Abstract
Purpose
The present study aimed to investigate the association between the human VDR gene and Helicobacter pylori infection.
Patients and methods
A cross-sectional study was conducted on 208 adult patients with upper gastrointestinal symptoms. Gastric biopsy specimens were obtained from each patient for molecular DNA and histological examination. Patients were genotyped for VDR gene polymorphisms using polymerase chain reaction and restriction fragment length polymorphism analysis.
Results
The allelic and genotypic distribution analyses of the FokI, ApaI and TaqI polymorphisms of the VDR gene did not show distribution differences between H. pylori-positive and -negative groups. The genotype distribution observed for polymorphism BsmI deviated significantly from what was expected in a Hardy–Weinberg equilibrium test in the H. pylori-positive group (χ2=29.048, p<0.001). The distribution of BsmI genotypes differed significantly between the H. pylori-negative and H. pylori-positive groups (p=0.0034), where the frequency of the bb genotype increased among H. pylori-positive individuals compared with those without infection (63.25% versus 50.55%, respectively). Conversely, the H. pylori-negative group showed a Bb frequency that was 20.27% higher than in the infected group.
Conclusion
We identified a possible association between the BsmI polymorphism and infection by H. pylori. However, further research is required to clarify this relationship.
Keywords: vitamin D, VDR, polymorphism, Helicobacter pylori virulence factors, infectious disease, VDR SNPs, admixed population
Introduction
Helicobacter pylori is a spiral-shaped Gram-negative bacterium that, despite the adverse conditions in the human stomach, shows many mechanisms and specific virulence factors that affect the properties of the gastric mucosa and determine its adhesion and survival in such an environment.1
Long-term colonization by H. pylori, with a posterior chronic gastritis induction, depends on the strains living in the gastric mucosa and how they interact with the host’s immune system.2 Moreover, human genetic polymorphisms also partly contribute to the susceptibility and clinical evolution of the infection. They can produce different expression levels of immune system mediators related to infection rates, H. pylori eradication, colonization by more virulent strains, gastric inflammation enhancement and cell damage.3
It was recently proposed that infection by H. pylori can induce an improvement in vitamin D receptor (VDR) expression in gastric epithelia. It was observed in vitro that the active form of vitamin D, 1α,25-dihydroxyvitamin D3, exhibits immune modulators properties against this pathogen, via stimulation of cathelicidin antimicrobial peptides and decreasing inflammatory cytokine and chemokine levels.4
The gene responsible for VDR expression is located on the 12q13.11 chromosome and comprises a region of >60 kb of DNA.5 From the single nucleotide polymorphisms (SNPs) of this gene that have been described so far, the restriction fragment length polymorphism (RFLP) for FokI, BsmI, ApaI and TaqI restriction enzymes is currently used in studies of genetic susceptibility to infectious diseases.6–8
Of these variants, the FokI polymorphism (rs2228570; C>T), characterized by the substitution of thymine to cytosine (ATG to ACG), changes the protein structure of the VDR receptor, which in the presence of the mutant allele C or F produces a protein with 424 amino acids instead of 427 amino acids.10 In contrast to the FokI variant, the TaqI polymorphism (rs731236; T>C) does not alter the amino acid composition of the resulting protein when thymine is changed to cytosine (ATT to ATC). The remaining polymorphisms are located in the noncoding regions of the VDR gene, and guanine is changed to adenine (GCG to GCA) in the BsmI polymorphism (rs1544410; G>A) and cytosine is changed to adenine in the ApaI polymorphism (rs7975232, A>C).9
Even though findings are inconclusive regarding the exact role of these VDR gene variants, studies suggest effects of the FokI, located in exon 2, on VDR functionality and transcriptional activity of immune-specific transcription factors.10,11 It is also hypothesized that possible alterations in mRNA expression levels of the VDR gene are attributed to a 3′ untranslated region (3′-UTR),12 which includes the genetic variants observed in intron 8 of BsmI and ApaI sites and in exon 9 of the TaqI restriction enzyme.13
In this regard, this study aimed to investigate the link between VDR gene polymorphism in H. pylori-infected patients with gastric symptomatology. The study was used as a basis for comparing the distribution of genic frequencies of VDR gene polymorphisms described in a Brazilian population14 with that identified in the ethnic groups of the International HapMap Project.15
Patients and methods
Study population
A cross-sectional study was conducted with 208 outpatients (126 females and 82 males) aged 18 years or older (mean age ± standard deviation 53.04±15.20 years) who underwent an upper gastrointestinal endoscopy between April 2011 to March 2012 at the University Hospital João de Barros Barreto (HUJBB), Belém, Para, Brazil.
After previous evaluations due to the presence of symptoms in the upper gastrointestinal tract, patients who showed dyspeptic disorders and who were subsequently diagnosed with chronic gastritis were considered for the study. Subjects who reported to have received anti-Helicobacter therapy within the last 6 months, used a proton pump inhibitor within the last 30 days, were pregnant, abused alcohol, used illicit drugs, or had an HIV infection were excluded.
Biological samples, epidemiological data and written informal consent were obtained from all patients in this study. All procedures performed in the study were approved by the Ethics Committee for Research Involving Human Subjects of HUJBB (protocol no. 1820/2010).
Description and laboratory evaluation
In total, two gastric antral biopsy specimens were taken from each patient. One fragment was fixed in 10% formalin and stained with hematoxylin–eosin for histological examination according to the Updated Sydney System.16 The other biopsy specimen was immersed in a saline solution (0.9%) ready for genomic DNA extraction using the phenol–chloroform method.17
A polymerase chain reaction (PCR) assay to detect H. pylori in gastric biopsy specimens was performed using gene-specific primers (P1 and P2) that amplified a 298 bp fragment present in all H. pylori strains.18 Expressions of cagA and vacA virulence genes were determined by multiplex PCR using a protocol adapted from the study of Chattopadhyay et al.19
VDR gene polymorphism genotyping was performed via PCR–RFLP using forward and reverse primers, DNA fragments, molecular size of PCR products and restriction endonucleases, as described previously.20 PCR products were prepared in a 25 μL reaction mixture containing 1 μL genomic DNA, 2.5 mM dNTP, 0.5 μL Taq DNA polymerase, 2.5 μL of a 10× buffer, 1.5 mM MgCl2 and 2.4 mM of each primer.
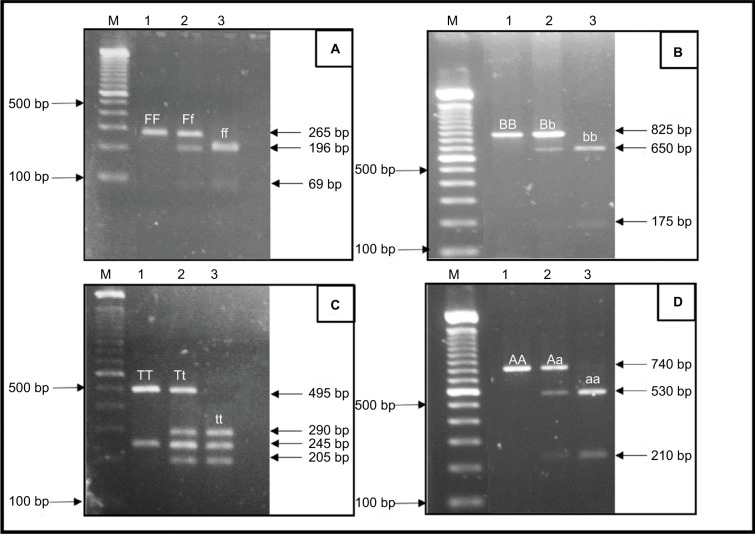
Figure 1 shows the cleavage patterns of the DNA fragments using a previously described technique. The genotype representation was denoted according to the presence (lower letter) or absence (capital letter) of the restriction site.
Figure 1.
Representative RFLP pattern for the detection of VDR polymorphisms in agarose gel.
Notes: (A) FokI: the FF (265 bp), Ff (265 bp, 196 bp and 69 bp) and ff (196 bp and 69 bp) genotypes. (B) BsmI: the BB (825 bp), Bb (825 bp, 650 bp and 175 bp) and bb (650 bp and 175 bp) genotypes. (C) TaqI: the TT (495 bp and 245 bp), Tt (495 bp, 290 bp, 245 bp and 205 bp) and tt (290 bp, 245 bp and 205 bp) genotypes. (D) ApaI: the AA (740 bp), Aa (740 bp, 530 bp and 210 bp) and aa (530 bp and 210 bp) genotypes. M, molecular marker 100 bp ladder.
Abbreviations: RFLP, restriction fragment length polymorphism; VDR, vitamin D receptor.
HapMap data
Allelic and haplotypic frequency comparisons of VDR polymorphisms based on individual ancestry were obtained from the HapMap database15 to represent three major continental populations: Asian, European and African. The study included a sample population of East Asian ancestries (ASN) comprising groups of Han Chinese in Beijing (CHB) and Japanese, Tokyo (JPT); Utah residents of Northern and Western European origin (CEU) and Sub-Saharan Africans from Yoruba, Ibadan, Nigeria (YRI).
Statistical analysis
Genotypic and allelic frequencies were compared between the H. pylori-negative and -positive groups using the Mantel–Haenszel chi-square test with an adjusted odds ratio (OR) for the covariates of sex and age. Hardy–Weinberg equilibrium (HWE) was also determined to the genotype frequencies of VDR polymorphisms. The variation in genotype proportions and a histopathological analysis of the BsmI polymorphism in the groups studied were compared by a binomial test. All analyses were performed using the statistical software program BioEstat 5.021 with a 5% level of statistical significance.
Pairwise linkage disequilibrium (LD) between VDR polymorphisms was computed using Haploview program v.4.2.22 Online SHEsi software was used to calculate the haplotype frequencies,23 and genetic distances were estimated for each population pair by Wright’s F statistics (FST) using Arlequin v. 3.01.24
Results
Of the 208 patients enrolled in the present study, the mean age at which H. pylori was diagnosed as positive was 51.38±15.22 years, while the mean age of the H. pylori-negative group was 55.18±14.98 years. Overall, 60.58% (126/208) of patients were females and 90.86% (189/208) were categorized as negroids (those with brown or black skin).
H. pylori infection was detected in 56.25% (117/208) of patients with upper gastrointestinal symptoms, 72.65% (85/117) of whom were colonized by CagA+/VacA s1m1 virulent strains. In relation to demographic and epidemiological aspects that may be considered a risk factor for H. pylori infection, no significant differences were observed between H. pylori-negative and H. pylori-positive groups in terms of sex, age, comorbidities and lifestyle (smoking, alcohol use and consumption of fruits).
It should be emphasized that in relation to ancestral family history based on their self-reported ethnicity, a similar distribution was observed between the study groups relative to the ethnic groups of African, European and Native American origin.
Analysis of allelic and genotypic distributions of the FokI, ApaI and TaqI polymorphisms in the VDR gene (Table 1) showed that there were no statistically significant differences between H. pylori-infected and -uninfected patients. However, concerning the BsmI polymorphism, the frequency of the bb genotype was higher in H. pylori-positive patients compared with those without an H. pylori infection (63.25% versus 50.55%, respectively). Conversely, the Bb genotype was more common in the H. pylori-negative group than in the H. pylori-positive group (37.26% versus 17.09%, respectively), and an OR of 0.28 suggests a protective effect of this genotype. No significant sex difference in genotypic distribution was found for polymorphism in the VDR gene.
Table 1.
Genotypic and allelic frequencies of VDR gene polymorphisms in H. pylori-infected and -uninfected patients from Belém, Pará, Brazil
| VDR/SNPs |
H. pylori infection
|
χ2 (p-value) | OR (CI 95%) | ORa (CI 95%) | |
|---|---|---|---|---|---|
| Positive
|
Negative
|
||||
| n=117 (%) | n=91 (%) | ||||
| FokI | |||||
| Genotypes | |||||
| FF | 56 (47.86) | 41 (45.05) | 0.2061 (0.9021) | 1.00b | 1.00b |
| Ff | 45 (38.46) | 36 (39.56) | 0.92 (0.50–1.66) | 0.81 (0.44–1.52) | |
| ff | 16 (13.68) | 14 (15.38) | 0.18 (0.37–1.90) | 1.04 (0.44–2.42) | |
| Alleles | |||||
| F | 157 (67.09) | 118 (64.84) | 0.2331 (0.6292) | 1.00b | |
| f | 77 (32.91) | 64 (35.16) | 0.90 (0.60–1.36) | ||
| BsmI | |||||
| Genotypes | |||||
| BB | 23 (19.66) | 11 (12.09) | 11.325 (0.0034) | 1.00b | 1.00b |
| Bb | 20 (17.09) | 34 (37.36) | 0.28 (0.11–0.70) | 0.29 (0.11–0.72) | |
| bb | 74 (63.25) | 46 (50.55) | 0.77 (0.34–1.72) | 0.70 (0.31–1.59) | |
| Alleles | |||||
| B | 66 (28.21) | 56 (30.77) | 0.3247 (0.5687) | 1.00b | |
| b | 168 (71.79) | 126 (69.23) | 1.13 (0.74–1.73) | ||
| ApaI | |||||
| Genotypes | |||||
| AA | 47 (40.17) | 37 (40.66) | 0.4525 (0.7974) | 1.00b | 1.00b |
| Aa | 48 (41.03) | 40 (43.96) | 0.94 (0.52–1.72) | 0.91 (0.49–1.68) | |
| aa | 22 (18.80) | 14 (15.38) | 1.24 (0.56–2.74) | 1.23 (0.53–2.83) | |
| Alleles | |||||
| A | 142 (60.68) | 114 (62.64) | 0.1650 (0.2331) | 1.00b | |
| a | 92 (39.32) | 68 (37.36) | 1.09 (0.73–1.62) | ||
| TaqI | |||||
| Genotypes | |||||
| TT | 70 (59.83) | 53 (58.24) | 0.0766 (0.9624) | 1.00b | 1.00b |
| Tt | 39 (33.33) | 32 (35.16) | 0.92 (0.51–1.66) | 0.95 (0.52–1.73) | |
| tt | 8 (6.84) | 6 (6.59) | 1.01 (0.33–3.09) | 1.00 (0.52–1.73) | |
| Alleles | |||||
| T | 179 (76.50) | 138 (75.82) | 0.0254 (0.8732) | 1.00b | |
| t | 55 (23.50) | 44 (24.18) | 0.96 (0.61–1.52) | ||
Notes:
Age and sex-adjusted OR.
1.00=reference group used in the analysis.
Abbreviations: VDR, vitamin D receptor; H. pylori, Helicobacter pylori; SNP, single nucleotide polymorphism; OR, odds ratio.
Additionally, the chi-square test of homogeneity revealed that the allelic frequencies of BsmI did not differ significantly between the H. pylori-negative and -positive groups and the b allele was the most prevalent in both samples, showing that these groups were extracted from the same population.
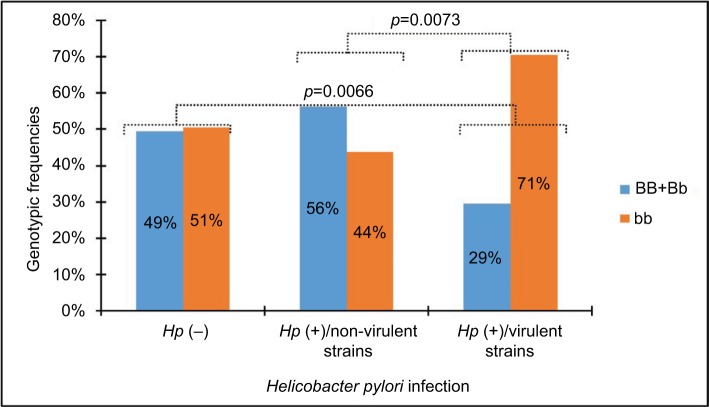
A significant deviation by HWE was found only in the genotypic distribution of the BsmI SNP identified in the H. pylori-positive group (χ2=29.048, p<0.001). In this sense, the results shown in Figure 2 indicate a very strong significant difference in the distribution of genotype frequencies of BsmI SNP between patients infected by virulent strains and those infected by non-virulent strains or uninfected patients. The frequency of the BB+Bb genotype was elevated in H. pylori-negative patients infected by non-virulent H. pylori strains. In contrast, the bb genotype was more prevalent in patients infected by virulent H. pylori strains.
Figure 2.
Genotypic frequencies of the BsmI VDR gene polymorphism in Helicobacter pylori-infected and -uninfected patients.
Abbreviations: VDR, vitamin D receptor; Hp, Helicobacter pylori.
As shown in Table 2, infection with virulent H. pylori strains and a high degree of neutrophil activity in the gastric mucosa were less frequently observed in patients with the bb genotype compared with subjects carrying the BB/Bb genotype, while no significant difference was observed among patients in the H. pylori-negative group and among those infected with non-virulent strains of H. pylori.
Table 2.
Genotypic distribution frequencies of the BsmI VDR gene polymorphism in H. pylori-infected and -uninfected patients according to the strong degree of neutrophil polymorphs activity in gastric mucosa
| H. pylori infection | Genotypes | Strong degree of neutrophil polymorphic activity
|
p-value | Z-value | |
|---|---|---|---|---|---|
| n | % | ||||
| Virulent strains | bb | 27/60 | 45.00 | 0.055 | 1.596 |
| BB+Bb | 16/25 | 64.00 | |||
| Non-virulent strains | bb | 6/14 | 42.86 | 0.187 | 0.891 |
| BB+Bb | 5/18 | 27.78 | |||
| Negative | bb | 3/46 | 6.25 | 0.220 | 0.773 |
| BB+Bb | 5/45 | 11.11 | |||
Abbreviations: VDR, vitamin D receptor; H. pylori, Helicobacter pylori.
The 3′-UTR VDR SNPs (BsmI, ApaI and TaqI) did not exhibit significant LD with each other. No LD was also observed between FokI and other polymorphisms tested.
Table 3 shows the allelic and haplotypic frequencies of VDR polymorphisms in infected and noninfected H. pylori patients in relation to data from the literature for the Brazilian population14 and HapMap population.15 Comparative analysis revealed differences in the allelic distribution of FokI polymorphism estimated according to relative populations from Europe and Africa. Indeed, the F allele of the FokI polymorphism was more common in the African group than in the Belém population. Similarly, differences were observed in the allelic frequencies of the 3′-UTR VDR polymorphisms (BsmI, ApaI and TaqI). The Belém population was genetically more distant from the Asian groups than the European and African populations, but the allelic frequencies of TaqI polymorphism in the studied population were distinguished from the Brazilian population.
Table 3.
Comparison of allelic frequencies of VDR gene polymorphisms between the study population and the HapMap populations
| Population | SNP (allele)
|
|||
|---|---|---|---|---|
| FokI (F) | BsmI (b) | ApaI (a) | TaqI (T) | |
| Belém | 0.661 | 0.707 | 0.385 | 0.762 |
| Hp+ | 0.671 | 0.718 | 0.393 | 0.765 |
| Hp− | 0.648 | 0.692 | 0.374 | 0.758 |
| BRZ | 0.674 | 0.605 | 0.460 | 0.627 |
| CEU | 0.525 | 0.525 | 0.424 | 0.526 |
| YRI | 0.833 | 0.712 | 0.375 | 0.750 |
| ASN | 0.646 | 0.921 | 0.645 | 0.933 |
Notes: Belém, total surveyed patients; Hp+, H. pylori positive; Hp−, H. pylori negative; Brazil, Brazilian population (BRZ) described by Lins et al;14 CEU, Utah residents of Northern and Western European origin; YRI, Sub-Saharan Africans from Yoruba, Ibadan, Nigeria; ASN, Han Chinese in Beijing (CHB) and Japanese, Tokyo (JPT).
Abbreviations: VDR, vitamin D receptor; SNP, single nucleotide polymorphism; H. pylori, Helicobacter pylori.
Table 4 summarizes the results of this comparative analysis among different interpopulation pairs, based on the frequencies of the four SNPs of the VDR gene. In general, significant differences in F-values indicate the level of genetic differentiation resulting from this comparison.
Table 4.
Locus-by-locus comparison of frequencies of fixation index (FST) values between different pairwise Brazilian and HapMap populations
| Pairwise population |
FokI
|
BsmI
|
ApaI
|
TaqI
|
||||
|---|---|---|---|---|---|---|---|---|
| FST | p | FST | p | FST | p | FST | p | |
| Hp+/Hp− | −0.009 | 0.782 | −0.008 | 0.709 | −0.009 | 0.754 | −0.009 | 0.718 |
| Hp+/BRZ | −0.007 | 0.909 | 0.021 | 0.045 | 0.002 | 0.218 | 0.040 | 0.009 |
| Hp+/CEU | 0.039 | 0.009 | 0.070 | 0.000 | −0.007 | 0.754 | 0.118 | 0.000 |
| Hp+/YRI | 0.062 | 0.000 | 0.008 | 1.000 | −0.007 | 0.800 | −0.006 | 0.636 |
| Hp+/ASN | −0.006 | 0.545 | 0.125 | 0.000 | 0.112 | 0.000 | 0.096 | 0.000 |
| Hp−/BRZ | −0.007 | 0.645 | −0.008 | 0.182 | 0.007 | 0.264 | 0.031 | 0.018 |
| Hp−/CEU | 0.026 | 0.045 | 0.049 | 0.018 | −0.004 | 0.527 | 0.103 | 0.000 |
| Hp−/YRI | 0.078 | 0.000 | −0.008 | 0.709 | −0.009 | 1.000 | −0.009 | 0.836 |
| Hp−/ASN | −0.009 | 0.727 | 0.158 | 0.000 | 0.129 | 0.000 | 0.112 | 0.000 |
Notes: Hp+, H. pylori positive; Hp−, H. pylori negative; Brazil, Brazilian population (BRZ) described by Lins et al;14 CEU, Utah residents of Northern and Western European origin; YRI, Sub-Saharan Africans from Yoruba, Ibadan, Nigeria; ASN, Han Chinese in Beijing (CHB) and Japanese, Tokyo (JPT).
Abbreviation: H. pylori, Helicobacter pylori.
Discussion
In the present study, a significant difference in genotypic distribution of the SNP BsmI between H. pylori-positive and -negative samples was observed. Basically, these findings showed a significant deviation in genotype frequency of SNP BsmI from that of the VDR gene, which was determined by a high bb genotype frequency in the H. pylori-positive group colonized by a virulent-type CagA/VacAs1m1 strain.
Evidence of an elevated frequency of the BsmI SNP bb in patients with gastritis, compared with that observed in patients carrying other genotypes (BB/Bb), suggests that this can be another additive factor in the interaction with H. pylori virulent strains, which encodes the CagA protein, conferring a higher risk for gastric cancer.25,26 In general, CagA shows the ability to bind several kinds of vital proteins, causing dysregulation of multiple classic signaling pathways by complex molecular mechanisms, especially target genes transcribed to promote tumorigenesis,26 with the possible involvement of VDR expression.
It is a fact that immune modulator properties of the active vitamin D are at least partially mediated by VDR,27 since the vitamin D metabolites are involved in antimicrobial activity induction and they also have anti-inflammatory effects,28,29 thereby inhibiting the Th1/Th17 immune response.30
Another relevant aspect is that H. pylori virulent strains can induce higher inflammatory reactions in the gastric mucosa. Previous studies have reported that virulent strains increased the secretion of proinflammatory cytokines that are chemotactic for neutrophils and mononuclear cells, resulting in intense infiltration of these immune cells. Therefore, they are related to more severe pathologies such as ulcer and gastric cancer.31,32
It is worth mentioning that in the present study, H. pylori infection diagnosed via molecular PCR showed 58% prevalence in patients with chronic gastritis. Similarly, a predominance of virulent type I (CagA+/s1m1) strains that reached 73% in the infected group was verified. Vinagre et al33 also reported similar findings using data from the same population.
During infection, H. pylori can persist for long periods in the mucosa, because it can neutralize gastric acid through strong activity of its urease enzymes, which hydrolyze urea and produce a large amount of ammonia and carbon dioxide that increase gastric pH34 and the enzyme γ-glutamyl transpeptidase, which supports its growth and survival in the gastric mucosa.35 It has been observed that both mechanisms can be inhibited by vitamin D administration.36
Particularly notable was the reduced neutrophil activity in the gastric mucosa observed among the patients infected by virulent strains of H. pylori and carrying the bb genotype, allowing bacterial development and persistent colonization of the stomach. This probably reflects the immunosuppressive role of vitamin D on the inhibition of neutrophil chemotaxis and host inflammatory response induced by CagA-positive H. pylori, suggesting that the BsmI polymorphism may be a marker of the cellular effect of vitamin D. These findings are consistent with the results reported by Guo et al,4 who observed a significant positive correlation between the chronic inflammation score, VDR mRNA expression and cathelicidin levels in H. pylori-infected patients.
It has been detected that expression of the cathelicidin–peptide antimicrobial complex,37 the product that is enzymatically cleaved by proteinase 3 to produce the antimicrobial agent cathelicidin (LL37),38 is more abundant in neutrophils and epithelial cells.39,40 The bactericidal activity of cathelicidin is mediated by its ability to bind to phosphatidylglycerol monolayer and disrupt the bacterial cell wall.41 The antibacterial activity of vitamin D3 and its metabolites was investigated recently in H. pylori cultures. In that experiment, the authors noticed that intact forms of 25-hydroxyvitamin D3 and 1α,25-dihydroxyvitamin D3 significantly inhibited the H. pylori proliferation in vitro.42
In this sense, our results showed an elevated frequency of the BsmI SNP BB/Bb genotypes in patients infected by CagA-negative H. pylori, supporting the hypothesis that there are VDR gene variants associated with different types of immunomodulatory effects. Furthermore, the BsmI B allele was reported to be significantly associated with the reduced risk of many types of cancer in a previous meta-analysis.43,44 Therefore, it is intriguing to speculate on the reason for the increased susceptibility to gastric risk presented by individuals infected by CagA-positive H. pylori and therefore having a less active VDR.
On the other hand, it is not clear whether the BsmI polymorphism has an effect on the expression level and activity of the translated VDR protein.45,46 In addition, there is controversy in the genetic association studies concerning the role of VDR polymorphism in diseases that can be assigned due to variation in LD patterns in different ethnic groups.12
These divergences strengthen the need to characterize the immunogenetic profile of patients carrying a set of variants that influence their genetic predisposition to H. pylori infection. In this respect, it is appropriate to evaluate the allelic and genotypic frequencies of VDR polymorphism in different ethnic groups, mainly because the population of Belém has a mixed racial origin and presents with a high prevalence of H. pylori infection. Thus, the F-statistics values revealed significant differences in the allele frequencies of VDR SNPs between the population studied and other ethnic groups reported in the literature.15 These differences might be the result of the mix of different racial groups, given that the population of Belém is highly distinctive in terms of genetic origin.47
On the other hand, it must be emphasized that infectious diseases, such as H. pylori infection, result from a complex interaction between genes and environmental and social factors. These interactions are crucial for the evolutionary processes and genetic composition of a population, which probably contributed (together with the ethnic influences) to the variation in the allelic frequencies observed in the study population.
In view of the reported results, there are some limitations, which should be considered with respect to the possible effects of population substructure, which can result in false-positive associations. The other critical point is that moderate sample sizes affect statistical power, making it difficult to evaluate the haplotypic interactions in a H. pylori infection. Contradictory data in genetic association studies might also be owing to differences in environmental exposure risk patterns and the combination of susceptibility variants, as previously reported.48
Thus, considering such limitations, a careful approach must be adopted in the interpretation of these findings, and more extensive investigations at the molecular level should be conducted to elucidate the relationship between VDR gene polymorphisms and pathogenesis of H. pylori infection, mainly because of the absence of data on this subject, making it impossible to perform a comparative analysis. Independent studies must be conducted to validate the relationship between VDR gene polymorphisms and the pathogenesis of H. pylori infection.
Conclusion
We identified a possible association between the BsmI polymorphism and infection by H. pylori. However, further research is required to clarify this relationship.
Acknowledgments
The authors thank all participating patients. The authors are grateful to the Brazilian National Research Council (CNPq) for financial support. They are grateful too to Marcelo Vieira, Rafael Silva, Isabella Oliveira and Camille Santos, members of the Immunogenetics Laboratory at Federal University of Pará.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yang I, Nell S, Suerbaum S. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev. 2013;37(5):736–761. doi: 10.1111/1574-6976.12027. [DOI] [PubMed] [Google Scholar]
- 2.Rhee KH, Park JS, Cho MJ. Helicobacter pylori: bacterial strategy for incipient stage and persistent colonization in human gastric niches. Yonsei Med J. 2014;55(6):1453–1466. doi: 10.3349/ymj.2014.55.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kusters JG, van Vliet AHM, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo LH, Chen WG, Zhu HT, et al. Helicobacter pylori induces increased expression of the vitamin D receptor in immune responses. Helicobacter. 2014;19(1):37–47. doi: 10.1111/hel.12102. [DOI] [PubMed] [Google Scholar]
- 5.Crofts LA, Hancock MS, Morrison NA, Eisman JA. Multiple promoters direct the tissue-specific expression of novel N-terminal variant human vitamin D receptor gene transcripts. Proc Natl Acad Sci U S A. 1998;95(18):10529–10534. doi: 10.1073/pnas.95.18.10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alagarasu K, Honap T, Mulay AP, Bachal RV, Shah PS, Cecilia D. Association of vitamin D receptor gene polymorphisms with clinical outcomes of dengue virus infection. Hum Immunol. 2012;73(11):1194–1199. doi: 10.1016/j.humimm.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Sortica VA, Cunha MG, Ohnishi MDO, et al. IL1B, IL4R, IL12RB1 and TNF gene polymorphisms are associated with Plasmodium vivax malaria in Brazil. Malar J. 2012;11:409. doi: 10.1186/1475-2875-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salimi S, Farajian-Mashhadi F, Alavi-Naini R, Talebian G, Narooie-Nejad M. Association between vitamin D receptor polymorphisms and haplotypes with pulmonary tuberculosis. Biomed Rep. 2015;3(2):189–194. doi: 10.3892/br.2014.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colin EM, Weel A, Uitterlinden AG, et al. Consequences of vitamin D receptor gene polymorphisms for growth inhibition of cultured human peripheral blood mononuclear cells by 1,25-dihydroxyvitamin D-3. Clin Endocrinol (Oxf) 2000;52(2):211–216. doi: 10.1046/j.1365-2265.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- 11.Jurutka PW, Remus LS, Whitfield GK, et al. The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol Endocrinol. 2000;14(3):401–420. doi: 10.1210/mend.14.3.0435. [DOI] [PubMed] [Google Scholar]
- 12.Morrison NA, Qi JC, Tokita A, et al. Prediction of bone-density from vitamin-d receptor alleles. Nature. 1994;367(6460):284–287. doi: 10.1038/367284a0. [DOI] [PubMed] [Google Scholar]
- 13.Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338(2):143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Lins TC, Vieira RG, Grattapaglia D, Pereira RW. Population analysis of vitamin D receptor polymorphisms and the role of genetic ancestry in an admixed population. Genet Mol Biol. 2011;34(3):377–385. doi: 10.1590/S1415-47572011000300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International HapMap Consortium The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 16.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis – the updated Sydney system. Am J Surg Pathol. 1996;20(10):1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammar M, Tyszkiewicz T, Wadstrom T, et al. Rapid detection of Helicobacter-pylori in gastric biopsy material by polymerase chain-reaction. J Clin Microbiol. 1992;30(1):54–58. doi: 10.1128/jcm.30.1.54-58.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chattopadhyay S, Patra R, Ramamurthy T, et al. Multiplex PCR assay for rapid detection and genotyping of Helicobacter pylori directly from biopsy specimens. J Clin Microbiol. 2004;42(6):2821–2824. doi: 10.1128/JCM.42.6.2821-2824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pani MA, Knapp M, Donner H, et al. Vitamin D receptor allele combinations influence genetic susceptibility to type 1 diabetes in Germans. Diabetes. 2000;49(3):504–507. doi: 10.2337/diabetes.49.3.504. [DOI] [PubMed] [Google Scholar]
- 21.Ayres M, Ayres M, Jr, Ayres DL, dos Santos AS. Bioestat 5.0: Aplicações estatísticas nas áreas das ciências biológicas e médicas. [Bioestat 5.0: Statistical applications in the areas of biological and medical sciences] 3th ed. Belém: MCT-CNPq; 2005. p. 364. Portuguese. [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 24.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 25.Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe. 2014;15(3):306–316. doi: 10.1016/j.chom.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Yong X, Tang B, Bo-Sheng L, et al. Helicobacter pylori virulence factor CagA promoter tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun Signal. 2015;13:30. doi: 10.1186/s12964-015-0111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White JH. Vitamin D metabolism and signaling in the immune system. Rev Endocr Metab Disord. 2012;13(1):21–29. doi: 10.1007/s11154-011-9195-z. [DOI] [PubMed] [Google Scholar]
- 28.Youssef DA, Miller CW, El-Abbassi AM, et al. Antimicrobial implications of vitamin D. Dermatoendocrinol. 2011;3(4):220–229. doi: 10.4161/derm.3.4.15027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangin M, Sinha R, Fincher K. Inflammation and vitamin D: the infection connection. Inflamm Res. 2014;63(10):803–819. doi: 10.1007/s00011-014-0755-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantorna MT, Snyder L, Lin YD, et al. Vitamin D and 1,25(OH)(2)D regulation of T cells. Nutrients. 2015;7(4):3011–3021. doi: 10.3390/nu7043011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montecucco C, Papini E, de Bernard M, Zoratti M. Molecular and cellular activities of Helicobacter pylori pathogenic factors. FEBS Lett. 1999;452(1–2):16–21. doi: 10.1016/s0014-5793(99)00652-3. [DOI] [PubMed] [Google Scholar]
- 32.Nogueira C, Figueiredo C, Carneiro F, et al. Helicobacter pylori genotypes may determine gastric histopathology. Am J Pathol. 2001;158(2):647–654. doi: 10.1016/s0002-9440(10)64006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinagre RM, Corvelo TC, Arnaud VC, Leite AC, Barile KA, Martins LC. Determination of strains of Helicobacter pylori and of polymorphism in the interleukin-8 gene in patients with stomach cancer. Arq Gastroenterol. 2011;48(1):46–51. doi: 10.1590/s0004-28032011000100010. [DOI] [PubMed] [Google Scholar]
- 34.Hazell SL, Lee A. Campylobacter pyloridis, urease, hydrogen-ion back diffusion, and gastric-ulcers. Lancet. 1986;2(8497):15–17. doi: 10.1016/s0140-6736(86)92561-4. [DOI] [PubMed] [Google Scholar]
- 35.Chevalier C, Thiberge JM, Ferrero RL, et al. Essential role of Helicobacter pylori gamma-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol Microbiol. 1999;31(5):1359–1372. doi: 10.1046/j.1365-2958.1999.01271.x. [DOI] [PubMed] [Google Scholar]
- 36.Kawaura A, Takeda E, Tanida N, et al. Inhibitory effect of long term 1 alpha-hydroxyvitamin D3 administration on Helicobacter pylori infection. J Clin Biochem Nutr. 2006;38(2):103–106. [Google Scholar]
- 37.Martineau AR, Wilkinson KA, Newton SM, et al. IFN-gamma- and TNF-independent vitamin D-inducible human suppression of mycobacteria: the role of cathelicidin LL-37. J Immunol. 2007;178(11):7190–7198. doi: 10.4049/jimmunol.178.11.7190. [DOI] [PubMed] [Google Scholar]
- 38.Sorensen OE, Follin P, Johnsen AH, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97(12):3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- 39.Risso A. Leukocyte antimicrobial peptides: multifunctional effector molecules of innate immunity. J Leukoc Biol. 2000;68(6):785–792. [PubMed] [Google Scholar]
- 40.van Wetering S, Tjabringa GS, Hiemstra PS. Interactions between neutrophil-derived antimicrobial peptides and airway epithelial cells. J Leukoc Biol. 2005;77(4):444–450. doi: 10.1189/jlb.0604367. [DOI] [PubMed] [Google Scholar]
- 41.Neville F, Cahuzac M, Konovalov O, et al. Lipid headgroup discrimination by antimicrobial peptide LL-37: insight into mechanism of action. Biophys J. 2006;90(4):1275–1287. doi: 10.1529/biophysj.105.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosoda K, Shimomura H, Wanibuchi K, et al. Identification and characterization of a vitamin D3 decomposition product bactericidal against Helicobacter pylori. Sci Rep. 2015;5:8860. doi: 10.1038/srep08860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raimondi S, Pasquali E, Gnagnarella P, et al. BsmI polymorphism of vitamin D receptor gene and cancer risk: a comprehensive meta-analysis. Mutat Res. 2014;769:17–34. doi: 10.1016/j.mrfmmm.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Xu YQ, He BS, Pan YQ, et al. Systematic review and meta-analysis on vitamin D receptor polymorphisms and cancer risk. Tumour Biol. 2014;35(5):4153–4169. doi: 10.1007/s13277-013-1544-y. [DOI] [PubMed] [Google Scholar]
- 45.Luo XY, Yang MH, Wu FX, et al. Vitamin D receptor gene BsmI polymorphism B allele, but not BB genotype, is associated with systemic lupus erythematosus in a Han Chinese population. Lupus. 2012;21(1):53–59. doi: 10.1177/0961203311422709. [DOI] [PubMed] [Google Scholar]
- 46.Selvaraj P, Prabhu AS, Harishankar M, Alagarasu K. Plasma 1,25 dihydroxy vitamin D3 level and expression of vitamin D receptor and cathelicidin in pulmonary tuberculosis. J Clin Immunol. 2009;29(4):4470–4478. doi: 10.1007/s10875-009-9277-9. [DOI] [PubMed] [Google Scholar]
- 47.Santos NPC, Ribeiro-Rodrigues EM, Ribeiro-dos-Santos AKC, et al. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31(2):184–190. doi: 10.1002/humu.21159. [DOI] [PubMed] [Google Scholar]
- 48.Kraft P, Hunter D. Integrating epidemiology and genetic association: the challenge of gene-environment interaction. Philos Trans R Soc Lond B Biol Sci. 2005;360(1460):1609–1616. doi: 10.1098/rstb.2005.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]