ABSTRACT
Survival of embryos largely depends on yolk processing during early development. Proteolytic enzymes, cathepsin B & D (ctsb & ctsd) are known to have some important roles in yolk processing of various fish species. Mature female red spotted groupers were injected with human chorionic gonadotropin (HCG) to induce ovulation. The fertilized eggs and embryos were sampled at 0, 4 and 24 HPF (hours post fertilization). Survivals of each groups of embryos were checked at 24 and 48 HPH (hours post hatching). Transcripts of ctsb & ctsd showed the highest level at 0 HPF and relatively high at 4 HPF, but greatly decreased at 24 HPF. In bad egg quality group (BE, embryos survived until 24 HPH), transcript level of ctsb at 4 HPF were significantly lower than the transcript level at the same stage in good egg quality group (GE, embryos survived until 48 HPH) while no significant change of ctsb transcript level was observed at 0 or 24 HPF between BE and GE. Transcript level of ctsd was decreased at 24 HPF, but the difference was not as strong as the case of ctsb transcript. These results suggest that maternal ctsb transcript rather than ctsd transcript is likely to be involved in egg quality resulting in the difference of survival rate of embryos at early developmental period in this species.
Keywords: Cathepsin B, Red spotted grouper, Egg quality, Embryo
INTRODUCTION
Grouper species such as red spotted grouper, Ephinephelus akaara have high economic value in Asian fish markets (Pierre et al., 2008; Wang et al., 2017). Production of these species, however, does not meet the high demand of markets. Evaluation of egg quality is important issue to improve the efficiency of artificial seed production. In a number of previous studies, survival of fish embryos has been suggested to be dependent on yolk processing during early development (Carnevali et al., 2001; Tingaud-Sequeira et al., 2011; Palomino et al., 2017). Several enzymes such as cathepsins and lipases are known to be involved in this process.
Cathepsins are members of the endosomal or lysosomal protease family and can be divided into several subgroups according to active-site amino acid such as cysteine group : cathepsin B, L, W and so on / aspartate group : cathepsin D and E / serine group : cathepsin G / metal ions proteases group (Duve, 1983; Roberts, 2005; Liaudet-Coopman et al., 2006; Tingaud-Sequeira et al., 2011). They have many biological functions associated with protein turnover, proenzyme activation, hormone maturation and epidermal homeostasis in animals, plants and also in microorganisms (Azaryan & Hook, 1994; Tobin et al., 2002; Yasothornsrkul et al., 2003; Callogari et al., 2005; Liaudet-Coopman et al., 2006). Cathepsins also play some important roles during the mobilization and hydrolysis of the stored yolk proteins in fish embryos, which is critical to embryonic development (Tingaud-Sequeira et al., 2011; Palomino et al., 2017).
The main molecules such as cathepsin B, D, L whose participation has been proposed during oocyte maturation were involved in egg yolk processing and early embryonic development in teleosts (Carnevali et al., 1999a, 2001, 2008; Kwon et al., 2001; Tingaud-Sequeira et al., 2011). A number of transcripts including cathepsin B and cathepsin D are produced and stored in fish oocytes during oogenesis (vitellogenesis) (Matsubara & Sawano, 1995; Carnevali et al., 1999b). After matured eggs fertilized with sperms, the stored transcripts in the eggs are translated into the respective enzymes and these enzymes mobilize and hydrolyze stored yolk proteins to supply necessary energy and basic materials for early development (Sire et al., 1994; Carnevali et al., 1999a; Kwon et al., 2001; Hiramatsu et al., 2002; Ohkubo & Matsubara, 2002; Matsubara et al., 2003; LaFleur et al., 2005; Finn, 2007; Amano et al., 2008; Tingaud-Sequeira et al., 2011). Recently, Palomino et al. (2017) demonstrated that cathepsins could determine egg quality in a pelagic fish species. These two enzymes were found to be important for early development of rainbow trout (Onchorhynchus mykiss), zebrafish (Danio rerio), killifish (Fundulus heteroclitus) and yellowtail kingfish (Seriola lalalndi) (Kwon et al., 2001; Tingaud-Sequeira & Cerda, 2007; Tingaud-Sequeira et al., 2011; Palomino et al., 2017). However, these two enzymes were not studied in red spotted grouper. In this study, we investigated a possible relationship of these enzymes with egg quality and the importance of these enzymes to the survival of embryos in red spotted grouper.
MATERIALS AND METHODS
1. Broodstocks and artificial fertilization
Red spotted grouper, E. akaara was obtained from Marine Science Institute of Jeju National University, Korea. Fish were reared at 22±1℃ with filtered seawater in an indoor tank (5 m × 5 m × 1.2 m). Photoperiod was maintained at 14 hours light: 10 hours dark (14L: 10D). Sex ratio of broodstocks (female: male) was almost 1:1 (42: 39). Fish were fed twice a day (Maruha nichiro, Japan). All broodstocks measured body length (BL, cm) and weight (BW, kg) before inducing ovulation. Mature female red spotted groupers were not fed one day before and after injection to minimize handling stress. Human chorionic gonadotropin (HCG, Sigma, USA) of 500 IU/Kg BW was administered to females to induce ovulation. Males also were injected with the same dose of HCG (Daesung biological labs, Korea). Eggs were collected one day after the injection by the method of abdominal pressure. Eggs from different female were fertilized with sperm from different male which was also injected with HCG (4 males were used to fertilize eggs from 9 females by random combination).
2. Incubation and sampling of embryos
After artificial fertilization, all fertilized eggs from different female were transferred immediately to a laboratory. Fertilized eggs were accommodated separately in 9 glass beakers at 22±1℃. Fertilization, hatching and survival rates were calculated as follows: fertilization rate = number of eggs – number of dead eggs / number of eggs × 100, hatching rate = number of fertilized eggs – number of unhatched eggs / number of fertilized eggs × 100, survival rate = number of hatched eggs – number of unhatched eggs / number of hatched eggs × 100. The fertilized eggs and developing embryos were sampled at 0 HPF (hours post fertilization, n=3), 4 HPF (n=3) and 24 HPF (n=3). Eggs and developing embryos were observed using a microscope (AMEX1000, Life technologies, USA). All samples were stored at –80℃ with RNAlater solution (Qiagen, Germany) until extraction. Survivals of each groups of embryos were checked at 24 HPH (hours post hatching) and 48 HPH. Fertilized eggs died before 24 HPH were grouped as ‘worst quality eggs, WE’, fertilized eggs survived only until 24 HPH were grouped as ‘bad quality eggs, BE’, fertilized eggs survived more than 48 HPH were grouped as ‘good quality eggs, GE’.
3. Identification of cathepsin B and cathepsin D
Sequences for cathepsin B (ctsb) and cathepsin D (ctsd) of red spotted grouper were not available in GenBank database since these enzymes have not been studied previously in this species. To identify these enzymes and obtain partial sequences, degenerate PCR was carried out using sequence information from orange spotted grouper. Total RNA was extracted from stored eggs using TRIzol® Reagent (ambion, USA) and treated chloroform twice and 0.8 M disodium citrate (Sigma, USA) with isopropanol for good quality of RNA purification. Extracted total RNA quantified using nanodrop-1000 (Thermo, USA). This RNA (1 μg) was reverse transcribed using TOPscrit™ RT Dry MIX (Enzynomics, Korea). The resultant cDNAs were used as a template for subsequent degenerate PCR. Electrophoresis of PCR products were conducted in 1% agarose gel. All primers were designed using the Primer 3 software (version 2.2.3) and showed in Table 1. The degenerate PCR was carried out using Takara Taq (Clontech, Japan). The condition for degenerate PCR was as follows: initial denaturation at 95℃ for 5 min, 40 cycles of denaturation at 95℃ for 15 seconds, annealing at 50℃ for 15 seconds and elongation at 72℃ for 1-2 min. PCR products were sequenced by 3730xl DNA Analyzer (Applied biosystems, USA).
Table 1. Primers for degenerate PCR to identify red spotted grouper.
| Genes | Primer sequence | |
|---|---|---|
| ctsb | Forward | 5’-CTGTTGATTAGTCCCGTGTAGAG-3’ |
| Reverse | 5’-GTGTCGCCCTCTAGTGATTATG-3’ | |
| ctsd | Forward | 5’-TCTTAGACTCCGTCGACTGTTA-3’ |
| Reverse | 5’-TGTCAAACACCACCGTGAA-3’ |
4. Transcripts level of ctsb and ctsd in eggs
Transcripts level of ctsb and ctsd in the egg of red spotted grouper during early development stages was investigated by quantitative real time PCR (qRT-PCR) using cDNAs as templates. Primers were designed by the Primer 3 software (version 2.2.3) and described in Table 2. The qRT-PCR was conducted using TOPreal™ qPCR 2× PreMIX SYBR Green (Enzymonics, Korea) and CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, USA). Abundance level of ctsb and ctsd transcripts was normalized against the level of β-actin transcript. β-actin primers were designed based on the sequence used by Wang et al. (2017). Relative abundance was determined through the comparative threshold cycle method, 2−△△Ct, along with CFX Manager™ Software (Bio-Rad, USA). Relative transcript levels of ctsb and ctsd were analyzed and compared for the differences between developmental stages and between egg quality groups. Data were presented as mean±SEM. Statistical difference of the expression levels between embryos of different development stages was analyzed by one-way ANOVA and Games-Howell range tests, independent two-sample t-test (P<0.05).
Table 2. Primers for qRT-PCR to identify ctsb, ctsd and β-actin.
| Genes | Primer sequence | Product size | |
|---|---|---|---|
| ctsb | Forward | 5’-CTGGTTTCTGGTGGTCTCTATG-3’ | 87 |
| Reverse | 5’-TCTGGTGCCATTCACATGAT-3’ | ||
| ctsd | Forward | 5’-CCCTTAAGTACAACTTTGGCTTTC-3’ | 98 |
| Reverse | 5’-CAGGCTGATCTCACCGTAATA-3’ | ||
| β-actin | Forward | 5’-GCGACCTCACAGACTACCT-3’ | 228 |
| Reverse | 5’-CTGGGCAACGGAACCT-3’ |
Results and Discussion
1. Fertilization, hatching and survival rates of eggs and embryos
Basic information on broodstocks used in the present study was shown in Table 3. Fertilization and hatching rates of eggs from each female was examined and calculated (Table 3). Survival of the hatched larvae from different female was monitored for 48 hours. After that, they were divided into three groups (GE, BE and WE) depending on the lengths of survival time as described in materials and methods. Fertilization rate of the three groups did not differ from each other, ranging 95.0-97.1%. Hatching rates of GE, BE and WE were 94%, 89% and 83 %, respectively. Eggs from one female in WE were dead before hatching. Survivals of GE and BE at 24 HPH was 72.6% and 67.6%, respectively. Survival of GE at 48 HPH was 57.5 %. Survival of WE was 0% at both 24 HPH and 48 HPH. Developmental speed and stages of the three groups observed were nearly the same. Egg size of these groups was around 700 μm in diameter at all stages examined. Developmental stages at the time of observations (0, 4 and 24 HPF) were fertilized egg, morula and Kupffer’s vesicle stages, respectively (Fig. 1).
Table 3. Fertilization, hatching and survival rates of eggs and embryos from different broodstocks.
| Female | Male | FR(%) | HR (%) | Survival at 24 HPH (%) | Survival at48 HPH (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID No. | BL | BW | ID No. | BL | BW | |||||
| 9503 | 43.6 | 1,200 | 8605 | 45.2 | 1600 | 95.0 | 95.0 | 75.0 | 62.5 | |
| GE | 2983 | 42.0 | 1,150 | 125A | 44.5 | 1328 | 98.8 | 94.0 | 71.4 | 55.0 |
| 9866 | 36.8 | 900 | 125A | 44.5 | 1328 | 97.0 | 94.0 | 71.4 | 55.0 | |
| 1114 | 41.5 | 1,000 | 4804 | 53.0 | 2450 | 97.5 | 82.0 | 64.4 | 0 | |
| BE | 9617 | 40.0 | 1,000 | 125A | 44.5 | 1328 | 94.0 | 90.4 | 69.2 | 0 |
| 6717 | 44.5 | 1,500 | 125A | 44.5 | 1328 | 98.0 | 94.0 | 69.2 | 0 | |
| 6885 | 39.0 | 898 | 4362 | 46.6 | 1540 | 95.0 | 81.0 | 0 | 0 | |
| WE | 6126 | 40.5 | 1,050 | 4362 | 46.6 | 1540 | 96.0 | - | 0 | 0 |
| 5322 | 38.2 | 960 | 8605 | 45.2 | 1600 | 95.0 | 85.0 | 0 | 0 | |
* GE : good quality egg group, BE : bad quality egg group, WE : worst quality egg group, ID : identification, BL : body length (cm), BW : body weight (kg), FR : fertilization rate, HR : hatching rate, Data were presented as mean±SEM.
Fig. 1. Developmental stages at the time of observation.
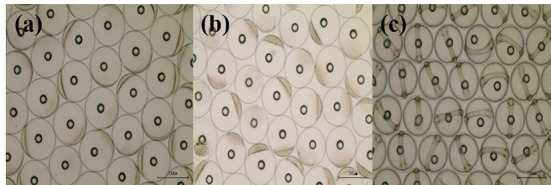
(a) fertilized eggs (0 HPF, hours post fertilization), (b) morula stage (4 HPF), (c) Kupffer’s vesicle stage (24 HPF).
2. Identification of ctsb & ctsd mRNA in eggs of red spotted grouper
Degenerate PCR successfully amplified a prominent band for ctsb and ctsd mRNA from the eggs of red spotted grouper. The sizes of PCR products for ctsb and ctsd mRNA were 950 and 248 base pairs (bp), respectively (Table 4). The partial sequences of these fragments exhibited 96-97% sequence identity to ctsb (accession number: KC832926.1), ctsd (accession number: GU988627.1) of orange spotted grouper, E. coioides.
Table 4. Red spotted grouper ctsb and ctsd mRNA partial sequences.
| Genes (size) | Sequence (5’ → 3’) |
|---|---|
| ctsb(950 bp) | TGGCGTGCAGCCCTCCTGGTGTTGGCTGCCAGCTTGTCAGTGAGCCTGGCCAGACCCCACCTCAA ACCACTGTCCGGTGACATGGTCAACTACATCAATAAGCTCAACACTACCTGGAGGGCTGGTCACA ACTTCCAAAATGTCGAATACAGTTACGTCCAGAAACTCTGCGGCACAATGCTGAAGGGACCCAAA CTGCCAGTCTTGGTTCAGTATTCTGGAGACATGAAGCTGCCTAAAAACTTTGACTCAAGAGAGCA GTGGCCCAACTGTCCCACTCTGAAGGAGATCAGAGACCAGGGCTCCTGTGGATCCTGCTGGGCGT TTGGTGCTGCAGAGGCCATCTCTGACCGTGTCTGTATCCACAGCAATGGCAAGGTCAGTGTGGAG ATCTCCTCGGAGGATCTGCTGACCTGCTGCGACAGCTGTGGCATGGGATGTAACGGTGGCTACCC TTCTGCTGCCTGGGACTTCTGGACCAATGCAGGACTGGTTTCTGGTGGTCTCTATGACTCTCACA TAGGTTGTCGGCCTTACACCATCCCTCCCTGTGAACATCATGTGAATGGCACCAGACCCCCCTGC ACTGGAGAGGGTGGAGACACACCCCAATGCGTCCTGCAGTGTGAACCTGGATACACACCTAGCTA CAAAGCAGACAAGCACTATGGTAAAACGTCCTACAGTGTGCTGTCAGATGAGGAGCAGATCCAGA GTGAGATATACAAGAATGGTCCAGTAGAGGGAGCCTTCACCGTCTTTGAAGACTTCTTGCTGTAT AAGACTGGTGTGTATCAGCACATGACTGGGTCTGCTGTTGGTGGCCATGCCATCAAGGTCCTGGG CTGGGGTGAGGAGGATGGTGTTCCCTACTGGCTCTGTGCCAACTCCTGGAACACTGACTGGGGTG ATAACGGTTTCTTTAAGTTCCTGCGTGGATCAGATCACTG |
| ctsd(248 bp) | TCGTGTTGGCGGCGGTAGCTCTCAGCACCGACGGACTGGTTCGAATTCCCTTAAAGAAATTCCGT TCCATCAGACGTGAGCTGACAGACTCGGGGAGAAGCGTTGAGGAGCTTCTGGCCGGCACACACTC CCTTAAGTACAACTTTGGCTTTCCCTCCAGCAGTGGACCCACTCCAGAAACCCTGAAGAACTACC TTGATGCCCAGTATTACGGTGAGATCAGCCTGGGGACCCCTCCTCAGACCTTC |
3. Relative quantification of ctsb & ctsd transcripts level in eggs of red spotted grouper
Degradation of yolk protein accumulated and stored in the eggs is important for early embryonic development. General lysosomal enzymes, phosphatases and proteinases have been suggested to be involved in yolk protein degradation of eggs in various animals including avians, amphibians, fish, crustaceans and insects (Perona & Vallejo, 1982; Lemanski & Aldoroty, 1977; Vogel & Gerster, 1997; Carnevali et al., 1999b). Products of maternal cathepsin transcripts including cathepsin B & D are associated with oocyte development in the ovary, and their RNA is depleted during embryo development (Carnevali et al., 2008; Follo et al., 2013; Fernandez et al., 2013; Langdon et al., 2016). It has been suggested that transcript level of cathepsin B & D can determine the quality of eggs and embryos in a pelagic fish species (Palomino et al., 2017).
The quantity of ctsb transcript in all egg groups quickly decreased at around 24 HPF regardless of egg quality (Fig. 2). This result is similar to the results from yellowtail kingfish (Palomino et al., 2017). In the present study, the quantity of ctsb transcript level decreased until kupffer’s vesicle stage. However, in killifish, ctsb transcript level decreased only until morula stage and increased after morula stage (Tingaud-Sequeira et al., 2011). In addition, rainbow trout showed a very low expression of ctsb transcript during all embryo development (Kwon et al., 2001). It seems that different fish species such as yellowtail kingfish, killifish and rainbow trout show different pattern of ctsb expression to each other during early development stages. This may be associated with the types of eggs such as floating eggs vs sinking eggs.
Fig. 2. Transcript level of cathepsin B (ctsb) at different time in three egg quality groups.
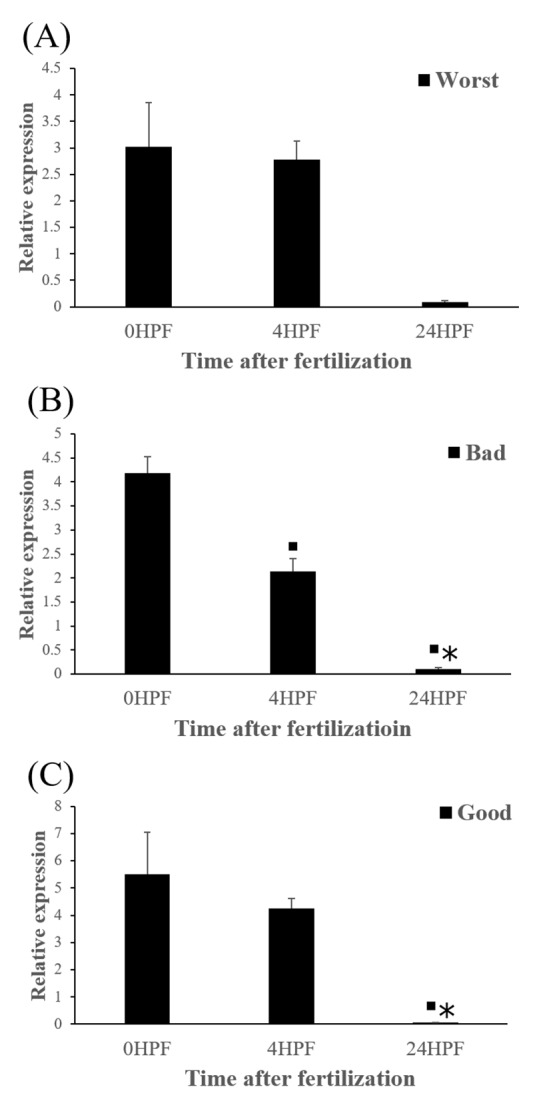
Worst egg quality group (A), Bad egg quality group (B), Good egg quality group (C). Each column represents mean± SEM (P<0.05). ▪ : differ from 0 HPF. * : differ from 4 HPF.
Level of ctsb transcript decreased during early embryo development in this species might be related to mobilization and hydrolyzation of stored yolk protein to supply necessary energy during early embryo development. In this study, transcript level of ctsb in GE was significantly higher at 4 HPF than that in BE (Fig. 3b). Transcript level of ctsb in GE seemed to be higher than other two groups at 0 HPF (Fig. 3a) and 4 HPF (Fig. 3b), but it decreased at 24 HPF (Fig. 3c). Good quality eggs could use ctsb transcript more and faster than bad quality eggs do in this species. Maternal factors could influence on several processes such as germ line establishment and pattern formation during embryonic development (Pelegri, 2003). Transcript of ctsb in the fertilized eggs of red spotted grouper could be one of the important maternal factors that affect survival and normal development.
Fig. 3. Transcript level of cathepsin B (ctsb) in the eggs of three egg quality groups at different time.
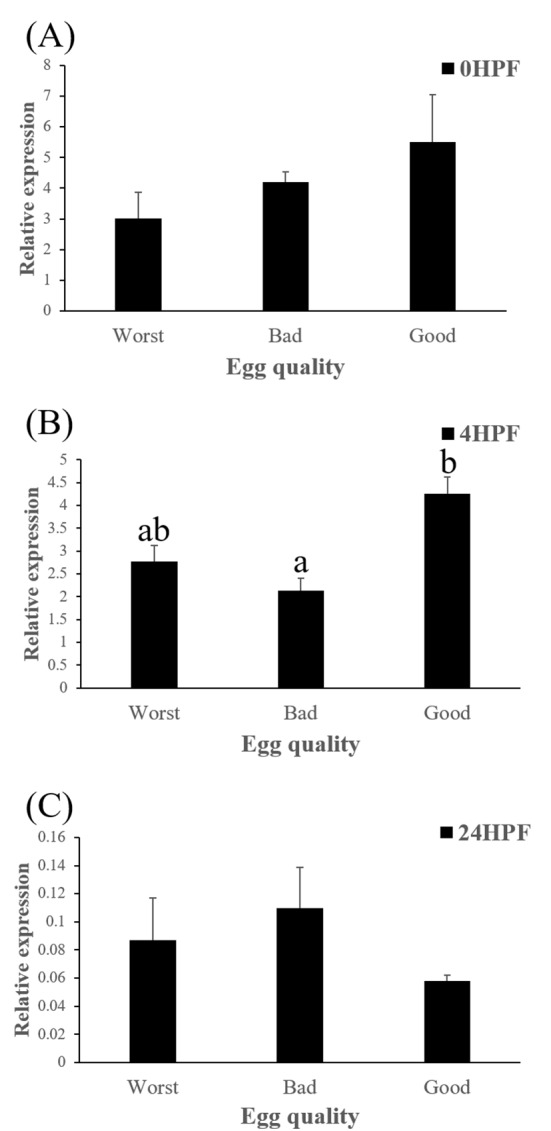
0 HPF (hours post fertilization) (a), 4 HPF (b), 24 HPF (c). Each column represents mean±SEM (P<0.05).
The quantity of ctsd transcript in all egg groups quickly decreased at around 24 HPF regardless of egg quality (Fig. 4) and this result was same pattern with ctsb transcript (Fig. 2) in the present study. Transcript level of ctsd also decreased until kupffer’s vesicle stage as ctsb transcript. Moreover, these pattern is similar to the results from rainbow trout and yellowtail kingfish (Kwon et al., 2001; Palomino et al., 2017). In the rainbow trout, ctsd transcript level was highest at 2 HPF and constantly decreased later until 44 days post-fertilization (won et al., 2001). The quantity of ctsd transcript also decreased until appearance of embryo in the yellowtail kingfish (Palomino et al., 2017). A similar result was also shown in grass carp, where ctsd activity by qRT-PCR was low during the early stages of the embryo (Dong et al., 2012). However, in gilthead sea bream, ctsd transcript level increased until morula stage but the level, thereafter, decreased until 50% epiboly (Fernandez et al., 2013). It seems that different fish species such as rainbow trout, yellowtail kingfish and gilthead sea bream show different pattern of ctsd expression to each other during early development stages. This may be also associated with the types of eggs such as floating eggs vs sinking eggs. The different transcript levels of ctsd during embryo development might be involved in the embryos’ energy requirements (Dong et al., 2012). In these species, ctsd transcript in the fertilized eggs is also associated with the process during early development.
Fig. 4. Transcript level of cathepsin D (ctsd) at different time in three egg quality groups.
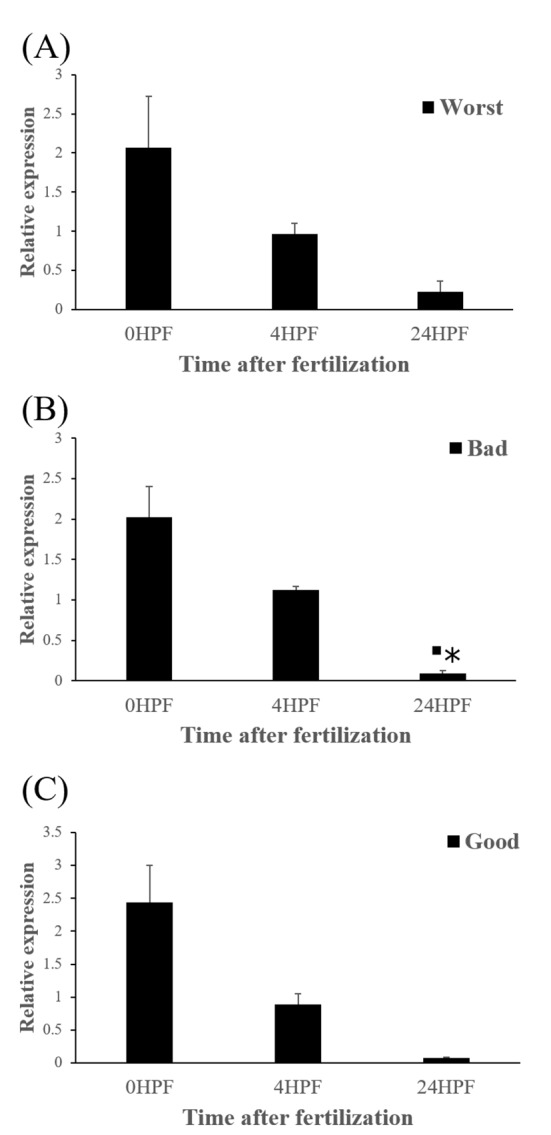
Worst egg quality group (A), Bad egg quality group (B), Good egg quality group (C). Each column represents the mean± SEM (P<0.05). ▪ : differ from 0 HPF. * : differ from 4 HPF.
The amount of ctsd enzymatic activity in poor quality eggs was showed to be much higher than that in good quality eggs, indicating that ctsd was suggested as a potential marker for bad egg quality in European seabass (Carnevali et al., 2001). In present study, however, transcript level of ctsd has no significant differences between good and bad quality egg groups at all time points studied (Fig 5). Therefore, ctsd seems not to be utilized for a potential mark of egg quality evaluation in this species.
Fig. 5. Transcript level of cathepsin D (ctsd) in the eggs of three egg quality groups at different time.
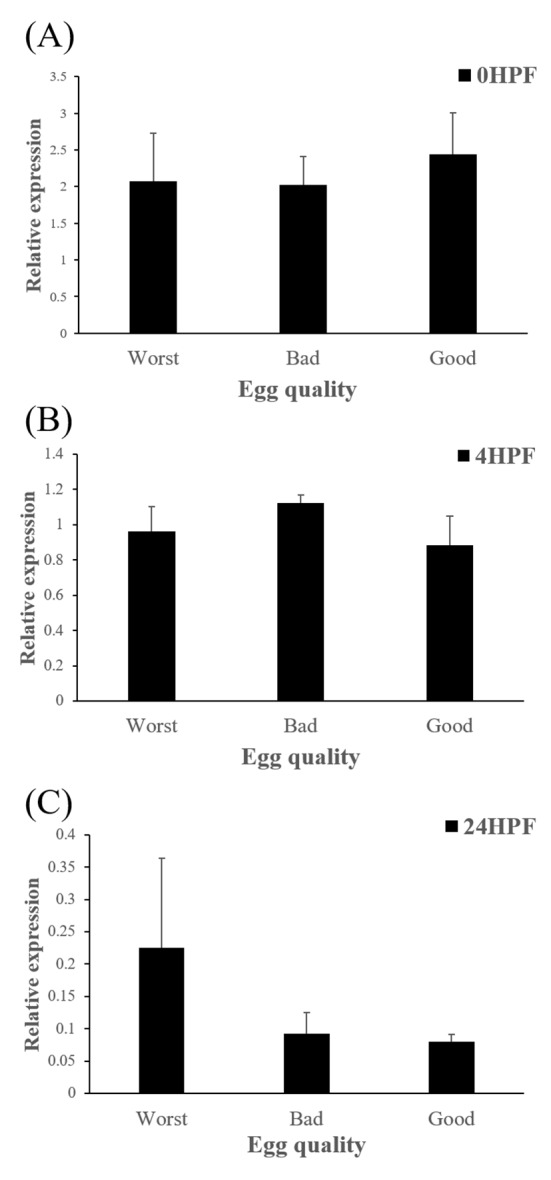
0 HPF (hours post fertilization) (a), 4 HPF (b), 24 HPF (c). Each column represents mean±SEM (P<0.05).
In summary, transcript level of ctsb in GE decreased more than the levels in other two groups (BE and WE) did, while transcript level of ctsd was not shown significant differences between three groups. Thus, the level of ctsb transcript in eggs could be considered as a potential marker for evaluating egg quality, but not the level of ctsd in red spotted grouper. In further studies, other enzymes such as cathepsin L (ctsl), lipoprotein lipase (lpl) which are also involved in yolk processing at early embryonic stages need to be investigated.
ACKNOWLEDGEMENTS
This research was supported by the Korean Ministry of Agriculture, Food and Rural Affairs (MAFRA), the Korean Ministry of Oceans and Fisheries (MOF), the Korean Rural Development Administration (RDA), and Korea Forest Service (KFS) (Grant number: 213008-05-1-WT511).
REFERENCES
- 1.Amano H, Fujita T, Hiramatsu N, Kagawa H, Matsubara T, Sullivan CV & Hara A. Multiple vitellogenin‐derived yolk proteins in gray mullet (Mugil cephalus): disparate proteolytic patterns associated with ovarian follicle maturation. Mol Reprod Dev. 2008;75:1307–1317. doi: 10.1002/mrd.20864. [DOI] [PubMed] [Google Scholar]
- 2.Azaryan AV, Hook VY. Unique cleavage specificity of 'prohormone thiol protease' related to proenkephalin processing. FEBS Lett. 1994;341:197–202. doi: 10.1016/0014-5793(94)80456-7. [DOI] [PubMed] [Google Scholar]
- 3.Callegari EA, Ferguson-Gottschall S, Gibori G. PGF2αinduced differential expression of genes involved in turnover of extracellular matrix in rat decidual cells. Reprod Biol Endocrinol. 2005;3:3. doi: 10.1186/1477-7827-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carnevali O, Carletta R, Cambi A, Vita A, Bromage N. Yolk formation and degradation during oocyte maturation in seabream Sparus aurata: involvement of two lysosomal proteinases. Biol Reprod. 1999a;60:140–146. doi: 10.1095/biolreprod60.1.140. [DOI] [PubMed] [Google Scholar]
- 5.Carnevali O, Centonze F, Brooks S, Marota I, Sumpter JP. Molecular cloning and expression of ovarian cathepsin D in seabream, Sparus aurata. Biol Reprod. 1999b;61:785–791. doi: 10.1095/biolreprod61.3.785. [DOI] [PubMed] [Google Scholar]
- 6.Carnevali O, Cionna C, Tosti L, Cerdà J, Gioacchini G. Changes in cathepsin gene expression and relative enzymatic activity during gilthead sea bream oogenesis. Mol Reprod Dev. 2008;75:97–104. doi: 10.1002/mrd.20768. [DOI] [PubMed] [Google Scholar]
- 7.Carnevali O, Mosconi G, Cambi A, Ridolfi S, Zanuy S, Polzonetti-Magni AM. Changes of lysosomal enzyme activities in sea bass (Dicentrarchus labrax) eggs and developing embryos. Aquaculture. 2001;202:249–256. doi: 10.1016/S0044-8486(01)00775-X. [DOI] [Google Scholar]
- 8.Dong ZD, Zhang J, Ji XS, Zhou FN, Fu Y, Chen W, Wang H. Molecular cloning, characterization and expression of cathepsin D from grass carp (Ctenopharyngodon idella) Fish Shellfish Immunol. 2012;33:1207–1214. doi: 10.1016/j.fsi.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Duve C. Lysosomes revisited. FEBS J. 1983;137:391–397. doi: 10.1111/j.1432-1033.1983.tb07841.x. [DOI] [PubMed] [Google Scholar]
- 10.Fernández CG, Roufidou C, Antonopoulou E, Sarropoulou E. Expression of developmental-stage-specific genes in the gilthead sea bream Sparus aurata L. Mar Biotechnol. 2013;15:313–320. doi: 10.1007/s10126-012-9486-8. [DOI] [PubMed] [Google Scholar]
- 11.Finn RN. The maturational disassembly and differential proteolysis of paralogous vitellogenins in a marine pelagophil teleost: a conserved mechanism of oocyte hydration. Biol Reprod. 2007;76:936–948. doi: 10.1095/biolreprod.106.055772. [DOI] [PubMed] [Google Scholar]
- 12.Follo C, Ozzano M, Montalenti C, Santoro MM, Isidoro C. Knockdown of cathepsin D in zebrafish fertilized eggs determines congenital myopathy. Biosci Rep. 2013;33:e00034. doi: 10.1042/BSR20120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiramatsu N, Ichikawa N, Fukada H, Fujita T, Sullivan CV, Hara A. Identification and characterization of proteases involved in specific proteolysis of vitellogenin and yolk proteins in salmonids. J Exp Zool A Ecol Genet Physiol. 2002;292:11–25. doi: 10.1002/jez.1138. [DOI] [PubMed] [Google Scholar]
- 14.Kwon JY, Prat F, Randall C, Tyler CR. Molecular characterization of putative yolk processing enzymes and their expression during oogenesis and embryogenesis in rainbow trout (Oncorhynchus mykiss) Biol Reprod. 2001;65:1701–1709. doi: 10.1095/biolreprod65.6.1701. [DOI] [PubMed] [Google Scholar]
- 15.LaFleur Jr GJ, Raldúa D, Fabra M, Carnevali O, Denslow N, Wallace RA, Cerda J. Derivation of major yolk proteins from parental vitellogenins and alternative processing during oocyte maturation in Fundulus heteroclitus. Biol Reprod. 2005;73:815–824. doi: 10.1095/biolreprod.105.041335. [DOI] [PubMed] [Google Scholar]
- 16.Langdon YG, Fuentes R, Zhang H, Abrams EW, Marlow FL, Mullins MC. Split top: a maternal cathepsin B that regulates dorsoventral patterning and morphogenesis. Development. 2016;143:1016–1028. doi: 10.1242/dev.128900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemanski LF, Aldoroty R. Role of acid phosphatase in the breakdown of yolk platelets in developing amphibian embryos. J Morphol. 1977;153:419–425. doi: 10.1002/jmor.1051530307. [DOI] [PubMed] [Google Scholar]
- 18.Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Vignon F. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237:167–179. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Matsubara T, Sawano K. Proteolytic cleavage of vitellogenin and yolk proteins during vitellogenin uptake and oocyte maturation in barfin flounder (Verasper moseri) J Exp Zool A Ecol Genet Physiol. 1995;272:34–45. [Google Scholar]
- 20.Matsubara T, Nagae M, Ohkubo N, Andoh T, Sawaguchi S, Hiramatsu N, Hara A. Multiple vitellogenins and their unique roles in marine teleosts. Fish Physiol Biochem. 2003;28:295–299. doi: 10.1023/B:FISH.0000030559.71954.37. [DOI] [Google Scholar]
- 21.Ohkubo N, Matsubara T. Sequential utilization of free amino acids, yolk proteins and lipids in developing eggs and yolk-sac larvae of barfin flounder Verasper moseri. Mar Biol. 2002;140:187–196. doi: 10.1007/s002270100647. [DOI] [Google Scholar]
- 22.Palomino J, Herrera G, Torres-Fuentes J, Dettleff P, Patel A, Martínez V. Assessment of cathepsin mRNA expression and enzymatic activity during early embryonic development in the yellowtail kingfish Seriola lalandi. Anim Reprod Sci. 2017;180:23–29. doi: 10.1016/j.anireprosci.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Pelegri F. Maternal factors in zebrafish development. Dev Dyn. 2003;228:535–554. doi: 10.1002/dvdy.10390. [DOI] [PubMed] [Google Scholar]
- 24.Perona R, Vallejo CG. The lysosomal proteinase of Artemia. FEBS J. 1982;124:357–362. doi: 10.1111/j.1432-1033.1982.tb06600.x. [DOI] [PubMed] [Google Scholar]
- 25.Pierre S, Gaillard S, Prévot‐D'Alvise N, Aubert J, Rostaing‐Capaillon O, Leung‐Tack D, Grillasca JP. Grouper aquaculture: Asian success and Mediterranean trials. Aquat Conserv. 2008;18:297–308. doi: 10.1002/aqc.840. [DOI] [Google Scholar]
- 26.Roberts R. Lysosomal cysteine proteases: structure, function and inhibition of cathepsins. Drug News Perspect. 2005;18:605–614. doi: 10.1358/dnp.2005.18.10.949485. [DOI] [PubMed] [Google Scholar]
- 27.Sire MF, Babin PJ, Vernier JM. Involvement of the lysosomal system in yolk protein deposit and degradation during vitellogenesis and embryonic development in trout. J Exp Zool A Ecol Genet Physiol. 1994;269:69–83. doi: 10.1002/jez.1402690109. [DOI] [Google Scholar]
- 28.Tingaud-Sequeira A, Cerdà J. Phylogenetic relationships and gene expression pattern of three different cathepsin L (Ctsl) isoforms in zebrafish: Ctsla is the putative yolk processing enzyme. Gene. 2007;386:98–106. doi: 10.1016/j.gene.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Tingaud-Sequeira A, Carnevali O, Cerdà J. Cathepsin B differential expression and enzyme processing and activity during Fundulus heteroclitus embryogenesis. Comp Biochem Physiol A Mol Integr Physiol. 2011;158:221–228. doi: 10.1016/j.cbpa.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Tobin DJ, Foitzik K, Reinheckel T, Mecklenburg L, Botchkarev VA, Peters C, Paus R. The lysosomal protease cathepsin L is an important regulator of keratinocyte and melanocyte differentiation during hair follicle morphogenesis and cycling. Am J Pathol. 2002;160:1807–1821. doi: 10.1016/S0002-9440(10)61127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel AM, Gerster T. Expression of a zebrafish cathepsin L gene in anterior mesendoderm and hatching gland. Dev Genes Evol. 1997;206:477–479. doi: 10.1007/s004270050078. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Zhang X, Liu Q, Liu X, Ding S. Selection and evaluation of new reference genes for RT-qPCR analysis in Epinephelus akaara based on transcriptome data. PloS one. 2017;12:e0171646. doi: 10.1371/journal.pone.0171646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Miller R, Goldsmith P. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc Natl Acad Sci. 2003;100:9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]


