Abstract
Changes in the expression of regulator of G protein signaling 2 (RGS2) are involved in the genesis and development of a number of malignancies. However, the association between changes in the expression of RGS2 and breast invasive carcinoma of no special type (BIC-NST) remains unknown. The present study found that, in comparison to normal breast tissue, BIC-NST exhibited low expression of RGS2 mRNA and protein, as detected using data mining and immunohistochemical analysis. The low expression of RGS2 was associated with the positive status of hormone receptor expression in BIC-NST. Kaplan-Meier analysis revealed that patients with low RGS2 expression had a significantly poorer overall survival rate. Furthermore, multivariate Cox regression analysis demonstrated that the RGS2 low expression was an independent high-risk factor. Gene set enrichment analysis using data from The Cancer Genome Atlas supported these results. In summary, the results of the current study indicate that RGS2 acts as a suppressor gene in the progression of BIC-NST. To the best of our knowledge, the present study is the first concerning the association between RGS2 and hormone receptors in BIC-NST, as well as that between RGS2 expression and the prognosis of patients with BIC-NST. However, the effect of RGS2 in breast cancer requires further investigation.
Keywords: breast invasive carcinoma of no special type, regulator of G protein signaling 2, prognosis, immunohistochemistry, data mining
Introduction
Disease-free survival in patients with breast cancer has significantly improved since the 1990s, owing to the combination of early breast cancer screening and a greater number of treatment approaches, including adjuvant chemotherapy, hormonal treatment and targeted therapy (1–3). However, there are certain patients who are only treated with maintenance therapy who receive an initial diagnosis of advanced-stage breast cancer or multidrug resistance (4). Breast cancer is the most commonly diagnosed cancer in women all over the world, and is the first and the second cause of cancer-associated mortality in women in developing and developed countries, respectively (5). Therefore, it is necessary to identify candidate biomarkers that are associated with the development of breast cancer and investigate therapeutic targets to provide patients with breast cancer with treatment options.
G-protein regulating factors negatively regulate the recycling of G-protein-coupled receptors; these factors accelerate GTP hydrolysis by binding to the α-subunit of a heterotrimeric G protein to deactivate G-protein signaling (6,7). With regard to malignant tumors, regulator of G-protein signaling 2 (RGS2) is one of the most well-characterized RGS genes. RGS2 can also inhibit the translation of mRNA into protein by binding to the ε-subunit of eukaryotic initiation factor 2B (8). Alteration of RGS2 expression is associated with a number of types of cancer, including prostate cancer (9), ovarian cancer (10), mantle cell lymphoma (11), acute myeloid leukemia (12) and fibrolamellar hepatocellular carcinoma (13). However, the association between RGS2 and breast cancer remains obscure. On the one hand, a large-sample study revealed that RGS2 is upregulated in breast cancer (14); on the other hand, another more recent study demonstrated that low expression of RGS2 is associated with breast cancer using MCF7 and MCF10A cells (15). The aim of the present study was to assess the association of RGS2 with breast cancer. Furthermore, according to the World Health Organization Classification of Tumors of the Breast (16), breast invasive carcinoma of no special type (BIC-NST) is the largest group among the different classifications of breast cancer. Thus, given the considerable heterogeneity of breast cancer, the present study focused on BIC-NST.
Materials and methods
Patients and samples
Paraffin-embedded breast cancer samples from patients who had undergone radical mastectomy were obtained from the archives of the Department of Pathology at the First Affiliated Hospital of Chongqing Medical University (CQMU) (Chongqing, China) between January and December 2011. Two pathologists screened 196 BIC-NST samples according to the criterion of 2012 WHO Classification of Tumors of the Breast and reached a consensus on the diagnosis. The patients enrolled in the present study were followed up every other year, and the last follow-up was in June 2016. All protocols in this study were approved by the Ethics Committee of the First Affiliated Hospital of CQMU, and each patient enrolled provided written informed consent.
RGS2 gene expression data in breast cancer and relevant clinical information were downloaded from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov). Data from 513 patients with breast cancer were analyzed, following the exclusion of all male patients and those female patients without BIC-NST, or those lacking complete information. Of the 513 patients, there were 93 paired datasets that included cancer tissue and adjacent tissue.
Immunohistochemistry (IHC) experimental procedures
RGS2 protein expression in 4-µm sections of 196 BIC-NST samples was detected by IHC. The polyclonal RGS2 antibody (cat. no. ab36561; dilution, 1:200) was purchased from Abcam (Cambridge, UK). Briefly, slides were deparaffinized using xylene, and rehydrating using a gradient ethanol series. Microwave antigen retrieval was performed using sodium citrate solution (10 mmol/l, pH 6.0) for 4 min in high heat (~100°C) and 10 min in low heat (~35°C), and the slides were cooled for 30 min. Slides were incubated with 3% hydrogen peroxide for 10 min at room temperature to block endogenous peroxidase activity. Samples were incubated with RGS2 antibody for 1 h at room temperature. Following incubation with Polymer Helper reagent and a poly-horseradish peroxidase-conjugated anti-mouse/rabbit IgG secondary antibody (cat. no. PV-9000; dilution ready-to-use; OriGene Technologies, Inc., Beijing, China) twice, for 20 min at room temperature each time, the sections were visualized using 3,3′-diaminobenzidine solution (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) and were counterstained with hematoxylin for 20 sec at room temperature. Image acquisition was performed using a light microscope (Olympus BX45; Olympus Corporation, Tokyo, Japan), and images were captured at magnification, ×200 and ×400.
IHC evaluation
The slides were independently reviewed by two pathologists who were blinded to any information on the clinical characteristics and follow-up data. RGS2 expression level scores were calculated according to the sum of IHC intensity (0, negative; 1, weak staining; 2, moderate staining; and 3, strong staining) and the rate of positive staining (0, 0%; 1, 1–50%; and 2, 51–100%). Differences in scoring were resolved by reaching a consensus. The total cumulative scores of 0–3 and 4–5 were defined as the low- and high-expression groups, respectively. Representative IHC staining images are shown in Fig. 1.
Figure 1.
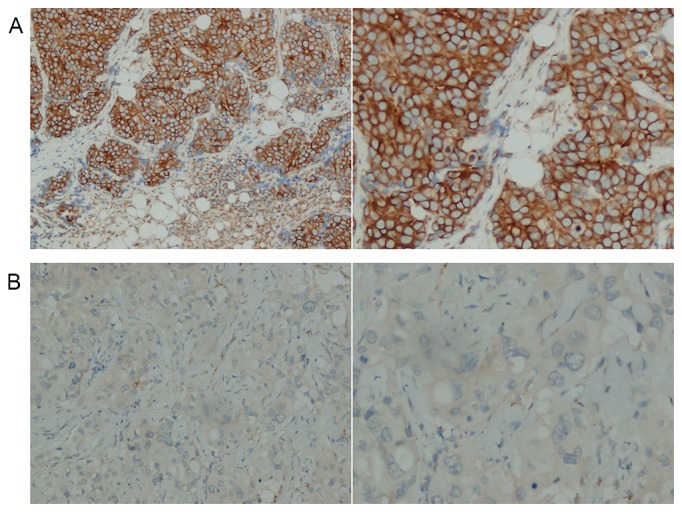
IHC staining of RGS2 protein in BIC-NST samples. (A) Representative IHC staining image showing high RGS2 expression in BIC-NST. (B) Representative IHC staining image showing low RGS2 expression in BIC-NST. Magnification: Left, ×200; right, ×400; IHC, immunohistochemistry; RGS2, regulator of G protein signaling 2; BIC-NST, breast invasive carcinoma of no special type.
Gene set enrichment analysis (GSEA)
GSEA was used to assess different enriched gene sets corresponding to each gene from microarray data. Every enriched gene set is sorted according to a common biological function. Thus, the association between the targeted gene and biological function could be indirectly shown by GSEA. GSEA software version 2.2.2 was downloaded from the Molecular Signatures Database, Broad Institute (http://software.broadinstitute.org/gsea/) (17). The enrichment of the C2 curated gene sets and the C5 Gene Ontology (GO) gene sets were analyzed by comparing low-expression and high-expression RGS2 subtypes. The number of permutations was set at 1,000.
Statistical analysis
Differences in expression between cancer tissue and adjacent tissue from the 93 paired TCGA datasets was analyzed by paired Student's t-test. The association between RGS2 expression and clinical pathological parameters was evaluated by the χ2 test. The association between RGS2 expression and overall survival was assessed using Kaplan-Meier method with log-rank test, and univariate and multivariate Cox regression analyses. P<0.05 was considered to indicate a statistically significant difference. Statistical analysis was conducted using SPSS version 19.0.0 (IBM Corp., Armonk, NY, USA) and statistical drawing was performed using GraphPad Prism version 6.01 (GraphPad Software Inc., La Jolla, CA, USA).
Results
mRNA and protein RGS2 expression is downregulated in BIC-NST
The expression of RGS2 mRNA and protein in tumor and adjacent normal breast tissue was detected by analysis of the TCGA datasets and IHC analysis of tissue samples. As shown in Fig. 2, the expression level of RGS2 mRNA in breast cancer tissue was significantly lower than that in adjacent normal breast tissue (P<0.001), as was the protein expression level of RGS2 (P<0.001).
Figure 2.
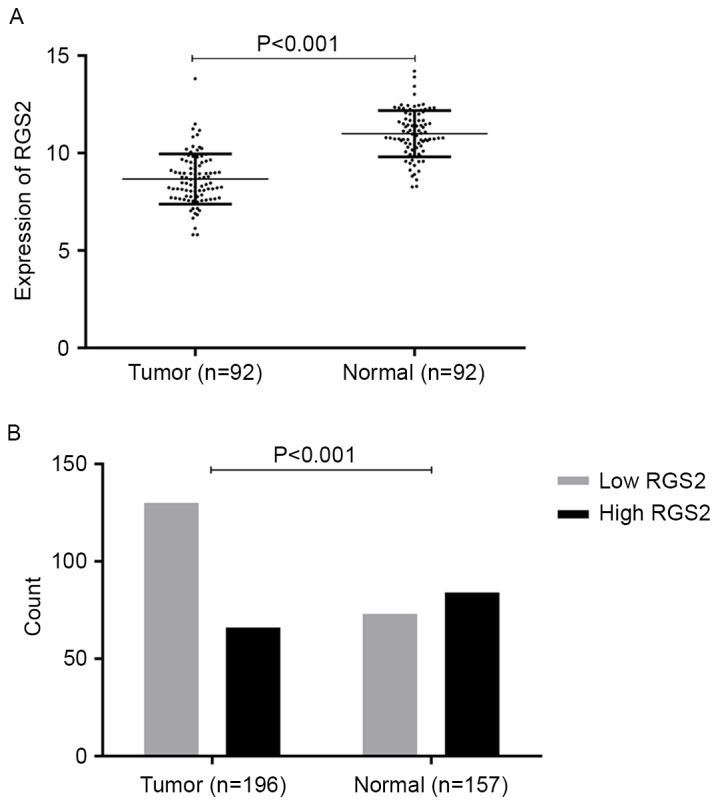
RGS2 expression in BIC-NST tissues and normal breast tissues. (A) In The Cancer Genome Atlas cohort, 92 paired mRNA expression datasets in patients with BIC-NST were analyzed using paired Student's t-test. (B) In the Chongqing Medical University cohort, protein expression levels in 196 tumor tissues and 157 normal tissues were analyzed using the χ2 test. RGS2, regulator of G protein signaling 2; BIC-NST, breast invasive carcinoma of no special type.
Association between RGS2 expression and clinical characteristics
Following IHC analysis, the relevant clinicopathological analysis of the 196 BIC-NST samples was performed (Table I). HER2 status is primarily determined by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) on formalin-fixed and paraffin-embedded samples. Definition of HER status is performed as follows: When the tumor is classified as 3+ by IHC staining, HER2 status considered positive. When the tumor is classified as 0 or 1+ by IHC staining, HER2 status is considered negative. If the tumor is classified as 2+ by IHC staining, HER2 status requires additional confirmation using FISH (18). In the present study, there were 14 cases of breast cancer which were classified as 2+ by IHC staining, but these samples were not examined using FISH as this protocol was not performed. Consequently, these cases were defined as ‘uncertain’ (Table I).
Table I.
Association between patient characteristics and expression status of RGS2 in the Chongqing Medical University cohort.
| Characteristic | Low RGS2, n (%) | High RGS2, n (%) | χ2 | P-value |
|---|---|---|---|---|
| Age, years | 3.300 | 0.069 | ||
| <60 | 112 (86.2) | 50 (75.8) | ||
| ≥60 | 18 (13.8) | 16 (24.2) | ||
| Histological gradea | 10.332 | 0.006 | ||
| Grade 1 | 30 (23.1) | 30 (45.5) | ||
| Grade 2 | 60 (46.2) | 22 (33.3) | ||
| Grade 3 | 40 (30.7) | 14 (21.2) | ||
| ER status | 8.790 | 0.003 | ||
| Negative | 32 (24.6) | 30 (45.5) | ||
| Positive | 98 (75.4) | 36 (54.5) | ||
| PR status | 5.085 | 0.024 | ||
| Negative | 38 (29.2) | 30 (45.5) | ||
| Positive | 92 (70.8) | 36 (54.5) | ||
| HER-2 status | 2.454 | 0.293 | ||
| Negative | 80 (61.5) | 48 (72.7) | ||
| Positive | 40 (30.8) | 14 (21.2) | ||
| Uncertainb | 10 (7.7) | 4 (6.1) | ||
| Ki-67, % | 1.807 | 0.179 | ||
| <14 | 50 (38.5) | 32 (48.5) | ||
| ≥14 | 80 (61.5) | 34 (51.5) | ||
| Distant metastasis | 3.142 | 0.076 | ||
| No | 124 (95.4) | 66 (100.0) | ||
| Yes | 6 (4.6) | 0 (0.0) | ||
| Tumor size, cm | 9.771 | 0.002 | ||
| ≤2 | 60 (46.2) | 46 (69.7) | ||
| >2 | 70 (53.8) | 20 (30.3) | ||
| Lymph node metastasis | 2.274 | 0.132 | ||
| No | 64 (49.2) | 40 (60.6) | ||
| Yes | 66 (50.8) | 26 (39.4) | ||
| Clinical stage | 1.143 | 0.285 | ||
| I–II | 102 (78.5) | 56 (84.8) | ||
| III–IV | 28 (21.5) | 10 (15.2) |
Histological grading was performed according to the World Health Organization Classification of Tumors of the Breast, 2012 (4th edition).
These samples were not examined using fluorescence in situhybridization. RGS2, regulator of G protein signaling 2; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2.
RGS2 protein expression was significantly negatively associated with the following clinical pathological characteristics: Histological grade (P=0.006), estrogen receptor (ER) status (P=0.003), progesterone receptor (PR) status (P=0.024) and tumor size (P=0.002). The TCGA BIC-NST sample was divided into two groups according to the expression of RGS2: The low-expression group (n=204, the lowest 35%), and the over-expression group (n=123, the highest 35%). As shown in Table II, age (P=0.002), ER status (P<0.001), PR status (P<0.001) and lymph node metastasis (P=0.002) were significantly negatively associated with RGS2 mRNA expression.
Table II.
Association between patient characteristics and expression status of RGS2 in The Cancer Genome Atlas cohort.
| Characteristic | Low RGS2, n (%) | High RGS2, n (%) | χ2 | P-value |
|---|---|---|---|---|
| Age, years | 9.307 | 0.002 | ||
| <60 | 99 (48.5) | 81 (65.9) | ||
| ≥60 | 105 (51.5) | 42 (34.1) | ||
| ER status | 27.172 | <0.001 | ||
| Negative | 33 (16.2) | 52 (42.3) | ||
| Positive | 171 (83.8) | 71 (57.7) | ||
| PR status | 22.132 | <0.001 | ||
| Negative | 55 (27.0) | 65 (52.8) | ||
| Positive | 149 (73.0) | 58 (47.2) | ||
| HER-2 status | 0.163 | 0.687 | ||
| Negative | 169 (82.8) | 104 (84.6) | ||
| Positive | 35 (17.2) | 19 (15.4) | ||
| Distant metastasis | 0.556 | 0.456 | ||
| No | 198 (97.1) | 121 (98.4) | ||
| Yes | 6 (2.9) | 2 (1.6) | ||
| Tumor size, cm | 0.029 | 0.866 | ||
| ≤2 | 53 (26.0) | 33 (26.8) | ||
| >2 | 151 (74.0) | 90 (73.2) | ||
| Lymph node metastasis | 9.469 | 0.002 | ||
| No | 82 (40.2) | 71 (57.7) | ||
| Yes | 122 (59.8) | 52 (42.3) | ||
| Clinical stage | 5.381 | 0.020 | ||
| I–II | 154 (75.5) | 106 (86.2) | ||
| III–IV | 50 (24.5) | 17 (13.8) |
RGS2, regulator of G protein signaling 2; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2.
Low expression levels of RGS2 are associated with poor survival rate in BIC-NST patients
Follow-up data from the TCGA cohort and the CQMU cohort were assessed, and the association between the RGS2 expression level and overall survival rate were analyzed using the Kaplan-Meier method and log-rank test. The median follow-up time in the TCGA and CQMU cohorts was 27 months (range, 0–238 months) and 44 months (range, 6–65 months), respectively. As shown in Fig. 3, the group with low expression of RGS2 mRNA had a significantly lower overall survival rate (P=0.002), as did the group with low expression of RGS2 protein (P=0.019). The results of univariate analysis in the TCGA and CQMU cohorts are presented in Tables III and IV, respectively. To evaluate the prognostic value of RGS2, the two groups of data were analyzed independently using multivariate Cox regression analysis. In the TCGA cohort, as demonstrated in Table V, the low expression of RGS2 [hazard ratio (HR), 0.435; 95% confidence interval (CI), 0.196–0.965; P=0.041], age (>60 years) (HR, 2.073; 95% CI, 1.013–4.243; P=0.046) and advanced disease stage (III–IV) (HR, 2.938; 95% CI, 1.390–6.208; P=0.005) were significantly associated with unfavorable survival in BIC-NST independently. In the CQMU cohort, as shown in Table VI, the low expression of RGS2 (HR, 4.602; 95% CI, 1.467–14.432; P=0.009), age (>60 years) (HR, 5.832; 95% CI, 2.149–15.830; P=0.001), advanced stage (III–IV) (HR, 5.021; 95% CI, 2.201–11.454; P<0.001) and lymph node metastasis (HR, 3.993; 95% CI, 1.406–11.340; P=0.009) were significantly independently associated with poor patient outcome in BIC-NST.
Figure 3.
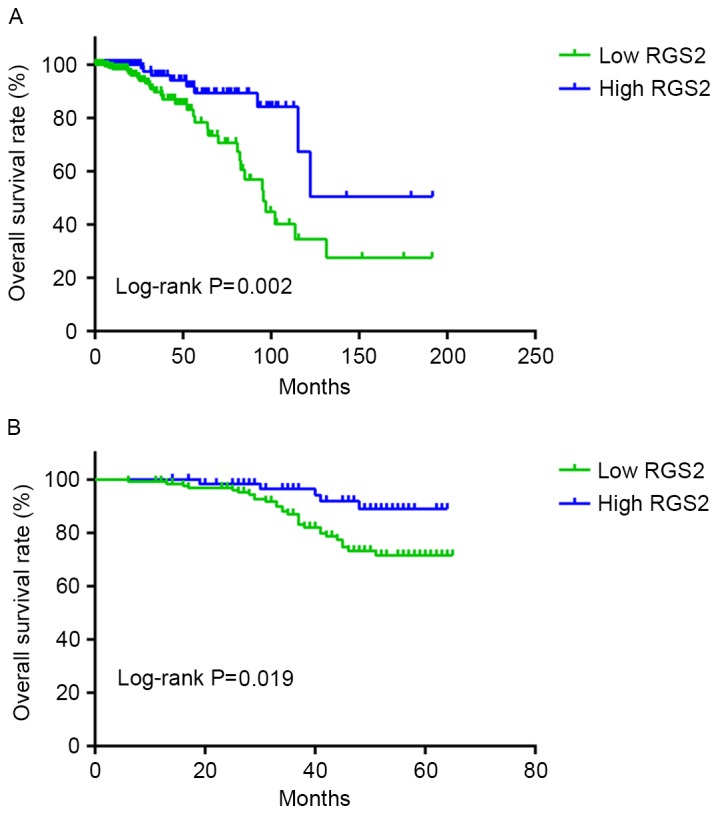
Kaplan-Meier analysis for different mRNA expression levels of RGS2 and overall survival in patients with breast invasive carcinoma of no special type. Analysis of (A) The Cancer Genome Atlas and (B) Chongqing Medical University cohorts showed that patients with low RGS2 expression exhibited a poor overall survival rate compared with those expressing high levels of RGS2.
Table III.
Univariate analysis for RGS2 and clinical characteristics in The Cancer Genome Atlas cohort.
| Characteristic | HR | 95% CI | P-value |
|---|---|---|---|
| RGS2 expression (low vs. high) | 0.324 | 0.154–0.683 | 0.003 |
| Age (≥60 vs. <60 years) | 2.014 | 1.213–3.342 | 0.007 |
| ER status (positive vs. negative) | 1.017 | 0.567–1.825 | 0.954 |
| PR status (positive vs. negative) | 0.799 | 0.477–1.338 | 0.393 |
| HER-2 status (positive vs. negative vs. uncertain) | 0.517 | 0.207–1.295 | 0.159 |
| Distant metastasis (presence vs. absence) | 3.642 | 1.770–7.495 | <0.001 |
| Tumor size (>2 vs. ≤2 cm) | 1.736 | 0.876–3.439 | 0.114 |
| Lymph node metastasis (presence vs. absence) | 1.676 | 0.970–2.894 | 0.064 |
| Stage (III–IV vs. I–II) | 2.277 | 1.363–3.803 | 0.002 |
HR, hazard ratio; 95% CI, 95% confidence interval; RGS2, regulator of G protein signaling 2; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2.
Table IV.
Univariate analysis for RGS2 and clinical characteristics in the Chongqing Medical University cohort.
| Characteristic | HR | 95% CI | P-value |
|---|---|---|---|
| RGS2 expression (low vs. high) | 0.337 | 0.130–0.874 | 0.025 |
| Age (≥60 vs. <60 years) | 4.036 | 1.777–9.169 | 0.001 |
| Histological gradea (grade 3 vs. 2 vs. 1) | 0.816 | 0.521–1.278 | 0.375 |
| ER status (positive vs. negative) | 0.989 | 0.470–2.080 | 0.977 |
| PR status (positive vs. negative) | 0.944 | 0.457–1.949 | 0.877 |
| HER-2 status (positive vs. negative vs. uncertain) | 0.835 | 0.464–1.503 | 0.547 |
| Ki-67 (≥14 vs. <14 %) | 0.544 | 0.274–1.080 | 0.082 |
| Distant metastasis (presence vs. absence) | 56.107 | 8.760–359.364 | <0.001 |
| Tumor size (>2 vs. ≤2 cm) | 2.276 | 1.119–4.629 | 0.023 |
| Lymph node metastasis (presence vs. absence) | 5.898 | 2.427–14.332 | <0.001 |
| Stage (III–IV vs. I–II) | 8.547 | 4.123–17.718 | <0.001 |
Histological grading was performed according to the World Health Organization Classification of Tumors of the Breast 2012 (4th edition). HR, hazard ratio; CI, confidence interval; RGS2, regulator of G protein signaling 2; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2.
Table V.
Multivariate Cox regression analysis for RGS2 and clinical characteristics in The Cancer Genome Atlas cohort.
| Characteristic | HR | 95% CI | P-value |
|---|---|---|---|
| Age (≥60 vs. <60 years) | 2.073 | 1.013–4.243 | 0.046 |
| RGS2 expression (low vs. high) | 0.435 | 0.196–0.965 | 0.041 |
| Stage (III–IV vs. I–II) | 2.938 | 1.390–6.208 | 0.005 |
| Distant metastasis (presence vs. absence) | 0.831 | 0.266–2.597 | 0.750 |
HR, hazard ratio; CI, confidence interval; RGS2, regulator of G protein signaling 2.
Table VI.
Multivariable Cox regression analysis for RGS2 and clinical characteristics in the Chongqing Medical University cohort.
| Characteristic | HR | 95% CI | P-value |
|---|---|---|---|
| Age (≥60 vs. <60 years) | 5.832 | 2.149–15.830 | 0.001 |
| RGS2 expression (low vs. high) | 4.602 | 1.467–14.432 | 0.009 |
| Lymph node metastasis (presence vs. absence) | 3.993 | 1.406–11.340 | 0.009 |
| Stage (III–IV vs. I–II) | 5.021 | 2.201–11.454 | <0.001 |
| Distant metastasis (presence vs. absence) | 7.517 | 0.773–73.114 | 0.082 |
| Tumor size (>2 vs. ≤2 cm) | 1.495 | 0.686–3.261 | 0.312 |
HR, hazard ratio; CI, confidence interval; RGS2, regulator of G protein signaling 2.
GSEA of RGS2 mRNA expression in TCGA microarray
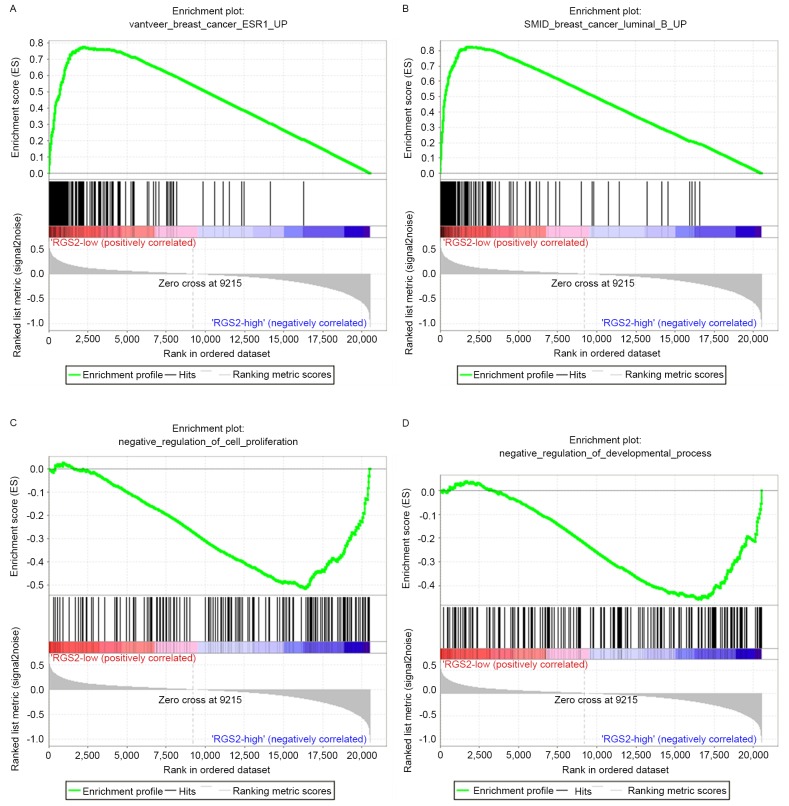
To investigate the biological function of RGS2, TCGA microarray data was analyzed using GSEA software. The C2 curated gene set results indicated that the significant gene set enrichment in the RGS2 low expression group contained ‘VANTVEER BREAST CANCER ESR1 UP’ (enrichment score, 1.8211; nominal P=0.002; false discovery rate, 0.1435; Fig. 4A) and ‘SMID BREAST CANCER LUMINAL B UP’ (enrichment score, 1.8208; nominal P<0.001; false discovery rate, 0.1295; Fig. 4B). C5 GO gene set results demonstrated that the significant gene set enrichment in the RGS2 overexpression group included ‘NEGATIVE REGULATION OF CELL PROLIFERATION’ (enrichment score, −1.8292; nominal P<0.001; false discovery rate, 0.2224; Fig. 4C) and ‘NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS’ (enrichment score, −1.8096; nominal P<0.001; false discovery rate, 0.2326; Fig. 4D).
Figure 4.
Gene set enrichment analysis of RGS2 expression in The Cancer Genome Atlas cohort. (A) Genes associated with upregulation of estrogen receptor 1 expression in breast cancer were significantly associated with low RGS2 expression. (B) Genes associated with upregulation in luminal B breast cancer were significantly associated with low RGS2 expression. (C) Genes associated with the negative regulation of cell proliferation were significantly associated with high RGS2 expression. (D) Genes associated with negative regulation of developmental process were significantly associated with high RGS2 expression. RGS2, regulator of G protein signaling 2.
Discussion
The present study was a large-sample comprehensive investigation into protein and mRNA levels, laboratory experiments and data mining; its workflow may become a trend of large-sample retrospective studies. The present study demonstrated that the RGS2 expression level was significantly downregulated in tumor tissues of BIC-NST compared with adjacent normal tissues. Although Smalley et al (14) revealed that RGS2 is overexpressed in the majority of breast cancer cases, no comparison was made with normal tissues. Additionally, the aforementioned study also demonstrated that RGS2 expression was lower in the breast cancer cell line compared with that in the normal cells. The results of the in vitro aspect of this study validated the results of the present study. Lyu et al (15) demonstrated that RGS2 expression was downregulated in breast cancer MCF-7 cells compared with normal mammary epithelium MCF-10A cells. The results of the present study were consistent with the results of Lyu et al (15) on the one hand; on the other hand, the present study expands these results from the cellular level to the level of human tissue, promoting the translation of the study from the laboratory bench to the clinic.
At the mRNA and protein levels, different results were obtained regarding the association between the clinicopathological features and RGS2 expression. There are a number of processes involved in the translation of mRNA to protein, including splicing, cleavage and modification. Thus, the expression of the gene is not always entirely consistent with that of the protein. However, RGS2 mRNA and protein expression was significantly downregulated in the ER-positive and PR-positive groups. GSEA revealed that the gene set enrichment in the RGS2 low-expression group was associated with estrogen receptor 1 and the luminal B subtype of breast cancer (19). According to the PAM50 Gene Expression Assay (20), the luminal B subtype of breast cancer is an ER-positive breast cancer. Therefore, the results of GSEA supported the results of the present study with regards to biological function. ER acts as a nuclear transcription factor when activated by estrogenic hormones and promotes the growth of the normal mammary epithelium (21). ER-positive cells in invasive breast cancer are frequently over-proliferative, resulting in harm to patients (22). The expression of PR is regulated by ER, so PR expression status is used to assess whether the estrogen-ER pathway is intact and functional (21). We hypothesize that the low expression of RGS2 may be a potential factor in the development of BIC-NST. To the best of our knowledge, the present study is the first to reveal that the low expression of RGS2 is associated with the positive status of hormone receptors in BIC-NST.
The Kaplan-Meier survival curve and Cox regression analysis demonstrated that low expression of RGS2 was significantly associated with the poor prognosis of patients with BIC-NST, and that it represents an independent risk factor. Through GSEA, the overexpression of RGS2 was found to be associated with the negative regulation of a number of biological processes. In the present study, the expression of RGS2 was negatively associated with pathological grade. Previous research revealed that there is an association between high pathological grade and poor survival outcome in patients with BIC-NST (23,24); this indirectly indicates that the low expression of RGS2 was associated with unfavorable prognosis in patients with BIC-NST. However, as TCGA data does not contain information regarding pathological grade, there was no way to validate the association between the expression of RGS2 mRNA and pathological grade in patients with BIC-NST. Taken together, these results indicate that RGS2 serves a role in the suppression of oncogene expression. To the best of our knowledge, the present study is also the first to report that low RGS2 expression was associated with poor outcome in patients with BIC-NST.
There are several limitations to the present study. The mechanism that underlies the association between the expression of RGS2 and the hormone receptors remains unknown. The subject of the present study focused solely on BIC-NST, among the various types of invasive breast cancer. This is the largest group and is thus the most likely to be experienced by patients; however, other types of invasive breast cancer exhibit different pathological features and gene profiles (25–29). Thus, the effect of RGS2 in invasive breast cancer requires further investigation.
In summary, the expression of RGS2 is under-regulated in patients with BIC-NST, and its low expression is closely associated with positive hormone receptor status. The low expression of RGS2 is predictive of poor prognosis in patients with BIC-NST, and is an independent risk factor. The results of the present study indicate that RGS2 may be a tumor suppressor gene in BIC-NST. Further study is required to investigate the precise mechanistic pathway underlying the effect of RGS2 in BIC-NST and to expand the study of RGS2 to other subtypes of invasive breast cancer.
Acknowledgements
The present study was funded by the Key Project of Chongqing Municipal Health Bureau, China (grant no. 2012-1-79).
Glossary
Abbreviations
- RGS2
regulator of G protein signaling 2
- BIC-NST
breast invasive carcinoma of no special type
- IHC
immunohistochemistry
- GSEA
gene set enrichment analysis
References
- 1.Early Breast Cancer Trialists' Collaborative Group (EBCTCG), corp-author Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Gianni L, Pienkowski T, Im YH, Tseng LM, Liu MC, Lluch A, Starosławska E, de la Haba-Rodriguez J, Im SA, Pedrini JL, et al. 5-year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): A multicentre, open-label, phase 2 randomised trial. Lancet Oncol. 2016;17:791–800. doi: 10.1016/S1470-2045(16)00163-7. [DOI] [PubMed] [Google Scholar]
- 3.Baum M. Modern concepts of the natural history of breast cancer: A guide to the design and publication of trials of the treatment of breast cancer. Eur J Cancer. 2013;49:60–64. doi: 10.1016/j.ejca.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Clarke R, Leonessa F, Trock B. Multidrug resistance/P-glycoprotein and breast cancer: Review and meta-analysis. Semin Oncol. 2005;32(Suppl 7):S9–S15. doi: 10.1053/j.seminoncol.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 6.Heximer SP, Lim H, Bernard JL, Blumer KJ. Mechanisms governing subcellular localization and function of human RGS2. J Biol Chem. 2001;276:14195–14203. doi: 10.1074/jbc.M009942200. [DOI] [PubMed] [Google Scholar]
- 7.Bosch DE, Zielinski T, Lowery RG, Siderovski DP. Evaluating modulators of ‘regulator of G-protein signaling’ (RGS) proteins. Curr Protoc Pharmacol: Chapter. 2012;2:Unit 2.8. doi: 10.1002/0471141755.ph0208s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen CH, Ming H, Zhao P, Hugendubler L, Gros R, Kimball SR, Chidiac P. Translational control by RGS2. J Cell Biol. 2009;186:755–765. doi: 10.1083/jcb.200811058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao X, Qin J, Xie Y, Khan O, Dowd F, Scofield M, Lin MF, Tu Y. Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene. 2006;25:3719–3734. doi: 10.1038/sj.onc.1209408. [DOI] [PubMed] [Google Scholar]
- 10.Hurst JH, Mendpara N, Hooks SB. Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cell Mol Biol Lett. 2009;14:153–174. doi: 10.2478/s11658-008-0040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, Hollmen J, Räty R, Aalto Y, Nagy B, Elonen E, Kere J, Mannila H, Franssila K, Knuutila S. Investigatory and analytical approaches to differential gene expression profiling in mantle cell lymphoma. Br J Haematol. 2002;119:905–915. doi: 10.1046/j.1365-2141.2002.03931.x. [DOI] [PubMed] [Google Scholar]
- 12.Schwable J, Choudhary C, Thiede C, Tickenbrock L, Sargin B, Steur C, Rehage M, Rudat A, Brandts C, Berdel WE, et al. RGS2 is an important target gene of Flt3-ITD mutations in AML and functions in myeloid differentiation and leukemic transformation. Blood. 2005;105:2107–2114. doi: 10.1182/blood-2004-03-0940. [DOI] [PubMed] [Google Scholar]
- 13.Kannangai R, Vivekanandan P, Martinez-Murillo F, Choti M, Torbenson M. Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3 and xenobiotic degradation pathways. Hum Pathol. 2007;38:639–644. doi: 10.1016/j.humpath.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Smalley MJ, Iravani M, Leao M, Grigoriadis A, Kendrick H, Dexter T, Fenwick K, Regan JL, Britt K, McDonald S, et al. Regulator of G-protein signalling 2 mRNA is differentially expressed in mammary epithelial subpopulations and over-expressed in the majority of breast cancers. Breast Cancer Res. 2007;9:R85. doi: 10.1186/bcr1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyu JH, Park DW, Huang B, Kang SH, Lee SJ, Lee C, Bae YS, Lee JG, Baek SH. RGS2 suppresses breast cancer cell growth via a MCPIP1-dependent pathway. J Cell Biochem. 2015;116:260–267. doi: 10.1002/jcb.24964. [DOI] [PubMed] [Google Scholar]
- 16.Ellis IO, Collins L, Ichihara S, et al. Invasive carcinoma of no special type. In: Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. WHO classification of tumours of the breast. Fourth. IARC 34; Lyon: 2012. [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles; Proc Natl Acad Sci USA; 2005; pp. 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colditz G, Chia KS. WHO classification of tumours of the breast. Fourth. IARC 23; Lyon: 2012. Invasive breast carcinoma: Introduction and general features. [Google Scholar]
- 19.Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H, Aimi J, Derynck MK, Chen M, Chan IT, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579. doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke RB. Steroid receptors and proliferation in the human breast. Steroids. 2003;68:789–794. doi: 10.1016/S0039-128X(03)00122-3. [DOI] [PubMed] [Google Scholar]
- 22.Schiff R, Osborne CK, Fuqua SAW. Clinical aspects of estrogen and prog-esterone receptors. Wolters Kluwer Lippincott Williams & Wilkins; Philadelphia: 2010. [Google Scholar]
- 23.Rakha EA, El-Sayed ME, Lee AH, Elston CW, Grainge MJ, Hodi Z, Blamey RW, Ellis IO. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J Clin Oncol. 2008;26:3153–3158. doi: 10.1200/JCO.2007.15.5986. [DOI] [PubMed] [Google Scholar]
- 24.Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, et al. Breast cancer prognostic classification in the molecular era: The role of histological grade. Breast Cancer Res. 2010;12:207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleton-Jansen AM. E-cadherin and loss of heterozygosity at chromosome 16 in breast carcinogenesis: Different genetic pathways in ductal and lobular breast cancer? Breast Cancer Res. 2002;4:5–8. doi: 10.1186/bcr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lacroix-Triki M, Suarez PH, MacKay A, Lambros MB, Natrajan R, Savage K, Geyer FC, Weigelt B, Ashworth A, Reis-Filho JS. Mucinous carcinoma of the breast is genomically distinct from invasive ductal carcinomas of no special type. J Pathol. 2010;222:282–298. doi: 10.1002/path.2763. [DOI] [PubMed] [Google Scholar]
- 27.Marchiò C, Iravani M, Natrajan R, Lambros MB, Geyer FC, Savage K, Parry S, Tamber N, Fenwick K, Mackay A, et al. Mixed micropapillary-ductal carcinomas of the breast: A genomic and immunohistochemical analysis of morphologically distinct components. J Pathol. 2009;218:301–315. doi: 10.1002/path.2572. [DOI] [PubMed] [Google Scholar]
- 28.Persson M, Andrén Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck; Proc Natl Acad Sci USA; 2009; pp. 18740–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer cell. 2002;2:367–376. doi: 10.1016/S1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]