Abstract
A set of pair-rule (PR) segmentation genes (PRGs) promotes the formation of alternate body segments in Drosophila melanogaster. Whereas Drosophila embryos are long-germ, with segments specified more or less simultaneously, most insects add segments sequentially as the germband elongates. The hide beetle Dermestes maculatus represents an intermediate between short- and long-germ development, ideal for comparative study of PRGs. We show that eight of nine Drosophila PRG orthologs are expressed in stripes in Dermestes. Functional results parse these genes into three groups: Dmac-eve, -odd and -run play roles in both germband elongation and PR patterning; Dmac-slp and -prd function exclusively as complementary, classic PRGs, supporting functional decoupling of elongation and segment formation; and orthologs of ftz, ftz-f1, h and opa show more variable function in Dermestes and other species. While extensive cell death generally prefigured Dermestes PRG RNAi-mediated cuticle defects, an organized region with high mitotic activity near the margin of the segment addition zone is likely to have contributed to truncation of eveRNAi embryos. Our results suggest general conservation of clock-like regulation of PR stripe addition in sequentially segmenting species while highlighting regulatory rewiring involving a subset of PRG orthologs.
KEY WORDS: Segmentation, Germband elongation, Pair-rule gene, Dermestes, Beetle, Evo-devo
Summary: Expression and functional studies in hide beetles suggest independent roles for pair-rule genes in elongation and segmentation of sequentially segmenting insects, and rewiring of a subset of these regulatory genes.
INTRODUCTION
Segmentation is a feature of all arthropods, and mechanisms controlling it would be expected to be largely shared among diverse members of this clade. The regulation of segmentation has been studied extensively in the model insect Drosophila melanogaster, where genes act hours before segments actually form, prefiguring and determining the patterns of segmentation that unfold during later stages of embryogenesis (Akam, 1987; Ingham, 1988; Lawrence, 1992). In recent years, the advent of molecular genetic approaches in diverse insects has enabled comparative studies of regulatory mechanisms in other species (Abzhanov et al., 1999; Angelini et al., 2005; Liu and Kaufman, 2005b; Peel et al., 2005; Williams and Nagy, 2017). Studies of orthologs of Drosophila segmentation genes have revealed that they are present in genomes throughout Insecta, as well as more distant arthropods, despite the fact that the morphological events leading to segment formation in most arthropods differ from those in Drosophila (Davis and Patel, 2002; Peel et al., 2005). Drosophila are long-germ insects (Krause, 1939) in which all segments are specified more or less simultaneously in the blastoderm [ʻsimultaneous segmentation' (Davis and Patel, 2002)]. By contrast, other arthropods and most insects add posterior segments sequentially after the blastoderm stage, during which only a small number of anterior segments are specified [ʻsequential segmentation' (Davis and Patel, 2002; Peel et al., 2005)]. Sequentially segmenting arthropods can be subdivided into short- and intermediate-germ developers, based on the number of segments specified in the blastoderm (Davis and Patel, 2002). Thus, the presence of morphological segments is ubiquitous but the way in which these segments develop varies, suggesting greater constraint on the morphological outcome – segment existence – than the developmental mechanisms leading to their formation, an example of ʻdevelopmental system drift' (True and Haag, 2001). Since the genes utilized in the most derived mode of segmentation, namely long-germ development seen in Drosophila, are present in animals that form segments by an alternate route, they must be, at least to some extent, repurposed to achieve this.
In Drosophila, nine pair-rule (PR) segmentation genes (PRGs) interact to establish repeated segments along the anterior-posterior axis (Gilbert, 2010; Nüsslein-Volhard et al., 1987; Nüsslein-Volhard and Wieschaus, 1980). Most Drosophila PRGs are expressed in striped patterns, termed PR stripes, at the blastoderm stage in the primordia of the alternate segmental regions, which are missing in mutant embryos (Akam, 1987; Lawrence, 1992). PR-like expression of orthologs of Drosophila PRGs has been reported in a number of arthropods, but their functions have been analyzed in only a few insects (Choe et al., 2006; Liu and Kaufman, 2005a; Mito et al., 2007; Nakao, 2015; Rosenberg et al., 2014; Wilson and Dearden, 2012). In the flour beetle Tribolium castaneum (Tc), which is a short-germ insect, most PRG orthologs are expressed in striped patterns but, unlike Drosophila, knockdown of Tc-even-skipped (eve), -odd-skipped (odd) and -runt (run) produced larvae consisting of only anterior structures (Choe and Brown, 2009; Choe et al., 2006). In addition, unlike Drosophila, Tc-odd and -eve are part of a wave-like clock mechanism that is responsible for the addition of segments sequentially during germband elongation, similar to the segmentation clock seen in vertebrates (El-Sherif et al., 2012; Sarrazin et al., 2012). In hemimetabolous Gryllus bimaculatus and Oncopeltus fasciatus, eveRNAi caused disorganized and/or truncated thorax and abdomen (Liu and Kaufman, 2005a; Mito et al., 2007), with PR-like defects detected in the anterior region in Gryllus (Mito et al., 2007). These findings revealed the differential function of some PRGs in these sequentially segmenting species, especially during posterior segment addition. Surprisingly, in two Hymenoptera, whose long-germ mode is thought to have evolved independently from that of Drosophila, PRG orthologs still show functional divergence: morpholino knockdown of eve, odd and hairy (h) resulted in loss of abdomen in Nasonia vitripennis, while eve, fushi tarazu (ftz) and run are required for maternal patterning in Apis mellifera (Rosenberg et al., 2014, 2009; Wilson and Dearden, 2012).
Coleoptera is the largest and perhaps most diverse insect order and, as first highlighted by Patel et al. (1994), this order includes species with short-, intermediate- and long-germ modes of segmentation, making it an ideal taxon for comparative studies of segmentation mechanisms (Davis and Patel, 2002; Sander, 1976). Here, we have investigated the expression and function of PRG orthologs in Dermestes maculatus (Dmac), an intermediate-germ beetle that diverged from T. castaneum ∼250 million years ago (Hedges et al., 2015). The twofold goal of this study was to provide comparative data on segmentation mechanisms in a second coleopteran model system, while providing insight into mechanisms underlying intermediate-germ development. Previous work by Patel et al. (1994) demonstrated that eve is expressed in PR stripes in a related intermediate-germ beetle, Dermestes frischi. Here we show that eight of the nine PRG orthologs are expressed in stripes arising sequentially from the posterior in both blastoderm stage embryos and in the elongating germband of D. maculatus embryos. PR-like defects were seen for sloppy paired (slp) RNAi knockdown, indicating exclusive roles in PR patterning, as previously reported for Dmac-paired (prd) (Xiang et al., 2015). Simultaneous knockdown of prd and slp resulted in elongated germbands with no Engrailed (En) stripes and lacking segment borders, indicating that germband elongation and patterning of segmental boundaries are decoupled processes. Knockdown of Dmac-eve, -odd and -run resulted in truncated embryos, indicating roles in germband elongation, as in Tribolium (Choe et al., 2006), while PR-like defects after moderate knockdown demonstrated that they also play roles in PR patterning similar to their Drosophila orthologs. Milder PR-like defects were observed for ftzRNAi, whereas Dmac-h knockdown impacted formation of the abdomen more broadly. Patterns of apoptosis seen after knockdown of each PRG ortholog generally prefigured morphological defects, although we also observed disruptions in mitotic patterns.
RESULTS
Dermestes PRG orthologs show PR-like expression with distinct features
D. maculatus PRG orthologs were isolated by degenerate PCR and 3′ RACE. Phylogenetic analysis demonstrated that each Dermestes PRG ortholog clustered closely with its Tribolium counterpart (Fig. S1). Expression was examined by in situ hybridization. Embryonic stages were determined using SYTOX Green nuclear staining, as described (Xiang et al., 2015).
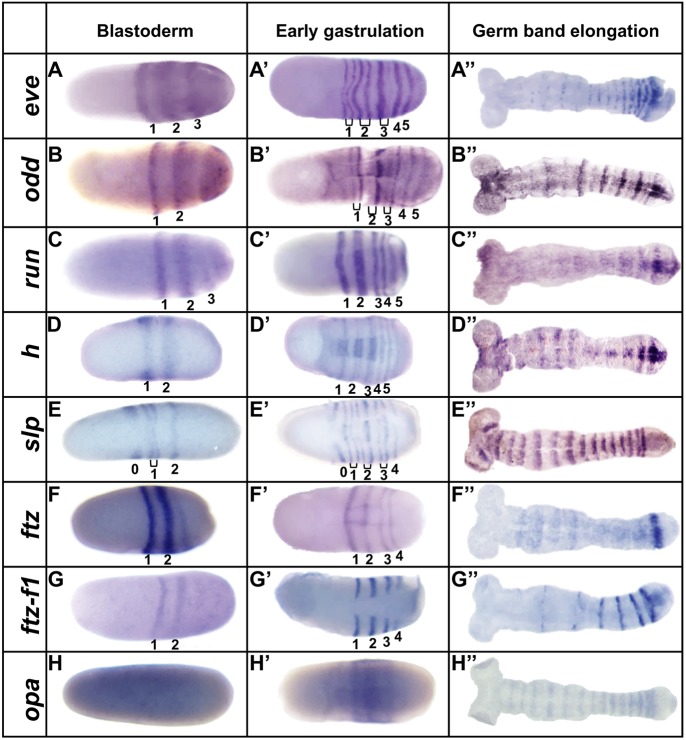
Dmac-eve, -odd and -run striped expression arose at the blastoderm stage, with primary stripes emerging sequentially from cap-like expression in the posterior region. Stripes arose in an anterior-to-posterior order (Fig. 1A-C, Fig. S2). As new stripes emerged posteriorly, the more anterior stripes refined (Fig. S2A-D,G-J,M-P). Prior to gastrulation, before formation of the ventral furrow, there were at least four primary stripes of each gene (Fig. S2D,J,P). During gastrulation, and as the germband elongated, posterior stripes were added sequentially while anterior stripes faded gradually (Fig. 1, Fig. S2).
Fig. 1.
Eight Dermestes PRG orthologs are expressed in PR stripes. Expression of Dermestes PRGs as assessed by in situ hybridization during early development at the stages indicated. Early gastrulation was defined by appearance of the ventral furrow. Note that Dermestes embryos vary in shape and size. (A-A″) Dmac-eve. (A) The first primary stripe has refined as a transverse stripe in the embryo's center. The second primary stripe is fuzzy and broad. A third stripe is resolving from cap-like expression in the posterior end. (A′) Five primary stripes have emerged. The anterior three have split to form secondary stripes (brackets). (A″) Stripes are fading in anterior segments but remain strong in posterior segments. (B-B″) Dmac-odd. (B) Two primary stripes are clear, along with cap-like expression in the posterior. (B′) Five primary stripes and three intercalated secondary stripes have emerged. (B″) Primary and secondary stripes alternate in intensity, with fading anterior expression. (C-C″) Dmac-run. (C) Three stripes have arisen with weak cap-like expression in the posterior end. (C′) In total, five stripes have emerged; the first and second are broader. (C″) Anterior expression has faded; weak stripes remain at the posterior end. (D-D″) Dmac-h. (D) Two Dmac-h stripes in the center of the embryo. (D′) At least five primary stripes are detectable. The second primary stripe has split into two thin stripes. (D″) Strong stripes in the most posterior two segments with faint expression in the anterior. (E-E″) Dmac-slp. (E) Three primary stripes with secondary stripes emerging for stripe 1 (bracket). The most anterior stripe (stripe 0) marks the future antennae. (E′) Stripe 0, four primary stripes and three secondary stripes are present. (E″) Segmental stripes show equal intensity, with fuzzy anterior borders. (F-F″) Dmac-ftz. (F) Two strong, central stripes. (F′) Four stripes at early gastrulation. (F″) Strong expression remains in the most posterior segment. (G-G″) Dmac-ftz-f1. (G) Lateral expression in two stripes. (G′) Four stripes are visible but Dmac-ftz-f1 is not expressed in the ventral furrow. (G″) Fading expression in the anterior; strong stripes in the posterior. (H-H″) Dmac-opa. (H) Uniform, weak expression at blastoderm. (H′) Ubiquitous expression in the trunk. (H″) Weak segmental stripes throughout the germband. Embryos are oriented with anterior, left.
For Dmac-eve, secondary segmental stripes appeared by splitting of the primary stripes (Fig. S2D, arrows). For Dmac-odd, one secondary stripe was added de novo anterior to each primary stripe (Fig. S2I,J, arrows). Throughout late blastoderm and germband elongation, Dmac-odd secondary stripes remained weaker than primary stripes (Fig. S2J-L). Secondary stripes were not observed for Dmac-run (Fig. 1C′,C″, Fig. S2O-R).
In contrast to eve, odd and run, Dmac-h was first observed broadly in the center of the embryo, with no obvious posterior cap-like expression (Fig. S3A). Two primary stripes resolved from this broad expression (Fig. S3B, Fig. 1D). Additional stripes were added sequentially from the posterior half; at least five primary stripes were detected at early gastrulation (Fig. S3D). Secondary stripes arose by splitting of most primary stripes (Fig. 1D′, stripes 2, 3). As the germband elongated, additional stripes emerged from the posterior end (Fig. S3E,F). The striped pattern was maintained through mid-germband stages, but then faded quickly (Fig. 1D″, Fig. S3G).
Dmac-slp was first expressed in a broad stripe in the center of the embryo, and later resolved into two stripes with stronger expression laterally (Fig. S3H,I). The first stripe remained strong only on the lateral sides, later contributing to the head lobes. Secondary striped expression appeared anterior to the first primary stripe, as a new primary stripe emerged posterior to it (Fig. 1E). Additional stripes appeared sequentially from the posterior region of the embryo and intercalated secondary stripes appeared between primary stripes, generating seven Dmac-slp stripes at early gastrulation (Fig. 1E′, Fig. S3J,K). Unlike many other D. maculatus PRG orthologs, anterior Dmac-slp stripes remained strong in each segment during germband elongation, although they became broader with slightly fuzzy anterior borders (Fig. 1E″, Fig. S3L-N). There was no obvious intensity difference among stripes in elongated germbands.
Dmac-ftz was first expressed as a single stripe in the center of the embryo (Fig. S3O). A second clear stripe then appeared posterior to the first stripe (Fig. 1F). Later, more stripes were added sequentially from the posterior half of the embryo. In total, there were four stripes at early gastrulation, with the first the most intense (Fig. 1F′). Posterior stripes appeared and anterior striped expression became weak and fuzzy as the germband elongated. In mid- to late-germband, strong striped expression was detectable only in the most posterior segments (Fig. S3P, Fig. 1F″). After segments were well established, Dmac-ftz was expressed segmentally, likely in the CNS (Fig. S3Q). No secondary segmental striped expression was detected throughout early embryogenesis. Dmac-ftz-f1, the Drosophila ortholog of which encodes a Ftz co-factor, was first expressed uniformly in pre-blastoderm stage embryos (Fig. S3R); this early ubiquitous expression of ftz-f1 is conserved in Drosophila and Tribolium (Heffer et al., 2013; Yussa et al., 2001). Maternally deposited Dmac-ftz-f1 was also confirmed by RT-PCR (not shown). Later, expression of Dmac-ftz-f1 was similar to that of Dmac-ftz, with one single stripe appearing first and posterior stripes arising sequentially from the posterior half of the embryo, as ubiquitous expression faded (Fig. S3S, Fig. 1G). Four Dmac-ftz-f1 stripes were present at early gastrulation (Fig. 1G′). These stripes differed from other PRG stripes in that no expression was detected on the dorsal side or in the ventral furrow region (Fig. 1G′). Striped expression persisted through gastrulation, but gradually faded in an anterior-to-posterior fashion as the germband elongated (Fig. S3T,U, Fig. 1G″). Dmac-ftz-f1 was also expressed in the distal leg tips and head appendages in late embryos (not shown). Dmac-odd-paired (opa) was ubiquitously expressed at blastoderm (Fig. 1H) and the onset of gastrulation (Fig. 1H′). Weak segmental striped expression resolved during gastrulation and germband elongation from broad expression in the posterior (Fig. S3W, Fig. 1H″). Expression of Dmac-opa during germband elongation appears similar to that in Tribolium (Choe et al., 2017; Clark and Peel, 2017 preprint).
In summary, except for opa, Dermestes PRG orthologs were expressed in primary PR-like patterns, with stripes added sequentially from the posterior. For each, there were four or five primary stripes established at the onset of gastrulation, consistent with an intermediate-germ designation. For several genes (eve, odd, h, slp), secondary stripes developed either by splitting (eve and h) or de novo (odd and slp). Additional stripes were added from the posterior during germband elongation, with more anterior stripes fading concurrently, except for Dmac-slp, which maintained segmental stripes throughout germband stages.
Knockdown of Dmac-eve, -odd or -run reveals dual roles in segment formation
Parental RNAi (pRNAi) was used to test the function of D. maculatus PRG orthologs. Knockdown (verified by qRT-PCR; Fig. S4) is expected to vary in individual embryos, generating phenocopies of an allelic series, allowing defects to be examined in both hatched (less severely affected) and unhatched (more severely affected) offspring. Average egg yield, hatch count, and penetrance after each knockdown are shown in Table S1.
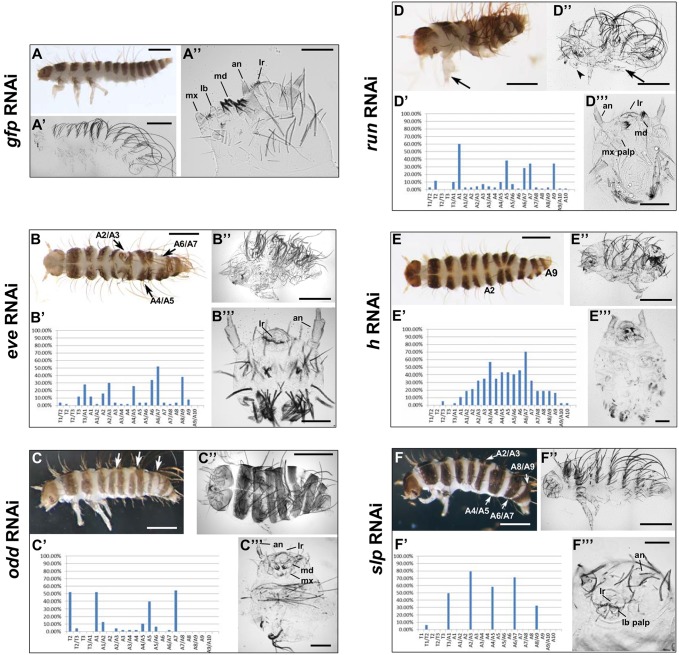
Dmac-eve, -odd and -run pRNAi did not affect egg yield; however, hatch counts were decreased compared with controls (gfp dsRNA injection). A graded series of effects was observed in hatched larvae after these knockdowns. Dmac-eveRNAi hatched larvae displayed defects including abnormalities within segment(s) and/or fusion at segmental boundaries for one or more segments (Fig. 2B). Defects occurred frequently at the boundaries between T3/A1, A2/A3, A4/A5, A6/A7, A8/A9 and neighboring regions, suggestive of PR-like patterning (Fig. 2B′). Similarly, defective segment(s) and/or fused adjacent segments were observed for Dmac-oddRNAi (Fig. 2C). Fusions were generally milder than for Dmac-eveRNAi, with loss of only portions of non-pigmented regions evident. Interestingly, pigmented stripes were sometimes disrupted by non-pigmented tissue (Fig. S5A, arrow). In other cases, the T2 leg was deformed with duplicated claw or thickened paddle-like structures at the tip (Fig. S5B). These are reminiscent of Drosophila odd mutants, which display mirror-image duplications, revealed by duplicated partial denticle belts with reversed polarity (Coulter and Wieschaus, 1988). Overall, for Dmac-odd, defects occurred most frequently in the T2, A1, A5, A7 segments (Fig. 2C′). Dmac-runRNAi produced defects including segment fusions and deletions in hatched larvae (Fig. 2D), sometimes accompanied by a deformed leg with duplicated claw or thickened structure (Fig. 2D, arrow). Extra pigmented regions were also observed, indicating partial segmental duplication (not shown). Defects occurred frequently at T2, A1, A5, A7, A9 and neighboring regions (Fig. 2D′).
Fig. 2.
Truncated embryos and PR defects after Dermestes PRG ortholog knockdown. Offspring of pRNAi for control gfp or for the indicated D. maculatus PRG orthologs. Bar charts indicate the frequency of defects (y-axis) in anterior to posterior segments (x-axis) of affected hatched larvae. Data were from hatched larvae with segmentation defects from three consecutive daily collections after 3′ dsRNA knockdowns. The patterns were very similar after 5′ dsRNA knockdowns. (A,A′) Control Dermestes larvae have three thoracic segments and ten abdominal segments. (A) Hatched larva with pigmented stripes on the head and every trunk segment. (A′) Cuticular preparation of unhatched larva without pigmentation. (A″) Head close-up shows antennae (an), labrum (lr), mandibles (md), maxillae (mx) and labium (lb). (B-B″) Dmac-evepRNAi. (B) Moderately affected hatched larva with multiple fused segments (arrows). (B′) T3/A1, A2/A3, A4/A5, A6/A7, A8/A9, and neighboring regions were most frequently affected (n=50). (B″) Unhatched larva with shortened body length. (B‴) Spherical cuticle of a severely affected unhatched larva consists of only head, with antenna and labrum. (C-C‴) Dmac-oddpRNAi. (C) Hatched larva with only two pairs of legs and several partially fused segments (arrows). (C′) T2, A1, A5 and A7 were most commonly affected (n=48). (C″) Unhatched larva with short body and fewer segments. (C‴) Severely affected unhatched offspring with head and some thoracic structure. (D-D‴) Dmac-runpRNAi. (D) Hatched larva has fewer segments and deformed leg with duplicated claw (arrow). (D′) T2, A1, A5, A7, A9, and adjacent regions most frequently affected (n=70). (D″) Unhatched, shortened larva with duplicated claw (arrow) and leg-like maxilla (arrowhead). (D‴) ‘Head-only’ cuticle lacking partial maxillary and labium structures. (E-E‴) Dmac-hpRNAi. (E) Hatched larva with irregular segmentation between A2 and A9. (E′) Regions between A1 and A9 most frequently affected (n=37). (E″,E‴) Unhatched larva with head and thoracic segments, but little or no segmented abdomen. No wild-type-like terminal structures are evident. (F-F‴) Dmac-slppRNAi. (F) Hatched larva with several fused adjacent segments (arrows). (F′) T3/A1, A2/A3, A4/A5, A6/A7 and A8/A9 are most frequently affected (n=101). (F″) Unhatched larvae with fewer trunk segments and only one pair of legs. (F‴) Unhatched larval head missing mandibles and maxillae. Scale bars: 500 µm, except 100 µm for A″,B‴,C‴,D‴,E‴,F‴.
Defects observed in unhatched offspring were more severe, as would be expected. For Dmac-eveRNAi, moderately affected unhatched larvae included some gnathal and trunk structures, but the overall body length was dramatically shortened (Fig. 2B″). The most severely affected were composed of only a small, spherical anterior structure with antennae and labrum, lacking mandible, maxilla, labium or posterior terminal structures, referred to as ‘head-only’ (Fig. 2B‴). Dmac-oddRNAi moderately affected unhatched larvae had significantly shortened body lengths with fewer trunk segments (Fig. 2C″). Severely affected unhatched larvae had head, one or two thoracic segments, and drastically shortened, asegmental posterior trunks (Fig. 2C‴). As in hatched larvae, deformed T2 legs or duplicated claws were sometimes observed (Fig. S5C). Moderately affected Dmac-runRNAi unhatched larvae had significantly shortened body length with fewer trunk segments (Fig. 2D″). Occasionally, maxillae-to-leg-like transformation was observed (Fig. 2D″, arrowhead). Duplicated claws were found (Fig. 2D″, arrow), as in hatched larvae. Severely affected offspring appeared to be ‘head-only’, similar to severe Dmac-eveRNAi, with wild-type-like antennae, labrum and mandible but missing the labium and some maxillary structures (Fig. 2D‴).
In summary, knockdown of Dmac-eve, -odd or -run revealed two classes of defects. In more severe cases, unhatched larvae were composed of ʻhead-only' (eve, run) or ʻanterior-only' (odd). In milder cases, larvae hatched, displaying PR-like defects (eve, run, odd) along with occasional mirror-image duplications (run, odd). This indicates that these three genes function in at least two phases of segmentation patterning in Dermestes: formation of an elongating germband, and subdivision of organized tissue into metameric units.
Loss of PR function for a subset of Dermestes PRG orthologs?
Unlike the two classes of defects described above, Dmac-h, -ftz or -ftz-f1 pRNAi produced distinct outcomes. Dmac-hRNAi disrupted segmentation throughout the abdominal region (Fig. 2E), with no evidence of PR-like or other specific register (Fig. 2E′), although these defects were only observed in ∼12% of hatched larvae. In unhatched larvae with more severe defects, 28% (27/98) developed to late stage with solid cuticular structures, with little (Fig. 2E″) or almost no (Fig. 2E‴) abdomen.
For Dmac-ftz pRNAi, only 2 of >500 hatched larvae showed fusion of segments, despite knockdown of Dmac-ftz to ∼6% of wild-type levels (Fig. S4). After injection of dsRNA directly into embryos [embryonic RNAi (eRNAi)], 5-11% of injected embryos had fused or missing segments (Fig. 3A,B). Fusion of segments was detected with a PR-like register, usually between A1/A2, A3/A4, A5/A6 and A7/A8, as revealed by anti-En staining (see below). The pattern of fusion was complementary to Dmac-eveRNAi mild effects (Fig. 2B′). However, the low penetrance of segmentation defects after eRNAi, and the absence of detectable defects after pRNAi, indicate that knockdown of Dmac-ftz is tolerated, perhaps due to redundancy with another PRG ortholog.
Fig. 3.
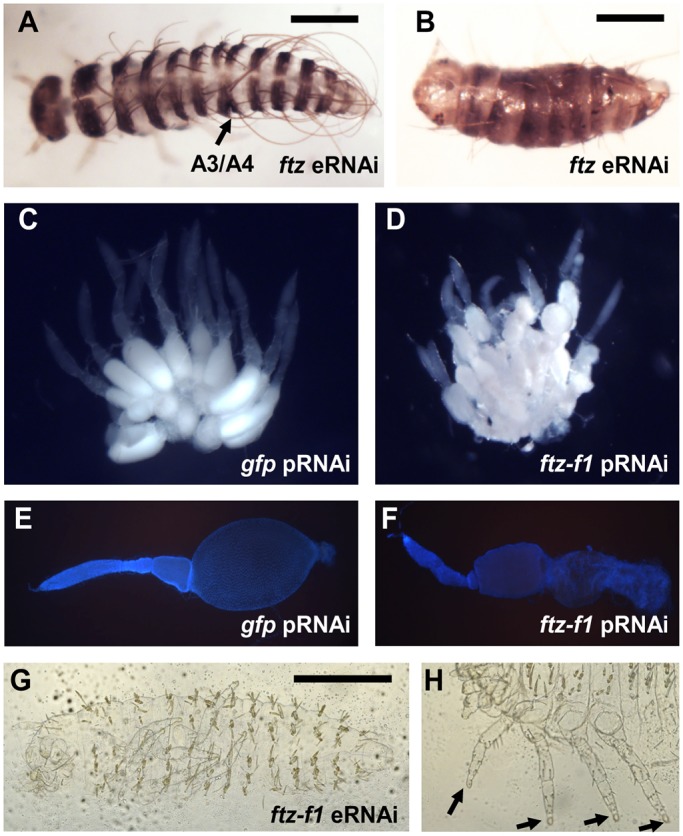
Dmac-ftz and ftz-f1 RNAi. (A,B) Hatched offspring after Dmac-ftz eRNAi. (A) Mildly affected embryo with single segmental fusion (arrow). (B) Severely affected larva with shortened body length and fewer segments. (C,E) Wild-type-like ovaries from gfp dsRNA-injected control female. (C) Large, oval-shaped, developing oocyte in each ovariole. (E) DAPI staining of dissected ovariole reveals large oocyte. (D,F) Ovaries from Dmac-ftz-f1 dsRNA-injected females. (D) Small primary oocytes clustered in each ovariole. No mature oocyte is visible. (F) Dissected ovariole with shrunken oocyte. (G) Unhatched larva from Dmac-ftz-f1 dsRNA-injected female has normal number of segments without any obvious segmentation defect. (H) Arrows indicate truncated distal ends of legs. Scale bars: 500 µm.
RNAi failed to provide convincing evidence for roles of Dmac-ftz-f1 in segmentation. As reported for Tribolium (Heffer et al., 2013; Xu et al., 2010), pRNAi for Dmac-ftz-f1 blocked oogenesis (Table S1, Fig. 3C-F); the small number of eggs laid hatched without any apparent defect. Surprisingly, segmentation defects were not observed after eRNAi either, although PR defects were observed in Tribolium using a similar approach (Heffer et al., 2013). Cuticle phenotypes of unhatched embryos showed well-patterned metameric segments with shortened setae all over the body (Fig. 3G). Claws and head appendages were blunt-ended, consistent with Dmac-ftz-f1 expression in tip primordia and demonstrating activity of the dsRNA (Fig. 3H).
No obvious segmentation defects were observed for Dmac-opa pRNAi, despite knockdown to ∼3% of wild-type levels (Fig. S4), similar to reports in Tribolium (Choe et al., 2006, 2017). We also performed Dmac-opa eRNAi. Although the survival rates were only ∼4% (n=70 and n=112 for two distinct dsRNAs targeting non-overlapping regions), significantly lower than that after gfp dsRNA injection (50%, n=46), specific segmentation defects were not observed. Moreover, >70% of embryos showed wild-type-like segmental striped En expression (not shown). The remaining embryos had overall weak expression or loss of posterior abdominal structures, which were also detected in >10% of control embryos and likely to be artifacts of directly injecting these embryos. Although analysis of genetic mutants for Dmac-h, -ftz-f1, -opa and -ftz will be necessary to definitively determine their roles, these RNAi results suggest that these four PRG orthologs are not required for PR patterning in Dermestes.
Expression of En suggests PR-like functions of Dermestes PRG orthologs
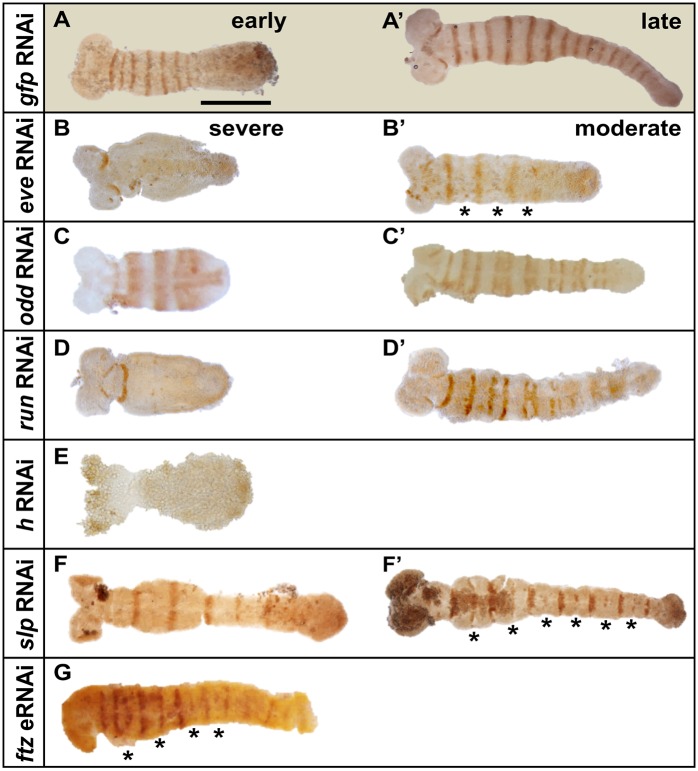
PRGs in Drosophila and Tribolium regulate segment formation in part by controlling the expression of en in segmental stripes (Choe and Brown, 2009; DiNardo and O'Farrell, 1987; Jaynes and Fujioka, 2004). We therefore examined En in early stage control (gfpRNAi) and PRG RNAi embryos. Striped En was detected in control embryos at early and late germband stages (Fig. 4A,A′). In severely affected Dmac-eveRNAi embryos, En was only expressed in antennal segments (Fig. 4B). In less severely affected embryos with relatively elongated germbands, En stripes were reduced or undetectable in alternate segments (Fig. 4B′, asterisks). Dmac-oddRNAi truncated embryos showed fuzzy and enlarged En stripes (Fig. 4C). Less severely affected embryos with relatively elongated germbands displayed abnormally spaced ‘paired’ En stripes (Fig. 4C′), similar to previous findings in Drosophila (Mullen and DiNardo, 1995). Extra En stripes were frequently detected between pairs of En stripes (Fig. S5D,E, arrows), and extra segmental furrows were observed in the region where ectopic En was expressed (Fig. S5D′, red arrow). Offspring of Dmac-run dsRNA-injected females included truncated embryos with En expression in only antenna and mandibular segments (Fig. 4D), while less severely affected embryos with relatively elongated germbands exhibited fused En stripes (Fig. 4D′). For Dmac-hRNAi, many embryos stopped developing at very early stages (∼25% of over 200 embryos; no divided nuclei in the center, not shown). No clear En stripes were detected in truncated embryos (Fig. 4E). For Dmac-slpRNAi, En expression was undetectable or decreased in alternate segments (Fig. 4F,F′, asterisks), consistent with PR cuticle defects. After Dmac-ftz eRNAi, ∼25% of embryos showed reduced En expression in a PR-like alternate segment register (Fig. 4G, asterisks), although the overall En expression at the posterior end of the germband was weak.
Fig. 4.
En expression is altered after Dermestes PRG knockdown. (A,A′) Control embryo (from gfp dsRNA-injected female) shows En stripes with equal intensity in every segment at early or late germband elongation stages. (B) Dmac-evepRNAi severely affected embryo with truncated, asegmental germband only has antennal En expression. (B′) A less severely affected embryo shows reduced En in alternate stripes (asterisks). (C) Dmac-oddpRNAi severely affected embryo with truncated germband has enlarged En stripes. (C′) ‘Paired’ En stripes detected in a moderately affected embryo. (D) Dmac-runpRNAi severely affected embryo. En is only detected in antennal and mandibular segments. (D′) Fused En stripes are visible in moderately affected offspring. (E) Dmac-hpRNAi. No striped En expression pattern is detectable. (F,F′) Dmac-slppRNAi. Alternate En stripes are absent (F) or reduced (F′, asterisks). (G) Dmac-ftzeRNAi. Alternate En stripes are weak or absent (asterisks). Scale bar: 500 µm.
In summary, these results indicate that Dmac-eve, -odd, -run, -slp and -prd (Xiang et al., 2015) are required for proper en expression, behaving similarly to their Drosophila counterparts. No effects on En expression were seen for Dmac-ftz-f1 or opa knockdown, and ftz knockdown had a mild effect.
Dmac-prd and -slp function exclusively in PR patterning in Dermestes
Dmac-slpRNAi produced a range of defects, with mildly affected larvae hatching with one fusion between neighboring segments (not shown) and moderately affected larvae displaying several fusions (Fig. 2F). Very often, T1 and/or T3 legs were missing or deformed (not shown). The fusions occurred between T1/T2, T3/A1, A2/A3, A4/A5, A6/A7 and A8/A9 segmental boundaries (Fig. 2F′), complementary to the pattern produced after Dmac-prdRNAi (Xiang et al., 2015, 2016). Unhatched larvae only had one pair of legs, with shorter body length and fewer segments (Fig. 2F″). The head also showed some defects, with missing mandibles, maxillae and partial labium (Fig. 2F‴). These results suggest a Drosophila-like PR function for Dmac-slp, similar to our previously reported Dmac-prdRNAi PR defects (Xiang et al., 2015), although defects were more severe for Dmac-slpRNAi.
To test whether these genes are required for patterning alternate sets of body segments, we simultaneously knocked down Dmac-slp and Dmac-prd (double pRNAi; Fig. 5, Fig. S7). As expected, the survival rates of gfp/slp and prd/slp double RNAi were significantly lower than for the gfp/gfp double-RNAi control (13.7% and 8.7% versus 75.2%, Table S1). Hatched prd/slp double-RNAi offspring had fusions at odd- or even-numbered or both sets of segmental boundaries, revealing a combination of the defects seen after Dmac-prd and Dmac-slp single knockdowns (Fig. S7). Examination of cuticle preparations of unhatched larvae revealed dramatically shortened larvae, lacking overt signs of segmentation (Fig. 5A,B). Remnants of segments were suggested by residual setae. There were no obvious leg structures, and all gnathal segments (mandible, maxillae, and labium) were missing; only antennae and labrum were evident in head regions. In severely affected embryos, there was no striped En expression in late germbands (Fig. 5C), and embryo morphology, as indicated by SYTOX Green staining, revealed no clear segmental furrows or grooves (Fig. 5C′). In some embryos, reduced En expression in every segment (Fig. 5D) together with fused adjacent segments (Fig. 5D′) indicated a milder phenotype. The defects were more obvious in the posterior region in even-numbered segments (Fig. 5D,D′, asterisks), which were affected in Dmac-slp single knockdown. This milder pattern is consistent with our findings that Dmac-slpRNAi knockdown produced more severe PR defects than Dmac-prdRNAi. In late stage embryos, after germband retraction, there was only residual En expression along the midline (Fig. 5F). No segmental grooves, gnathal, or leg appendages were evident (Fig. 5F′). Taken together, Dmac-prd/slp double RNAi resulted in failure to establish both odd- and even-numbered segments, whereas germband development per se appeared to be largely unaffected.
Fig. 5.
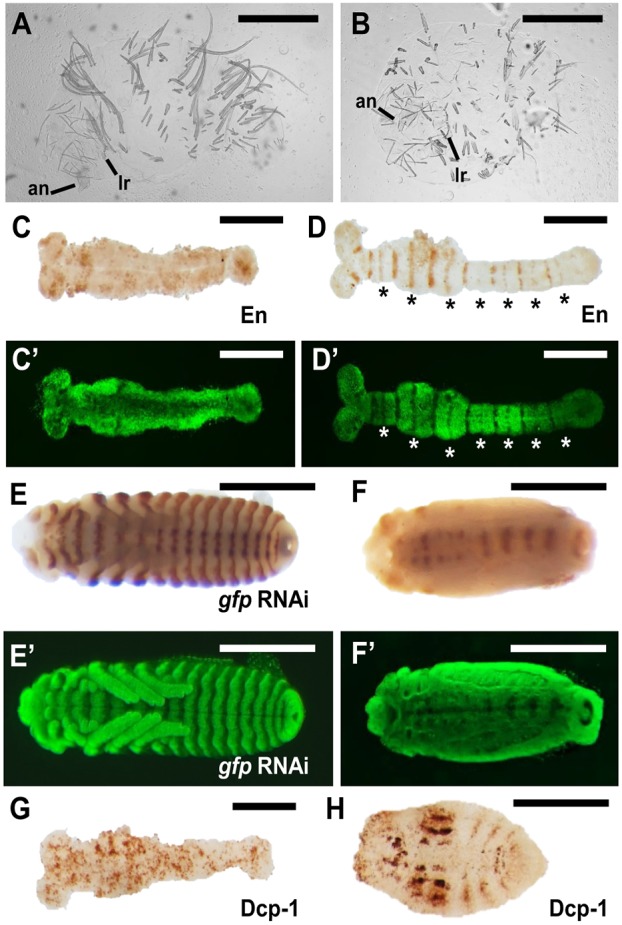
Dmac-prd and -slp double RNAi produces asegmental embryos. (A,B) Significantly shortened unhatched larvae after Dmac-prd, -slp double pRNAi. Only antenna and labrum are visible. No gnathal appendages or legs are present. (C) No striped-like En expression is detected in elongated germband in a severely affected embryo. (D) Moderately affected embryo shows reduced En expression. Reduction is more obvious for even-numbered En stripes (asterisks). (C′,D′) SYTOX Green staining of embryos in C and D for visualization of embryo morphology. Note that there are no obvious segmental grooves (C′) or fused adjacent segments (D′), indicating overt lack of segmentation or milder segmentation defects, respectively. (E) Striped En expression at the posterior border of each segment in control embryos. (F) Only residual En expression is detected along the midline after double knockdown. (E′,F′) SYTOX Green staining of embryos in E and F for visualization of embryo morphology. (G,H) Dcp-1 antibody staining for apoptosis at elongated germband stage (G) and in a later stage embryo (H). There is almost uniform apoptosis signal in the elongated germband (G). Note that we previously observed stronger effects for Dmac-prd using eRNAi than pRNAi (Xiang et al., 2015). Scale bars: 500 µm.
Knockdown of Dermestes PRGs results in increased apoptosis
In Drosophila, cell death contributes to patterning defects in PRG mutants (Hughes and Krause, 2001; Ingham et al., 1985; Magrassi and Lawrence, 1988). To further characterize the role of D. maculatus PRG orthologs, we examined cell death patterns in control and pRNAi embryos by performing antibody staining against the apoptosis marker cleaved Dcp-1 (Fig. 6A-F). Very little apoptotic activity was detected in gfpRNAi offspring from early stages to late germband elongation (not shown). When segmental grooves were clear, only weak apoptotic signal was detected in the head lobes and posterior germband ends (Fig. 6A).
Fig. 6.
Increased apoptosis and disrupted mitosis after Dermestes PRG knockdown. (A-F) Dcp-1 (top) and SYTOX Green nuclear staining (bottom) in pRNAi offspring. (A) gfpRNAi control. Apoptosis is detected in the head and posterior at low levels after germband elongation. Primordial appendages are visible. (B) Dmac-evepRNAi. Increased apoptosis is seen in the trunk. Antennae primordia, but no appendages, are visible. (C) Dmac-oddpRNAi. Increased apoptosis in the trunk of the early embryo. Antennal, mandible, maxillary and some thoracic primordia are present. (D) Dmac-runpRNAi. Extensive apoptosis in the center of the embryo; only antennal and mandible appendages are present. (E) Dmac-hpRNAi. High apoptotic activity at the posterior end. (F) Dmac-slppRNAi. Striped apoptosis concentrated in alternate compartments is apparent in an extended germband stage embryo. (G-O) PH3 staining to monitor mitosis. (G-J) Control gfppRNAi. (G) Mitotic cells are clustered in head lobes at early germband elongation. (H) Later, there is increased mitosis signal in the head and along the anterior and central trunk. In the posterior, mitotic cells are arranged in a stripe (arrow). (I) Concentrated mitotic activity persists in the posterior at late germband elongation (arrow). (J) High mitotic activity all along the embryo at later stage. (K-M) Dmac-slppRNAi. PH3 staining very similar to control. Concentrated mitosis is seen in the posterior (arrows in K,L). (N,O) Dmac-evepRNAi. (N) Embryo during germband elongation has fewer mitotic cells and without a clear striped-like arrangement in the posterior. (O) Later stage embryo with extensive mitotic activity throughout. Among the 200 0-1 day AEL embryos that were examined for every staining, over 25% developed to appropriate developmental stages for the examination of apoptosis or mitosis pattern. Scale bars: 500 µm.
Dmac-eve, -odd and -run RNAi produced truncated embryos with extensive apoptosis. Severely affected Dmac-eveRNAi embryos showed high apoptotic activity almost uniformly throughout the embryo (Fig. 6B). Dmac-oddRNAi caused less apoptosis in the trunk, and striped apoptotic signals were detected in the anterior (Fig. 6C). These results are consistent with the finding that Dmac-oddRNAi cuticle defects were milder than Dmac-eveRNAi. For Dmac-runRNAi, extensive apoptosis was detected uniformly in the trunk (Fig. 6D). A concentrated region with apoptotic activity in the posterior end was detected for Dmac-hRNAi (Fig. 6E). For Dmac-slpRNAi, apoptosis was detected in alternate compartments, at the time when segmental furrows appeared, in a PR-like pattern (Fig. 6F). Finally, for Dmac-slp/prd double RNAi, strong apoptotic signals were detected almost uniformly in elongated germbands (Fig. 5G) and extensive apoptotic activity persisted after germband retraction (Fig. 5H). Overall, apoptosis patterns prefigured morphological defects (Fig. 2), indicating that cell death is involved in causing the defects seen after Dermestes PRG knockdown.
Knockdown of Dmac-eve causes disruption of posterior mitotic activity
Cell proliferation has been documented in the posterior growth zone in annelids and some arthropods (Scholtz, 1993; Shankland and Seaver, 2000). Although more recent work has highlighted the importance of cell rearrangement in contributing to germband elongation in sequentially segmenting insects (Nakamoto et al., 2015), cell proliferation is also likely to play a role, and the relative contributions of these processes may vary in different species (Auman et al., 2017; Sarrazin et al., 2012). We examined mitotic figures in Dermestes embryos using anti-phospho-Histone H3 antibody (PH3; Fig. 6G-O, Fig. S6). In gfpRNAi control embryos, mitosis was detected as nuclei divided in the syncytial blastoderm (Fig. S6A′). No mitotic activity was detected in the embryonic rudiment at late blastoderm stages (Fig. S6B′). However, the possibility of transient cell division cannot be excluded. During gastrulation and early germband elongation, mitosis was detected in patches in the head lobes, and in some regions of the trunk (Fig. S6C′-E′, Fig. 6G). At mid-germband stages, mitotic activity expanded and a localized region with high mitotic activity was evident in the segment addition zone (SAZ) (Fig. 6H, arrow). This region retained mitotic activity throughout germband elongation (Fig. 6I, arrow). In fully elongated germband embryos with segmental furrows, mitosis was detected throughout the embryo, with stronger activity in the posterior end (Fig. 6J).
To test whether Dermestes PRGs impact cell proliferation in addition to cell viability, post-knockdown mitotic activity was compared between a gene that functions exclusively in PR patterning (Dmac-slp) and one that functions in both PR patterning and germband elongation (Dmac-eve). Patterns of mitosis after Dmac-slp knockdown did not appear different from gfpRNAi controls. Localized mitotically active regions were detected during germband elongation in patterns similar to controls (Fig. 6K,L,M, arrow). Mitotic activity was normal in the head and trunk regions, and the SAZ that shows high levels of mitosis in controls appeared unaffected. By contrast, these regions showed little mitotic activity after knockdown of Dmac-eve, with only a few mitotic cells evident (Fig. 6N, arrow). These cells were also not as well organized as those in gfpRNAi control or Dmac-slpRNAi embryos at comparable stages. Extensive mitosis was detected almost uniformly in later stage, shortened Dmac-eveRNAi embryos (Fig. 6O).
In summary, these results indicate that there is a localized region within the SAZ with high mitotic activity during Dermestes germband elongation. Whereas there was little effect on mitosis of Dmac-slp knockdown, mitosis in the SAZ was greatly reduced and mitotic cells were not well organized in Dmac-eveRNAi embryos, suggesting that failure of cell proliferation contributes to the truncation of the body axis after Dmac-eve knockdown.
DISCUSSION
Dermestes has fewer posterior segments added after gastrulation than its well-studied beetle counterpart, Tribolium. It might represent an intermediate state between ancestral short-germ and derived long-germ modes. Here, we examined the expression and function of PRG orthologs in Dermestes. Dmac-eve, -run and -odd function in both posterior elongation and PR segmentation; slp and prd function exclusively in PR segmentation; h has a role in posterior patterning; ftz has mild PR-like function; and no roles in segmentation were found for ftz-f1 or opa. As in Drosophila, PRGs were shown to promote cell viability. Knockdown of Dmac-eve, but not Dmac-slp, disrupted mitotic activity in the SAZ. Together, these studies revealed overall similarities in the PR patterning process between Dermestes and Tribolium while also highlighting subtle changes in expression pattern and the divergence of function of individual PRGs during insect evolution.
PRG expression in holometabolous insects
PRG expression has been examined in both long-germ and sequentially segmenting species in recent years (El-Sherif et al., 2012; Liu and Kaufman, 2005a; Mito et al., 2007; Rosenberg et al., 2014; Sarrazin et al., 2012; Wilson and Dearden, 2012). In short-germ Tribolium, sequential addition of PR stripes from the posterior end of the embryo suggested that a clock-based oscillating mechanism regulates PRG expression at both blastoderm and germband stages (El-Sherif et al., 2012; Sarrazin et al., 2012). In Nasonia, in which long-germ development was derived independently from that of Drosophila, a Drosophila-like, gap gene-dependent regulation of PRG expression was proposed for anterior segments, whereas posterior PR stripes arise in an anterior-to-posterior order, similar to sequentially segmenting species (Keller et al., 2010; Rosenberg et al., 2014). By contrast, in Drosophila, while stripe 1 is often the first stripe to arise for several PRGs, the remaining stripes arise in different orders for different PRGs and even in different individual embryos, with no anterior-to-posterior bias (Clark and Akam, 2016; Surkova et al., 2008; Yu and Pick, 1995). Dermestes display posterior cap-like expression of eve, odd and run, and stripes arose at both blastoderm and germband stages from the posterior, likely reflecting clock mechanisms shared with Tribolium. Severe ʻhead-only' or ʻanterior-only' defects confirmed that their function in generating a growing germband is conserved between these two beetles with distinct germ modes. Patel et al. (1994) previously observed sequential eve expression in Callosobruchus maculatus, suggesting that all segments are patterned sequentially in this long-germ beetle species, in contrast to long-germ Drosophila and Nasonia. This supports the perspective that long-germ segmentation evolved independently in insects (Davis and Patel, 2002; Liu and Kaufman, 2005b). Future studies will investigate the function of PRG orthologs in Callosobruchus.
In all species examined to date, a subset of PRGs generate secondary stripes. For Dermestes, secondary stripes were observed for eve, odd, h, prd and slp, emerging either by splitting (eve, h and prd) or de novo (odd and slp). Secondary h stripes do not form in Drosophila but do in Tribolium (Eckert et al., 2004). For eve, secondary stripes are added de novo in Drosophila but by splitting in Dermestes and Tribolium (Brown et al., 1997; Frasch and Levine, 1987; Macdonald et al., 1986; Patel et al., 1994). Larger differences in expression were seen for opa and ftz-f1. D. melanogaster opa is expressed ubiquitously but mutations cause a PR phenotype (Benedyk et al., 1994; Clark and Akam, 2016; Jürgens et al., 1984). In Dermestes, marginal segmental stripes of opa resolved from a broad expression domain during gastrulation and germband elongation, whereas in Tribolium clear segmental stripes arise from the posterior. In Drosophila, Ftz-F1 is expressed ubiquitously (Yussa et al., 2001), whereas in Dermestes and Tribolium (Heffer et al., 2013) PR-like stripes were observed. Finally, intensities of primary and secondary stripes alternated only for Dmac-odd, while secondary stripes exhibit weaker expression in Drosophila for run, odd, slp, and prd (Clark and Akam, 2016; Surkova et al., 2008). Dmac-slp differed from other PRGs in Dermestes in that anterior stripes did not fade in elongating germbands, similar to the situation in Tribolium (Choe and Brown, 2007). Together, these results reveal extensive fine-tuning of regulatory mechanisms controlling PRG expression in multiple lineages.
Two distinct modes of PRG function in segmentation
Knockdown of either Dmac-slp or -prd caused PR-like segmentation defects exclusively, consistent with their functions as ʻcore' PRGs. Similar PR-like defects were seen after RNAi knockdown of these genes in Tribolium, and slp and prd were the only PRGs identified in Tribolium mutant screens (Choe and Brown, 2007; Choe et al., 2006; Maderspacher et al., 1998). Knockdown of a second group of genes comprising Dmac-eve, -odd and -run produced two distinct classes of defect: mildly affected offspring usually hatched, showing PR-like defects, whereas severely affected unhatched offspring displayed defects in elongation and cuticles were composed of head or anterior regions only. These findings suggest that two independent modes of PRG action are required for segment formation in sequentially segmenting insects, which are likely to require the regulation of different sets of downstream target genes: (1) eve, odd and run promote the formation of an elongating germband; (2) eve, odd and run, as well as the core PRGs slp and prd, promote the subdivision of organized tissue into metameric units. This ‘classic’ PR patterning role for eve, odd and run in sequentially segmenting species can be concealed by severely truncated phenotypes and/or if the identity of lost segments cannot be easily determined, as the case in the cricket Gryllus (Mito et al., 2005) but was also observed for Tribolium (S. J. Brown, personal communication). Here, we circumvented these issues by quantitative examination of moderate cuticle defects in hatched larvae after Dmac-eve, -odd and -run knockdown. The PR-like register of defects was revealed by compiling results from individual affected larvae (Fig. 7), and confirmed by the observation of irregular En expression in young embryos. Simultaneous knockdown of Dmac-prd and -slp produced embryos with no segmental boundaries across gnathal, thoracic and abdominal regions, but truncated embryos retaining only anterior structures were never observed, supporting the notion that germband elongation and patterning of segmental boundaries are decoupled modes. Similarly, elongated but unsegmented posterior tissue was reported in crustaceans when Notch signaling was disrupted pharmacologically (Williams et al., 2012). It is likely that these two modes of PRG action also occur in long-germ Drosophila, but the ‘classic’ PR-based assignment of tissue to segmental units occurs for the entire embryo at blastoderm, prior to elongation of the germband. This ‘classic’ PR function requires nine PRGs in Drosophila and involves direct regulation of other regulatory genes such as en. Similar to sequentially segmenting species, Drosophila germband elongation involves run- and eve-dependent cell intercalation (Irvine and Wieschaus, 1994; Zallen and Wieschaus, 2004), which occurs, at least in part, by PRG regulation of Toll-like cell surface proteins (Paré et al., 2014), targets that are likely to be shared by Tribolium PRG orthologs (Benton et al., 2016).
Fig. 7.
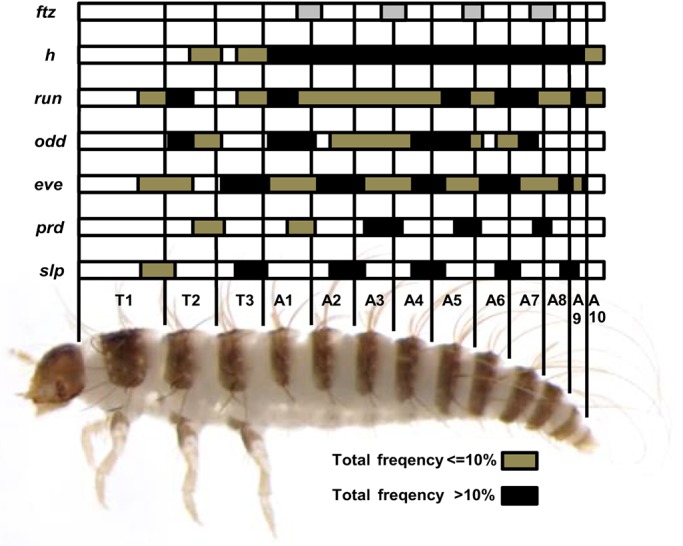
Dermestes PRG orthologs are required for elongation and PR-like segmentation. Schematic representation of defective patterns after PRG RNAi. Results are based on defective patterns in affected hatched larvae after pRNAi, except for ftz (eRNAi, gray bars). Black bars indicate regions affected at high frequency (>10%); tan bars show regions affected at low frequency (≤10%). For genes playing dual roles in Dermestes segmentation (eve, odd, run), black bars alone show PR-like phenotypes in moderately affected offspring (hatched), whereas black and tan bars together indicate severely affected, truncated offspring (unhatched). PR-like defects seen for slppRNAi versus prdpRNAi or for ftzeRNAi versus evepRNAi are complementary.
Divergent function of PRG orthologs
Even though the outcome of PRG regulation appears highly similar across species – germband elongation and subdivision of tissue into metameric units – the function of individual PRGs varies, even within the beetle clade (Table S2). Perhaps the most drastic case is ftz, which arose as a homeotic gene, but is required for en expression in Drosophila (Florence et al., 1997; Heffer et al., 2011; Lohr et al., 2001; Pick, 2016; Telford, 2000). Although expressed in PR stripes, Ftz is not required for en expression in Tribolium (Choe et al., 2006), but appears to maintain a marginal role in this process in Dermestes. A second variable PRG is opa, a PRG in Drosophila expressed ubiquitously in the trunk, where it modulates the function of other PRGs (Clark and Akam, 2016). Although expressed in stripes in Tribolium (Clark, 2017 preprint; Clark and Akam, 2016; Clark and Peel, 2017 preprint), segmentation defects were not produced after RNAi knockdown (Choe et al., 2006, 2017). Similarly, in Dermestes, neither Dmac-opapRNAi nor Dmac-opaeRNAi, using two non-overlapping dsRNAs, produced segmentation defects. The PRG ftz-f1, which is expressed ubiquitously in Drosophila, is striped in both beetle species examined and has PR function in Tribolium (Heffer et al., 2013). In Dermestes, we did not detect a PR-like segmentation function. However, we cannot rule out the possibility that maternally deposited transcript was ineffectively targeted in injected embryos, masking a role in segmentation.
Whereas h is a typical PRG in Drosophila, Dmac-hRNAi disrupted abdominal segmentation with variable and low penetrance but without PR register, and no trunk segmentation defects were reported in Tribolium (Aranda et al., 2008; Choe et al., 2006). Dmac-ftzRNAi also resulted in PR defects with low penetrance. We propose that the Dmac-ftzRNAi and Dmac-hRNAi low penetrance effects reflect marginal requirements in segmentation, perhaps due to partial redundancy with other PRGs. Their situation in beetles might represent an intermediate evolutionary stage, in which they retain the freedom to functionally diverge. In Apis, ftz has a role in head patterning (Wilson and Dearden, 2012), whereas outside of holometabolous insects ftz is unlikely to play a role in segmentation (Dawes et al., 1994) and h may be restricted to a role in Notch/Delta signaling (Pueyo et al., 2008; Stollewerk et al., 2003). In Drosophila, each of these genes is essential for PR patterning. We note that a major role of h in Drosophila is to regulate ftz expression (Carroll and Scott, 1986; Howard and Ingham, 1986; Ish-Horowicz and Pinchin, 1987). Thus, these two genes became interdependent in a lineage in which they are both absolutely required for segmentation. Future studies will examine the rewiring underlying these types of changes in PRG function during evolution.
MATERIALS AND METHODS
Animal rearing and PRG ortholog isolation
A D. maculatus colony has been maintained in our lab for over 4 years, as described (Xiang et al., 2015, 2016). RNA was extracted from Dermestes embryos 0-1 day after egg laying (AEL) and reverse transcription was performed to prepare cDNA. Forward and reverse degenerate primers targeting conserved protein motifs designed by ourselves or from previous literature (Damen et al., 2005; Damen et al., 2000) were used for amplification, followed by 3′ RACE. To ensure that the separately isolated regions correspond to a single gene, gene-specific primers including the regions isolated by both degenerate PCR and 3′ RACE were used to amplify cDNA and the products were sequenced. See Fig. S8 and supplementary Materials and Methods for further details of gene isolation.
Phylogenetic analysis
To determine orthologs of PRGs from other species, phylogenetic analysis was performed as described (Rosenberg et al., 2014). Homologous protein sequences from other species were trimmed to align with the partial D. maculatus PRG ortholog sequence that we isolated. Multiple sequence alignment was carried out using MUSCLE (Edgar, 2004). Phylogenetic analysis was carried out using RAxML at CIPRES Science Gateway (http://www.phylo.org/sub_sections/portal/) (Miller et al., 2010; Stamatakis, 2006; Stamatakis et al., 2008). Trees were visualized using Dendroscope (Huson and Scornavacca, 2012).
Gene knockdown by RNAi
RNAi was carried out according to Xiang et al. (2016). Two non-overlapping target regions (5′ or 3′) were separately tested for each gene to ensure that effects were specific (Fig. S8, dark blue boxes). gfp dsRNA was used as a negative control. See supplementary Materials and Methods for further details of dsRNA synthesis. For parental RNAi, ∼2 µl at 3 µg/µl was injected into 8-10 females for each dsRNA. After ∼24 h recovery at 30°C, injected females were mated with uninjected young males for 1 day before embryo collection. For eRNAi, pre-blastoderm stage embryos were injected; 3 µg/µl dsRNA was used for eRNAi and 50-100 ng was typically injected into each embryo. After injection, embryos were held at 30°C until hatching or appropriate developmental stage.
Gene expression and phenotypic analysis
Analysis of gene expression patterns was carried out following previously established protocols (Xiang et al., 2015) using overnight embryo collections. Early Dermestes embryogenesis has been characterized using SYTOX Green nuclear staining (Xiang et al., 2015). Thus, SYTOX Green nuclear staining was performed after in situ hybridization for visualizing embryo morphology to determine developmental stage, e.g. a wide shallow ventral furrow is evident in embryos at early gastrulation. Phenotypic analysis of embryos and larvae was carried out as described (Xiang et al., 2016). Daily embryo collections were incubated for 72 h at 30°C (wild-type embryos hatch within 60 h). Hatched first instar larvae or unhatched embryos were collected for analysis of cuticle defects. 4D9 anti-Engrailed, anti-phospho-Histone H3 (mitosis marker) and anti-cleaved Drosophila Dcp-1 (apoptosis marker) antibodies were used for immunohistochemistry. For details of antibody staining and analysis of ovarian morphology, see the supplementary Materials and Methods. Embryos were visualized by microscopy using an Olympus SZX12, Leica SP5X, Leica 501007, Zeiss SteREO Discovery V12 or Zeiss Axio Imager M1. Image stacking used CombineZP and merged images were prepared using Photoshop (Adobe) if necessary.
qPCR for knockdown validation
To verify gene knockdown after RNAi, 0-6 h AEL embryos from 3′ dsRNA-injected females were collected on the third day after injection and aged for another 2 h at 30°C to reach blastoderm to germband stages (Xiang et al., 2015) when PRGs are relatively highly expressed. To calculate relative expression of the gene of interest in pRNAi offspring compared with gfp pRNAi offspring, we used the 2−ΔΔCt method. For each RNAi knockdown, three biological replicates were performed and mean and standard error were calculated (Fig. S4). For details of qPCR, see the supplementary Materials and Methods.
Supplementary Material
Acknowledgements
We thank Alys Cheatle Jarvela and Hsiao-Ling Lu for sharing expertise on qPCR; Cassandra Extavour, Seth Donoughe and Reinhard Schröder for sharing protocols on apoptosis; Eric Haag, Steve Mount, Alexa Bely for advice; and L.P. lab members for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: L.P.; Methodology: J.X., K.R., A.H.; Data curation: J.X., K.R., A.H., L.P.; Writing - original draft: J.X., L.P.; Writing - review & editing: J.X., K.R., A.H., L.P.; Supervision: L.P.; Project administration: L.P.; Funding acquisition: L.P.
Funding
This work was supported by the National Institutes of Health (R01GM113230 to L.P.). Deposited in PMC for release after 12 months.
Data availability
Dmac gene sequences are available at GenBank under the following accession numbers: MG437053, fushi tarazu (ftz) mRNA; MG437054, ftz transcription factor 1 (ftz-f1) mRNA; MG437055, even-skipped (eve) mRNA, partial; MG437056, odd-skipped (odd) mRNA, partial; MG437057, runt (run) mRNA, partial; MG437058, hairy (h) mRNA, partial; MG437059, sloppy paired (slp) mRNA, partial; and MG437060, odd paired (opa) mRNA, partial.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.154039.supplemental
References
- Abzhanov A., Popadic A. and Kaufman T. C. (1999). Chelicerate Hox genes and the homology of arthropod segments. Evol. Dev. 1, 77-89. 10.1046/j.1525-142x.1999.99014.x [DOI] [PubMed] [Google Scholar]
- Akam M. (1987). The molecular basis for metameric pattern in the Drosophila embryo. Development 101, 1-22. [PubMed] [Google Scholar]
- Angelini D. R., Liu P. Z., Hughes C. L. and Kaufman T. C. (2005). Hox gene function and interaction in the milkweed bug Oncopeltus fasciatus (Hemiptera). Dev. Biol. 287, 440-455. 10.1016/j.ydbio.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Aranda M., Marques-Souza H., Bayer T. and Tautz D. (2008). The role of the segmentation gene hairy in Tribolium. Dev. Genes Evol. 218, 465-477. 10.1007/s00427-008-0240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman T., Vreede B. M. I., Weiss A., Hester S. D., Williams T. A., Nagy L. M. and Chipman A. D. (2017). Dynamics of growth zone patterning in the milkweed bug Oncopeltus fasciatus. Development 144, 1896-1905. 10.1242/dev.142091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk M. J., Mullen J. R. and DiNardo S. (1994). odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes Dev. 8, 105-117. 10.1101/gad.8.1.105 [DOI] [PubMed] [Google Scholar]
- Benton M. A., Pechmann M., Frey N., Stappert D., Conrads K. H., Chen Y.-T., Stamataki E., Pavlopoulos A. and Roth S. (2016). Toll genes have an ancestral role in axis elongation. Curr. Biol. 26, 1609-1615. 10.1016/j.cub.2016.04.055 [DOI] [PubMed] [Google Scholar]
- Brown S. J., Parrish J. K., Beeman R. W. and Denell R. E. (1997). Molecular characterization and embryonic expression of the even-skipped ortholog of Tribolium castaneum. Mech. Dev. 61, 165-173. 10.1016/S0925-4773(96)00642-9 [DOI] [PubMed] [Google Scholar]
- Carroll S. B. and Scott M. P. (1986). Zygotically active genes that affect the spatial expression of the fushi tarazu segmentation gene during early Drosophila embryogenesis. Cell 45, 113-126. 10.1016/0092-8674(86)90543-X [DOI] [PubMed] [Google Scholar]
- Choe C. P. and Brown S. J. (2007). Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev. Biol. 302, 281-294. 10.1016/j.ydbio.2006.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe C. P. and Brown S. J. (2009). Genetic regulation of engrailed and wingless in Tribolium segmentation and the evolution of pair-rule segmentation. Dev. Biol. 325, 482-491. 10.1016/j.ydbio.2008.10.037 [DOI] [PubMed] [Google Scholar]
- Choe C. P., Miller S. C. and Brown S. J. (2006). A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc. Natl. Acad. Sci. USA 103, 6560-6564. 10.1073/pnas.0510440103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe C. P., Stellabotte F. and Brown S. J. (2017). Regulation and function of odd-paired in Tribolium segmentation. Dev. Genes Evol. 227, 309-317. 10.1007/s00427-017-0590-7 [DOI] [PubMed] [Google Scholar]
- Clark E. (2017). Dynamic patterning by the Drosophila pair-rule network reconciles long-germ and short-germ segmentation. PLoS Biol. 15, e2002439 10.1371/journal.pbio.2002439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E. and Akam M. (2016). Odd-paired controls frequency doubling in Drosophila segmentation by altering the pair-rule gene regulatory network. Elife 5, e18215 10.1101/052241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E. and Peel A. (2017). Evidence for the temporal regulation of insect segmentation by a conserved set of developmental transcription factors. bioRxiv , 145151 10.1101/145151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter D. E. and Wieschaus E. (1988). Gene activities and segmental patterning in Drosophila: analysis of odd-skipped and pair-rule double mutants. Genes Dev. 2, 1812-1823. 10.1101/gad.2.12b.1812 [DOI] [PubMed] [Google Scholar]
- Damen W. G., Weller M. and Tautz D. (2000). Expression patterns of hairy, even-skipped, and runt in the spider Cupiennius salei imply that these genes were segmentation genes in a basal arthropod. Proc. Natl. Acad. Sci. USA 97, 4515-4519. 10.1073/pnas.97.9.4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen W. G., Janssen R. and Prpic N. M. (2005). Pair rule gene orthologs in spider segmentation. Evol. Dev. 7, 618-628. 10.1111/j.1525-142X.2005.05065.x [DOI] [PubMed] [Google Scholar]
- Davis G. K. and Patel N. H. (2002). Short, long, and beyond: molecular and embryological approaches to insect segmentation. Annu. Rev. Entomol. 47, 669-699. 10.1146/annurev.ento.47.091201.145251 [DOI] [PubMed] [Google Scholar]
- Dawes R., Dawson I., Falciani F., Tear G. and Akam M. (1994). Dax, a locust Hox gene related to fushi-tarazu but showing no pair-rule expression. Development 120, 1561-1572. [DOI] [PubMed] [Google Scholar]
- DiNardo S. and O'Farrell P. H. (1987). Establishment and refinement of segmental pattern in the Drosophila embryo: spatial control of engrailed expression by pair-rule genes. Genes Dev. 1, 1212-1225. 10.1101/gad.1.10.1212 [DOI] [PubMed] [Google Scholar]
- Eckert C., Aranda M., Wolff C. and Tautz D. (2004). Separable stripe enhancer elements for the pair-rule gene hairy in the beetle Tribolium. EMBO Rep. 5, 638-642. 10.1038/sj.embor.7400148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792-1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherif E., Averof M. and Brown S. J. (2012). A segmentation clock operating in blastoderm and germband stages of Tribolium development. Development 139, 4341-4346. 10.1242/dev.085126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence B., Guichet A., Ephrussi A. and Laughon A. (1997). Ftz-F1 is a cofactor in Ftz activation of the Drosophila engrailed gene. Development 124, 839-847. [DOI] [PubMed] [Google Scholar]
- Frasch M. and Levine M. (1987). Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1, 981-995. 10.1101/gad.1.9.981 [DOI] [PubMed] [Google Scholar]
- Gilbert S. F. (2010). Developmental Biology, 9th edn. Sunderland, MA: Sinauer. [Google Scholar]
- Hedges S. B., Marin J., Suleski M., Paymer M. and Kumar S. (2015). Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835-845. 10.1093/molbev/msv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffer A., Löhr U. and Pick L. (2011). ftz evolution: findings, hypotheses and speculations (response to DOI 10.1002/bies.201100019). BioEssays 33, 910-918. 10.1002/bies.201100112 [DOI] [PubMed] [Google Scholar]
- Heffer A., Grubbs N., Mahaffey J. and Pick L. (2013). The evolving role of the orphan nuclear receptor ftz-f1, a pair-rule segmentation gene. Evol. Dev. 15, 406-417. 10.1111/ede.12050 [DOI] [PubMed] [Google Scholar]
- Howard K. and Ingham P. (1986). Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell 44, 949-957. 10.1016/0092-8674(86)90018-8 [DOI] [PubMed] [Google Scholar]
- Hughes S. C. and Krause H. M. (2001). Establishment and maintenance of parasegmental compartments. Development 128, 1109-1118. [DOI] [PubMed] [Google Scholar]
- Huson D. H. and Scornavacca C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061-1067. 10.1093/sysbio/sys062 [DOI] [PubMed] [Google Scholar]
- Ingham P. W. (1988). The molecular genetics of embryonic pattern formation in Drosophila. Nature 335, 25-34. 10.1038/335025a0 [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Howard K. R. and Ish-Horowicz D. (1985). Transcription pattern of the Drosophila segmentation gene hairy. Nature 318, 439-445. 10.1038/318439a0 [DOI] [Google Scholar]
- Irvine K. D. and Wieschaus E. (1994). Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827-841. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D. and Pinchin S. M. (1987). Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell 51, 405-415. 10.1016/0092-8674(87)90636-2 [DOI] [PubMed] [Google Scholar]
- Jaynes J. B., Fujioka M. (2004). Drawing lines in the sand: even skipped et al. and parasegment boundaries. Dev. Biol. 269, 609-622. 10.1016/j.ydbio.2004.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G., Wieschaus E., Nüsslein-Volhard C. and Kluding H. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome . Wilhelm Roux's Arch. Dev. Biol. 193, 283-295. 10.1007/BF00848157 [DOI] [PubMed] [Google Scholar]
- Keller R. G., Desplan C. and Rosenberg M. I. (2010). Identification and characterization of Nasonia Pax genes. Insect Mol. Biol. 19 Suppl. 1, 109-120. 10.1111/j.1365-2583.2009.00921.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause G. (1939). Die Eitypen der Insekten. Biol. Zentralbl. 59, 495-536. [Google Scholar]
- Lawrence P. A. (1992). The Making of a Fly: The Genetics of Animal Design. Oxford, UK: Blackwell Scientific. [Google Scholar]
- Liu P. Z. and Kaufman T. C. (2005a). even-skipped is not a pair-rule gene but has segmental and gap-like functions in Oncopeltus fasciatus, an intermediate germband insect. Development 132, 2081-2092. 10.1242/dev.01807 [DOI] [PubMed] [Google Scholar]
- Liu P. Z. and Kaufman T. C. (2005b). Short and long germ segmentation: unanswered questions in the evolution of a developmental mode. Evol. Dev. 7, 629-646. 10.1111/j.1525-142X.2005.05066.x [DOI] [PubMed] [Google Scholar]
- Lohr U., Yussa M. and Pick L. (2001). Drosophila fushi tarazu. a gene on the border of homeotic function. Curr. Biol. 11, 1403-1412. 10.1016/S0960-9822(01)00443-2 [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Ingham P. and Struhl G. (1986). Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell 47, 721-734. 10.1016/0092-8674(86)90515-5 [DOI] [PubMed] [Google Scholar]
- Maderspacher F., Bucher G. and Klingler M. (1998). Pair-rule and gap gene mutants in the flour beetle Tribolium castaneum. Dev. Genes Evol. 208, 558-568. 10.1007/s004270050215 [DOI] [PubMed] [Google Scholar]
- Magrassi L. and Lawrence P. A. (1988). The pattern of cell death in fushi tarazu, a segmentation gene of Drosophila. Development 104, 447-451. [DOI] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W. and Schwartz T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), pp. 1-8. New Orleans: IEEE; 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Mito T., Sarashina I., Zhang H., Iwahashi A., Okamoto H., Miyawaki K., Shinmyo Y., Ohuchi H. and Noji S. (2005). Non-canonical functions of hunchback in segment patterning of the intermediate germ cricket Gryllus bimaculatus. Development 132, 2069-2079. 10.1242/dev.01784 [DOI] [PubMed] [Google Scholar]
- Mito T., Kobayashi C., Sarashina I., Zhang H., Shinahara W., Miyawaki K., Shinmyo Y., Ohuchi H. and Noji S. (2007). even-skipped has gap-like, pair-rule-like, and segmental functions in the cricket Gryllus bimaculatus, a basal, intermediate germ insect (Orthoptera). Dev. Biol. 303, 202-213. 10.1016/j.ydbio.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Mullen J. R. and DiNardo S. (1995). Establishing parasegments in Drosophila embryos: roles of the odd-skipped and naked genes. Dev. Biol. 169, 295-308. 10.1006/dbio.1995.1145 [DOI] [PubMed] [Google Scholar]
- Nakamoto A., Hester S. D., Constantinou S. J., Blaine W. G., Tewksbury A. B., Matei M. T., Nagy L. M. and Williams T. A. (2015). Changing cell behaviours during beetle embryogenesis correlates with slowing of segmentation. Nat. Commun. 6, 6635 10.1038/ncomms7635 [DOI] [PubMed] [Google Scholar]
- Nakao H. (2015). Analyses of interactions among pair-rule genes and the gap gene Krüppel in Bombyx segmentation. Dev. Biol. 405, 149-157. 10.1016/j.ydbio.2015.06.012 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C. and Wieschaus E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795-801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Frohnhöfer H. G. and Lehmann R. (1987). Determination of anteroposterior polarity in Drosophila. Science 238, 1675-1681. 10.1126/science.3686007 [DOI] [PubMed] [Google Scholar]
- Paré A. C., Vichas A., Fincher C. T., Mirman Z., Farrell D. L., Mainieri A. and Zallen J. A. (2014). A positional Toll receptor code directs convergent extension in Drosophila. Nature 515, 523-527. 10.1038/nature13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N. H., Condron B. G. and Zinn K. (1994). Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature 367, 429-434. 10.1038/367429a0 [DOI] [PubMed] [Google Scholar]
- Peel A. D., Chipman A. D. and Akam M. (2005). Arthropod segmentation: beyond the Drosophila paradigm. Nat. Rev. Genet. 6, 905-916. 10.1038/nrg1724 [DOI] [PubMed] [Google Scholar]
- Pick L. (2016). Hox genes, evo-devo, and the case of the ftz gene. Chromosoma 125, 535-551. 10.1007/s00412-015-0553-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueyo J. I., Lanfear R. and Couso J. P. (2008). Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. Proc. Natl. Acad. Sci. USA 105, 16614-16619. 10.1073/pnas.0804093105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. I., Lynch J. A. and Desplan C. (2009). Heads and tails: evolution of antero-posterior patterning in insects. Biochim. Biophys. Acta 1789, 333-342. 10.1016/j.bbagrm.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. I., Brent A. E., Payre F. and Desplan C. (2014). Dual mode of embryonic development is highlighted by expression and function of Nasonia pair-rule genes. Elife 3, e01440 10.7554/eLife.01440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander K. (1976). Specification of the basic body pattern in insect embryogenesis. Adv. Insect Physiol. 12, 125-238. 10.1016/S0065-2806(08)60255-6 [DOI] [Google Scholar]
- Sarrazin A. F., Peel A. D. and Averof M. (2012). A segmentation clock with two-segment periodicity in insects. Science 336, 338-341. 10.1126/science.1218256 [DOI] [PubMed] [Google Scholar]
- Scholtz G. (1993). Teloblasts in decapod embryos: an embryonic character reveals the monophyletic origin of freshwater crayfishes (Crustacea, Decapoda). Zool. Anz. 230, 45-54. [Google Scholar]
- Shankland M. and Seaver E. C. (2000). Evolution of the bilaterian body plan: what have we learned from annelids? Proc. Natl. Acad. Sci. USA 97, 4434-4437. 10.1073/pnas.97.9.4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688-2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P. and Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758-771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stollewerk A., Schoppmeier M. and Damen W. G. M. (2003). Involvement of Notch and Delta genes in spider segmentation. Nature 423, 863-865. 10.1038/nature01682 [DOI] [PubMed] [Google Scholar]
- Surkova S., Kosman D., Kozlov K., Manu, Myasnikova E., Samsonova A. A., Spirov A., Vanario-Alonso C. E., Samsonova M. and Reinitz J. (2008). Characterization of the Drosophila segment determination morphome. Dev. Biol. 313, 844-862. 10.1016/j.ydbio.2007.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford M. J. (2000). Evidence for the derivation of the Drosophila fushi tarazu gene from a Hox gene orthologous to lophotrochozoan Lox5. Curr. Biol. 10, 349-352. 10.1016/S0960-9822(00)00387-0 [DOI] [PubMed] [Google Scholar]
- True J. R. and Haag E. S. (2001). Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3, 109-119. 10.1046/j.1525-142x.2001.003002109.x [DOI] [PubMed] [Google Scholar]
- Williams T. A. and Nagy L. M. (2017). Linking gene regulation to cell behaviors in the posterior growth zone of sequentially segmenting arthropods. Arthropod Struct. Dev. 46, 380-394. 10.1016/j.asd.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Williams T., Blachuta B., Hegna T. A. and Nagy L. M. (2012). Decoupling elongation and segmentation: notch involvement in anostracan crustacean segmentation. Evol. Dev. 14, 372-382. 10.1111/j.1525-142X.2012.00555.x [DOI] [PubMed] [Google Scholar]
- Wilson M. J. and Dearden P. K. (2012). Pair-rule gene orthologues have unexpected maternal roles in the honeybee (Apis mellifera). PLoS ONE 7, e46490 10.1371/journal.pone.0046490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Forrest I. S. and Pick L. (2015). Dermestes maculatus: an intermediate-germ beetle model system for evo-devo. Evodevo 6, 32 10.1186/s13227-015-0028-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Reding K. and Pick L. (2016). Rearing and double-stranded RNA-mediated gene knockdown in the hide beetle, Dermestes maculatus. J. Vis. Exp. 118, e54976 10.3791/54976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Tan A. and Palli S. R. (2010). The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J. Insect Physiol. 56, 1471-1480. 10.1016/j.jinsphys.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. and Pick L. (1995). Non-periodic cues generate seven ftz stripes in the Drosophila embryo. Mech. Dev. 50, 163-175. 10.1016/0925-4773(94)00333-I [DOI] [PubMed] [Google Scholar]
- Yussa M., Löhr U., Su K. and Pick L. (2001). The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains. Mech. Dev. 107, 39-53. 10.1016/S0925-4773(01)00448-8 [DOI] [PubMed] [Google Scholar]
- Zallen J. A. and Wieschaus E. (2004). Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343-355. 10.1016/S1534-5807(04)00060-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.