ABSTRACT
Inflammasomes are multimeric protein complexes that typically comprise a sensor, an adaptor and the zymogen procaspase-1. An inflammasome assembles in response to a diverse range of pathogen-associated or danger-associated molecular patterns (PAMPs or DAMPs). The inflammasome platform leads to activation of caspase-1 through proximity-induced self-cleavage, which further induces maturation of interleukins 1β and 18 (IL-1β and IL-18) through proteolytic cleavage of pro-IL-1β and pro-IL-18. Activated caspase-1 also cleaves gasdermin D, which leads to a particular form of cell death called pyroptosis. Mutations in genes that encode inflammasome components are associated with many inflammatory disorders, and studies in the past decade have highlighted the importance of appropriate activation of the inflammasome in homeostasis and disease pathogenesis. Therefore, much attention is being paid to uncover the modulators and regulators of inflammasome assembly and pyroptosis. This Cell Science at a Glance article and accompanying poster outlines the concepts in the activation of inflammasome sensors and assembly of the inflammasome platform. We also discuss recent insights into the mechanisms of regulation of inflammasome activity and the induction of cell death by pyroptosis.
KEY WORDS: Inflammasome, Pyroptosis, Cell death, IL-1 cytokines
Summary: This article and accompanying poster provides an outline of the concepts in the assembly of inflammasomes and cell death by pyroptosis.
Introduction
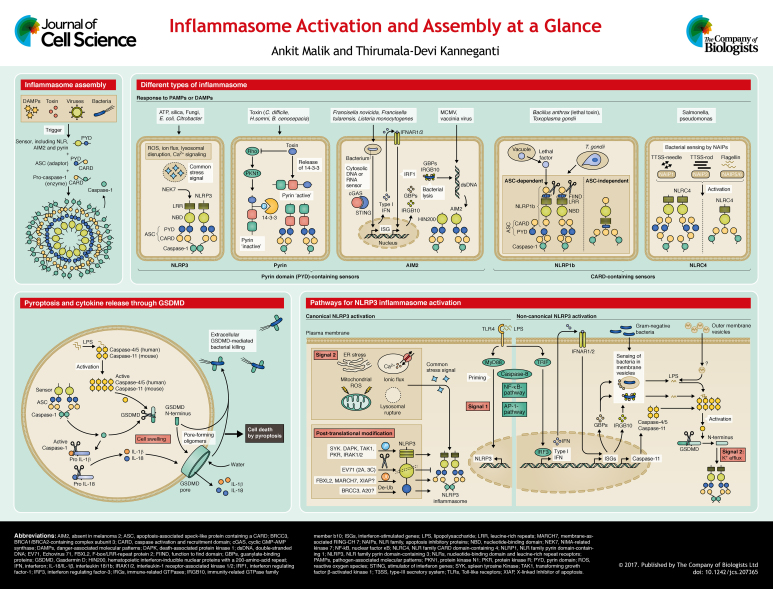
Sensors of PAMPs and DAMPs in the cytoplasm include nucleotide-binding oligomerization domain (NOD)- and leucine-rich repeat (LRR)-containing receptors (NLRs), the absent in melanoma-2 (AIM2)-like receptors (ALRs) and proteins that contain a tripartite motif (TRIM), including pyrin. Among these sensors, members of the evolutionary conserved NLRs, together with AIM2 and pyrin, can assemble into a multimeric protein complex that is called the inflammasome (see poster). Inflammasomes are typically composed of a sensor, the adaptor molecule ASC [apoptosis-associated speck-like protein containing a caspase-activation and recruitment domain (CARD); also known as PYCARD] and the cysteine protease procaspase-1. Upon detection of specific stimuli, the sensor, which includes NLRs, AIM2 or pyrin, recruits ASC to form a multimeric complex that is referred to as ‘speck’. Oligomerized or ‘nucleated’ ASC recruits procaspase-1 into the complex, which then is converted into bioactive caspase-1 by proximity-induced self-cleavage. Following this, the active caspase-1 subunits p20 and p10 cleave pro-IL-1β and pro-IL-18 to yield bioactive cytokines and activate pore-forming gasdermin D (GSDMD) to induce a form of cell death called ‘pyroptosis’ (see poster and Box 1). Inflammasome activation serves critical functions in pathogen defense; it helps to remove damaged and transformed host cells and stimulates an adaptive immune response. Conversely, aberrant inflammasome activation is linked to many inflammatory disorders, infectious diseases and cancer. Therefore, inflammasome activation is a tightly regulated event that encompasses many molecular and cellular signals. Here, we summarize the mechanisms that regulate the activation of the inflammasome sensors and the assembly of the inflammasome platform. We also discuss recent insights on the induction of pyroptosis and open questions in inflammasome biology.
Box 1. Pyroptosis.
Pyroptosis is an inflammatory form of cell death that is induced by inflammasome activation; it is characterized by swelling of the cell that is followed by lysis and release of intracellular contents. Loss of osmotic potential had been previously implicated as a critical event in cell death that is induced by pyroptosis (Fink et al., 2008; Fink and Cookson, 2006), but how inflammasome activation induced loss of osmotic potential was not known. Gasdermin D (GSDMD) was recently identified as the missing link between cell death and caspase-1 and/or caspase-11 activation (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015b). The N-terminal domain of GSDMD exhibits robust and specific binding to membrane lipids, phosphoinositides and cardiolipin. It is held in an inhibited form by its C-terminal domain (Aglietti et al., 2016; Ding et al., 2016; Liu et al., 2016; Sborgi et al., 2016). Pyroptotic caspases 1 and 11 (caspase-4 and caspase-5 in humans) cleave GSDMD at D276 in the linker region, thereby relieving the intramolecular inhibitory effect of the C-terminal domain (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015b). Following the binding to membrane lipids, this N-terminal region of GSDMD (GSDMD-N) oligomerizes to form pores with an inner diameter of 10 to 14 nm (see poster) (Ding et al., 2016). The formation of these pores disrupts the osmotic potential of the cell, thereby causing its swelling and lysis. Therefore, whereas IL-1β is processed normally in GSDMD-deficient cells, its release is abrogated. Expression of the N-terminal fragment is also sufficient for killing bacterial cells (He et al., 2015; Shi et al., 2015b). GSDMD belongs to the gasdermin family that shares 45% sequence homology amongst its six members. Except for autosomal recessive deafness type 59 protein (DFNB59), all other GSDM proteins have a GSDMD-like two-domain structure. In addition, similar to GSDMD, their N-terminus can induce pyroptosis (Ding et al., 2016; Shi et al., 2015b). However, unlike GSDMD, other members lack the pyroptotic caspase cleavage site (Shi et al., 2015b), which underscores its importance in pyroptosis. Although GSDMD can be cleaved by caspase-3 (Wang et al., 2017), if and how other GSDM molecules are also cleaved to release the pore-forming N-terminus is not yet known. It is also not clear why pyroptosis follows inflammasome activation in macrophages (Fink et al., 2008), dendritic cells (Edgeworth et al., 2002) and epithelial cells (Sellin et al., 2014), but not in neutrophils or monocytes (Chen et al., 2014; Gaidt et al., 2016; Miao et al., 2010a). Whether these cell type-specific differences are due to differences in GSDMD expression and regulation is another unanswered question.
Inflammasome assembly
All members of the NLR family of proteins contain a central nucleotide-binding domain (NBD), and most also have a variable N-terminal domain and a C-terminal LRR domain. Based on the presence of an N-terminal pyrin domain (PYD) or CARD, this family is further divided into NLRP or NLRC receptors. The human and mouse genomes encode 22 and 34 NLRs, respectively (Harton et al., 2002). Among them, NLRP1, NLRP3 and NLRC4 are the NLRs capable of inducing the formation of the inflammasome as a platform to activate caspase-1. NLRP12, NLRP6 and NLRP9b have also been suggested to form inflammasomes, but their role as inflammasome sensors is not well defined (Anand et al., 2012; Elinav et al., 2011; Mamantopoulos et al., 2017; Vladimer et al., 2012; Zhu et al., 2017).
Inflammasome assembly requires homotypic CARD–CARD or PYD–PYD interactions between its components, and both the PYD and CARD domains can induce oligomerization, which is the basis of inflammasome assembly (Cai et al., 2014; Lu et al., 2014; Sborgi et al., 2015). When the ligand is detected, the sensor is relieved from an inhibitory state and oligomerizes or nucleates ASCs by inducing homotypic interactions between their PYDs. Next, ASC recruits pro-caspase-1 through interactions between its CARD domain with that of the caspase. The resultant multimeric inflammasome complex contains the sensor, the adaptor and the enzyme at increasing stoichiometric ratios (Lu et al., 2014) (see poster). The assembly of NLRP3, AIM2 and pyrin inflammasomes is strictly dependent on the adaptor ASC. In contrast, NLRP1 and NLRC4 possess a CARD domain and can recruit caspase-1 directly (Jin et al., 2013a; Nour et al., 2009; Ponomareva et al., 2013). Therefore, NLRP1 and NLRC4 can induce inflammasome assembly and pyroptosis independently of ASC. However, ASC recruitment into the complex still promotes the efficient processing of IL-1β and IL-18 (Broz et al., 2010; Guey et al., 2014; Mariathasan et al., 2004; Van Opdenbosch et al., 2014). As inflammasome assembly requires homotypic CARD–CARD and PYD–PYD interactions, certain PYD-only proteins (POPs) and CARD-only proteins (COPs) can act as dominant-negative regulators of inflammasome assembly (Matusiak et al., 2015).
Inflammasome sensors
NLRP1
The first sensor that was identified as being capable of forming the inflammasome was NLRP1 (Martinon et al., 2002). Human NLRP1 contains a PYD, NBD and LRR domain, a ‘function-to find’ domain (FIIND) and a C-terminal CARD. However, the mouse genome encodes for three NLRP1 paralogs (a–c), all of which lack the PYD. NLRP1b is activated by cleavage at its N-terminal and FIIND domains by lethal factor (LF), a component of the anthrax lethal toxin (LeTx) produced by Bacillus anthracis (Boyden and Dietrich, 2006; Chavarría-Smith and Vance, 2013; Finger et al., 2012; Frew et al., 2012). This NLRP1b activation by LeTx has been shown to protect mice from Bacillus anthracis infection (Moayeri et al., 2010; Terra et al., 2010). Single-nucleotide polymorphisms (SNPs) in NLRP1 are associated with congenital toxoplasmosis, and, furthermore, NLRP1 activation is required to limit Toxoplasma infectivity (Witola et al., 2011). Although the NLRP1b inflammasome is activated by T. gondii, no cleavage product for NLRP1b is detectable in response to the parasitic stimuli (Cavailles et al., 2014; Ewald et al., 2014; Gorfu et al., 2014), which suggests that NLRP1b proteolysis, or cleavage, may not be a global mechanism for its inflammasome assembly. As mentioned above, NLRP1 can also induce ASC-independent inflammasome assembly (see poster).
Whereas the specific trigger(s) for NLRP1a-mediated inflammasomes are unknown, mice homozygous for a dominant-negative Q593P mutation in NLRP1a develop an autoinflammatory disease that is dependent on caspase-1 and IL-1β. These mice (Nlrp1aQ593P/Q593P) have aberrant myelopoiesis and display an increased susceptibility to chemoablation and infection with lymphocytic choriomenigitis virus (LCMV) (Masters et al., 2012); in contrast, Nlrp1a−/− mice demonstrate enhanced hematopoietic recovery in response to the same stresses. However, how NLRP1 affects these processes is largely unknown.
NLRP3
NLRP3 is composed of an N-terminal PYD, a central NBD and a C-terminal LRR domain. Mutations in NLRP3 have been observed in autoinflammatory disorders such as cryopyrin-associated periodic syndromes (CAPS) that are characterized by episodic skin rashes and fever (Feldmann et al., 2002; Hoffman et al., 2001; Neven et al., 2004). NLRP3 was eventually discovered to function as a NLR that forms inflammasomes and senses a vast array of infectious and endogenous DAMPs. These include microbial cell wall components, nucleic acids, pore-forming toxins, environmental crystalline agents such as silica and endogenous molecules, including ATP and uric acid crystals (Cassel et al., 2008; Cruz et al., 2007; Dostert et al., 2008; Hornung et al., 2008; Kanneganti et al., 2006a,b). As the NLRP3 inflammasome assembles in response to a vast range of DAMPs, it is likely that it senses a common cellular distress signal that is induced by these molecules, instead of undergoing a direct interaction with all of these triggers. Changes in cell volume, rupture of lysosomes, production of reactive oxygen species (ROS), K+ efflux and Ca2+ signaling have all been proposed as the distress signals that are sensed by NLRP3 (Compan et al., 2012; Halle et al., 2008; Kanneganti et al., 2007; Munoz-Planillo et al., 2013; Schorn et al., 2011; Zhou et al., 2011).
Two distinct steps – priming (denoted signal 1 on poster) and inflammasome assembly (signal 2 on poster) – are required to activate the NLRP3 inflammasome (see the poster section on pathways for NLRP3 inflammasome activation). Priming involves activation of myeloid differentiation primary response protein (MyD88) or of other nuclear factor (NF)-κB- or activator protein 1 (AP-1)-activating pathways, which upregulate the expression of Nlrp3 and other inflammasome components. Apoptotic caspase-8 has also been implicated in inflammasome priming (Gurung et al., 2014; Lemmers et al., 2007), its assembly (Gringhuis et al., 2012; Gurung et al., 2014) and IL-1β processing (Maelfait et al., 2008). In human monocytes, an axis comprising toll-like receptor 4 (TLR4), the adaptor TIR-domain-containing adapter-inducing interferon-β (TRIF; also known as TICAM1), receptor-interacting serine/threonine-protein kinase 1 (RIPK1), FAS-associated death domain protein (FADD) and caspase-8 has been described for NLRP3 inflammasome activation that is independent of K+ efflux and ASC (Gaidt et al., 2016). In addition to promoting the activity of the NLRP3 inflammasome, caspase-8 can act redundantly with caspase-1 to process IL-1β in a model of osteomyelitis (Gurung et al., 2016; Lukens et al., 2014). Whereas this pathway suggests an integration of inflammasome activation with extracellular ligand recognition signaling, further studies are required to define the molecular basis of the assembly of the NLRP3 inflammasome.
Caspase-11 and the NLRP3 inflammasome
Caspase-11 (which has two orthologs in humans, caspase-4 and caspase-5, and only the ortholog caspase-4 in mice) is an inflammatory caspase that can bind to caspase-1 (Wang et al., 1998). The genes encoding caspase-1 (Casp1) and caspase-11 display linkage disequilibrium, and the caspase-11 gene (Casp4) was lost when a primary knockout for caspase-1 was generated (in a 129 mouse strain background) (Kayagaki et al., 2011). Therefore, the functional significance of caspase-11 was initially assigned to caspase-1. Caspase-11 was eventually identified to have a non-redundant role in the NLRP3 inflammasome activation and pyroptosis after infection of mice with Gram-negative bacteria E. coli and Citrobacter rodentium (Kayagaki et al., 2011; Rathinam et al., 2012). Further studies described caspase-11 as a sensor of endotoxin [i.e. lipopolysaccharide (LPS)] in the cytoplasm and a critical regulator of susceptibility to endotoxemia (Kayagaki et al., 2011). Caspase-11 directly binds to the penta- and hexa-acylated lipid A-component of LPS (Hagar et al., 2013) through its CARD domain (Shi et al., 2014). This interaction of LPS and caspase-11 in the cytoplasm is independent of the extracellular detection of LPS, which is mediated by TLR4, MD2 (also known as LY96) and CD14 (Hagar et al., 2013; Kayagaki et al., 2013). In contrast to the upregulation of NLRP3 and ASC by MyD88 and NF-κB-activating signals, expression of caspase-11 is upregulated by TRIF and interferon (IFN)-mediated signaling (Gurung et al., 2012; Rathinam et al., 2012).
The mechanism by which caspase-11 promotes the activation of NLRP3 is, however, unclear. Whereas caspase-11 can bind to caspase-1 (Wang et al., 1998), it is dispensable for inflammasome assembly following the activation of NLRP3 with canonical triggers, such as silica and uric acid crystals. One hypothesis is that caspase-11 instigates an effector molecule that induces NLRP3 inflammasome assembly. For example, caspase-11 can cleave pannexin-1, leading to a drop in intracellular K+ levels, which then induces activation of the NLRP3 inflammasome (Rühl and Broz, 2015; Schmid-Burgk et al., 2015; Yang et al., 2015). However, K+ efflux is also associated with NLRP1b activation (Pétrilli et al., 2007). Therefore, a specific link between caspase-11 and the NLRP3 inflammasome is currently unknown. The human genome encodes two orthologs of caspase-11: caspase-4 and caspase-5, either of which can functionally provide the caspase-11 function in murine cells (Shi et al., 2014). However, caspase-4 and caspase-5 may not be redundant, as both are required for cell death and IL-1β release following Salmonella infection. In contrast, only caspase-4 is required for inflammasome activation by cytosolic LPS (Baker et al., 2015).
The mechanism by which LPS is delivered into the cytoplasm for recognition by caspase-11 is one of the major questions in the field. A recent study demonstrated that LPS that is contained within the bacterial outer membrane vesicles (OMVs) is detected by caspase-11 (Vanaja et al., 2016) (see poster). However, how OMV-associated LPS is transported into the cytoplasm is unclear. Another study identified a role for interferon-induced molecules in the liberation of LPS from bacteria. The signal transducer and activator of transcription 1 (STAT1)–IRF9 signaling axis downstream of IFNs upregulates the transcription factor IRF1, which in turn promotes the expression of guanylate-binding proteins (GBPs) and immunity-related GTPase family member B10 (IRGB10; UniProt U5NFV2) (Man et al., 2016). These proteins target intracellular bacteria for lysis and enhance the release of LPS into the cytosol, which increases its detection by caspase-11 and induces subsequent activation of the NLRP3 inflammasome (Man et al., 2016; Pilla et al., 2014) (see poster). Therefore, IFN signaling promotes both caspase-11 upregulation and liberation of LPS into the cytoplasm. Consistent with its role as a cytosolic LPS sensor and an inducer of the NLRP3 inflammasome, caspase-11 is particularly important in the defense against intracellular bacterial infections (Aachoui et al., 2013; Gurung et al., 2012).
In addition to the above mechanisms of activation, the NLRP3 inflammasome is also regulated by many different types of post-translational modifications and several other cellular mechanisms (Box 2).
Box 2. Regulation of the NLRP3 inflammasome.
Post-translational modifications are integral to the assembly of the NLRP3 inflammasome, including deubiquitylation of NLRP3 by BRCA1/BRCA2-containing complex subunit 3 (BRCC3) (Py et al., 2013), dephosphorylation of PYD of NLRP3 by phosphatase 2A (Stutz et al., 2017), linear ubiquitin chain assembly complex (LUBAC)-induced ubiquitylation of ASC (Rodgers et al., 2014) and the phosphorylation of ASC by Syk kinase (Hara et al., 2013; Lin et al., 2015). Interleukin-1 receptor-associated kinase 1 (IRAK1) was also identified to have a role in the transcription-independent, early activation of the NLRP3 inflammasome (Fernandes-Alnemri et al., 2013; Lin et al., 2014). Other kinases such as death-associated protein kinase (DAPK) (Chuang et al., 2011) and double stranded RNA-dependent protein kinase (PKR) (Lu et al., 2012) promote NLRP3 assembly through unknown mechanisms. On the other hand, ubiquitylation of NLRP3 by the E3 ligases FBXL2 (Han et al., 2015), MARCH7 (Yan et al., 2015) and XIAP (Yabal et al., 2014) restricts the level and activation of the NLRP3 inflammasome. NLRP3 is also targeted and inhibited by various host factors including Enterovirus 71 (EV71) proteases 2A and 3C that cleave NLRP3 and dampen the innate immune response (see poster). Inflammasome activation is tightly regulated by multiple cellular mechanisms. Under homeostatic conditions, ASC localizes in the cytosol and the nucleus, whereas NLRP3 is localized on the membrane of the endoplasmic reticulum (ER) (Zhou et al., 2011). Therefore, changes in subcellular localization of NLRP3 and ASC are essential for inflammasome nucleation. Dynein-mediated transport of the mitochondria towards the ER has been proposed as a mechanism for bringing ASC and NLRP3 into close proximity (Misawa et al., 2013). Recent studies have also identified the serine-threonine kinase NEK7, a protein involved in mitotic spindle formation and cytokinesis, as a critical regulator of the assembly of NLRP3 inflammasome (He et al., 2016; Schmid-Burgk et al., 2016; Shi et al., 2015a). NEK7 interacts with the LRR domain of NLRP3 independently of its kinase activity and promotes nucleation (He et al., 2016; Shi et al., 2015a). During cell division, NEK7 is localized on mitotic spindles, which prevents NLRP3 inflammasome assembly (Shi et al., 2015a). It can therefore be hypothesized that NEK7 functions to safeguard dividing cells from the activation of the NLRP3 inflammasome because during cell division, the cytoplasm is enriched with endogenous ligands that could be misrepresented as stress signals. Autoinflammatory disorders associated with NLRP3 often involve mutations in or around the NBD (Dodé et al., 2002; Feldmann et al., 2002; Hoffman et al., 2001; Neven et al., 2004; Touitou et al., 2004) and display NLRP3 auto-activation (Brydges et al., 2013, 2009; Meng et al., 2009; Mortimer et al., 2016), which suggests an auto-inhibitory function of the NBD in NLRP3 activation. NLRP3 inflammasome, therefore, has multiple levels of regulation including post-translational modifications and structural restriction. Understanding and employing strategies to modulate NLRP3 activation would likely be beneficial in multiple inflammatory disorders.
NLRC4
NLRC4 was characterized as an NLR capable of binding to and activating caspase-1 through its CARD-domain (Poyet et al., 2001). Subsequently, NLRC4 was identified as inducing inflammasome formation following Salmonella infection (Mariathasan et al., 2004). NLRC4 can be activated by the bacterial flagellin (Franchi et al., 2006; Miao et al., 2006; Molofsky et al., 2006; Ren et al., 2006) and components of the flagellin-associated secretion systems (Miao et al., 2010b; Yang et al., 2013; Zhao et al., 2011). However, NLRC4 is not a direct sensor of these ligands. Instead, NLR family apoptosis inhibitory proteins (NAIPs) are the sensors of NLRC4 ligands and are therefore critical for NLRC4 inflammasome activation. Whereas only a single NAIP has been identified in humans (Endrizzi et al., 2000), the mouse genome encodes seven NAIP proteins. Mouse NAIP1 and NAIP2 bind to the bacterial needle and inner rod proteins of the type 3 secretion system (T3SS), whereas NAIP5 and NAIP6 recognize flagellin (Kofoed and Vance, 2011; Rayamajhi et al., 2013; Yang et al., 2013; Zhao et al., 2011) (see poster). The sole human NAIP can sense both flagellin and the components of bacterial T3SS (Kortmann et al., 2015; Yang et al., 2013). The function of other mouse NAIPs is currently unknown.
Inflammasome assembly is initiated by a single activated NAIP molecule that provides a platform for NLRC4 self-oligomerization (Hu et al., 2015; Zhang et al., 2015). In the absence of a ligand, NLRC4 is maintained in an inactive state by its NBD and the winged helix domain (WHD) that stabilize its closed conformation, while the LRR domain provides steric hindrance to its oligomerization (Diebolder et al., 2015; Tenthorey et al., 2014). The importance of these auto-inhibitory mechanisms is underscored by the description of auto-inflammation and spontaneous colitis in humans and mouse models with gain-of-function mutations in NLRC4 (Canna et al., 2014; Kitamura et al., 2014; Romberg et al., 2014).
Unsurprisingly, NAIPs and NLRC4 are critical components of the host defense against flagellated bacteria. In addition to maturation of IL-1β and IL-18, NLRC4-mediated pyroptosis promotes shedding of infected epithelial cells, which help to control the pathogen load during Salmonella infection (Sellin et al., 2014). Apart from pyroptosis and maturation of IL-1β and IL-18, NLRC4 activation also affects other aspects of cell biology such as the release of eicosanoids, including prostaglandins and leukotrienes (von Moltke et al., 2012). Whether the function of NLRC4 in eicosanoid production can be extended to other inflammasomes and cell death pathways is currently unclear.
AIM2
AIM2 is a highly conserved member of the ALR family that contains an N-terminal PYD and a C-terminal hematopoietic interferon-inducible nuclear protein with a 200-amino-acid repeat (HIN200) domain (Cridland et al., 2012). The PYD and HIN200 domain, when they occur together, are referred to as the PYHIN domain. The ALR family has 14 members in mice and four in humans. Unlike other members of the ALR family, AIM2 lacks a nuclear localization domain and interacts with ASC through its PYD (Hornung et al., 2009).
Although AIM2 was initially seen as an IFN-inducible tumor suppressor, subsequent studies identified it as a cytosolic sensor of double-stranded (ds)DNA that can assemble into inflammasomes (Bürckstümmer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009). The AIM2 inflammasome assembles during viral and bacterial infections of, for example, vaccinia virus, mouse cytomegalovirus (MCMV) (Rathinam et al., 2010), Francisella tularensis (Fernandes-Alnemri et al., 2010; Jones et al., 2010) and Listeria monocytogenes (Kim et al., 2010; Rathinam et al., 2010) (see poster). In the absence of dsDNA, AIM2 exists in the cytoplasm in an auto-inhibitory state with the HIN200 domain bound to the PYD. Binding to dsDNA by the HIN200 domain relieves this auto-inhibition, which allows the PYD to undergo homotypic interaction with ASC (Jin et al., 2013b). The HIN200 domain of AIM2 binds to dsDNA independently of its sequence identity; however, it does require the DNA to be of a certain length (about 80 bp) (Jin et al., 2012; Li et al., 2014). Therefore, AIM2 inflammasome activation is induced by cytoplasmic dsDNA from both pathogenic and host sources.
Interferon signaling is required to induce the activation of AIM2 after infection with the bacterium Francisella, but not the DNA virus MCMV. Whereas MCMV actively releases its genomic DNA into the cytoplasm, cGAS-STING-mediated IFN signaling IFN signaling is required to induce the expression of GBPs and IRGB10, which mediate bacterial lysis and release of genomic DNA into the cytoplasm (Man et al., 2015a, 2016; Meunier et al., 2015) (see poster). Intriguingly, AIM2 deficiency actually results in increased type-I IFN production (Corrales et al., 2016; Fernandes-Alnemri et al., 2010; Hornung et al., 2009). Whether this is because of a direct inhibition of interferon-producing pathways by AIM2 is not clear. As AIM2 binds to dsDNA in a sequence-independent manner, it has also been implicated in recognition of host DNA during inflammatory diseases (Choubey, 2012; Dihlmann et al., 2014a; Dombrowski et al., 2011) and cancer (Dihlmann et al., 2014b; Ponomareva et al., 2013). However, the function of AIM2 as a tumor suppressor was found to be independent of its inflammasome activity, but instead dependent on its ability to regulate Akt kinase signaling and cellular proliferation (Man et al., 2015b; Wilson et al., 2015).
Other members of the ALR family can inhibit AIM2: p202 (also known as IFI202) lacks the PYD but has two HIN domains (HIN1 and HIN2). HIN1 can bind to and sequester dsDNA from AIM2, whereas HIN2 can bind to the HIN domain of AIM2 and inhibit inflammasome assembly (Roberts et al., 2009; Yin et al., 2013). In contrast, POP3 lacks a HIN domain, but does have a PYD through which it can bind to AIM2 and so block ASC binding and oligomerization, which blunts the immune response to DNA viruses (Khare et al., 2014). Furthermore, alternatively spliced isoforms of AIM2 that lack the PYD or HIN domain have been predicted (Choubey et al., 2010). These putative, truncated proteins may similarly inhibit AIM2 activation and associated immune responses.
Pyrin
Pyrin was first described as the protein associated with familial Mediterranean fevers (FMF), an autoinflammatory disorder characterized by episodic fever and joint inflammation (Bernot et al., 1998). Recently, pyrin was characterized as an inflammasome sensor (Gavrilin et al., 2012; Xu et al., 2014). In humans, pyrin is composed of an N-terminal PYD, central B-box and coiled-coil domain, and a C-terminal B30.2/SPRY domain. In mice, the C-terminal B30.2/SPRY domain is absent. A mouse model in which the human B30.2 domain was spliced into mouse pyrin to generate a mouse–human chimeric protein develops a severe autoinflammatory disorder (Chae et al., 2011). As evidence for pyrin forming an inflammasome, the autoinflammatory disorder in these mice could be rescued by deletion of caspase-1, ASC and IL-1β, but not NLRP3, caspase-8 or IL-1α (Chae et al., 2011; Sharma et al., 2017).
Recent studies have demonstrated that the pyrin inflammasome assembles upon the modification of cytoskeletal proteins. Toxins produced by various bacterial species, such as Clostridium difficile (TcdB), Vibrio parahemolyticus (VopS), Clostridium botulinum (C3), Burkholderia cenocepacia and Bordetella pertussis (PT), induce covalent modifications within the switch I region of Rho family members. These modifications include glycosylation, adenylylation and ADP-ribosylation and lead to assembly of the pyrin inflammasome (Xu et al., 2014). The modification of Rho was found to be essential for pyrin inflammasome activation, even though a direct interaction between Rho and pyrin was not detected (Dumas et al., 2014; Xu et al., 2014). Mice with an inactive mutant of the actin-associated WD repeat-containing protein 1 (Wdr1) also develop a pyrin- and IL-18-mediated autoimmune disorder (Kim et al., 2015). Furthermore, defects in the mevalonate kinase pathway (MVK), which is required for synthesis of geranyl pyrophosphate, a substrate involved in geranylgeranylation and function of small cellular GTPases, promotes pyrin inflammasome activation (Akula et al., 2016; Park et al., 2016). The pyrin inflammasome is inhibited by microtubule disruption in response to pyrin-activating stimuli (Gao et al., 2016; Park et al., 2016), although peripheral blood mononuclear cells (PBMCs) from human patients with FMF are recalcitrant to colchicine-mediated inhibition of IL-1β release (Van Gorp et al., 2016). Whereas it has been posited that microtubule disruption reverses the inactivation of Rho, the exact mechanism of colchicine-mediated pyrin inhibition is unknown. These findings suggest that pyrin senses a common stress signal induced by cytoskeletal modifications instead of interacting with the inducers directly, which is similar to the proposed way of action for NLRP3.
Similar to other inflammasomes, cellular machinery exists to prevent pyrin activation under resting conditions. In homeostasis, pyrin is phosphorylated at S242 and bound by 14-3-3 proteins (Masters et al., 2016). TcdB induces dephosphorylation at S242, which relieves the binding of 14-3-3 and allows inflammasome assembly (Akula et al., 2016; Gao et al., 2016; Masters et al., 2016; Park et al., 2016) (see poster). However, the molecular events that control the phosphorylation, dephosphorylation and subsequent nucleation events are unknown. Furthermore, whether auto-activating mutations that are associated with pyrin bypass this regulatory inhibition has also not been studied.
Conclusion and future directions
Inflammasomes play a critical role in health and disease. They are required for maturation and release of IL-1β, IL-18 and other cellular contents, removal of malignant or damaged cells and host defense from infectious agents. Inflammasome activation intersects with many critical cellular processes that include inflammatory signaling, metabolism, cell division and cell death pathways. Given their high inflammatory potential, many cellular processes are also devoted to the regulation of inflammasome activation. In spite of recent progress in elucidating these mechanisms, many fundamental questions still remain. For example, the exact ligand and mechanisms that activate NLRP3- and pyrin-mediated inflammasomes are not entirely known. Furthermore, the mechanisms that regulate sensing of DAMPs and/or PAMPs present within membrane vesicles by sensors that are localized in the cytoplasm are still unclear. Investigating these and other fundamental questions highlighted in this article will help to significantly improve our understanding of the inflammasome biology, and likely yield novel therapeutic options for the associated diseases and conditions.
Supplementary Material
Acknowledgements
We would like to thank Deepika Sharma and Teneema Kuriakose for helpful editing of the manuscript. We would like to apologize to our colleagues whose work could not be cited due to space limitations.
Footnotes
Funding
This work was supported by grants from the National Institutes of Health (AI101935, AI124346, AR056296 and CA163507) and the ALSAC to T.-D.K. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.207365.supplemental.
Conflict of interest
The authors declare no competing or financial interests.
References
- Aachoui Y., Leaf I. A., Hagar J. A., Fontana M. F., Campos C. G., Zak D. E., Tan M. H., Cotter P. A., Vance R. E., Aderem A. et al. (2013). Caspase-11 protects against bacteria that escape the vacuole. Science (New York, N.Y.) 339, 975-978. 10.1126/science.1230751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglietti R. A., Estevez A., Gupta A., Ramirez M. G., Liu P. S., Kayagaki N., Ciferri C., Dixit V. M. and Dueber E. C. (2016). GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. USA 113, 7858-7863. 10.1073/pnas.1607769113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akula M. K., Shi M., Jiang Z., Foster C. E., Miao D., Li A. S., Zhang X., Gavin R. M., Forde S. D., Germain G. et al. (2016). Control of the innate immune response by the mevalonate pathway. Nat. Immunol. 17, 922-929. 10.1038/ni.3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P. K., Malireddi R. K. S., Lukens J. R., Vogel P., Bertin J., Lamkanfi M. and Kanneganti T.-D. (2012). NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 488, 389-393. 10.1038/nature11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. J., Boucher D., Bierschenk D., Tebartz C., Whitney P. G., D'Silva D. B., Tanzer M. C., Monteleone M., Robertson A. A. B., Cooper M. A. et al. (2015). NLRP3 inflammasome activation downstream of cytoplasmic LPS recognition by both caspase-4 and caspase-5. Eur. J. Immunol. 45, 2918-2926. 10.1002/eji.201545655 [DOI] [PubMed] [Google Scholar]
- Bernot A., Da Silva C., Petit J.-L., Cruaud C., Caloustian C., Castet V., Ahmed-Arab M., Dross C., Dupont M. and Cattan D. (1998). Non-founder mutations in the MEFV gene establish this gene as the cause of familial Mediterranean fever (FMF). Hum. Mol. Genet. 7, 1317-1325. 10.1093/hmg/7.8.1317 [DOI] [PubMed] [Google Scholar]
- Boyden E. D. and Dietrich W. F. (2006). Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38, 240-244. 10.1038/ng1724 [DOI] [PubMed] [Google Scholar]
- Broz P., von Moltke J., Jones J. W., Vance R. E. and Monack D. M. (2010). Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 8, 471-483. 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brydges S. D., Mueller J. L., McGeough M. D., Pena C. A., Misaghi A., Gandhi C., Putnam C. D., Boyle D. L., Firestein G. S., Horner A. A. et al. (2009). Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity 30, 875-887. 10.1016/j.immuni.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brydges S. D., Broderick L., McGeough M. D., Pena C. A., Mueller J. L. and Hoffman H. M. (2013). Divergence of IL-1, IL-18, and cell death in NLRP3 inflammasomopathies. J. Clin. Invest. 123, 4695-4705. 10.1172/JCI71543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K. L. et al. (2009). An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10, 266-272. 10.1038/ni.1702 [DOI] [PubMed] [Google Scholar]
- Cai X., Chen J., Xu H., Liu S., Jiang Q.-X., Halfmann R. and Chen Z. J. (2014). Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207-1222. 10.1016/j.cell.2014.01.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canna S. W., de Jesus A. A., Gouni S., Brooks S. R., Marrero B., Liu Y., DiMattia M. A., Zaal K. J., Sanchez G. A., Kim H. et al. (2014). An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46, 1140-1146. 10.1038/ng.3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassel S. L., Eisenbarth S. C., Iyer S. S., Sadler J. J., Colegio O. R., Tephly L. A., Carter A. B., Rothman P. B., Flavell R. A. and Sutterwala F. S. (2008). The Nalp3 inflammasome is essential for the development of silicosis. Proc. Natl. Acad. Sci. USA 105, 9035-9040. 10.1073/pnas.0803933105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavailles P., Flori P., Papapietro O., Bisanz C., Lagrange D., Pilloux L., Massera C., Cristinelli S., Jublot D., Bastien O. et al. (2014). A highly conserved Toxo1 haplotype directs resistance to toxoplasmosis and its associated caspase-1 dependent killing of parasite and host macrophage. PLoS Pathog. 10, e1004005 10.1371/journal.ppat.1004005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae J. J., Cho Y.-H., Lee G.-S., Cheng J., Liu P. P., Feigenbaum L., Katz S. I. and Kastner D. L. (2011). Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1beta activation and severe autoinflammation in mice. Immunity 34, 755-768. 10.1016/j.immuni.2011.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarría-Smith J. and Vance R. E. (2013). Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog. 9, e1003452 10.1371/journal.ppat.1003452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. W., Groß C. J., Sotomayor F. V., Stacey K. J., Tschopp J., Sweet M. J. and Schroder K. (2014). The neutrophil NLRC4 inflammasome selectively promotes IL-1beta maturation without pyroptosis during acute Salmonella challenge. Cell Rep. 8, 570-582. 10.1016/j.celrep.2014.06.028 [DOI] [PubMed] [Google Scholar]
- Choubey D. (2012). Interferon-inducible Ifi200-family genes as modifiers of lupus susceptibility. Immunol. Lett. 147, 10-17. 10.1016/j.imlet.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D., Duan X., Dickerson E., Ponomareva L., Panchanathan R., Shen H. and Srivastava R. (2010). Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J. Interferon Cytokine Res. 30, 371-380. 10.1089/jir.2009.0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y. T., Lin Y. C., Lin K. H., Chou T. F., Kuo W. C., Yang K. T., Wu P. R., Chen R. H., Kimchi A. and Lai M. Z. (2011). Tumor suppressor death-associated protein kinase is required for full IL-1beta production. Blood. 117, 960-970. 10.1182/blood-2010-08-303115 [DOI] [PubMed] [Google Scholar]
- Compan V., Baroja-Mazo A., López-Castejón G., Gomez A. I., Martínez C. M., Angosto D., Montero M. T., Herranz A. S., Bazán E., Reimers D. et al. (2012). Cell volume regulation modulates NLRP3 inflammasome activation. Immunity 37, 487-500. 10.1016/j.immuni.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Corrales L., Woo S.-R., Williams J. B., McWhirter S. M., Dubensky T. W. and Gajewski T. F. (2016). Antagonism of the STING pathway via activation of the AIM2 inflammasome by intracellular DNA. J. Immunol. 196, 3191-3198. 10.4049/jimmunol.1502538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. A., Curley E. Z., Wykes M. N., Schroder K., Sweet M. J., Roberts T. L., Ragan M. A., Kassahn K. S. and Stacey K. J. (2012). The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol. Biol. 12, 140 10.1186/1471-2148-12-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz C. M., Rinna A., Forman H. J., Ventura A. L. M., Persechini P. M. and Ojcius D. M. (2007). ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J. Biol. Chem. 282, 2871-2879. 10.1074/jbc.M608083200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebolder C. A., Halff E. F., Koster A. J., Huizinga E. G. and Koning R. I. (2015). Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: implications for NLR activation. Structure 23, 2349-2357. 10.1016/j.str.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Dihlmann S., Erhart P., Mehrabi A., Nickkholgh A., Lasitschka F., Bockler D. and Hakimi M. (2014a). Increased expression and activation of absent in melanoma 2 inflammasome components in lymphocytic infiltrates of abdominal aortic aneurysms. Mol. Med. 20, 230-237. 10.2119/molmed.2013.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihlmann S., Tao S., Echterdiek F., Herpel E., Jansen L., Chang-Claude J., Brenner H., Hoffmeister M. and Kloor M. (2014b). Lack of Absent in Melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients. Int. J. Cancer 135, 2387-2396. 10.1002/ijc.28891 [DOI] [PubMed] [Google Scholar]
- Ding J., Wang K., Liu W., She Y., Sun Q., Shi J., Sun H., Wang D.-C. and Shao F. (2016). Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111-116. 10.1038/nature18590 [DOI] [PubMed] [Google Scholar]
- Dodé C., Le Dû N., Cuisset L., Letourneur F., Berthelot J.-M., Vaudour G., Meyrier A., Watts R. A., David Scott G. I., Nicholls A. et al. (2002). New mutations of CIAS1 that are responsible for muckle-wells syndrome and familial cold urticaria: a novel mutation underlies both syndromes. Am. J. Hum. Genet. 70, 1498-1506. 10.1086/340786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski Y., Peric M., Koglin S., Kammerbauer C., Goss C., Anz D., Simanski M., Glaser R., Harder J., Hornung V. et al. (2011). Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 3, 82ra38 10.1126/scitranslmed.3002001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C., Petrilli V., Van Bruggen R., Steele C., Mossman B. T. and Tschopp J. (2008). Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science (New York, N.Y.) 320, 674-677. 10.1126/science.1156995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas A., Amiable N., de Rivero Vaccari J. P., Chae J. J., Keane R. W., Lacroix S. and Vallières L. (2014). The inflammasome pyrin contributes to pertussis toxin-induced IL-1beta synthesis, neutrophil intravascular crawling and autoimmune encephalomyelitis. PLoS Pathog. 10, e1004150 10.1371/journal.ppat.1004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgeworth J. D., Spencer J., Phalipon A., Griffin G. E. and Sansonetti P. J. (2002). Cytotoxicity and interleukin-1beta processing following Shigella flexneri infection of human monocyte-derived dendritic cells. Eur. J. Immunol. 32, 1464-1471. [DOI] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A. L., Henao-Mejia J., Thaiss C. A., Booth C. J., Peaper D. R., Bertin J., Eisenbarth S. C., Gordon J. I. et al. (2011). NLRP6 inflammasome regulator colonic microbial ecology and risk for colitis. Cell 145, 745-757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi M. G., Hadinoto V., Growney J. D., Miller W. and Dietrich W. F. (2000). Genomic sequence analysis of the mouse Naip gene array. Genome Res. 10, 1095-1102. 10.1101/gr.10.8.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald S. E., Chavarria-Smith J. and Boothroyd J. C. (2014). NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect. Immun. 82, 460-468. 10.1128/IAI.01170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann J., Prieur A.-M., Quartier P., Berquin P., Certain S., Cortis E., Teillac-Hamel D., Fischer A. and de Saint Basile G. (2002). Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am. J. Hum. Genet. 71, 198-203. 10.1086/341357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.-W., Datta P., Wu J. and Alnemri E. S. (2009). AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509-513. 10.1038/nature07710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.-W., Juliana C., Solorzano L., Kang S., Wu J., Datta P., McCormick M., Huang L., McDermott E. et al. (2010). The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11, 385-393. 10.1038/ni.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Kang S., Anderson C., Sagara J., Fitzgerald K. A. and Alnemri E. S. (2013). Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. J. Immunol. 191, 3995-3999. 10.4049/jimmunol.1301681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger J. N., Lich J. D., Dare L. C., Cook M. N., Brown K. K., Duraiswami C., Bertin J. and Gough P. J. (2012). Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem. 287, 25030-25037. 10.1074/jbc.M112.378323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S. L. and Cookson B. T. (2006). Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell. Microbiol. 8, 1812-1825. 10.1111/j.1462-5822.2006.00751.x [DOI] [PubMed] [Google Scholar]
- Fink S. L., Bergsbaken T. and Cookson B. T. (2008). Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl Acad. Sci. USA 105, 4312-4317. 10.1073/pnas.0707370105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.-D., Özören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A. et al. (2006). Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7, 576-582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Frew B. C., Joag V. R. and Mogridge J. (2012). Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 8, e1002659 10.1371/journal.ppat.1002659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt M. M., Ebert T. S., Chauhan D., Schmidt T., Schmid-Burgk J. L., Rapino F., Robertson A. A. B., Cooper M. A., Graf T. and Hornung V. (2016). Human monocytes engage an alternative inflammasome pathway. Immunity 44, 833-846. 10.1016/j.immuni.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Gao W., Yang J., Liu W., Wang Y. and Shao F. (2016). Site-specific phosphorylation and microtubule dynamics control Pyrin inflammasome activation. Proc. Natl. Acad. Sci. USA 113, E4857-E4866. 10.1073/pnas.1601700113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilin M. A., Abdelaziz D. H. A., Mostafa M., Abdulrahman B. A., Grandhi J., Akhter A., Abu Khweek A., Aubert D. F., Valvano M. A., Wewers M. D. et al. (2012). Activation of the pyrin inflammasome by intracellular Burkholderia cenocepacia. J. Immunol. 188, 3469-3477. 10.4049/jimmunol.1102272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfu G., Cirelli K. M., Melo M. B., Mayer-Barber K., Crown D., Koller B. H., Masters S., Sher A., Leppla S. H., Moayeri M., (2014). Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. MBio 5, e01117-13 10.1128/mBio.01117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis S. I., Kaptein T. M., Wevers B. A., Theelen B., van der Vlist M., Boekhout T. and Geijtenbeek T. B. H. (2012). Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1beta via a noncanonical caspase-8 inflammasome. Nat. Immunol. 13, 246-254. 10.1038/ni.2222 [DOI] [PubMed] [Google Scholar]
- Guey B., Bodnar M., Manié S. N., Tardivel A. and Petrilli V. (2014). Caspase-1 autoproteolysis is differentially required for NLRP1b and NLRP3 inflammasome function. Proc. Natl. Acad. Sci. USA 111, 17254-17259. 10.1073/pnas.1415756111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung P., Malireddi R. K. S., Anand P. K., Demon D., Vande Walle L., Liu Z., Vogel P., Lamkanfi M. and Kanneganti T. D. (2012). Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-beta (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J. Biol. Chem. 287, 34474-34483. 10.1074/jbc.M112.401406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung P., Anand P. K., Malireddi R. K. S., Vande Walle L., Van Opdenbosch N., Dillon C. P., Weinlich R., Green D. R., Lamkanfi M. and Kanneganti T. D. (2014). FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J. Immunol. 192, 1835-1846. 10.4049/jimmunol.1302839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung P., Burton A. and Kanneganti T.-D. (2016). NLRP3 inflammasome plays a redundant role with caspase 8 to promote IL-1beta-mediated osteomyelitis. Proc. Natl. Acad. Sci. USA 113, 4452-4457. 10.1073/pnas.1601636113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar J. A., Powell D. A., Aachoui Y., Ernst R. K. and Miao E. A. (2013). Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science (New York, N.Y.) 341, 1250-1253. 10.1126/science.1240988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle A., Hornung V., Petzold G. C., Stewart C. R., Monks B. G., Reinheckel T., Fitzgerald K. A., Latz E., Moore K. J. and Golenbock D. T. (2008). The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat. Immunol. 9, 857-865. 10.1038/ni.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Lear T. B., Jerome J. A., Rajbhandari S., Snavely C. A., Gulick D. L., Gibson K. F., Zou C., Chen B. B. and Mallampalli R. K. (2015). Lipopolysaccharide Primes the NALP3 Inflammasome by Inhibiting Its Ubiquitination and Degradation Mediated by the SCFFBXL2 E3 Ligase. J. Biol. Chem. 290, 18124-18133. 10.1074/jbc.M115.645549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H., Tsuchiya K., Kawamura I., Fang R., Hernandez-Cuellar E., Shen Y., Mizuguchi J., Schweighoffer E., Tybulewicz V. and Mitsuyama M. (2013). Phosphorylation of the adaptor ASC acts as a molecular switch that controls the formation of speck-like aggregates and inflammasome activity. Nat. Immunol. 14, 1247-1255. 10.1038/ni.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harton J. A., Linhoff M. W., Zhang J. and Ting J.-P. (2002). Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J. Immunol. 169, 4088-4093. 10.4049/jimmunol.169.8.4088 [DOI] [PubMed] [Google Scholar]
- He W.-T., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z.-H., Zhong C.-Q. and Han J. (2015). Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 25, 1285-1298. 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zeng M. Y., Yang D., Motro B. and Nunez G. (2016). NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354-357. 10.1038/nature16959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H. M., Mueller J. L., Broide D. H., Wanderer A. A. and Kolodner R. D. (2001). Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29, 301-305. 10.1038/ng756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A. and Latz E. (2008). Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847-856. 10.1038/ni.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E. and Fitzgerald K. A. (2009). AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514-518. 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhou Q., Zhang C., Fan S., Cheng W., Zhao Y., Shao F., Wang H.-W., Sui S.-F. and Chai J. (2015). Structural and biochemical basis for induced self-propagation of NLRC4. Science (New York, N.Y.) 350, 399-404. 10.1126/science.aac5489 [DOI] [PubMed] [Google Scholar]
- Jin T., Perry A., Jiang J., Smith P., Curry J. A., Unterholzner L., Jiang Z., Horvath G., Rathinam V. A., Johnstone R. W. et al. (2012). Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36, 561-571. 10.1016/j.immuni.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., Curry J., Smith P., Jiang J. and Xiao T. S. (2013a). Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1. Proteins 81, 1266-1270. 10.1002/prot.24287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., Perry A., Smith P., Jiang J. and Xiao T. S. (2013b). Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J. Biol. Chem. 288, 13225-13235. 10.1074/jbc.M113.468033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. W., Kayagaki N., Broz P., Henry T., Newton K., O'Rourke K., Chan S., Dong J., Qu Y., Roose-Girma M. et al. (2010). Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. USA 107, 9771-9776. 10.1073/pnas.1003738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti T.-D., Body-Malapel M., Amer A., Park J.-H., Whitfield J., Franchi L., Taraporewala Z. F., Miller D., Patton J. T. and Inohara N. (2006a). Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281, 36560-36568. 10.1074/jbc.M607594200 [DOI] [PubMed] [Google Scholar]
- Kanneganti T.-D., Özören N., Body-Malapel M., Amer A., Park J.-H., Franchi L., Whitfield J., Barchet W., Colonna M. and Vandenabeele P. (2006b). Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440, 233 10.1038/nature04517 [DOI] [PubMed] [Google Scholar]
- Kanneganti T.-D., Lamkanfi M. and Núñez G. (2007). Intracellular NOD-like receptors in host defense and disease. Immunity 27, 549-559. 10.1016/j.immuni.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S. et al. (2011). Non-canonical inflammasome activation targets caspase-11. Nature 479, 117-121. 10.1038/nature10558 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Wong M. T., Stowe I. B., Ramani S. R., Gonzalez L. C., Akashi-Takamura S., Miyake K., Zhang J., Lee W. P., Muszynski A. et al. (2013). Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science (New York, N.Y.) 341, 1246-1249. 10.1126/science.1240248 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Stowe I. B., Lee B. L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T. et al. (2015). Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666-671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Khare S., Ratsimandresy R. A., de Almeida L., Cuda C. M., Rellick S. L., Misharin A. V., Wallin M. C., Gangopadhyay A., Forte E., Gottwein E. et al. (2014). The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat. Immunol. 15, 343-353. 10.1038/ni.2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Bauernfeind F., Ablasser A., Hartmann G., Fitzgerald K. A., Latz E. and Hornung V. (2010). Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 40, 1545-1551. 10.1002/eji.201040425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. L., Chae J. J., Park Y. H., De Nardo D., Stirzaker R. A., Ko H.-J., Tye H., Cengia L., DiRago L., Metcalf D. et al. (2015). Aberrant actin depolymerization triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1beta. J. Exp. Med. 212, 927-938. 10.1084/jem.20142384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A., Sasaki Y., Abe T., Kano H. and Yasutomo K. (2014). An inherited mutation in NLRC4 causes autoinflammation in human and mice. J. Exp. Med. 211, 2385-2396. 10.1084/jem.20141091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed E. M. and Vance R. E. (2011). Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592-595. 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J., Brubaker S. W. and Monack D. M. (2015). Cutting edge: inflammasome activation in primary human macrophages is dependent on flagellin. J. Immunol. 195, 815-819. 10.4049/jimmunol.1403100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers B., Salmena L., Bidère N., Su H., Matysiak-Zablocki E., Murakami K., Ohashi P. S., Jurisicova A., Lenardo M., Hakem R. et al. (2007). Essential role for caspase-8 in Toll-like receptors and NFkappaB signaling. J. Biol. Chem. 282, 7416-7423. 10.1074/jbc.M606721200 [DOI] [PubMed] [Google Scholar]
- Li H., Wang J., Wang J., Cao L.-S., Wang Z.-X. and Wu J.-W. (2014). Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling. Acta Crystallogr. F Struct. Biol. Commun. 70, 21-29. 10.1107/S2053230X1303135X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K.-M., Hu W., Troutman T. D., Jennings M., Brewer T., Li X., Nanda S., Cohen P., Thomas J. A. and Pasare C. (2014). IRAK-1 bypasses priming and directly links TLRs to rapid NLRP3 inflammasome activation. Proc. Natl. Acad. Sci. USA 111, 775-780. 10.1073/pnas.1320294111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. C., Huang D. Y., Wang J. S., Lin Y. L., Hsieh S. L., Huang K. C. and Lin W. W. (2015). Syk is involved in NLRP3 inflammasome-mediated caspase-1 activation through adaptor ASC phosphorylation and enhanced oligomerization. J. Leukoc. Biol. 97, 825-835. 10.1189/jlb.3HI0814-371RR [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V. G., Wu H. and Lieberman J. (2016). Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153-158. 10.1038/nature18629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Magupalli V. G., Ruan J., Yin Q., Atianand M. K., Vos M. R., Schröder G. F., Fitzgerald K. A., Wu H. and Egelman E. H. (2014). Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193-1206. 10.1016/j.cell.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Nakamura T., Inouye K., Li J., Tang Y., Lundbäck P., Valdes-Ferrer S. I., Olofsson P. S., Kalb T., Roth J. et al. (2012). Novel role of PKR in inflammasome activation and HMGB1 release. Nature 488, 670-674. 10.1038/nature11290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens J. R., Gurung P., Vogel P., Johnson G. R., Carter R. A., McGoldrick D. J., Bandi S. R., Calabrese C. R., Vande Walle L., Lamkanfi M. et al. (2014). Dietary modulation of the microbiome affects autoinflammatory disease. Nature 516, 246-249. 10.1038/nature13788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait J., Vercammen E., Janssens S., Schotte P., Haegman M., Magez S. and Beyaert R. (2008). Stimulation of Toll-like receptor 3 and 4 induces interleukin-1beta maturation by caspase-8. J. Exp. Med. 205, 1967-1973. 10.1084/jem.20071632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamantopoulos M., Ronchi F., Van Hauwermeiren F., Vieira-Silva S., Yilmaz B., Martens L., Saeys Y., Drexler S. K., Yazdi A. S. and Raes J. (2017). Nlrp6-and ASC-dependent inflammasomes do not shape the commensal gut microbiota composition. Immunity 47, 339-348, e4. 10.1016/j.immuni.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Man S. M., Karki R., Malireddi R. K. S., Neale G., Vogel P., Yamamoto M., Lamkanfi M. and Kanneganti T.-D. (2015a). The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16, 467-475. 10.1038/ni.3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S. M., Zhu Q., Zhu L., Liu Z., Karki R., Malik A., Sharma D., Li L., Malireddi R. K. S., Gurung P. et al. (2015b). Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell 162, 45-58. 10.1016/j.cell.2015.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S. M., Karki R., Sasai M., Place D. E., Kesavardhana S., Temirov J., Frase S., Zhu Q., Malireddi R. K. S. and Kuriakose T. (2016). IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell 167, 382-396.e317. 10.1016/j.cell.2016.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S. and Dixit V. M. (2004). Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213-218. 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Martinon F., Burns K. and Tschopp J. (2002). The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417-426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- Masters S. L., Gerlic M., Metcalf D., Preston S., Pellegrini M., O'Donnell J. A., McArthur K., Baldwin T. M., Chevrier S., Nowell C. J. et al. (2012). NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity 37, 1009-1023. 10.1016/j.immuni.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters S. L., Lagou V., Jeru I., Baker P. J., Van Eyck L., Parry D. A., Lawless D., De Nardo D., Garcia-Perez J. E., Dagley L. F. et al. (2016). Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Sci. Transl. Med. 8, 332ra345 10.1126/scitranslmed.aaf1471 [DOI] [PubMed] [Google Scholar]
- Matusiak M., Van Opdenbosch N. and Lamkanfi M. (2015). CARD- and pyrin-only proteins regulating inflammasome activation and immunity. Immunol. Rev. 265, 217-230. 10.1111/imr.12282 [DOI] [PubMed] [Google Scholar]
- Meng G., Zhang F., Fuss I., Kitani A. and Strober W. (2009). A mutation in the Nlrp3 gene causing inflammasome hyperactivation potentiates Th17 cell-dominant immune responses. Immunity 30, 860-874. 10.1016/j.immuni.2009.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E., Wallet P., Dreier R. F., Costanzo S., Anton L., Rühl S., Dussurgey S., Dick M. S., Kistner A., Rigard M. et al. (2015). Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16, 476-484. 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E. A., Alpuche-Aranda C. M., Dors M., Clark A. E., Bader M. W., Miller S. I. and Aderem A. (2006). Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7, 569-575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Miao E. A., Leaf I. A., Treuting P. M., Mao D. P., Dors M., Sarkar A., Warren S. E., Wewers M. D. and Aderem A. (2010a). Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11, 1136-1142. 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E. A., Mao D. P., Yudkovsky N., Bonneau R., Lorang C. G., Warren S. E., Leaf I. A. and Aderem A. (2010b). Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA 107, 3076-3080. 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa T., Takahama M., Kozaki T., Lee H., Zou J., Saitoh T. and Akira S. (2013). Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14, 454-460. 10.1038/ni.2550 [DOI] [PubMed] [Google Scholar]
- Moayeri M., Crown D., Newman Z. L., Okugawa S., Eckhaus M., Cataisson C., Liu S., Sastalla I. and Leppla S. H. (2010). Inflammasome sensor Nlrp1b-dependent resistance to anthrax is mediated by caspase-1, IL-1 signaling and neutrophil recruitment. PLoS Pathog. 6, e1001222 10.1371/journal.ppat.1001222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A. B., Byrne B. G., Whitfield N. N., Madigan C. A., Fuse E. T., Tateda K. and Swanson M. S. (2006). Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203, 1093-1104. 10.1084/jem.20051659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer L., Moreau F., MacDonald J. A. and Chadee K. (2016). NLRP3 inflammasome inhibition is disrupted in a group of auto-inflammatory disease CAPS mutations. Nat. Immunol. 17, 1176-1186. 10.1038/ni.3538 [DOI] [PubMed] [Google Scholar]
- Munoz-Planillo R., Kuffa P., Martínez-Colón G., Smith B. L., Rajendiran T. M. and Nunez G. (2013). K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142-1153. 10.1016/j.immuni.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neven B., Callebaut I., Prieur A. M., Feldmann J., Bodemer C., Lepore L., Derfalvi B., Benjaponpitak S., Vesely R., Sauvain M. J. et al. (2004). Molecular basis of the spectral expression of CIAS1 mutations associated with phagocytic cell-mediated autoinflammatory disorders CINCA/NOMID, MWS, and FCU. Blood 103, 2809-2815. 10.1182/blood-2003-07-2531 [DOI] [PubMed] [Google Scholar]
- Nour A. M., Yeung Y.-G., Santambrogio L., Boyden E. D., Stanley E. R. and Brojatsch J. (2009). Anthrax lethal toxin triggers the formation of a membrane-associated inflammasome complex in murine macrophages. Infect. Immun. 77, 1262-1271. 10.1128/IAI.01032-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. H., Wood G., Kastner D. L. and Chae J. J. (2016). Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat. Immunol. 17, 914-921. 10.1038/ni.3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F. and Tschopp J. (2007). Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 14, 1583-1589. 10.1038/sj.cdd.4402195 [DOI] [PubMed] [Google Scholar]
- Pilla D. M., Hagar J. A., Haldar A. K., Mason A. K., Degrandi D., Pfeffer K., Ernst R. K., Yamamoto M., Miao E. A. and Coers J. (2014). Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. USA 111, 6046-6051. 10.1073/pnas.1321700111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomareva L., Liu H., Duan X., Dickerson E., Shen H., Panchanathan R. and Choubey D. (2013). AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Mol. Cancer Res. 11, 1193-1202. 10.1158/1541-7786.MCR-13-0145 [DOI] [PubMed] [Google Scholar]
- Poyet J.-L., Srinivasula S. M., Tnani M., Razmara M., Fernandes-Alnemri T. and Alnemri E. S. (2001). Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276, 28309-28313. 10.1074/jbc.C100250200 [DOI] [PubMed] [Google Scholar]
- Py B. F., Kim M.-S., Vakifahmetoglu-Norberg H. and Yuan J. (2013). Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 49, 331-338. 10.1016/j.molcel.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Rathinam V. A. K., Jiang Z., Waggoner S. N., Sharma S., Cole L. E., Waggoner L., Vanaja S. K., Monks B. G., Ganesan S., Latz E. et al. (2010). The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395-402. 10.1038/ni.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V. A. K., Vanaja S. K., Waggoner L., Sokolovska A., Becker C., Stuart L. M., Leong J. M. and Fitzgerald K. A. (2012). TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell 150, 606-619. 10.1016/j.cell.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Zak D. E., Chavarria-Smith J., Vance R. E. and Miao E. A. (2013). Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191, 3986-3989. 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T., Zamboni D. S., Roy C. R., Dietrich W. F. and Vance R. E. (2006). Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2, e18 10.1371/journal.ppat.0020018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. L., Idris A., Dunn J. A., Kelly G. M., Burnton C. M., Hodgson S., Hardy L. L., Garceau V., Sweet M. J., Ross I. L. et al. (2009). HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science (New York, N.Y.) 323, 1057-1060. 10.1126/science.1169841 [DOI] [PubMed] [Google Scholar]
- Rodgers M. A., Bowman J. W., Fujita H., Orazio N., Shi M., Liang Q., Amatya R., Kelly T. J., Iwai K., Ting J. et al. (2014). The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J. Exp. Med. 211, 1333-1347. 10.1084/jem.20132486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg N., Al Moussawi K., Nelson-Williams C., Stiegler A. L., Loring E., Choi M., Overton J., Meffre E., Khokha M. K., Huttner A. J. et al. (2014). Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat. Genet. 46, 1135-1139. 10.1038/ng.3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl S. and Broz P. (2015). Caspase-11 activates a canonical NLRP3 inflammasome by promoting K(+) efflux. Eur. J. Immunol. 45, 2927-2936. 10.1002/eji.201545772 [DOI] [PubMed] [Google Scholar]
- Sborgi L., Ravotti F., Dandey V. P., Dick M. S., Mazur A., Reckel S., Chami M., Scherer S., Huber M., Böckmann A. et al. (2015). Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc. Natl. Acad. Sci. USA 112, 13237-13242. 10.1073/pnas.1507579112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sborgi L., Rühl S., Mulvihill E., Pipercevic J., Heilig R., Stahlberg H., Farady C. J., Müller D. J., Broz P. and Hiller S. (2016). GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 35, 1766-1778. 10.15252/embj.201694696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk J. L., Gaidt M. M., Schmidt T., Ebert T. S., Bartok E. and Hornung V. (2015). Caspase-4 mediates non-canonical activation of the NLRP3 inflammasome in human myeloid cells. Eur. J. Immunol. 45, 2911-2917. 10.1002/eji.201545523 [DOI] [PubMed] [Google Scholar]
- Schmid-Burgk J. L., Chauhan D., Schmidt T., Ebert T. S., Reinhardt J., Endl E. and Hornung V. (2016). A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 291, 103-109. 10.1074/jbc.C115.700492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn C., Frey B., Lauber K., Janko C., Strysio M., Keppeler H., Gaipl U. S., Voll R. E., Springer E., Munoz L. E. et al. (2011). Sodium overload and water influx activate the NALP3 inflammasome. J. Biol. Chem. 286, 35-41. 10.1074/jbc.M110.139048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M. E., Müller A. A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K. M. and Hardt W.-D. (2014). Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe 16, 237-248. 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Sharma D., Sharma B. R., Vogel P. and Kanneganti T.-D. (2017). IL-1β and caspase-1 drive autoinflammatory disease independently of IL-1α or caspase-8 in a mouse model of familial mediterranean fever. Am. J. Pathol. 187, 236-244. 10.1016/j.ajpath.2016.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L. and Shao F. (2014). Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187-192. 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- Shi H., Wang Y., Li X., Zhan X., Tang M., Fina M., Su L., Pratt D., Bu C. H., Hildebrand S. et al. (2015a). NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17, 250-258. 10.1038/ni.3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F. and Shao F. (2015b). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660-665. 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- Stutz A., Kolbe C.-C., Stahl R., Horvath G. L., Franklin B. S., van Ray O., Brinkschulte R., Geyer M., Meissner F. and Latz E. (2017). NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J. Exp. Med. 214, 1725-1736. 10.1084/jem.20160933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenthorey J. L., Kofoed E. M., Daugherty M. D., Malik H. S. and Vance R. E. (2014). Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell 54, 17-29. 10.1016/j.molcel.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra J. K., Cote C. K., France B., Jenkins A. L., Bozue J. A., Welkos S. L., LeVine S. M. and Bradley K. A. (2010). Cutting edge: resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. J. Immunol. 184, 17-20. 10.4049/jimmunol.0903114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touitou I., Lesage S., McDermott M., Cuisset L., Hoffman H., Dode C., Shoham N., Aganna E., Hugot J.-P., Wise C. et al. (2004). Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum. Mutat. 24, 194-198. 10.1002/humu.20080 [DOI] [PubMed] [Google Scholar]
- Vanaja S. K., Russo A. J., Behl B., Banerjee I., Yankova M., Deshmukh S. D. and Rathinam V. A. K. (2016). Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell 165, 1106-1119. 10.1016/j.cell.2016.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gorp H., Saavedra P. H., de Vasconcelos N. M., Van Opdenbosch N., Vande Walle L., Matusiak M., Prencipe G., Insalaco A., Van Hauwermeiren F., Demon D. et al. (2016). Familial Mediterranean fever mutations lift the obligatory requirement for microtubules in Pyrin inflammasome activation. Proc. Natl. Acad. Sci. USA 113, 14384-14389. 10.1073/pnas.1613156113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opdenbosch N., Gurung P., Vande Walle L., Fossoul A., Kanneganti T. D. and Lamkanfi M. (2014). Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat. Commun. 5, 3209 10.1038/ncomms4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimer G. I., Weng D., Paquette S. W. M., Vanaja S. K., Rathinam V. A., Aune M. H., Conlon J. E., Burbage J. J., Proulx M. K. and Liu Q. (2012). The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37, 96-107. 10.1016/j.immuni.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Trinidad N. J., Moayeri M., Kintzer A. F., Wang S. B., van Rooijen N., Brown C. R., Krantz B. A., Leppla S. H., Gronert K. et al. (2012). Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature 490, 107-111. 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Miura M., Jung Y.-K., Zhu H., Li E. and Yuan J. (1998). Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell 92, 501-509. 10.1016/S0092-8674(00)80943-5 [DOI] [PubMed] [Google Scholar]
- Wang Y., Gao W., Shi X., Ding J., Liu W., He H., Wang K. and Shao F. (2017). Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a Gasdermin. Nature 547, 99-103. advance online publication 10.1038/nature22393 [DOI] [PubMed] [Google Scholar]
- Wilson J. E., Petrucelli A. S., Chen L., Koblansky A. A., Truax A. D., Oyama Y., Rogers A. B., Brickey W. J., Wang Y., Schneider M. et al. (2015). Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat. Med. 21, 906-913. 10.1038/nm.3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witola W. H., Mui E., Hargrave A., Liu S., Hypolite M., Montpetit A., Cavailles P., Bisanz C., Cesbron-Delauw M.-F., Fournie G. J. et al. (2011). NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect. Immun. 79, 756-766. 10.1128/IAI.00898-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yang J., Gao W., Li L., Li P., Zhang L., Gong Y.-N., Peng X., Xi J. J., Chen S. et al. (2014). Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513, 237-241. 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- Yabal M., Muller N., Adler H., Knies N., Gross C. J., Damgaard R. B., Kanegane H., Ringelhan M., Kaufmann T., Heikenwalder M., Strasser A., Gross O., Ruland J., Peschel C., Gyrd-Hansen M. and Jost P. J. (2014). XIAP restricts TNF- and RIP3-dependent cell death and inflammasome activation. Cell Rep. 7, 1796-1808. 10.1016/j.celrep.2014.05.008 [DOI] [PubMed] [Google Scholar]
- Yan Y., Jiang W., Liu L. Wang X., Ding C., Tian Z. and Zhou R. (2015). Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 160, 62-73. 10.1016/j.cell.2014.11.047 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J. and Shao F. (2013). Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA 110, 14408-14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., He Y., Muñoz-Planillo R., Liu Q. and Núñez G. (2015). Caspase-11 requires the pannexin-1 channel and the purinergic P2X7 pore to mediate pyroptosis and endotoxic shock. Immunity 43, 923-932. 10.1016/j.immuni.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q., Sester D. P., Tian Y., Hsiao Y.-S., Lu A., Cridland J. A., Sagulenko V., Thygesen S. J., Choubey D., Hornung V. et al. (2013). Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 4, 327-339. 10.1016/j.celrep.2013.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen S., Ruan J., Wu J., Tong A. B., Yin Q., Li Y., David L., Lu A., Wang W. L. et al. (2015). Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science (New York, N.Y.) 350, 404-409. 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.-N., Lu Q., Xu H., Liu L. and Shao F. (2011). The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596-600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- Zhou R., Yazdi A. S., Menu P. and Tschopp J. (2011). A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221-225. 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
- Zhu S., Ding S., Wang P., Wei Z., Pan W., Palm N. W., Yang Y., Yu H., Li H.-B. and Wang G. (2017). Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 546, 667 10.1038/nature22967 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.