Abstract
Pluripotent stem cells have broad utility in biomedical research and their molecular regulation has thus garnered substantial interest. While the principles that establish and regulate pluripotency have been well defined in the mouse, it has been difficult to extrapolate these insights to the human system due to species-specific differences and the distinct developmental identities of mouse versus human embryonic stem cells. In this Review, we examine genome-wide approaches to elucidate the regulatory principles of pluripotency in human embryos and stem cells, and highlight where differences exist in the regulation of pluripotency in mice and humans. We review recent insights into the nature of human pluripotent cells in vivo, obtained by the deep sequencing of pre-implantation embryos. We also present an integrated overview of the principal layers of global gene regulation in human pluripotent stem cells. Finally, we discuss the transcriptional and epigenomic remodeling events associated with cell fate transitions into and out of human pluripotency.
KEY WORDS: Chromatin, Embryogenesis, Epigenetics, Pluripotency, Reprogramming, Transcription
Summary: This Review examines genome-wide approaches to elucidate the regulatory principles of pluripotency in human embryos and stem cells, and highlights differences in the regulation of pluripotency between mice and humans.
Introduction
Pluripotent cells reside naturally in the blastocyst of early mouse and human embryos and constitute the founder tissue of the embryo proper. Pluripotent stem cells (PSCs) that are capable of long-term self-renewal can be derived from the blastocyst (Evans and Kaufman, 1981; Martin, 1981; Thomson et al., 1998) or can be induced from somatic cells by direct reprogramming (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007). Given appropriate cues, embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) can differentiate into virtually any cell type found in the adult body. Elucidating the molecular nature of pluripotency is thereby essential for designing enhanced strategies to reprogram somatic cells and for understanding how PSCs can be directed into lineages of interest in vitro.
Owing to their high proliferation rate and amenability to genetic manipulation, mouse ESCs (mESCs) provide a robust in vitro system with which to dissect the molecular regulation of pluripotency. The genome-wide DNA targets and protein-protein interaction partners of core pluripotency regulators have been mapped in mESCs (Young, 2011; Huang and Wang, 2014), and an essential transcription factor network that can explain mESC behavior has been defined using computational methods (Dunn et al., 2014). Furthermore, transcriptomic studies and functional experiments have shown that, under chemically defined conditions, mESCs closely resemble the epiblast compartment of the embryonic day (E) 4.5 mouse blastocyst (Boroviak et al., 2014). Thus, mESCs recapitulate key features of in vivo pluripotency.
It is becoming increasingly clear, however, that the regulatory principles of pluripotency cannot simply be extrapolated from mouse to human, but must be interrogated in human cells directly. Evidence obtained in recent years has revealed that extensive differences exist between mouse and human early embryogenesis, including the timing of zygotic genome activation (ZGA; see Glossary, Box 1) (Blakeley et al., 2015), divergent responses of mouse and human embryos to signal inhibitors (Kuijk et al., 2012; Roode et al., 2012), differences in the expression of key developmental regulators (Blakeley et al., 2015; Petropoulos et al., 2016), and different mechanisms to accomplish X-chromosome dosage compensation (Okamoto et al., 2011; Petropoulos et al., 2016; Vallot et al., 2017). Adding further complexity, human ESCs (hESCs) are considered to be developmentally more mature than mESCs, and to more closely resemble mouse epiblast stem cells (mEpiSCs) that are derived from the post-implantation epiblast (Brons et al., 2007; Tesar et al., 2007). Therefore, the molecular mechanisms that regulate human pluripotency are not easily inferred from studies in mice.
Box 1. Glossary.
ChIP-Chip. A method to identify the genome-wide DNA targets of a protein of interest by chromatin immunoprecipitation followed by DNA microarray analysis.
ChIP-Seq. A method to identify the genome-wide DNA targets of a protein of interest by chromatin immunoprecipitation followed by massively parallel DNA sequencing.
CpG methylation. The addition of a methyl group to the fifth carbon of a cytosine base in a cytosine-phosphate-guanine (CpG) dinucleotide. The methylation of CpG-dense promoter regions is associated with gene repression.
Epiblast (EPI). The lineage of the blastocyst that gives rise to all somatic lineages and the germ line.
Expression quantitative trait loci (eQTL). Regions of the genome that contain variations in DNA sequence that correlate with the expression of one or more genes.
Extended pluripotent stem (EPS) cells. Pluripotent stem cells that can contribute to embryonic as well as to extraembryonic tissues upon injection into early mouse embryos.
Fluorescence ubiquitin cell cycle indicator (FUCCI). A system to track cell cycle progression in live cells based on cell cycle-dependent proteolysis of fluorescent ubiquitylation oscillators.
Inner cell mass (ICM). A cellular mass on the inside of the blastocyst containing the epiblast and primitive endoderm (hypoblast) lineages.
Insulated neighborhoods. Chromosomal loop structures that are formed by CTCF homodimers and co-occupied by the Cohesin complex. Such neighborhoods function to insulate genes and their regulatory elements within the loop.
Mesendoderm. A bipotential embryonic tissue layer that arises during gastrulation and gives rise to both mesoderm and endoderm.
Naive pluripotency. A state of pluripotency associated with the pre-implantation epiblast, which is characterized by an unbiased developmental potential and depletion of repressive chromatin features. Naive pluripotency is recapitulated in vitro in the form of mESCs. Recently, a number of studies have attempted to derive hESCs in a naive state.
Pioneer factors. Factors that can engage target sequences on nucleosomes or in compacted chromatin and facilitate the binding of other transcription factors.
Polycomb repressive complex 2 (PRC2). A complex of Polycomb group proteins that di- and tri-methylates lysine 27 of histone H3 (H3K27me2/3). The PRC2 complex consists of four subunits: EED, SUZ12, EZH1/2 and RBAP46/48 (RBBP7/4).
Primed pluripotency. A state of pluripotency associated with the late post-implantation epiblast, which is characterized by lineage priming and enrichment in repressive chromatin features. Primed pluripotency is recapitulated in vitro in the form of mEpiSCs. Conventional hESCs also display defining features of primed pluripotency.
Primitive endoderm (PE, or hypoblast). The lineage of the blastocyst that gives rise to the extraembryonic endoderm of the yolk sac.
Primordial germ cells (PGCs). Precursors to sperm and egg, which are specified from pluripotent epiblast and subsequently migrate to the gonads.
RNA fluorescence in situ hybridization (RNA FISH). A method to detect the presence of RNA molecules in fixed cells using complementary fluorescent probes.
Trophectoderm (TE). Outer layer of cells in the blastocyst, which gives rise to a large part of the placenta.
Zygotic genome activation (ZGA). Time point during cleavage when the embryonic genome becomes transcriptionally active and takes over control of cellular functions from maternal products.
2i/L. A serum-free medium that includes inhibitors of MEK and GSK3 together with the cytokine leukemia inhibitory factor (LIF), which maintains mESCs in a naive pluripotent state.
t2i/L+Gö. A medium for maintaining hESCs in a naive pluripotent state that includes a MEK inhibitor, a reduced dose of GSK3 inhibitor, human LIF and a protein kinase C inhibitor.
5i/L/A. A medium for maintaining hESCs in a naive pluripotent state that comprises 2i/L together with inhibitors of BRAF, SRC and ROCK, and recombinant activin A. FGF2 is added in some formulations to improve the efficiency of naive conversion.
In this Review, we provide a broad overview of the global regulatory mechanisms that operate specifically in human pluripotent cells. We first review our current understanding of the major transcriptional and epigenomic events that lead to human blastocyst formation. This provides essential background for the rest of the article, in which we discuss the key determinants of global gene expression in hESCs and the molecular changes that occur during cell fate transitions around the human pluripotent state. Where relevant, we highlight notable differences in gene regulatory networks between mouse and human pluripotent cells.
Deep sequencing of early human embryos: a molecular blueprint for pluripotency in vivo
What are the key molecular properties of human pluripotent cells in their in vivo environment? Owing to limited access to human embryos, molecular profiling of the human blastocyst has lagged behind comparable studies in mice. However, in the past 5 years, several studies have characterized the major changes that take place in the transcriptome and DNA methylome during human pre-implantation development (summarized in Fig. 1).
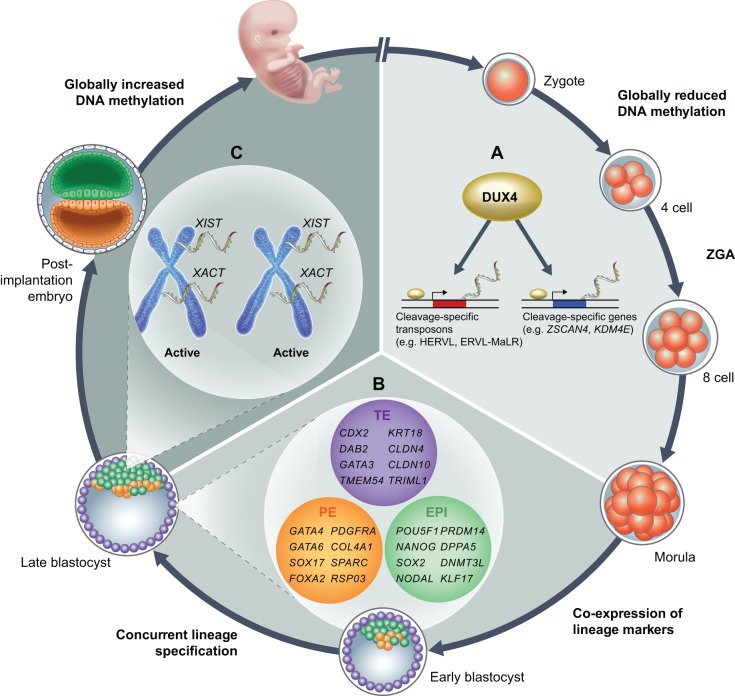
Fig. 1.
Insights into the transcriptional and epigenetic properties of human pluripotent cells in vivo. An overview of molecular events occurring during human pre-implantation development. (A) The transcription factor DUX4 activates cleavage-specific gene and transposon expression during zygotic genome activation (ZGA) (Hendrickson et al., 2017; De Iaco et al., 2017; Whiddon et al., 2017). (B) The three lineages of the human blastocyst – epiblast (EPI), primitive endoderm (PE) and trophectoderm (TE) – form concurrently and are associated with specific markers, as inferred from single-cell RNA-Seq analyses (Blakeley et al., 2015). (C) Both X chromosomes are actively transcribed in female blastocysts and show co-expression of the lncRNAs XIST and XACT in mutually exclusive nuclear domains (Okamoto et al., 2011; Petropoulos et al., 2016; Vallot et al., 2017). Human pre-implantation development is also marked by globally reduced levels of DNA methylation (Smith et al., 2014; Guo et al., 2014).
The transcriptome of human pre-implantation embryos
The initial transcriptional profiling of whole human embryos was performed on microarray platforms (Vassena et al., 2011; Xie et al., 2010; Zhang et al., 2009; Dobson et al., 2004; Madissoon et al., 2014). However, owing to the heterogeneous nature of these samples, it was challenging to derive lineage-specific gene expression patterns from these studies. The advent of single-cell transcriptome analysis has allowed lineage-specific gene expression in human pre-implantation embryos to be delineated more precisely. For example, Tang and colleagues analyzed global gene expression in 90 single human cells from the oocyte to late blastocyst stage, and in 34 single hESCs at early and late passages (Yan et al., 2013). The expression profiles of single cells obtained from late blastocysts clustered into three distinct groups, representing the epiblast (EPI), primitive endoderm (PE) and trophectoderm (TE) lineages (see Glossary, Box 1). Dramatic transcriptional changes were observed between the EPI cells and hESCs derived in culture (Yan et al., 2013). Thus, conventional hESCs differ substantially from the pluripotent cells that are present in the human blastocyst in terms of their global gene expression patterns.
By comparing single-cell RNA-Seq data from early human embryos (Yan et al., 2013) with that of stage-matched mouse embryos (Deng et al., 2014), Niakan and colleagues reported that a single wave of ZGA occurs between the human 4-cell and 8-cell stages (Blakeley et al., 2015). This contrasts with prior studies that had reported that a minor wave of ZGA occurs before the human 4-cell stage (Dobson et al., 2004; Xue et al., 2013), and indicates a delay in the onset of ZGA in human embryos compared with mouse embryos (Flach et al., 1982). Three groups recently identified the double homeobox transcription factor 4 (DUX4) as a global regulator of ZGA in mouse and human embryos (Hendrickson et al., 2017; De Iaco et al., 2017; Whiddon et al., 2017). Mutations in DUX4 are associated with facioscapulohumeral dystrophy, a common muscular dystrophy, and forced DUX4 expression activates an early embryonic transcriptional program in primary human myoblasts (Geng et al., 2012). DUX4 mRNA is expressed from the oocyte to 4-cell stage of human embryos, and when overexpressed in hESCs, DUX4 induces the derepression of hundreds of genes, including ZSCAN4 and KDM4E, and HERVL retrotransposons. In addition, it was shown that a mouse ortholog, Dux, can induce a ʻ2 cell-like' state in mESCs (De Iaco et al., 2017; Hendrickson et al., 2017). Hence, DUX family proteins are potent transcriptional activators of cleavage-specific gene and transposon expression (Fig. 1A).
In the mouse, lineage segregation in pre-implantation embryos occurs in two distinct waves: the TE first segregates from the inner cell mass (ICM; see Glossary, Box 1) as Cdx2 becomes restricted to outer cells of the morula, and this is then followed by segregation of the EPI and PE. In human embryos, however, CDX2 is not detected until after blastocyst formation (Niakan and Eggan, 2013). In fact, a single-cell transcriptome analysis of 1529 cells from 88 human embryos suggested that human lineage specification occurs simultaneously during blastocyst formation at around E5 (Petropoulos et al., 2016). An intermediate state in which EPI, PE and TE markers are co-expressed appears to precede the onset of concurrent lineage commitment. It is worth noting that human blastocysts seem to undergo an additional round of cell division prior to implantation (Niakan et al., 2012), which may be linked to the differences in the timing of lineage segregation. Human blastocysts also differ from their mouse counterparts in the spatial distribution of emerging EPI and PE cells. While cells expressing Nanog and Gata4/6 are scattered around the mouse ICM in a mutually exclusive ʻsalt-and-pepper' pattern (Chazaud et al., 2006), NANOG-positive cells assume a more tightly clustered organization in the early human ICM (Durruthy-Durruthy et al., 2016b).
How similar are the mouse and human blastocysts at the transcriptional level? Blakeley et al. (2015) identified conserved and human-specific gene expression patterns in the three lineages of the blastocyst (Fig. 1B). Conserved lineage-specific genes include NANOG, OCT4 (POU5F1) and SOX2 (in the EPI), CDX2, GATA3 and KRT18 (in the TE), and GATA4, PDGFRA and SOX17 (in the PE). Several genes are exclusively expressed in the human EPI, such as KLF17 (confirmed at the protein level), whereas the mouse EPI factors Esrrb, Klf2 and Bmp4 are not expressed in the human EPI. Another notable species-specific difference was the expression of TE-associated genes: Elf5 and Eomes are specific to mouse TE, while CLDN10, PLAC8 and TRIML1 are specific to human TE (Blakeley et al., 2015). Hence, there are fundamental differences in the expression of key developmental regulators between mouse and human blastocysts. These data indicate that the transcriptional regulation of human pre-implantation development cannot easily be inferred from studies in mice. This conclusion is further underscored by the recent finding that OCT4 may be required at an earlier stage of pre-implantation development in human embryos compared with mouse embryos (Fogarty et al., 2017).
The epigenome of human pre-implantation embryos
The DNA methylation landscape of early human embryos has been charted by whole-genome bisulfite sequencing from the oocyte to post-implantation stages (Okae et al., 2014; Smith et al., 2014; Guo et al., 2014). Genome-wide cytosine DNA demethylation occurs shortly after fertilization, but with different kinetics in the maternal and paternal genomes. The paternal genome is actively demethylated in the zygote, while the maternal genome undergoes progressive passive demethylation during cleavage. CpG methylation (see Glossary, Box 1) is restored overall to somatic levels after implantation, although the precise timing of global remethylation in human embryos has not been established. Hence, pluripotent cells in human blastocysts are hypomethylated. Meissner and colleagues have also shown that hESCs undergo rapid remethylation upon derivation from the blastocyst (Smith et al., 2014). In addition to the aforementioned transcriptional differences, this highlights a second molecular difference between hESCs in vitro and the pluripotent cells of the blastocyst.
Changes in the epigenetic state of the X chromosome have also been studied in mouse and human embryos. The paternally inherited X chromosome is inactivated in female mouse embryos at the 4-cell stage, and this is followed by X-chromosome reactivation in the EPI at E4.5 (reviewed by Pasque and Plath, 2015). This process is mediated by the long non-coding RNA (lncRNA) Xist, which coats the inactive X chromosome. RNA fluorescence in situ hybridization (RNA FISH; see Glossary, Box 1) studies have indicated that both X chromosomes are also actively transcribed in human female blastocysts but, surprisingly, XIST is co-expressed (Okamoto et al., 2011). These observations were confirmed by allele-specific gene expression analyses from Lanner and colleagues, who proposed that a progressive dampening of X-linked gene expression occurs on both X chromosomes during human pre-implantation development (Petropoulos et al., 2016). Such a model of dosage compensation is reminiscent of that observed in hermaphrodite worms, in which expression of both X chromosomes is reduced by half (reviewed by Meyer, 2010). Why does XIST not induce X-chromosome silencing in human blastocysts? Rougeulle and colleagues showed that XIST assumes a dispersed organization in human blastocysts, which might explain why it does not induce X inactivation (Vallot et al., 2017). This study also suggested that the repressive effect of XIST in human embryos is antagonized by another X-linked lncRNA, XACT, which co-accumulates with XIST on the same actively transcribed X chromosomes (Fig. 1C).
Layers of gene regulation in hPSCs
Having summarized the pivotal molecular events occurring during human pre-implantation development, we now turn our focus to the regulation of gene expression in hPSCs. Although our understanding of gene control in human embryos still remains fragmentary, the ability to map the genome-wide location of crucial nuclear regulators and perturb their expression has identified complex interactions among multiple layers of gene regulation in hPSCs (summarized in Fig. 2). Below, we focus on those regulatory mechanisms that have been shown to be functionally significant by either genetic or pharmacological manipulation.
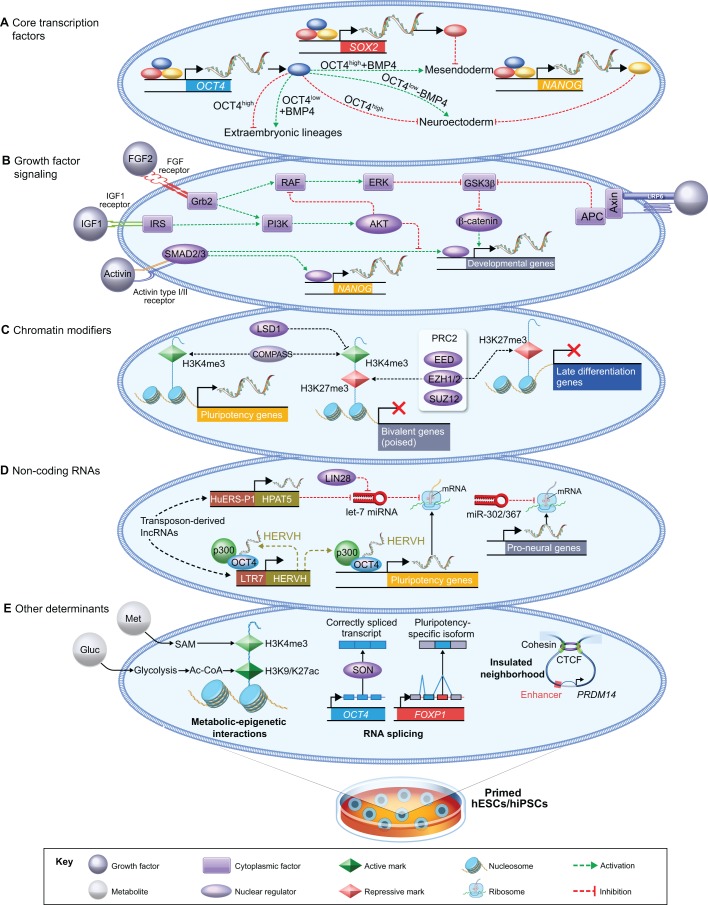
Fig. 2.
Layers of gene expression control in primed human PSCs. Overview of the major determinants of global gene expression in hPSCs cultured under conventional (primed) culture conditions. (A) The core transcription factors OCT4, SOX2 and NANOG form an autoregulatory network and repress distinct lineage fates in hESCs (Boyer et al., 2005; Wang et al., 2012). (B) Crosstalk among the major signaling pathways in hESCs as proposed by Singh et al. (2012). According to this model, activation of the PI3K/AKT pathway by IGF1 or FGF2 promotes the self-renewal of hESCs via two mechanisms. First, PI3K/AKT modulates the threshold of SMAD2/3 activity, allowing for the activation of NANOG but not mesendoderm-associated genes. As shown in A, the activation of NANOG stimulates the expression of core pluripotency genes and blocks neuroectoderm differentiation. Active PI3K/AKT also inhibits MEK/ERK and maintains GSK3β activity, which blocks β-catenin-mediated stimulation of pro-differentiation genes. Note that Wnt/β-catenin signaling has a distinct role in mESCs, where it functions to promote self-renewal instead of differentiation (ten Berge et al., 2011; Wray et al., 2011; Yi et al., 2011). (C) Chromatin-modifying enzymes with functional significance in hESC self-renewal include EZH1/2, which deposit H3K27me3 (Collinson et al., 2016; Shan et al., 2017), and enzymes that control the levels of H3K4me3 (Adamo et al., 2011; Bertero et al., 2015). (D) Various non-coding RNAs regulate the human pluripotent state. Transposon-derived lncRNAs, including HERVH and HPAT5, contribute to the self-renewal of hESCs (Lu et al., 2014; Durruthy-Durruthy et al., 2016a), as does miR-302/367 (Rosa et al., 2009; Lipchina et al., 2011; Rosa and Brivanlou, 2011). However, let-7 miRNA blocks the processing of pluripotency transcripts and is inhibited by LIN28 (Viswanathan et al., 2008; Newman et al., 2008; Rybak et al., 2008). (E) Other major determinants of gene expression in hPSCs. (Left) Metabolites, such as methionine (Met) and glucose (Gluc), generate substrates for histone modifications in hPSCs (Shiraki et al., 2014; Moussaieff et al., 2015). (Middle) The spliceosome produces a pluripotency-specific isoform of the transcription factor FOXP1, while SON ensures the accurate splicing of OCT4 and PRDM14 (Gabut et al., 2011; Lu et al., 2013). (Right) Insulated neighborhoods established by cohesion-associated CTCF loops constrain enhancer-promoter interactions at human pluripotency loci (Ji et al., 2016).
Core transcriptional circuitry
A core set of master transcription factors is considered to be essential for the maintenance of hESCs. Boyer and colleagues mapped the genome-wide binding of OCT4, SOX2 and NANOG (collectively OSN) by chromatin immunoprecipitation (ChIP) coupled with DNA microarrays (ChIP-Chip; see Glossary, Box 1) and made three major observations: (1) OSN co-occupy a large fraction of promoters in their target genes; (2) roughly half of the genes bound by OSN are actively transcribed in hESCs; and (3) OSN collaborate in a regulatory circuitry that involves extensive autoregulatory and feedforward loops (Boyer et al., 2005). The target genes of OSN are poorly conserved between mouse and human ESCs (Loh et al., 2006); however, this is partially explained by the evolution of new transcription factor binding sites by species-specific transposable elements (Kunarso et al., 2010). As we will discuss, the core pluripotency transcription factors are directly plugged into multiple layers of gene expression control in hESCs (Fig. 2A-E).
Do OSN act collectively on shared target genes or do they act individually to repress distinct cell fates? Ivanova and colleagues performed loss- and gain-of-function studies in multiple independent hESC lines and reported that OSN exercise distinct roles in suppressing lineage commitment (Wang et al., 2012). Specifically, OCT4 interacts with the BMP4 pathway to specify four distinct cell fates in a dose-dependent manner (Fig. 2A). High levels of OCT4 induce mesendoderm (see Glossary, Box 1) in the presence of BMP4, but maintain hESC self-renewal in the absence of BMP4. By contrast, low levels of OCT4 induce extraembryonic genes in the presence of BMP4, but stimulate neuroectoderm genes in the absence of BMP4. In addition, NANOG represses neuroectoderm differentiation, whereas SOX2 represses mesendoderm differentiation (Fig. 2A). Furthermore, the overexpression of OSN does not trigger hESC differentiation, indicating that OSN act primarily as differentiation repressors in hESCs (Wang et al., 2012).
Other transcription factors that cooperate with the core transcription factors in sustaining the human pluripotent state have also been identified. For instance, Ng and colleagues performed a high-throughput RNA interference (RNAi) screen in OCT4-GFP reporter hESCs, and identified an essential role for the transcription factor PRDM14 (Chia et al., 2010), the ortholog of which is required for the establishment of primordial germ cells (PGCs; see Glossary, Box 1) in mice (Yamaji et al., 2008). PRDM14 regulates the expression of OCT4 by binding to its proximal enhancer and colocalizes with OSN across the genome in hESCs. Several Forkhead box (FOX) transcription factors have also been implicated in the control of human pluripotency. FOXO1 binds and regulates the promoters of OCT4 and SOX2 (Zhang et al., 2011). In addition, a specific level of FOXD3 is required for hESC self-renewal: overexpression of FOXD3 induces differentiation to paraxial mesoderm, whereas depletion of FOXD3 generates mesodermal and endodermal derivatives (Arduini and Brivanlou, 2012).
Growth factor signaling
There has been a concerted effort to define the signaling requirements of hESCs since their initial derivation on mouse embryonic fibroblasts (MEFs) in serum-containing media (Thomson et al., 1998). Two growth factors have been identified that can replace the requirement for MEFs or for MEF-conditioned medium: transforming growth factor β (TGFβ)/Activin/Nodal, which signals through the downstream effectors SMAD2/3 (James et al., 2005; Beattie et al., 2005); and basic fibroblast growth factor (FGF2) (Xu et al., 2005). Recombinant provision of Activin and FGF2 allows hESCs to be maintained in the absence of both MEFs and serum (Vallier et al., 2005). But how do these signaling pathways contribute to maintenance of the pluripotent state?
FGF/ERK signaling was reported to induce the expression of NANOG in hESCs, but the underlying mechanism remains unclear (Greber et al., 2010; Yu et al., 2011). On the other hand, direct binding of SMAD2/3 to the NANOG promoter suggests that the TGFβ/Activin/Nodal pathway directly regulates the expression of this core transcription factor (Vallier et al., 2009; Xu et al., 2008; Greber et al., 2008) (Fig. 2B). Interestingly, the transcription of components of the TGFβ signaling pathway is enriched in human EPI cells (Blakeley et al., 2015). Moreover, NANOG protein expression is reduced in human blastocysts treated with a TGFβ inhibitor, but this is not seen in treated mouse blastocysts (Blakeley et al., 2015). Thus, the TGFβ-mediated regulation of NANOG appears common to human pluripotent cells in vitro and in vivo. Inhibition of TGFβ/Activin/Nodal signaling in hESCs rapidly suppresses NANOG expression and induces neural differentiation (Chambers et al., 2009). This is consistent with the aforementioned role of NANOG in repressing neuroectoderm-associated genes.
Wnt signaling has also been implicated in regulating hESCs, although its role has been harder to define due to its dose-dependent effects. It was initially reported that hESC self-renewal can be maintained by providing recombinant Wnt3a or by inhibiting glycogen synthase kinase 3 (GSK3), the enzyme that targets β-catenin for degradation (Sato et al., 2004). However, Moon and colleagues showed that the long-term treatment of hESCs with Wnt3a actually compromises their self-renewal (Davidson et al., 2012). A similar phenotype was observed by increasing the concentration of GSK3 inhibitor, which resulted in a more robust induction of β-catenin transcriptional activity. Variable levels of endogenous Wnt pathway activity have been linked to the heterogeneous differentiation propensity of hESCs: whereas Wnthigh cells predominantly form mesendoderm, Wntlow cells are biased towards neuroectoderm (Blauwkamp et al., 2012). A function for endogenous Wnt signals in driving exit from pluripotency was also observed in mEpiSCs (Kurek et al., 2015), and this thus appears to be a conserved feature of PSCs that resemble the post-implantation EPI. It should be noted, however, that the pro-differentiation role of Wnt/β-catenin signaling in mEpiSCs/hESCs is distinct from its effects in mESCs, where Wnt signals and GSK3 inhibition are known to reinforce the pluripotency network and suppress differentiation (Ying et al., 2008; ten Berge et al., 2011; Wray et al., 2011; Yi et al., 2011).
Extensive crosstalk has been proposed to occur among the major signaling pathways in hESCs (Singh et al., 2012). This model of Singh et al. centers on the role of PI3K/AKT signaling as a node that modulates inputs from FGF/ERK, Activin/Nodal and Wnt/β-catenin signaling (Fig. 2B). Activation of PI3K/AKT by growth factors such as FGF2, IGF1 or heregulin (neuregulin 1) keeps phosphorylated SMAD2/3 levels within a tight range that allows for the activation of pluripotency genes, including NANOG. Conversely, the withdrawal of PI3K/AKT stimulation leads to elevated SMAD2/3 activity and to the induction of mesendoderm genes. According to this model, PI3K/AKT also inhibits MEK/ERK signaling, which in turn blocks β-catenin-mediated stimulation of developmental genes (Singh et al., 2012).
Chromatin-modifying enzymes
The chromatin landscape of pluripotent cells was first charted in mESCs. This work surprisingly revealed the genome-wide colocalization of active and repressive chromatin marks: tri-methylation of lysine 4 on histone 3 (H3K4me3) and tri-methylation of lysine 27 on histone 3 (H3K27me3), respectively (Bernstein et al., 2006; Azuara et al., 2006). Such a ʻbivalent' chromatin signature is thought to mark genes that are repressed in ESCs but poised to allow for alternative fates. H3K4me3 and H3K27me3 also overlap at thousands of genes in hESCs, but few genes exhibit H3K27me3 alone (Pan et al., 2007; Zhao et al., 2007). This indicates that bivalency is the default chromatin state at key developmental control genes marked by H3K27me3 in hESCs (Fig. 2C).
Deposition of H3K27me3 is mediated by Polycomb repressive complex 2 (PRC2; see Glossary, Box 1) via its catalytic subunit EZH1/2. Chromatin immunoprecipitation followed by DNA sequencing (ChIP-Seq; see Glossary, Box 1) studies by the Bernstein lab have revealed that many bivalent domains in mouse and human ESCs are also occupied by the PRC1 complex, and that such types of bivalent domains more efficiently retain repressive chromatin marks during differentiation (Ku et al., 2008). The precise mechanisms governing Polycomb recruitment to specific genomic targets remain unknown (reviewed by Holoch and Margueron, 2017), but a recent study in mESCs showed that Polycomb-associated proteins have a unique DNA recognition motif that specifically binds to unmethylated CpG sites (Li et al., 2017). In addition, PRDM14 has been reported to direct PRC2 to specific developmental regulators in hESCs (Chan et al., 2013a), and a significant fraction of PRC2 targets in mouse and human ESCs is co-occupied by OSN (Boyer et al., 2005; Lee et al., 2006).
The genetic ablation of various enzymes belonging to the PRC2 complex causes the removal of repressive H3K27me3 marks, reduced self-renewal and upregulation of mesendoderm genes in hESCs (Collinson et al., 2016; Shan et al., 2017). By contrast, the loss of Eed (which encodes a PRC2 component) has little impact on the transcriptome of mESCs grown in chemically defined 2i/L conditions (see Glossary, Box 1) (Galonska et al., 2015), indicating that mESCs can self-renew in the complete absence of H3K27me3. Enzymes that regulate the levels of H3K4me3 are also required for the self-renewal of hESCs. Lysine-specific demethylase 1 (LSD1; also known as KDM1A) keeps the levels of H3K4 methylation in check at the promoters of developmental genes marked by bivalent domains (Adamo et al., 2011). By contrast, the DPY30-COMPASS methyltransferase promotes H3K4me3 deposition at a subset of pluripotency genes and bivalent genes involved in mesendoderm specification (Bertero et al., 2015). Hence, enzymes controlling both active and repressive histone marks are essential for hESC identity (Fig. 2C).
In addition to their dependence on histone-modifying enzymes, hESCs are exquisitely sensitive to the loss of DNMT1, the DNA methyltransferase responsible for maintaining CpG methylation during DNA replication. Meissner and colleagues have shown that ablating the de novo methyltransferases DNMT3A and/or DNMT3B in hESCs causes only a modest reduction in DNA methylation content, and does not compromise hESC self-renewal (Liao et al., 2015). However, the deletion of DNMT1 in hESCs causes global DNA demethylation and cell death. This dependence of hESCs on DNMT1 contrasts with the role of DNMTs in mESCs; mESCs tolerate the combined removal of all three DNMT enzymes and the complete loss of DNA methylation (Tsumura et al., 2006). Together, these examples demonstrate that certain gene-silencing mechanisms, while dispensable for the self-renewal of mESCs, are functionally required in hESCs.
Functional roles of non-coding RNAs
MicroRNAs (miRNAs) are small RNAs (∼22 nt) that mediate the post-transcriptional repression of mRNA targets (Bartel, 2009). The miR-302/367 cluster comprises the most abundantly expressed miRNA family in hESCs (Suh et al., 2004; Morin et al., 2008; Bar et al., 2008). miR-302/367 is thought to repress neural differentiation in hESCs by modulating Nodal and BMP signaling (Rosa et al., 2009; Lipchina et al., 2011) and by downregulating NR2F2 (Coup-TFII), a transcription factor that induces neural differentiation (Rosa and Brivanlou, 2011) (Fig. 2D). The expression of miR-302/367 is positively influenced by OCT4, which binds directly to the miR-302/367 promoter in mouse and human ESCs (Marson et al., 2008). Other families of miRNAs counteract the pluripotency network. For example, miR-145 post-transcriptionally silences the expression of OCT4, SOX2 and KLF4 during hESC differentiation (Xu et al., 2009). Another well-studied example of an miRNA that antagonizes pluripotency is let-7, the processing of which into mature miRNAs is blocked by the RNA-binding protein LIN28 in mouse and human ESCs (Viswanathan et al., 2008; Rybak et al., 2008; Newman et al., 2008).
Long non-coding RNAs (lncRNAs) are another class of non-coding RNAs that contribute to the transcriptional regulation of hESCs, and are associated with endogenous retroviral elements. One such lncRNA derives from retrovirally encoded HERVH transcripts and functions by recruiting OCT4 and co-activators, such as p300 (EP300), to activate the expression of nearby genes (Lu et al., 2014) (Fig. 2D). Reijo Pera and colleagues (Durruthy-Durruthy et al., 2016a) also identified three transposon-derived lncRNAs, called HPAT2, HPAT3 and HPAT5, that are found in hESCs and are also highly expressed in the human blastocyst. The injection of siRNAs that target HPAT2, HPAT3 and HPAT5 into human embryos at the 2-cell stage prevents the injected blastomeres from contributing to the ICM. Hence, these lncRNAs are required for the establishment of human pluripotent cells in vivo (Durruthy-Durruthy et al., 2016a). The exogenous expression of HPAT5 inhibits hESC differentiation by interfering with the expression and activity of let-7 miRNAs, which reveals an interesting example of functional crosstalk between miRNAs and lncRNAs (Fig. 2D). Like the miRNA families discussed above, many transposon-derived lncRNAs are under the direct transcriptional regulation of core pluripotency factors in hESCs (Santoni et al., 2012; Durruthy-Durruthy et al., 2016a; Fort et al., 2014).
Other determinants
Several additional layers of gene expression control have been implicated in hPSCs. First, numerous links have been identified between the levels of metabolites and epigenetic marks in hESCs (reviewed by Mathieu and Ruohola-Baker, 2017). Notably, methionine metabolism provides a substrate for histone methylation (Shiraki et al., 2014), while glycolysis provides a substrate for histone acetylation in hESCs (Moussaieff et al., 2015) (Fig. 2E, left). Second, it has also been shown that the spliceosome ensures the accurate splicing of pluripotency-associated transcripts (Lu et al., 2013) and is known to produce a pluripotency-specific isoform of the transcription factor FOXP1 (Gabut et al., 2011) (Fig. 2E, middle). Third, cohesin-associated chromatin loops are known to establish boundaries between insulated neighborhoods (see Glossary, Box 1), which constrain enhancer-promoter interactions at loci such as PRDM14 (Ji et al., 2016) (Fig. 2E, right). Thus, global gene expression in hESCs is regulated by a complex web of interactions among transcription factors, chromatin modifiers, metabolites and 3D chromatin structure.
Transcriptome and epigenome remodeling during the transitions into and out of pluripotency
In this final section, we examine how the regulatory network that governs human pluripotency is established and dynamically remodeled during three pivotal cell fate transitions: the induction of pluripotency by defined factors, the interconversion between distinct pluripotent states, and the exit from pluripotency. A graphical summary of the major molecular changes during these three cell fate transitions is provided in Fig. 3.
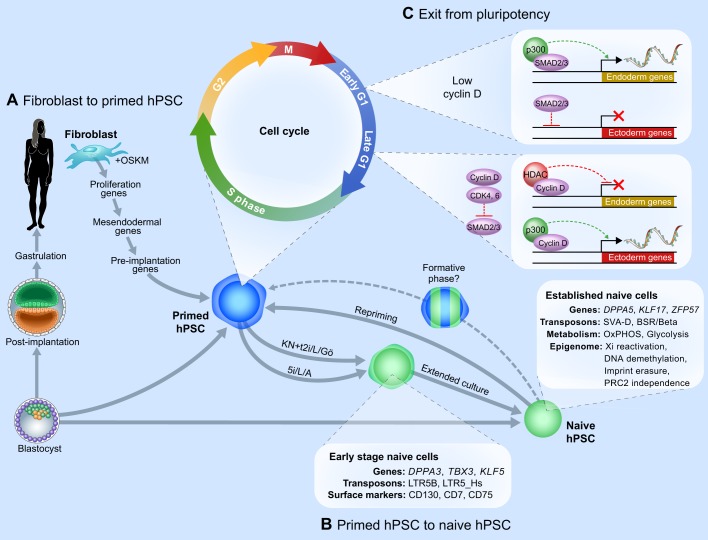
Fig. 3.
Molecular dynamics during the transitions into and out of human pluripotency. Summary of the transcriptional and epigenetic changes during cell fate transitions involving hPSCs. (A) Sequence of events occurring during the reprogramming of human somatic cells to pluripotency upon overexpression of OCT4, SOX2, KLF4 and c-MYC (OSKM) (Cacchiarelli et al., 2015). (B) Events occurring during the primed-to-naive PSC transition. Collier et al. (2017) assessed transcriptional dynamics at early and late stages of naive resetting upon overexpression of KLF2 and NANOG (KN) transgenes in primed hESCs and transfer to t2i/L+Gö medium (Takashima et al., 2014) or by switching primed hESCs to 5i/L/A medium (Theunissen et al., 2014). Note that the Smith laboratory recently showed that hPSCs can also be reset to naive pluripotency by transient histone deacetylase inhibition and transfer to t2i/L+Gö medium (Guo et al., 2017). The dynamics of X-chromosome reactivation during naive resetting were analyzed by Sahakyan et al. (2017). Naive hPSCs derived in t2i/L+Gö or 5i/L/A display globally reduced DNA methylation levels and erasure of most imprinted regions (Pastor et al., 2016; Theunissen et al., 2016). (C) The exit of human pluripotency, specifically the relationship between the cell cycle and lineage commitment of hPSCs (Pauklin and Vallier, 2013; Pauklin et al., 2016). The existence of a formative phase, during which PSCs acquire competence for multilineage and germ cell induction, has recently been proposed (Smith, 2017). OxPHOS, oxidative phosphorylation; Xi, inactive X.
The induction of pluripotency in human somatic cells
Since the discovery that somatic cells can be reprogrammed to pluripotency by exogenous transcription factors (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007), there has been significant interest in elucidating how reprogramming factors act. Zaret and colleagues performed ChIP-Seq analyses of OCT4, SOX2, KLF4 and c-MYC (collectively OSKM) in human fibroblasts to examine their initial engagement with chromatin (Soufi et al., 2012). They reported that OSKM predominantly bind to distal regulatory regions after 48 h of reprogramming factor expression. While OCT4, SOX2 and KLF4 (OSK) act as ʻpioneer factors' (see Glossary, Box 1) that open up closed chromatin, c-MYC facilitates the engagement of OSK with chromatin. Collectively, OSKM activate genes in fibroblasts that are known to promote reprogramming, such as the transcription factor GLIS1 (Maekawa et al., 2011) and miR-302/367 (Anokye-Danso et al., 2011). Early OSKM binding was also enriched at genes with roles in apoptosis and senescence, including TP53 (Soufi et al., 2012). This is consistent with prior reports that the cell death pathway is induced rapidly after expression of the reprogramming factors in human fibroblasts (Kawamura et al., 2009). A recent analysis of OSK activity during mouse reprogramming indicates that these reprogramming factors do not act in isolation, however, but rely on collaborative interactions with stage-specific transcription factors to silence somatic enhancers and to activate pluripotency-specific enhancers (Chronis et al., 2017).
Analyzing the changes in gene expression and chromatin state during reprogramming is an arduous task due to the heterogeneous and inefficient nature of the process. Mikkelsen and colleagues (Cacchiarelli et al., 2015) took advantage of immortalized ʻsecondary' fibroblasts to characterize the global transcriptional and epigenomic events that occur during human reprogramming (Fig. 3A). Secondary fibroblasts are derived from a clonal line of human iPSCs that was generated with drug-inducible transcription factors, and therefore contain a uniform stoichiometry of the reprogramming factors (Hockemeyer et al., 2008). Using these cells, it was shown that, shortly after factor induction, genes associated with fibroblast identity are downregulated, whereas genes important for proliferation and metabolic reprogramming are activated by day 5 (Cacchiarelli et al., 2015). Subsequently, the fibroblasts transiently acquire a mesendodermal gene expression signature, in agreement with a prior study (Takahashi et al., 2014). As fibroblasts are of mesodermal origin, this intermediate state marks a reversal of the normal order of development. Another transient wave of gene expression was observed at the later stages of reprogramming when certain mRNAs (e.g. DPPA3, DNMT3L, TFCP2L1) and miRNAs (miR371) that are associated with human pre-implantation development are upregulated. However, this pre-implantation gene expression signature is lost again as the hiPSCs acquire a post-implantation identity typical of hESCs. This final step is also accompanied by a gain of DNA hypermethylation and of bivalent chromatin domains (Cacchiarelli et al., 2015). It will be of interest to investigate whether passage through these transient mesendodermal and pre-implantation states is required for successful reprogramming.
Understanding the sources of gene expression variability between iPSC lines is imperative for establishing well-controlled models of human disease. The NextGen Consortium and HipSci Initiative combined whole-genome DNA and RNA sequencing in hundreds of iPSC lines to quantify the fraction of transcriptional variability caused by genetic background (Carcamo-Orive et al., 2017; DeBoever et al., 2017; Kilpinen et al., 2017). These analyses revealed that differences between individuals contribute the largest proportion of variation between iPSC lines (50-53% of variation at the single-gene level, depending on the study). In addition, a significant fraction of inter-individual variability is driven by expression quantitative trait loci (eQTL; see Glossary, Box 1) specific to iPSCs, which are enriched at the binding sites for OCT4, NANOG and other pluripotency regulators (Carcamo-Orive et al., 2017; DeBoever et al., 2017; Kilpinen et al., 2017). By contrast, non-genetic background-associated transcriptional variability is largely driven by the skewed deposition of H3K27me3 silencing marks at Polycomb target genes (Carcamo-Orive et al., 2017). This is consistent with the findings from an RNAi screen that revealed that Polycomb enzymes are important regulators of human somatic cell reprogramming (Onder et al., 2012).
An accompanying study from the NextGen Consortium examined variability in DNA methylation between iPSC lines obtained from multiple pairs of monozygotic twins (Panopoulos et al., 2017). This work revealed that aberrant DNA methylation is not primarily driven by genetic variation, but seems to occur at regulatory regions that contain MYC or MYC-like binding sites in a clone-specific manner. The process of epigenetic reprogramming – whether by defined factors or by somatic cell nuclear transfer – also renders hPSCs vulnerable to the loss of genomic imprinting, especially at paternally methylated imprints (Bar et al., 2017). These large-scale analyses offer a cautionary note about the extent of variability between hiPSC lines and provide a wealth of data to further dissect the molecular drivers of gene expression variability during reprogramming.
Interconversions between distinct human pluripotent states
Two distinct pluripotent stem cell states can be derived from mouse embryos: mESCs derived from the blastocyst represent an uncommitted ʻnaive' state, whereas mEpiSCs derived from the post-implantation EPI are designated as ʻprimed' (see Glossary, Box 1) (Nichols and Smith, 2009). Evidence has accumulated that hESCs and hiPSCs are in a primed pluripotent state analogous to that observed in mEpiSCs. hPSCs share basic biological properties with mEpiSCs, including a flat morphology, dependence on FGF/Activin and propensity for X-chromosome inactivation (XCI) (Brons et al., 2007; Tesar et al., 2007). Like mEpiSC lines, hPSC lines also frequently display lineage bias during differentiation (Osafune et al., 2008; Bock et al., 2011; Nishizawa et al., 2016; Blauwkamp et al., 2012). In addition, global gene expression in hPSCs correlates most strongly with that of the primate late post-implantation EPI (Nakamura et al., 2016), reaffirming their primed identity. These observations have generated widespread interest in methods to isolate naive hPSCs (Hanna et al., 2010; Gafni et al., 2013; Chan et al., 2013b; Ware et al., 2014; Takashima et al., 2014; Theunissen et al., 2014; Guo et al., 2016; Qin et al., 2016; Zimmerlin et al., 2016; Guo et al., 2017; Liu et al., 2017).
Recent studies have begun to explore the transcriptional and epigenetic dynamics that occur during the naive ʻresetting' of conventional hPSCs and their subsequent ʻrepriming'. For example, Sahakyan et al. (2017) examined the status of the X chromosome in female naive hESCs derived using two alternative culture conditions: t2i/L+Gö (Takashima et al., 2014) and 5i/L/A (see Glossary, Box 1) (Theunissen et al., 2014). These two conditions induce a global transcriptional profile that correlates strongly with that of human and monkey pre-implantation embryos (Huang et al., 2014; Nakamura et al., 2016). The conversion of primed hESCs to the naive state results in X-chromosome reactivation together with the expression of XIST, although XIST is predominantly expressed from a single X chromosome (Sahakyan et al., 2017). Unlike primed hESCs that undergo irreversible ʻerosion' of the inactive X chromosome, naive hESCs can initiate de novo XCI upon differentiation, albeit in a non-random manner (Sahakyan et al., 2017). Lanner and colleagues defined combinations of cell-surface markers that could distinguish primed from naive hESCs (Collier et al., 2017). Using these markers, these authors tracked temporal changes in gene expression during the primed-to-naive conversion (Fig. 3B). Whereas some naive-specific genes (such as DPPA3 and TBX3) are activated relatively early, others (such as DPPA5, KLF17 and ZFP57) are fully induced only upon subsequent passaging (Collier et al., 2017). Naive resetting of hPSCs also involves profound metabolic changes, including increased oxidative phosphorylation (Takashima et al., 2014; Sperber et al., 2015) and glycolytic metabolism (Gu et al., 2016), and confers tolerance to the removal or inhibition of PRC2 enzymes (Moody et al., 2017; Shan et al., 2017).
Given their stage-specific patterns of expression during early human development, it has been proposed that transposable elements might serve as biomarkers to distinguish between distinct human pluripotent states. Initial work based on the culture of hESCs in 2i/L suggested that HERVH might be a marker of naive human pluripotency (Wang et al., 2014). However, it was reported that naive cells cultured in t2i/L+Gö or 5i/L/A show more specific upregulation of LTR5-HERVK and SVA-D integrants (Theunissen et al., 2016; Collier et al., 2017; Guo et al., 2017) (Fig. 3B). HERVK is actively transcribed during human embryogenesis from the 8-cell stage, and encodes viral proteins that are expressed in the human blastocyst (Goke et al., 2015; Grow et al., 2015). SVA-D elements are highly expressed at the human morula and blastocyst stages and are part of a hominid-specific family of retroelements (Hancks and Kazazian, 2010). The full repertoire of naive-specific transposable elements was not activated until after passaging and subsequent maturation of the naive phenotype in the course of primed-to-naive resetting (Collier et al., 2017). Whether these transposon families simply provide biomarkers of distinct human pluripotent states or also have functional significance remains to be elucidated.
The above studies indicate that naive hESCs offer a window into understanding the mechanisms of human pre-implantation development that are difficult to model in conventional hESCs, such as X-chromosome reactivation and inactivation, and the role of early embryonic transposons. However, current naive hESCs do not perfectly resemble pluripotent cells in the human blastocyst, as exemplified by the loss of imprint methylation in t2i/L+Gö or 5i/L/A (Pastor et al., 2016; Theunissen et al., 2016; Guo et al., 2017), and the observation that naive hESCs maintained in 5i/L/A are prone to genetic instability (Theunissen et al., 2014; Pastor et al., 2016; Liu et al., 2017). Thus, it remains to be determined whether strategies can be devised to capture naive hESCs that are as robust as their mouse counterparts, an advance that could facilitate a wide range of applications in regenerative medicine. While the contribution of currently available naive hESCs to interspecies chimeras has been inefficient (reviewed by Wu et al., 2016a), a recent study reported robust interspecies chimerism upon injection of human extended pluripotent stem cells (EPS cells; see Glossary, Box 1), which were derived in a newly defined chemical culture condition (Yang et al., 2017). The exact developmental identity of EPS cells remains unclear, but these data suggest that alternative pluripotent states that have enhanced proliferative capacity could enable human cells to more efficiently contribute to interspecies chimeras. Finally, the generation of hPSCs with distinct developmental identities might also facilitate other research applications, such as the induction of human PGC-like cells (von Meyenn et al., 2016; Irie et al., 2015), and more efficient genome editing (Yang et al., 2016).
Exiting the human pluripotency program
How is the human pluripotency network disassembled during the onset of lineage commitment and what are the accompanying transcriptional and epigenetic changes? In attempt to address this, Ng and colleagues performed a high-throughput RNAi screen to identify the mechanisms that promote exit from the human pluripotent state (Gonzales et al., 2015). hESCs containing a NANOG-GFP reporter transgene were transfected with siRNAs 24 h before induction of differentiation. As the expression of NANOG is tightly linked to pluripotency, the depletion of genes that promote dissolution of pluripotency is expected to delay the downregulation of NANOG-GFP reporter activity during differentiation. This screen identified cell cycle regulators that are involved in DNA replication during S phase or in the G2-to-M transition as regulators of pluripotent state dissolution (Gonzales et al., 2015). As depletion of these factors effectively arrests the cells in S or G2, these cell cycle phases appear to be intrinsically inclined towards maintenance of pluripotency. The underlying mechanism rests on the activation of the ATM/ATR-p53 axis during S phase and of cyclin B1 during G2 phase, which stimulate the expression of TGFβ-related genes (Gonzales et al., 2015).
By contrast, the G1 phase of the cell cycle has been identified as being favorable to hESC differentiation. Using the fluorescence ubiquitin cell cycle indicator (FUCCI; see Glossary, Box 1) reporter system to isolate hESCs in different stages of the cell cycle, Pauklin and Vallier demonstrated that hESCs in early G1 can only initiate differentiation into endoderm, while late G1 is permissive for ectoderm differentiation. This partitioning of G1 is the result of increasing levels of Cyclin D-CDK4/6 in the course of G1, which has two major consequences: (1) the removal of SMAD2/3 from target genes by Cyclin D-CDK4/6 (Pauklin and Vallier, 2013); and (2) the direct transcriptional repression of endoderm genes and activation of ectoderm genes by Cyclin D (Pauklin et al., 2016) (Fig. 3C). The G1 phase also has a unique epigenetic configuration that renders it transcriptionally competent to exit from pluripotency: many developmental genes transiently gain bivalent domains during G1, but are only marked by H3K27me3 during the remainder of the cell cycle (Singh et al., 2015). Collectively, these studies demonstrate that the phase of the cell cycle contributes to the decision of whether to maintain or dissolve the pluripotent state in response to lineage-inductive cues.
Genome-wide studies have revealed that the lineage specification and differentiation of hESCs involve broad changes in DNA methylation, in active and repressive histone marks, and in transcription factor binding (Xie et al., 2013; Gifford et al., 2013; Tsankov et al., 2015). An important focus for future work will be to define the order in which these regulatory dynamics occur during exit from pluripotency. In view of their heterogeneity, lineage priming and post-implantation identity, conventional (primed) hESCs are an imperfect model for addressing these questions. By contrast, naive hESCs may provide a more uniform and unbiased starting point to investigate how the pluripotent state becomes disassembled. It should be noted, however, that naive hESCs derived in t2i/L+Gö or 5i/L/A appear to be less responsive to lineage-inductive cues, although they can differentiate efficiently after re-exposure to primed culture conditions (Takashima et al., 2014; Sahakyan et al., 2017; Guo et al., 2017; Liu et al., 2017). This might reflect a developmental requirement for transition through a formative phase in which pluripotent cells acquire increased capacity to respond to specification cues (Smith, 2017). This formative period may last longer in humans, as the primate post-implantation EPI maintains a relatively stable transcriptome for one week (Nakamura et al., 2016). Hence, an important objective will be to better understand the transition between naive and formative pluripotency, and determine whether it is possible to capture formative hPSCs in culture.
Future perspectives
Research in the past decade has illuminated fundamental aspects of genome regulation in early human embryos and PSCs, but some significant gaps in our understanding remain. First, while gene expression dynamics during human pre-implantation development have been studied in depth by whole-embryo and single-cell RNA sequencing, information about the post-implantation stages remains limited. The ability to culture human embryos in vitro beyond implantation paves the way for a detailed investigation of this critical period of human development (Deglincerti et al., 2016; Shahbazi et al., 2016). Second, studies of chromatin accessibility and histone modifications using limited numbers of cells have revealed that dramatic chromatin reorganization occurs during mouse pre-implantation development (Liu et al., 2016; Zhang et al., 2016; Dahl et al., 2016; Wu et al., 2016b). We anticipate that such approaches will also offer key insights into epigenetic aspects of early human embryogenesis. Third, recent advances in genome editing present an opportunity to examine the function of human pluripotency regulators in their in vivo context, as recently shown for OCT4 (Fogarty et al., 2017). Fourth, whereas the composition of pluripotency-associated protein complexes has been analyzed in mESCs (Wang et al., 2006; Rafiee et al., 2016; Costa et al., 2013; Ding et al., 2015; Gagliardi et al., 2013), comparable studies have not been performed in hESCs. Such biochemical approaches might reveal connections among seemingly disparate regulatory modules and could identify novel reprogramming factors. Finally, there remains a strong interest in deriving novel types of human PSCs that reflect distinct stages of human embryogenesis. Efforts in this regard are guided by emerging insights into the transcriptional and epigenetic status of pluripotent cells in early primate embryos, which provide a set of rigorous benchmarks to assess the developmental identity of hPSCs (Boroviak and Nichols, 2017). The ability to capture hPSCs with distinct embryonic identities will enable researchers to more precisely reconstruct the earliest steps of human development in vitro.
Acknowledgements
We thank Steven Lee for assistance with illustrations.
Footnotes
Competing interests
R.J. is co-founder of Fate Therapeutics, Fulcrum Therapeutics and Omega Therapeutics.
Funding
T.W.T. is supported by a Sir Henry Wellcome Postdoctoral Fellowship from the Wellcome Trust (098889/Z/12/Z) and R.J. is supported by grants from the National Institutes of Health. Deposited in PMC for release after 12 months.
References
- Adamo A., Sesé B., Boue S., Castaño J., Paramonov I., Barrero M. J. and Izpisua Belmonte J. C. (2011). LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 13, 652-659. 10.1038/ncb2246 [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F., Trivedi C. M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P. J., Epstein J. A. et al. (2011). Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376-388. 10.1016/j.stem.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduini B. L. and Brivanlou A. H. (2012). Modulation of FOXD3 activity in human embryonic stem cells directs pluripotency and paraxial mesoderm fates. Stem Cells 30, 2188-2198. 10.1002/stem.1200 [DOI] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H. F., John R. M., Gouti M., Casanova M., Warnes G., Merkenschlager M. et al. (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532-538. 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- Bar M., Wyman S. K., Fritz B. R., Qi J., Garg K. S., Parkin R. K., Kroh E. M., Bendoraite A., Mitchell P. S., Nelson A. M. et al. (2008). MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26, 2496-2505. 10.1634/stemcells.2008-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar S., Schachter M., Eldar-Geva T. and Benvenisty N. (2017). Large-scale analysis of loss of imprinting in human pluripotent stem cells. Cell Rep. 19, 957-968. 10.1016/j.celrep.2017.04.020 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215-233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie G. M., Lopez A. D., Bucay N., Hinton A., Firpo M. T., King C. C. and Hayek A. (2005). Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells 23, 489-495. 10.1634/stemcells.2004-0279 [DOI] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315-326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Bertero A., Madrigal P., Galli A., Hubner N. C., Moreno I., Burks D., Brown S., Pedersen R. A., Gaffney D., Mendjan S. et al. (2015). Activin/nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes Dev. 29, 702-717. 10.1101/gad.255984.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeley P., Fogarty N. M. E., Del Valle I., Wamaitha S. E., Hu T. X., Elder K., Snell P., Christie L., Robson P. and Niakan K. K. (2015). Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3151-3165. 10.1242/dev.123547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauwkamp T. A., Nigam S., Ardehali R., Weissman I. L. and Nusse R. (2012). Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat. Commun. 3, 1070 10.1038/ncomms2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z. D., Ziller M., Croft G. F., Amoroso M. W., Oakley D. H. et al. (2011). Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439-452. 10.1016/j.cell.2010.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T. and Nichols J. (2017). Primate embryogenesis predicts the hallmarks of human naïve pluripotency. Development 144, 175-186. 10.1242/dev.145177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Bertone P., Smith A. and Nichols J. (2014). The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 16, 516-528. 10.1038/ncb2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G. et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947-956. 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I. G. M., Smithers L. E., Trotter M. W. B., Rugg-Gunn P., Sun B., Chuva De Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A. et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191-195. 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D., Trapnell C., Ziller M. J., Soumillon M., Cesana M., Karnik R., Donaghey J., Smith Z. D., Ratanasirintrawoot S., Zhang X. et al. (2015). Integrative analyses of human reprogramming reveal dynamic nature of induced pluripotency. Cell 162, 412-424. 10.1016/j.cell.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo-Orive I., Hoffman G. E., Cundiff P., Beckmann N. D., D'souza S. L., Knowles J. W., Patel A., Papatsenko D., Abbasi F. Reaven G. M. et al. (2017). Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell 20, 518-532 e9. 10.1016/j.stem.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M. and Studer L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275-280. 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.-S., Göke J., Lu X., Venkatesan N., Feng B., Su I.-H. and Ng H.-H. (2013a). A PRC2-dependent repressive role of PRDM14 in human embryonic stem cells and induced pluripotent stem cell reprogramming. Stem Cells 31, 682-692. 10.1002/stem.1307 [DOI] [PubMed] [Google Scholar]
- Chan Y.-S., Göke J., Ng J.-H., Lu X., Gonzales K. A. U., Tan C.-P., Tng W.-Q., Hong Z.-Z., Lim Y.-S. and Ng H.-H. (2013b). Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13, 663-675. 10.1016/j.stem.2013.11.015 [DOI] [PubMed] [Google Scholar]
- Chazaud C., Yamanaka Y., Pawson T. and Rossant J. (2006). Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 10, 615-624. 10.1016/j.devcel.2006.02.020 [DOI] [PubMed] [Google Scholar]
- Chia N.-Y., Chan Y.-S., Feng B., Lu X., Orlov Y. L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.-S. et al. (2010). A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468, 316-320. 10.1038/nature09531 [DOI] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J. and Plath K. (2017). Cooperative binding of transcription factors orchestrates reprogramming. Cell 168, 442-459.e20. 10.1016/j.cell.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier A. J., Panula S. P., Schell J. P., Chovanec P., Plaza Reyes A., Petropoulos S., Corcoran A. E., Walker R., Douagi I., Lanner F. et al. (2017). Comprehensive cell surface protein profiling identifies specific markers of human naive and primed pluripotent states. Cell Stem Cell 20, 874-890.e7. 10.1016/j.stem.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson A., Collier A. J., Morgan N. P., Sienerth A. R., Chandra T., Andrews S. and Rugg-Gunn P. J. (2016). Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in human embryonic stem cells. Cell Rep. 17, 2700-2714. 10.1016/j.celrep.2016.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y., Ding J., Theunissen T. W., Faiola F., Hore T. A., Shliaha P. V., Fidalgo M., Saunders A., Lawrence M., Dietmann S. et al. (2013). NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370-374. 10.1038/nature11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J. A., Jung I., Aanes H., Greggains G. D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A. Y. et al. (2016). Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548-552. 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson K. C., Adams A. M., Goodson J. M., Mcdonald C. E., Potter J. C., Berndt J. D., Biechele T. L., Taylor R. J. and Moon R. T. (2012). Wnt/beta-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. USA 109, 4485-4490. 10.1073/pnas.1118777109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A., Planet E., Coluccio A., Verp S., Duc J. and Trono D. (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49, 941-945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboever C., Li H., Jakubosky D., Benaglio P., Reyna J., Olson K. M., Huang H., Biggs W., Sandoval E., D'antonio M. et al. (2017). Large-scale profiling reveals the influence of genetic variation on gene expression in human induced pluripotent stem cells. Cell Stem Cell 20, 533-546.e7. 10.1016/j.stem.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A., Croft G. F., Pietila L. N., Zernicka-Goetz M., Siggia E. D. and Brivanlou A. H. (2016). Self-organization of the in vitro attached human embryo. Nature 533, 251-254. 10.1038/nature17948 [DOI] [PubMed] [Google Scholar]
- Deng Q., Ramskold D., Reinius B. and Sandberg R. (2014). Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193-196. 10.1126/science.1245316 [DOI] [PubMed] [Google Scholar]
- Ding J., Huang X., Shao N., Zhou H., Lee D. F., Faiola F., Fidalgo M., Guallar D., Saunders A., Shliaha P. V. et al. (2015). Tex10 coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming. Cell Stem Cell 16, 653-668. 10.1016/j.stem.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A. T., Raja R., Abeyta M. J., Taylor T., Shen S., Haqq C. and Pera R. A. R. (2004). The unique transcriptome through day 3 of human preimplantation development. Hum. Mol. Genet. 13, 1461-1470. 10.1093/hmg/ddh157 [DOI] [PubMed] [Google Scholar]
- Dunn S.-J., Martello G., Yordanov B., Emmott S. and Smith A. G. (2014). Defining an essential transcription factor program for naive pluripotency. Science 344, 1156-1160. 10.1126/science.1248882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durruthy-Durruthy J., Sebastiano V., Wossidlo M., Cepeda D., Cui J., Grow E. J., Davila J., Mall M., Wong W. H., Wysocka J. et al. (2016a). The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat. Genet. 48, 44-52. 10.1038/ng.3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durruthy-Durruthy J., Wossidlo M., Pai S., Takahashi Y., Kang G., Omberg L., Chen B., Nakauchi H., Reijo Pera R. and Sebastiano V. (2016b). Spatiotemporal reconstruction of the human blastocyst by single-cell gene-expression analysis informs induction of naive pluripotency. Dev. Cell 38, 100-115. 10.1016/j.devcel.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Evans M. J. and Kaufman M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154-156. 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- Flach G., Johnson M. H., Braude P. R., Taylor R. A. and Bolton V. N. (1982). The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1, 681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty N. M. E., McCarthy A., Snijders K. E., Powell B. E., Kubikova N., Blakeley P., Lea R., Elder K., Wamaitha S. E., Kim D. et al. (2017). Genome editing reveals a role for OCT4 in human embryogenesis. Nature 550, 67-73. 10.1038/nature24033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort A., Hashimoto K., Yamada D., Salimullah M., Keya C. A., Saxena A., Bonetti A., Voineagu I., Bertin N., Kratz A. et al. (2014). Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat. Genet. 46, 558-566. 10.1038/ng.2965 [DOI] [PubMed] [Google Scholar]
- Gabut M., Samavarchi-Tehrani P., Wang X., Slobodeniuc V., O'hanlon D., Sung H. K., Alvarez M., Talukder S., Pan Q., Mazzoni E. O. et al. (2011). An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147, 132-146. 10.1016/j.cell.2011.08.023 [DOI] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A. A. F., Manor Y. S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282-286. 10.1038/nature12745 [DOI] [PubMed] [Google Scholar]
- Gagliardi A., Mullin N. P., Ying Tan Z., Colby D., Kousa A. I., Halbritter F., Weiss J. T., Felker A., Bezstarosti K., Favaro R. et al. (2013). A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal. EMBO J. 32, 2231-2247. 10.1038/emboj.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galonska C., Ziller M. J., Karnik R. and Meissner A. (2015). Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell 17, 462-470. 10.1016/j.stem.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L. N., Yao Z., Snider L., Fong A. P., Cech J. N., Young J. M., Van Der Maarel S. M., Ruzzo W. L., Gentleman R. C., Tawil R. et al. (2012). DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38-51. 10.1016/j.devcel.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford C. A., Ziller M. J., Gu H., Trapnell C., Donaghey J., Tsankov A., Shalek A. K., Kelley D. R., Shishkin A. A., Issner R. et al. (2013). Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149-1163. 10.1016/j.cell.2013.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke J., Lu X., Chan Y.-S., Ng H.-H., Ly L.-H., Sachs F. and Szczerbinska I. (2015). Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16, 135-141. 10.1016/j.stem.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Gonzales K. A. U., Liang H., Lim Y.-S., Chan Y.-S., Yeo J.-C., Tan C.-P., Gao B., Le B., Tan Z.-Y., Low K.-Y. et al. (2015). Deterministic restriction on pluripotent state dissolution by cell-cycle pathways. Cell 162, 564-579. 10.1016/j.cell.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Greber B., Lehrach H. and Adjaye J. (2008). Control of early fate decisions in human ES cells by distinct states of TGFbeta pathway activity. Stem Cells Dev. 17, 1065-1077. 10.1089/scd.2008.0035 [DOI] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J. Y., Han D. W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P. et al. (2010). Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6, 215-226. 10.1016/j.stem.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Grow E. J., Flynn R. A., Chavez S. L., Bayless N. L., Wossidlo M., Wesche D. J., Martin L., Ware C. B., Blish C. A., Chang H. Y. et al. (2015). Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522, 221-225. 10.1038/nature14308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Gaeta X., Sahakyan A., Chan A. B., Hong C. S., Kim R., Braas D., Plath K., Lowry W. E. and Christofk H. R. (2016). Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell 19, 476-490. 10.1016/j.stem.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., Yan J., Ren X., Lin S., Li J. et al. (2014). The DNA methylation landscape of human early embryos. Nature 511, 606-610. 10.1038/nature13544 [DOI] [PubMed] [Google Scholar]
- Guo G., Von Meyenn F., Santos F., Chen Y., Reik W., Bertone P., Smith A. and Nichols J. (2016). Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep. 6, 437-446. 10.1016/j.stemcr.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Von Meyenn F., Rostovskaya M., Clarke J., Dietmann S., Baker D., Sahakyan A., Myers S., Bertone P., Reik W. et al. (2017). Epigenetic resetting of human pluripotency. Development 144, 2748-2763. 10.1242/dev.146811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks D. C. and Kazazian H. H. Jr (2010). SVA retrotransposons: Evolution and genetic instability. Semin. Cancer Biol. 20, 234-245. 10.1016/j.semcancer.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Cheng A. W., Saha K., Kim J., Lengner C. J., Soldner F., Cassady J. P., Muffat J., Carey B. W. and Jaenisch R. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. USA 107, 9222-9227. 10.1073/pnas.1004584107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson P. G., Dorais J. A., Grow E. J., Whiddon J. L., Lim J. W., Wike C. L., Weaver B. D., Pflueger C., Emery B. R., Wilcox A. L. et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925-934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Cook E. G., Gao Q., Mitalipova M. and Jaenisch R. (2008). A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3, 346-353. 10.1016/j.stem.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D. and Margueron R. (2017). Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci. 42, 531-542. 10.1016/j.tibs.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Huang X. and Wang J. (2014). The extended pluripotency protein interactome and its links to reprogramming. Curr. Opin. Genet. Dev. 28, 16-24. 10.1016/j.gde.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Maruyama T. and Fan G. (2014). The naive state of human pluripotent stem cells: a synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell 15, 410-415. 10.1016/j.stem.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W. W. C., Kobayashi T., Viukov S., Manor Y. S., Dietmann S., Hanna J. H. and Surani M. A. (2015). SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253-268. 10.1016/j.cell.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D., Levine A. J., Besser D. and Hemmati-Brivanlou A. (2005). TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132, 1273-1282. 10.1242/dev.01706 [DOI] [PubMed] [Google Scholar]
- Ji X., Dadon D. B., Powell B. E., Fan Z. P., Borges-Rivera D., Shachar S., Weintraub A. S., Hnisz D., Pegoraro G., Lee T. I. et al. (2016). 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell 18, 262-275. 10.1016/j.stem.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y. V., Menendez S., Morera L. B., Raya A., Wahl G. M. and Izpisua Belmonte J. C. (2009). Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140-1144. 10.1038/nature08311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F. P., Culley O. J. et al. (2017). Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370-375. 10.1038/nature22403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M., Koche R. P., Rheinbay E., Mendenhall E. M., Endoh M., Mikkelsen T. S., Presser A., Nusbaum C., Xie X., Chi A. S. et al. (2008). Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijk E. W., Van Tol L. T., Van de Velde H., Wubbolts R., Welling M., Geijsen N. and Roelen B. A. (2012). The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 139, 871-882. 10.1242/dev.071688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G., Chia N.-Y., Jeyakani J., Hwang C., Lu X., Chan Y.-S., Ng H.-H. and Bourque G. (2010). Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 42, 631-634. 10.1038/ng.600 [DOI] [PubMed] [Google Scholar]
- Kurek D., Neagu A., Tastemel M., Tuysuz N., Lehmann J., Van De Werken H. J., Philipsen S., Van Der Linden R., Maas A., Van I. W. F. et al. (2015). Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Rep. 4, 114-128. 10.1016/j.stemcr.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K. et al. (2006). Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301-313. 10.1016/j.cell.2006.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liefke R., Jiang J., Kurland J. V., Tian W., Deng P., Zhang W., He Q., Patel D. J., Bulyk M. L. et al. (2017). Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287-291. 10.1038/nature23881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Karnik R., Gu H., Ziller M. J., Clement K., Tsankov A. M., Akopian V., Gifford C. A., Donaghey J., Galonska C. et al. (2015). Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 47, 469-478. 10.1038/ng.3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipchina I., Elkabetz Y., Hafner M., Sheridan R., Mihailovic A., Tuschl T., Sander C., Studer L. and Betel D. (2011). Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 25, 2173-2186. 10.1101/gad.17221311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H. et al. (2016). Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558-562. 10.1038/nature19362 [DOI] [PubMed] [Google Scholar]
- Liu X., Nefzger C. M., Rossello F. J., Chen J., Knaupp A. S., Firas J., Ford E., Pflueger J., Paynter J. M., Chy H. S. et al. (2017). Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat. Methods 14, 1055-1062. 10.1038/nmeth.4436 [DOI] [PubMed] [Google Scholar]
- Loh Y.-H., Wu Q., Chew J.-L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J. et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431-440. 10.1038/ng1760 [DOI] [PubMed] [Google Scholar]
- Lu X., Göke J., Sachs F., Jacques P. E., Liang H., Feng B., Bourque G., Bubulya P. A. and Ng H.-H. (2013). SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat. Cell Biol. 15, 1141-1152. 10.1038/ncb2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Sachs F., Ramsay L., Jacques P. E., Göke J., Bourque G. and Ng H.-H. (2014). The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol. 21, 423-425. 10.1038/nsmb.2799 [DOI] [PubMed] [Google Scholar]
- Madissoon E., Töhönen V., Vesterlund L., Katayama S., Unneberg P., Inzunza J., Hovatta O. and Kere J. (2014). Differences in gene expression between mouse and human for dynamically regulated genes in early embryo. PLoS ONE 9, e102949 10.1371/journal.pone.0102949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T., Kawamura Y., Mochizuki H., Goshima N. and Yamanaka S. (2011). Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 474, 225-229. 10.1038/nature10106 [DOI] [PubMed] [Google Scholar]
- Marson A., Levine S. S., Cole M. F., Frampton G. M., Brambrink T., Johnstone S., Guenther M. G., Johnston W. K., Wernig M., Newman J. et al. (2008). Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521-533. 10.1016/j.cell.2008.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634-7638. 10.1073/pnas.78.12.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J. and Ruohola-Baker H. (2017). Metabolic remodeling during the loss and acquisition of pluripotency. Development 144, 541-551. 10.1242/dev.128389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J. (2010). Targeting X chromosomes for repression. Curr. Opin. Genet. Dev. 20, 179-189. 10.1016/j.gde.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody J. D., Levy S., Mathieu J., Xing Y., Kim W., Dong C., Tempel W., Robitaille A. M., Dang L. T., Ferreccio A. et al. (2017). First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor. Proc. Natl. Acad. Sci. USA 114, 10125-10130. 10.1073/pnas.1706907114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R. D., O'connor M. D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.-L., Zhao Y., Mcdonald H., Zeng T., Hirst M. et al. (2008). Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18, 610-621. 10.1101/gr.7179508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. et al. (2015). Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 21, 392-402. 10.1016/j.cmet.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Okamoto I., Sasaki K., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Yamamoto T. and Saitou M. (2016). A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537, 57-62. 10.1038/nature19096 [DOI] [PubMed] [Google Scholar]
- Newman M. A., Thomson J. M. and Hammond S. M. (2008). Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539-1549. 10.1261/rna.1155108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan K. K. and Eggan K. (2013). Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev. Biol. 375, 54-64. 10.1016/j.ydbio.2012.12.008 [DOI] [PubMed] [Google Scholar]
- Niakan K. K., Han J., Pedersen R. A., Simon C. and Pera R. A. R. (2012). Human pre-implantation embryo development. Development 139, 829-841. 10.1242/dev.060426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J. and Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487-492. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Chonabayashi K., Nomura M., Tanaka A., Nakamura M., Inagaki A., Nishikawa M., Takei I., Oishi A., Tanabe K. et al. (2016). Epigenetic variation between human induced pluripotent stem cell lines is an indicator of differentiation capacity. Cell Stem Cell 19, 341-354. 10.1016/j.stem.2016.06.019 [DOI] [PubMed] [Google Scholar]
- Okae H., Chiba H., Hiura H., Hamada H., Sato A., Utsunomiya T., Kikuchi H., Yoshida H., Tanaka A., Suyama M. et al. (2014). Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 10, e1004868 10.1371/journal.pgen.1004868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I., Patrat C., Thépot D., Peynot N., Fauque P., Daniel N., Diabangouaya P., Wolf J.-P., Renard J.-P., Duranthon V. et al. (2011). Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472, 370-374. 10.1038/nature09872 [DOI] [PubMed] [Google Scholar]
- Onder T. T., Kara N., Cherry A., Sinha A. U., Zhu N., Bernt K. M., Cahan P., Marcarci B. O., Unternaehrer J., Gupta P. B. et al. (2012). Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598-602. 10.1038/nature10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R. J., Fitz-Gerald C. S., Sato Y., Cowan C. A., Chien K. R. and Melton D. A. (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26, 313-315. 10.1038/nbt1383 [DOI] [PubMed] [Google Scholar]
- Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G. A., Stewart R. and Thomson J. A. (2007). Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1, 299-312. 10.1016/j.stem.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Panopoulos A. D., Smith E. N., Arias A. D., Shepard P. J., Hishida Y., Modesto V., Diffenderfer K. E., Conner C., Biggs W., Sandoval E. et al. (2017). Aberrant DNA methylation in human iPSCs associates with MYC-binding motifs in a clone-specific manner independent of genetics. Cell Stem Cell 20, 505-517.e6. 10.1016/j.stem.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V. and Plath K. (2015). X chromosome reactivation in reprogramming and in development. Curr. Opin. Cell Biol. 37, 75-83. 10.1016/j.ceb.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W. A., Chen D., Liu W., Kim R., Sahakyan A., Lukianchikov A., Plath K., Jacobsen S. E. and Clark A. T. (2016). Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell 18, 323-329. 10.1016/j.stem.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S. and Vallier L. (2013). The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135-147. 10.1016/j.cell.2013.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., Madrigal P., Bertero A. and Vallier L. (2016). Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev. 30, 421-433. 10.1101/gad.271452.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S., Edsgard D., Reinius B., Deng Q., Panula S. P., Codeluppi S., Plaza Reyes A., Linnarsson S., Sandberg R. and Lanner F. (2016). Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012-1026. 10.1016/j.cell.2016.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Hejna M., Liu Y., Percharde M., Wossidlo M., Blouin L., Durruthy-Durruthy J., Wong P., Qi Z., Yu J. et al. (2016). YAP induces human naive pluripotency. Cell Rep. 14, 2301-2312. 10.1016/j.celrep.2016.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiee M.-R., Girardot C., Sigismondo G. and Krijgsveld J. (2016). Expanding the circuitry of pluripotency by selective isolation of chromatin-associated proteins. Mol. Cell 64, 624-635. 10.1016/j.molcel.2016.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roode M., Blair K., Snell P., Elder K., Marchant S., Smith A. and Nichols J. (2012). Human hypoblast formation is not dependent on FGF signalling. Dev. Biol. 361, 358-363. 10.1016/j.ydbio.2011.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A. and Brivanlou A. H. (2011). A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 30, 237-248. 10.1038/emboj.2010.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Spagnoli F. M. and Brivanlou A. H. (2009). The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 16, 517-527. 10.1016/j.devcel.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E. E., Nitsch R. and Wulczyn F. G. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10, 987-993. 10.1038/ncb1759 [DOI] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T. W., Kuoy E., Langerman J., Clark A. T., Jaenisch R. et al. (2017). Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 20, 87-101. 10.1016/j.stem.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni F. A., Guerra J. and Luban J. (2012). HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 9, 111 10.1186/1742-4690-9-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P. and Brivanlou A. H. (2004). Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55-63. 10.1038/nm979 [DOI] [PubMed] [Google Scholar]
- Shahbazi M. N., Jedrusik A., Vuoristo S., Recher G., Hupalowska A., Bolton V., Fogarty N. M., Campbell A., Devito L. G., Ilic D. et al. (2016). Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 18, 700-708. 10.1038/ncb3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y., Liang Z., Xing Q., Zhang T., Wang B., Tian S., Huang W., Zhang Y., Yao J., Zhu Y. et al. (2017). PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naive ESCs. Nat. Commun. 8, 672 10.1038/s41467-017-00668-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki N., Shiraki Y., Tsuyama T., Obata F., Miura M., Nagae G., Aburatani H., Kume K., Endo F. and Kume S. (2014). Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19, 780-794. 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Singh A. M., Reynolds D., Cliff T., Ohtsuka S., Mattheyses A. L., Sun Y., Menendez L., Kulik M. and Dalton S. (2012). Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell 10, 312-326. 10.1016/j.stem.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. M., Sun Y., Li L., Zhang W., Wu T., Zhao S., Qin Z. and Dalton S. (2015). Cell-cycle control of bivalent epigenetic domains regulates the exit from pluripotency. Stem Cell Rep. 5, 323-336. 10.1016/j.stemcr.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. (2017). Formative pluripotency: the executive phase in a developmental continuum. Development 144, 365-373. 10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z. D., Chan M. M., Humm K. C., Karnik R., Mekhoubad S., Regev A., Eggan K. and Meissner A. (2014). DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611-615. 10.1038/nature13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G. and Zaret K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 151, 994-1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z., Fischer K. A., Devi A., Detraux D., Gu H. et al. (2015). The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 17, 1523-1535. 10.1038/ncb3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M.-R., Lee Y., Kim J. Y., Kim S.-K., Moon S.-H., Lee J. Y., Cha K.-Y., Chung H. M., Yoon H. S., Moon S. Y. et al. (2004). Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270, 488-498. 10.1016/j.ydbio.2004.02.019 [DOI] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861-872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Sasaki A., Yamamoto M., Nakamura M., Sutou K., Osafune K. and Yamanaka S. (2014). Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun. 5, 3678 10.1038/ncomms4678 [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. et al. (2014). Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254-1269. 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R. K. and Nusse R. (2011). Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 13, 1070-1075. 10.1038/ncb2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L. and Mckay R. D. G. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196-199. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Theunissen T. W., Powell B. E., Wang H., Mitalipova M., Faddah D. A., Reddy J., Fan Z. P., Maetzel D., Ganz K., Shi L. et al. (2014). Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 471-487. 10.1016/j.stem.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T. W., Friedli M., He Y., Planet E., O'neil R. C., Markoulaki S., Pontis J., Wang H., Iouranova A., Imbeault M. et al. (2016). Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19, 502-515. 10.1016/j.stem.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S. and Jones J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145-1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Tsankov A. M., Gu H., Akopian V., Ziller M. J., Donaghey J., Amit I., Gnirke A. and Meissner A. (2015). Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344-349. 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H. R. et al. (2006). Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805-814. 10.1111/j.1365-2443.2006.00984.x [DOI] [PubMed] [Google Scholar]
- Vallier L., Alexander M. and Pedersen R. A. (2005). Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495-4509. 10.1242/jcs.02553 [DOI] [PubMed] [Google Scholar]
- Vallier L., Mendjan S., Brown S., Chng Z., Teo A., Smithers L. E., Trotter M. W. B., Cho C. H.-H., Martinez A., Rugg-Gunn P. et al. (2009). Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 136, 1339-1349. 10.1242/dev.033951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallot C., Patrat C., Collier A. J., Huret C., Casanova M., Liyakat Ali T. M., Tosolini M., Frydman N., Heard E., Rugg-Gunn P. J. et al. (2017). XACT noncoding RNA competes with XIST in the control of X chromosome activity during human early development. Cell Stem Cell 20, 102-111. 10.1016/j.stem.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R., Boue S., Gonzalez-Roca E., Aran B., Auer H., Veiga A. and Izpisua Belmonte J. C. I. (2011). Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 138, 3699-3709. 10.1242/dev.064741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S. R., Daley G. Q. and Gregory R. I. (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97-100. 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Meyenn F., Berrens R. V., Andrews S., Santos F., Collier A. J., Krueger F., Osorno R., Dean W., Rugg-Gunn P. J. and Reik W. (2016). Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Dev. Cell 39, 104-115. 10.1016/j.devcel.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W. and Orkin S. H. (2006). A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364-368. 10.1038/nature05284 [DOI] [PubMed] [Google Scholar]
- Wang Z., Oron E., Nelson B., Razis S. and Ivanova N. (2012). Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440-454. 10.1016/j.stem.2012.02.016 [DOI] [PubMed] [Google Scholar]
- Wang J., Xie G., Singh M., Ghanbarian A. T., Raskó T., Szvetnik A., Cai H., Besser D., Prigione A., Fuchs N. V. et al. (2014). Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 516, 405-409. 10.1038/nature13804 [DOI] [PubMed] [Google Scholar]
- Ware C. B., Nelson A. M., Mecham B., Hesson J., Zhou W., Jonlin E. C., Jimenez-Caliani A. J., Deng X., Cavanaugh C., Cook S. et al. (2014). Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA 111, 4484-4489. 10.1073/pnas.1319738111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon J. L., Langford A. T., Wong C.-J., Zhong J. W. and Tapscott S. J. (2017). Conservation and innovation in the DUX4-family gene network. Nat. Genet. 49, 935-940. 10.1038/ng.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R. and Smith A. (2011). Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 13, 838-845. 10.1038/ncb2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Greely H. T., Jaenisch R., Nakauchi H., Rossant J. and Belmonte J. C. I. (2016a). Stem cells and interspecies chimaeras. Nature 540, 51-59. 10.1038/nature20573 [DOI] [PubMed] [Google Scholar]
- Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W. et al. (2016b). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652-657. 10.1038/nature18606 [DOI] [PubMed] [Google Scholar]
- Xie D., Chen C.-C., Ptaszek L. M., Xiao S., Cao X., Fang F., Ng H. H., Lewin H. A., Cowan C. and Zhong S. (2010). Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 20, 804-815. 10.1101/gr.100594.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Schultz M. D., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J. W., Tian S., Hawkins R. D., Leung D. et al. (2013). Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134-1148. 10.1016/j.cell.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Rosler E., Jiang J., Lebkowski J. S., Gold J. D., O'sullivan C., Delavan-Boorsma K., Mok M., Bronstein A., Carpenter M. K. (2005). Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem Cells 23, 315-323. 10.1634/stemcells.2004-0211 [DOI] [PubMed] [Google Scholar]
- Xu R.-H., Sampsell-Barron T. L., Gu F., Root S., Peck R. M., Pan G., Yu J., Antosiewicz-Bourget J., Tian S., Stewart R. et al. (2008). NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3, 196-206. 10.1016/j.stem.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Papagiannakopoulos T., Pan G., Thomson J. A. and Kosik K. S. (2009). MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647-658. 10.1016/j.cell.2009.02.038 [DOI] [PubMed] [Google Scholar]
- Xue Z., Huang K., Cai C., Cai L., Jiang C. Y., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y. E. et al. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593-597. 10.1038/nature12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M., Seki Y., Kurimoto K., Yabuta Y., Yuasa M., Shigeta M., Yamanaka K., Ohinata Y. and Saitou M. (2008). Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 40, 1016-1022. 10.1038/ng.186 [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J. et al. (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131-1139. 10.1038/nsmb.2660 [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhang X., Yi L., Hou Z., Chen J., Kou X., Zhao Y., Wang H., Sun X.-F., Jiang C. et al. (2016). Naive induced pluripotent stem cells generated from beta-thalassemia fibroblasts allow efficient gene correction with CRISPR/Cas9. Stem Cells Transl. Med. 5, 8-19. 10.5966/sctm.2015-0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liu B., Xu J., Wang J., Wu J., Shi C., Xu Y., Dong J., Wang C., Lai W. et al. (2017). Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 169, 243-257.e25. 10.1016/j.cell.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Pereira L., Hoffman J. A., Shy B. R., Yuen C. M., Liu D. R. and Merrill B. J. (2011). Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat. Cell Biol. 13, 762-770. 10.1038/ncb2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P. and Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519-523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A. (2011). Control of the embryonic stem cell state. Cell 144, 940-954. 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R. et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917-1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Yu P., Pan G., Yu J. and Thomson J. A. (2011). FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 8, 326-334. 10.1016/j.stem.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zucchelli M., Bruce S., Hambiliki F., Stavreus-Evers A., Levkov L., Skottman H., Kerkelä E., Kere J. and Hovatta O. (2009). Transcriptome profiling of human pre-implantation development. PLoS ONE 4, e7844 10.1371/journal.pone.0007844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yalcin S., Lee D.-F., Yeh T.-Y. J., Lee S.-M., Su J., Mungamuri S. K., Rimmelé P., Kennedy M., Sellers R. et al. (2011). FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat. Cell Biol. 13, 1092-1099. 10.1038/ncb2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zheng H., Huang B., Li W., Xiang Y., Peng X., Ming J., Wu X., Zhang Y., Xu Q. et al. (2016). Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553-557. 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- Zhao X. D., Han X., Chew J. L., Liu J., Chiu K. P., Choo A., Orlov Y. L., Sung W. K., Shahab A., Kuznetsov V. A. et al. (2007). Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1, 286-298. 10.1016/j.stem.2007.08.004 [DOI] [PubMed] [Google Scholar]
- Zimmerlin L., Park T. S., Huo J. S., Verma K., Pather S. R., Talbot C. C. Jr, Agarwal J., Steppan D., Zhang Y. W., Considine M. et al. (2016). Tankyrase inhibition promotes a stable human naive pluripotent state with improved functionality. Development 143, 4368-4380. 10.1242/dev.138982 [DOI] [PMC free article] [PubMed] [Google Scholar]