Abstract
Drosophila neuroblasts are an excellent model for investigating how neuronal diversity is generated. Most brain neuroblasts generate a series of ganglion mother cells (GMCs) that each make two neurons (type I lineage), but 16 brain neuroblasts generate a series of intermediate neural progenitors (INPs) that each produce 4-6 GMCs and 8-12 neurons (type II lineage). Thus, type II lineages are similar to primate cortical lineages, and may serve as models for understanding cortical expansion. Yet the origin of type II neuroblasts remains mysterious: do they form in the embryo or larva? If they form in the embryo, do their progeny populate the adult central complex, as do the larval type II neuroblast progeny? Here, we present molecular and clonal data showing that all type II neuroblasts form in the embryo, produce INPs and express known temporal transcription factors. Embryonic type II neuroblasts and INPs undergo quiescence, and produce embryonic-born progeny that contribute to the adult central complex. Our results provide a foundation for investigating the development of the central complex, and tools for characterizing early-born neurons in central complex function.
KEY WORDS: Neurogenesis, Type II neuroblast, Intermediate neural progenitors, INPs, Temporal patterning, Pdm, Castor, Grainy head, Dichaete, Central complex
Summary: This study shows that embryonic Drosophila type II neuroblasts produce INPs, express late temporal transcription factors, undergo quiescence, and contribute to the central complex of the adult brain.
INTRODUCTION
Drosophila neural progenitors, called neuroblasts, are a model system for investigating stem cell self-renewal versus differentiation (Doe, 2008; Reichert, 2011), as well as how a single progenitor generates different types of neurons and glia over time (Alsio et al., 2013; Kohwi et al., 2013). Drosophila type I neuroblasts have a relatively simple cell lineage: they undergo a series of asymmetric cell divisions to produce a series of smaller ganglion mother cells (GMCs) that typically differentiate into a pair of neurons. There are about 100 type I neuroblasts in each larval brain lobe; they generate progeny during embryogenesis, undergo a period of quiescence, and then resume their lineage in the larva (Truman and Bate, 1988; Datta, 1995; Maurange and Gould, 2005; Sousa-Nunes et al., 2010). Type I neuroblasts have a molecular profile that is Deadpan (Dpn)+, Asense (Ase)+ and Pointed P1 (PntP1; Pnt)− and produce GMCs that are Dpn− Prospero (Pros)+ (Zhu et al., 2011; Xie et al., 2016). Moreover, many embryonic type I neuroblasts can transition to a simpler ‘type 0’ lineage, in which each neuroblast daughter cell directly differentiates into a neuron (Karcavich and Doe, 2005; Baumgardt et al., 2014; Bertet et al., 2014).
In contrast, Drosophila larval type II neuroblasts have a more elaborate cell lineage: they divide asymmetrically to bud off smaller intermediate neural progenitors (INPs), which themselves produce a series of four to six GMCs that each make a pair of neurons or glia (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Izergina et al., 2009). Type II neuroblasts have a molecular profile that is Dpn+ Ase− PntP1+; they produce INPs with the molecular profile Dpn+ Ase+ and these INPs each produce approximately six GMCs that are Dpn− Ase+ (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Izergina et al., 2009; Zhu et al., 2011). Although there are only eight type II neuroblasts per larval brain lobe, they generate a major portion of the intrinsic neurons of the adult central complex (Bayraktar et al., 2010; Ito et al., 2013; Riebli et al., 2013; Yu et al., 2013), a neuropil devoted to multimodal sensory processing and locomotion (Martin et al., 1999; Renn et al., 1999; Strauss, 2002; Wessnitzer and Webb, 2006; Poeck et al., 2008; Wang et al., 2008; Pan et al., 2009; Bender et al., 2010; Boyan and Reichert, 2011; Ofstad et al., 2011; Seelig and Jayaraman, 2011, 2013, 2015).
A large amount of work over the past two decades has illuminated the general principles for how type I neuroblasts generate neuronal diversity. First, dorsoventral, anterior-posterior, and Hox spatial patterning cues generate unique neuroblast identities (Chu-LaGraff and Doe, 1993; Prokop and Technau, 1994; Skeath et al., 1995; McDonald et al., 1998; Weiss et al., 1998; Skeath and Thor, 2003; Marin et al., 2012; Estacio-Gómez and Díaz-Benjumea, 2014; Moris-Sanz et al., 2015). Second, the temporal transcription factors Hunchback (Hb), Krüppel, Nubbin/Pdm2 (referred to here as Pdm), Castor (Cas) and Grainy head (Grh) specify unique GMC identities within each neuroblast lineage (Brody and Odenwald, 2000; Berger et al., 2001; Isshiki et al., 2001; Novotny et al., 2002; Cenci and Gould, 2005; Kanai et al., 2005; Grosskortenhaus et al., 2006; Mettler et al., 2006; Urban and Mettler, 2006; Maurange et al., 2008; Tran and Doe, 2008; Tsuji et al., 2008; Ulvklo et al., 2012; Herrero et al., 2014; Moris-Sanz et al., 2014).
In contrast, much less is known about the eight type II neuroblasts in each brain lobe. Larval type II neuroblasts undergo a temporal transcription factor cascade (Syed et al., 2017), and, similarly, the shorter larval INP lineages undergo a three-factor temporal transcription cascade of Dichaete, Grh, and Eyeless (Ey) (Bayraktar and Doe, 2013). In the embryo, only one of the eight type II neuroblasts has been identified (Hwang and Rulifson, 2011); the origin of the other type II neuroblasts has not been reported in existing embryonic brain neuroblast maps (Urbach and Technau, 2003). It remains unknown whether type II neuroblasts arise de novo from the neuroectoderm, similar to type I neuroblasts, or whether they arise from a type I→type II transition similar to the type I→type 0 neuroblast transitions (Baumgardt et al., 2014; Bertet et al., 2014). If type II neuroblasts form during embryogenesis, it is unknown whether they utilize the same Hb→Krüppel→Pdm→Cas→Grh temporal transcription factor cascade to generate neuronal diversity, or whether they make embryonic-born INPs that sequentially express Dichaete→Grh→Ey, similar to larval INPs (Bayraktar and Doe, 2013). Furthermore, if type II neuroblast lineages are initiated in the embryo, it would be interesting to know if their INPs undergo quiescence, similar to type I and II neuroblasts; if so, they would be the only cell type beyond neuroblasts known to enter quiescence at the embryo/larval transition. Perhaps most importantly, identifying embryonic type II neuroblasts is essential for subsequent characterization of their early-born progeny, which are likely to generate pioneer neurons that are crucially important for establishing larval or adult brain architecture.
Here, we address all of these open questions. We show that all eight type II neuroblasts form during embryogenesis. We use molecular markers and clonal data to show that embryonic type II neuroblasts give rise to INPs that produce multiple GMCs and neurons during embryogenesis, and that INPs undergo quiescence during the embryo-larval transition. We find that embryonic type II neuroblasts sequentially express a subset of neuroblast temporal transcription factors (Pdm→Cas→Grh), and embryonic INPs express a subset of the known larval INP temporal transcription factors (Dichaete). Finally, we show that embryonic INPs give rise to neurons that survive to populate the adult central complex.
RESULTS
All type II neuroblasts arise during embryogenesis
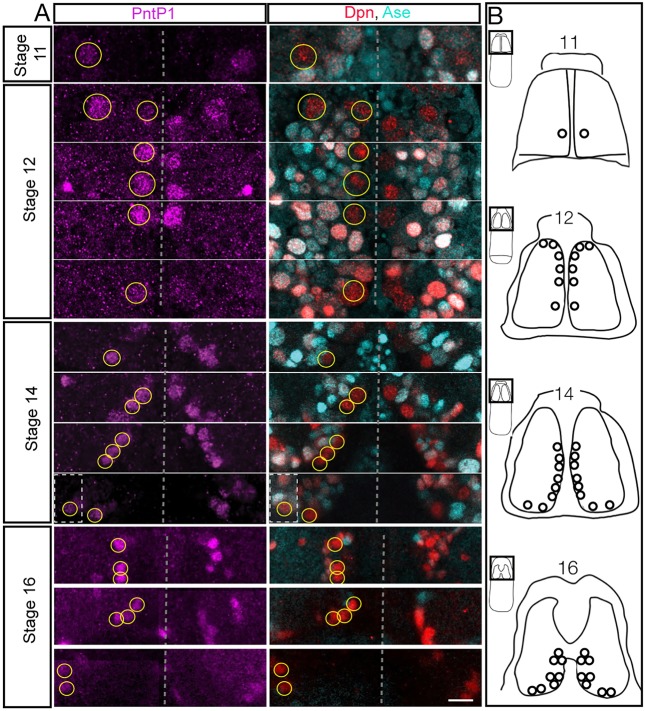
Larval type II neuroblasts are PntP1+ Dpn+ Ase− and here we used these markers to determine whether type II neuroblasts exist in the embryo. We found that type II neuroblasts formed internal to the dorsal cephalic neuroectoderm beginning at late stage 11. At this stage, there is one PntP1+ Dpn+ Ase− type II neuroblast in a stereotyped dorsal posteromedial location; this is always the first type II neuroblast to appear (Fig. 1). By stage 12, the number of type II neuroblasts along the dorsomedial region of the brain increased from four (8 h) to six (9.5 h), and from stage 15 (12 h) to the end of embryogenesis there were reliably eight type II neuroblasts per lobe (Fig. 1), the same number previously observed at all stages of larval development (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Izergina et al., 2009). We reliably found three clusters of type II neuroblasts: an anteromedial group of three neuroblasts, a medial group of three neuroblasts, and a posterior ventrolateral group of two neuroblasts (Fig. 1A; summarized in Fig. 1B). Owing to the dynamic morphogenetic movements of head involution, and the close positioning of the type II neuroblasts, we could not reliably identify individual neuroblasts within each cluster.
Fig. 1.
Eight type II neuroblasts arise during embryogenesis. (A) Embryonic type II neuroblasts (yellow circles on left brain lobe; unlabeled on right brain lobe) are PntP1+ (magenta) Dpn+ (red) Ase− (cyan). Each stage shows multiple focal planes from anterior to posterior (top to bottom in the figure) to clearly visualize each type II neuroblast, except for stage 11 where there is a single type II neuroblast. n>10 for each stage. Dashed boxes are from a different focal plane. (B) Summary of type II neuroblast formation; owing to rapid morphogenetic movements it is not possible to identify individual type II neuroblasts from stage to stage, but beginning at stage 14 it is possible to recognize three clusters of neuroblasts. All panels are dorsal views with the dorsal midline in the center of the panel, anterior up. Scale bar: 10 μm.
We tried to link the embryonic type II neuroblasts to the map of embryonic brain neuroblasts (Urbach and Technau, 2003), but were unsuccessful, probably because most type II neuroblasts arise later than the stages described in that study. Based on molecular marker analysis, we conclude that all eight known type II neuroblasts form during embryogenesis and they are among the last neuroblasts to form during embryogenesis.
Embryonic type II neuroblasts generate INPs, GMCs and neurons during embryogenesis
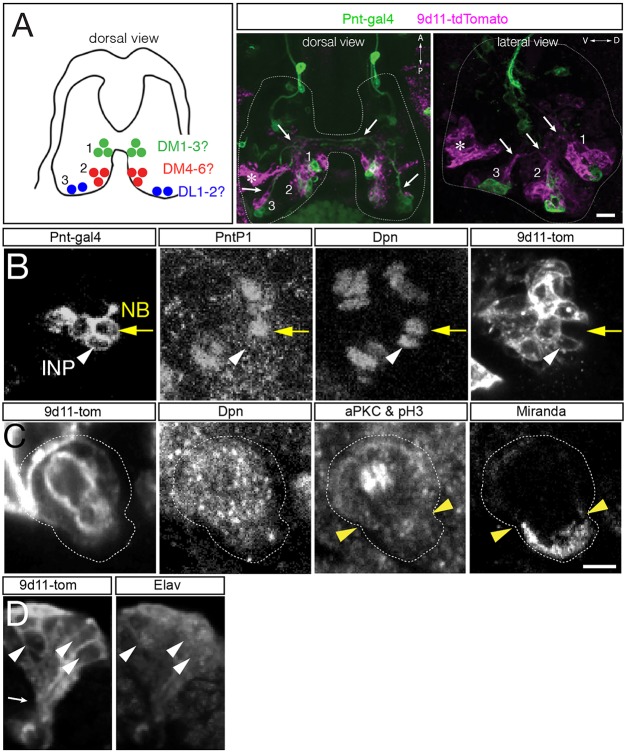
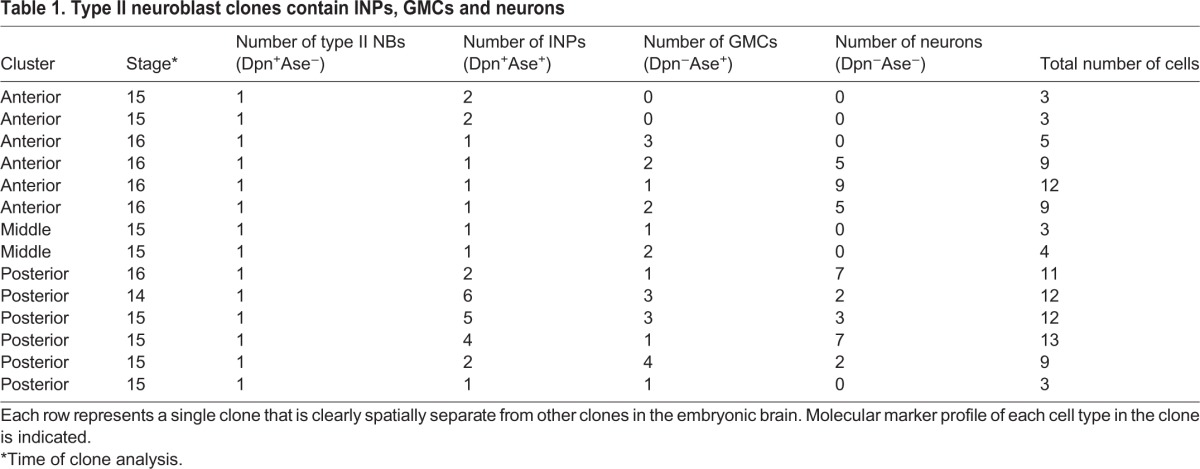
Here, we use molecular markers and clonal analysis to determine whether embryonic type II lineages produce INPs, GMCs or neurons. We used a Pnt-gal4 line to make clones; to validate the type II lineage-specific expression of this line, we stained for Pnt-gal4 and type II neuroblast and INP markers (Fig. 2A). We found that Pnt-gal4 is expressed in the parental type II neuroblast, the maturing INPs and their GMC progeny (Fig. 2B). We define INPs as Dpn+ Ase+ Pros− cells adjacent to type II neuroblasts; the presence of Dpn was sufficient to distinguish these cells from GMCs (Fig. 2B). We did not detect any type I neuroblasts expressing this marker. Next, we generated ‘flip-out’ clones using the heat shock-inducible multicolor flip-out method (Nern et al., 2015) crossed to the Pnt-gal4 line. When we assayed clones relatively early in embryogenesis (stage 13) we detected small clones containing a single type II neuroblast and one or more INPs (Fig. 2C; Table 1). Allowing the embryos to develop further resulted in larger clones that additionally contained GMCs and neurons (Fig. 2D). We found clones containing one type II neuroblast with up to five INPs at the latest stages of embryogenesis (Table 1). Taken together, these data show that embryonic type II neuroblasts generate multiple INPs, which themselves produce GMCs and neurons prior to larval hatching.
Fig. 2.
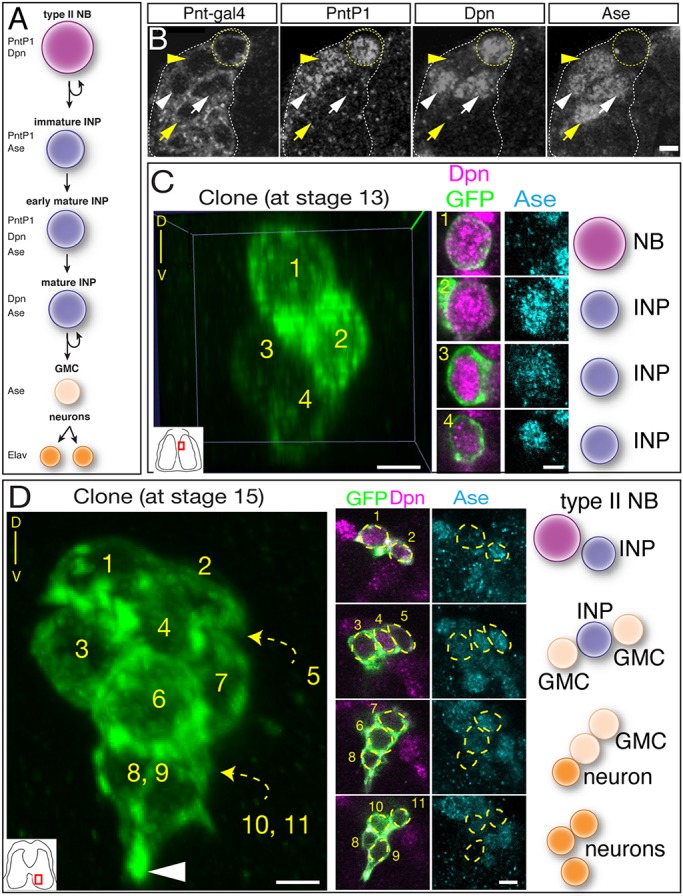
Clonal analysis shows that type II neuroblasts make INPs, GMCs and neurons during embryogenesis. (A) Molecular markers used to identify cell types within type II lineages. (B) Embryonic type II neuroblasts generate embryonic-born INPs and GMCs. Dorsomedial view of a type II neuroblast cluster in a stage 16 embryo showing a type II neuroblast (Pnt-gal4+ PntP1+ Dpn+ and Ase−; yellow circle); an immature INP (Pnt-gal4+ PntP1+ Dpn− and Ase+; yellow arrowhead); mature INP (Pnt-gal4+ PntP1+ Dpn+ and Ase+; white arrowhead); a mature INP that has lost PntP1 expression (Pnt-gal4+ PntP1− Dpn+ and Ase+; white arrow); and a GMC (Pnt-gal4+ PntP1− Dpn− and Ase+; yellow arrow). (C) Single neuroblast clone assayed at stage 13 (location shown in inset): four-cell clone containing a type II neuroblast and three INPs. Orientation is dorsal up, with the neuroblast closest to the dorsal surface of the brain. (D) Single neuroblast clone assayed at stage 15 (location shown in inset): eleven-cell clone containing a type II neuroblast, two INPs, four GMCs and four neurons. Orientation is dorsal up, showing that the neurons are sending projections ventrally (arrowhead). Scale bars: 5 μm (B); 10 μm (C,D, clone projection); 5 μm (C,D, insets). n=1 for each clone shown; n>20 for total clone number analyzed. NB, neuroblast.
Table 1.
Type II neuroblast clones contain INPs, GMCs and neurons
A defining feature of type II neuroblasts is their ability to make INPs, which undergo a molecularly asymmetric cell division to self-renew and generate a GMC (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Izergina et al., 2009). Here, we determine whether embryonic INPs undergo asymmetric cell division. To identify INPs and their progeny, we used the INP marker R9D11-tdTomato (henceforth 9D11-tom) (Bayraktar and Doe, 2013) and confirmed that it is expressed in embryonic INPs (Fig. 3A,B). We also detected a deep ventral cluster of unrelated cells that expressed 9D11-tom but not Dpn, but these can be excluded from analysis owing to their distinct position (Fig. 3A, asterisk). Using this marker, we found that 9D11-tom+ Dpn+ embryonic INPs undergo asymmetric cell division: they partition aPKC and Miranda to opposite cortical domains (Fig. 3C). To confirm that these GMCs generate post-mitotic neurons during embryogenesis, we stained for the neuronal marker Elav, and found that 9D11-tom clusters contained Elav+ neurons (Fig. 3D). Additionally, axon fascicles from single type II neuroblast lineage clones were visible during embryogenesis (Fig. 3A,D, arrows), confirming the production of embryonic-born neurons from type II lineages. We conclude that embryonic type II neuroblasts generate asymmetrically dividing INPs that produce GMCs and neurons during embryogenesis.
Fig. 3.
Embryonic INPs undergo asymmetric cell division. (A,B) R9D11-tdTomato (9D11-tom) labels embryonic INPs and their progeny, but not type II neuroblasts. (A) Left: summary of type II neuroblast positions (dorsal view). Center and right panels: dorsal or lateral view of the three type II neuroblast clusters labeled with Pnt-gal4 (green; type II neuroblasts and progeny) and 9D11-tom (magenta; INPs and progeny). Axon fascicles are visible in dorsal and lateral views (white arrows). Note there is 9D11-tom expression at a deep ventral location that is not near any type II lineage (asterisk). (B) Type II neuroblast (Pnt-gal4+ PntP1+ Dpn+ 9D11-tom−; yellow arrow); INP (Pnt-gal4+ PntP1− Dpn+ 9D11-tom+; white arrowhead) at stage 16. n>10 for experiment shown. (C) Embryonic INPs undergo asymmetric cell division. INPs were identified as 9D11-tom+ Dpn+ and positioned within the middle cluster of neuroblasts in the dorsal posterior medial brain lobe. aPKC and pH3 are co-stained: aPKC is localized to the larger apical cell cortex (white cortex above arrowheads; future INP daughter cell) whereas pH3 decorates the mitotic chromosomes in the middle of the INP. Miranda is localized to the smaller basal cell cortex (cortex below arrowheads; future GMC daughter cell). n>5 for mitotic INPs. (D) Embryonic INPs generate embryonic-born neurons. Lateral view of a 9D11-tom+ cluster in a stage 16 embryo. The post-mitotic neuronal marker Elav is detected in a subset of the 9D11-tom+ cluster (white arrowheads), and axon projections can be observed (white arrow, bottom left). Scale bars: 15 μm (A); in C: 10 μm (B), 5 μm (C,D). n>10 for the experiment shown.
Embryonic type II neuroblasts and INPs undergo quiescence
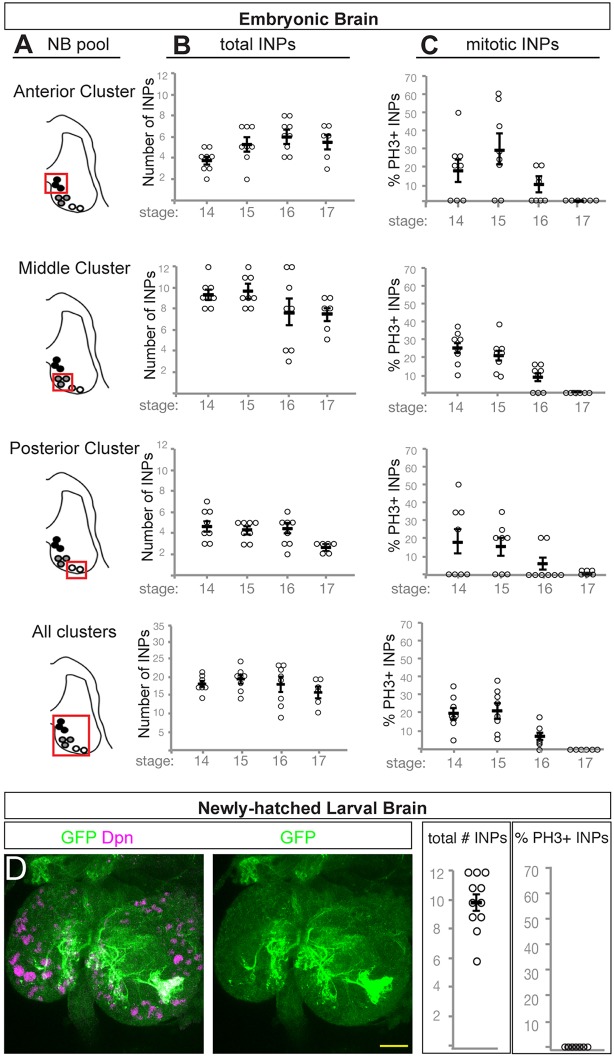
Type I central brain and thoracic neuroblasts have been shown to undergo quiescence at the embryo-larval transition (Truman and Bate, 1988). Type II neuroblasts also undergo quiescence, because only the four mushroom body neuroblasts and a single lateral neuroblast maintain proliferation during the embryo-larval transition (Egger et al., 2008). In contrast, nothing is known about whether INPs undergo quiescence. To address this question, we counted the total number of INPs over time, as well as the number of mitotic INPs. We identified INPs as 9D11-tom+ Dpn+ and mitotic INPs by immunoreactivity for phospho-histone H3 (pH3). We quantified INPs in each cluster independently as well as all INPs in each brain lobe (Fig. 4A). We observed a fairly constant number of INPs in each cluster from embryonic stage 14 to stage 17 (Fig. 4B), yet the number of proliferating INPs declined significantly over time, reaching zero by stage 17 (Fig. 4C). We conclude that the INPs enter quiescence by embryonic stage 17.
Fig. 4.
INPs undergo quiescence across the embryo-larval transition. (A) Schematic outlining the three pools of type II neuroblast INP progeny assayed in B and C (red box). (B) Total number of INPs per pool at the indicated stages; INPs identified as 9D11-tom+ Dpn+ cells. Each circle represents the number of INPs per brain lobe. (C) Number of phospho-histone H3 (pH3)-positive mitotic INPs per pool at the indicated stages; INPs are identified as 9D11-tom+ Dpn+ cells. Each circle represents the number of INPs in the cluster of neuroblasts shown in A; black bar represents the average, shown with s.e.m. (D) Quiescent INPs are present in the newly hatched larva. INPs marked with 9D11-gal4 UAS-tdTomato (green); brain neuroblasts and INPs marked with Dpn (magenta). Anterior up, left and right brain lobes shown. Scale bar: 15 μm.
If INPs enter quiescence in the late embryo, we should be able to detect them in the newly hatched larvae, prior to production of larval-born INPs made from type II neuroblasts that have re-entered the cell cycle. We assayed 0-4 h newly hatched larvae for Dpn and 9D11-tom to mark the small quiescent INPs (Fig. 4D). We observed an average of 10±2 9D11-tom+ Dpn+ cells in each brain lobe, and none of these INPs was mitotic (n=11; Fig. 4D). We conclude that INPs undergo quiescence in the late embryo and can persist into the larvae. The fate of these quiescent INPs – whether they resume proliferation, differentiate or die – remains to be determined.
Embryonic type II neuroblasts undergo a late temporal transcription factor cascade
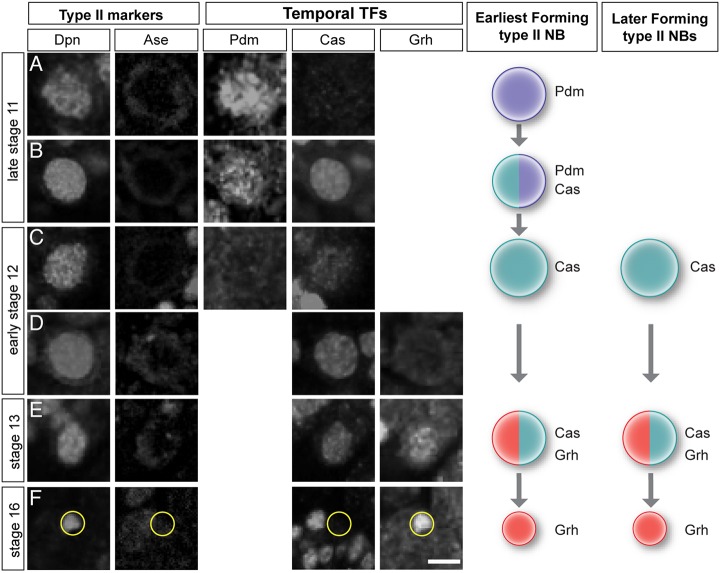
Embryonic type I neuroblasts undergo a well-characterized temporal transcription factor cascade that generates GMC diversity and, ultimately, neuronal diversity. Most type I neuroblasts sequentially express Hb→Krüppel→Pdm→Cas→Grh (Kohwi and Doe, 2013), although late-forming neuroblasts can skip some of the early factors: neuroblast 3-3 begins the series with Krüppel (Tsuji et al., 2008) and NB6-1 begins the series with Cas (Cui and Doe, 1992). As type II neuroblasts are among the latest to form, it is possible that they do not express any known temporal transcription factors.
We stained embryos for type II neuroblast markers (Dpn+ Ase–) and individual temporal identity transcription factors. We did not observe the first two temporal transcription factors, Hb or Krüppel, in any type II neuroblasts at any stage of development (Fig. S1). We next focused on the first type II neuroblast to form, which can be uniquely identified at late stage 11 (see Fig. 1). This early-forming neuroblast showed the temporal cascade of Pdm→Pdm/Cas→Cas→Cas/Grh→Grh (Fig. 5A-F). All later-forming type II neuroblasts exhibited a more truncated temporal cascade of Cas→Cas/Grh→Grh (Fig. 5D-F). We conclude that embryonic type II neuroblasts undergo a late temporal transcription factor cascade.
Fig. 5.
Embryonic type II neuroblasts express late temporal transcription factors. (A-F) Temporal transcription factor expression in the earliest type II neuroblast to form (posterior-most, see Fig. 1). Type II neuroblasts are identified as Dpn+ Ase− (left columns); temporal transcription factor expression reveals sequential expression of Pdm+→Pdm+/Cas+→Cas+→Cas+/Grh+→Grh+. Schematics on the right summarize this cascade. Later-forming type II neuroblasts start the cascade with Cas. Yellow circles indicate the small Grh+ neuroblast. Scale bar: 10 μm. n>10 for the experiment shown.
Embryonic INPs do not express the full temporal transcription factor cascade during embryogenesis
Larval INPs undergo a temporal transcription factor cascade of Dichaete→Grh→Ey over their ∼12 h lifespan (Bayraktar and Doe, 2013). We wondered whether the shorter time frame of embryogenesis might result in shorter temporal transcription factor expression windows, a truncated temporal cascade, or perhaps a lack of all temporal transcription factor expression.
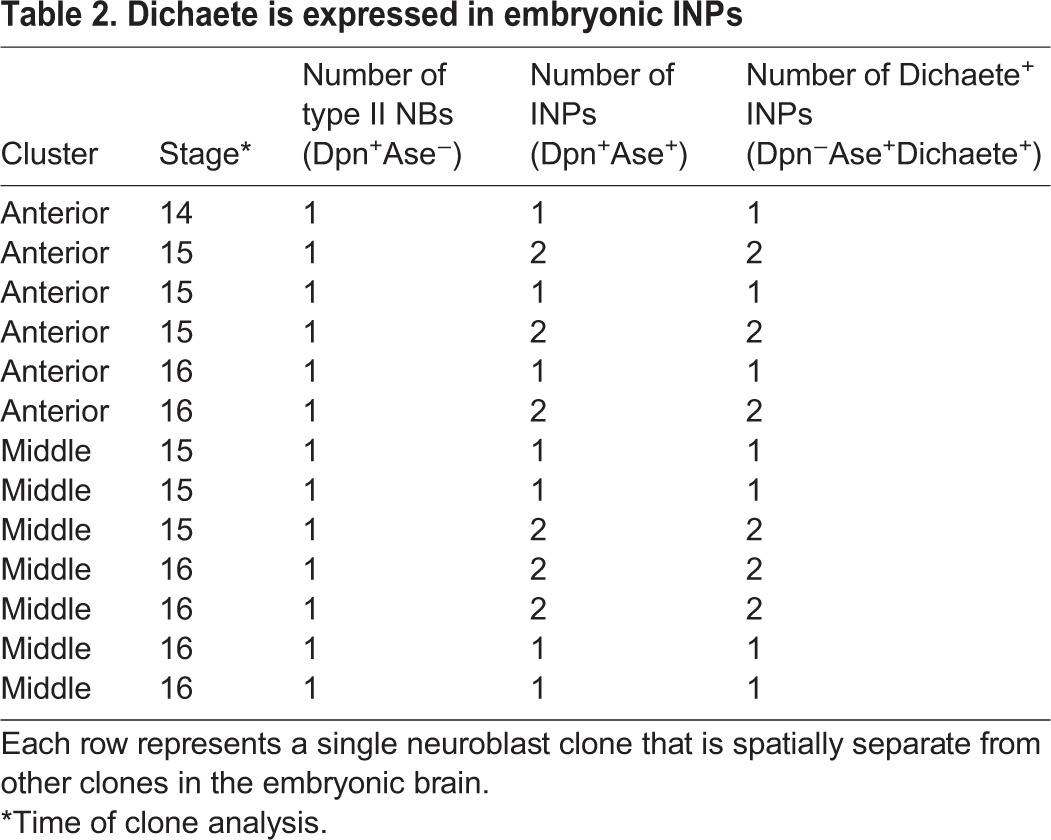
To identify embryonic INPs expressing known INP temporal transcription factors, we generated FLP-out clones using a heat-shock FLP in mid-embryogenesis (4 h-9 h) and assayed brains containing a single type II neuroblast clone. We stained embryos for the clone marker, Dpn, and Ase to identify the neuroblast (Dpn+ Ase−) and INPs (Dpn+ Ase+), and for one of the larval INP temporal transcription factors (Dichaete, Grh or Ey). We found that the early temporal factor Dichaete was detected in all INPs within the presumptive DM1-6 anterior and middle clusters (n=15 clones, anterior; n=12 clones, middle) (Fig. 6A,B; quantified in Table 2), but the presumptive DL1-2 posterior cluster contained no Dichaete+ INPs (n=9 clones) (Fig. 6C). Interestingly, the larval DL1-2 neuroblasts also fail to produce Dichaete+ or Grh+ INPs (Fig. S2). Grh was commonly detected in a single INP next to Grh+ neuroblasts, but not next to Grh− neuroblasts (Fig. S3), suggesting that it is transiently inherited from the parental neuroblast, as is also observed in larval INP lineages (Bayraktar and Doe, 2013); we never detected Grh in INPs distant from the neuroblasts, as would be expected for a middle temporal transcription factor. The late temporal factor Ey was never detected in INPs during embryogenesis (data not shown). We conclude that embryonic INPs undergo a temporal cascade that is truncated during the Dichaete window by entry into quiescence (Fig. 6D). It would be interesting to determine whether embryonic-born INPs express the later temporal factors Grh and Ey in the larvae, if they re-enter the cell cycle (Bayraktar and Doe, 2013).
Fig. 6.
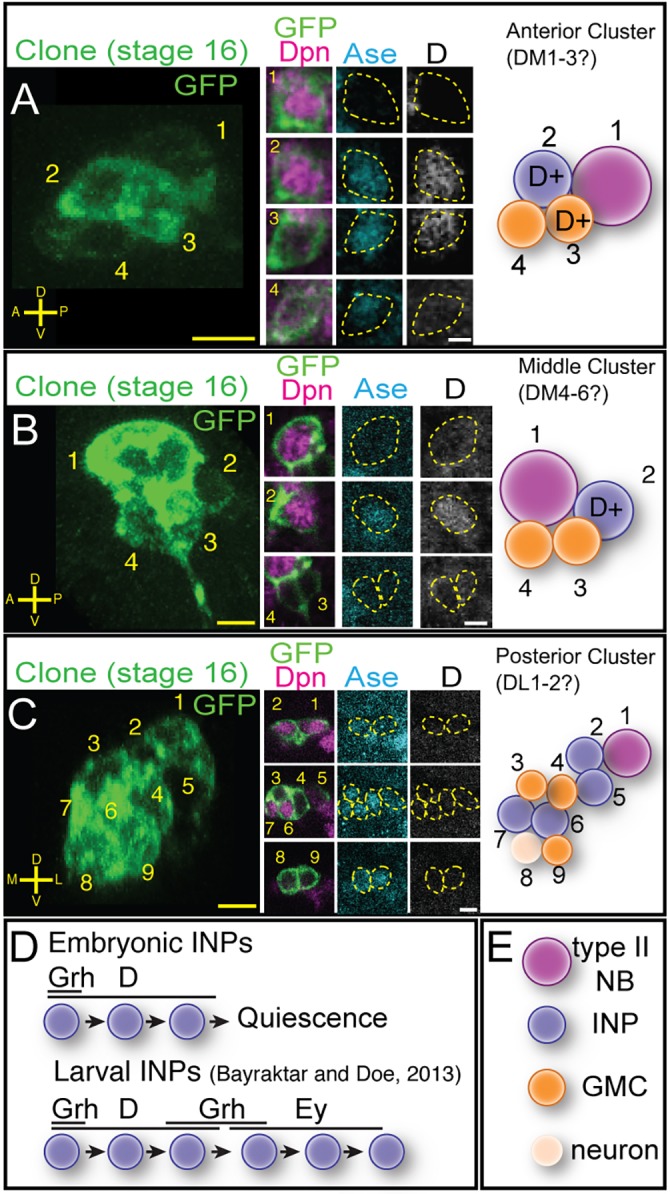
Embryonic INPs express the temporal transcription factor Dichaete. (A) Anterior cluster clone containing Dichaete (D)+ INPs. Four-cell FLP-out clone at stage 16 (left) stained for the clone marker (GFP, green), Dpn (magenta), Ase (cyan) and D (white). The clone contains a type II neuroblast (1), a D+ INP (2) and two GMCs, one D+ and one D− (3,4). (B) Middle cluster clone containing D+ INPs. Four-cell FLP-out clone at stage 16 stained as in A containing a type II neuroblast (1), one D+ INP (4) and two D− GMCs (2,3). (C) Posterior cluster clone lacking D+ INPs. Nine-cell FLP-out clone at stage 16 (left) stained as in A containing a type II neuroblast (1), four D− INPs (2,5-7), three D− GMCs (3,4,9) and one D− neuron (8). Scale bars: 7 μm (clonal projections); 5 μm (insets). (D) Model for INP temporal factor expression; top, embryonic INPs from anterior and middle clusters; bottom, larval INP temporal factor expression (Bayraktar and Doe, 2013). (E) Cell type key for panels above. n>20 for stage 16 clones analyzed.
Table 2.
Dichaete is expressed in embryonic INPs
Embryonic-born INPs contribute to the adult central complex
Embryonic type II neuroblasts produce neurons with contralateral projections, where they have been proposed to pioneer the fan-shaped body neuropil of the central complex (Riebli et al., 2013). To determine whether embryonic-born INP progeny persist into adulthood, we used the FLEXAMP system (Bertet et al., 2014) to permanently mark embryonic INPs and their progeny and trace them into the adult brain. FLEXAMP uses a brief inactivation of temperature-sensitive Gal80 protein (by shifting to 29°C) to allow transient expression of Gal4, which induces FLP expression and the permanent expression of nsyb-LexA LexAop-myr:GFP (Fig. 7A). We crossed R9D11-gal4 (expressed in embryonic INPs) to the FLEXAMP stock and raised the flies at 18°C (negative control), 29°C (positive control), or with a 10 h pulse of 29°C at late embryogenesis followed by 18°C for the rest of the fly's life (‘immortalization of embryonic progeny’ experiment).
Fig. 7.

Embryonic INP progeny contribute to the adult central complex. (A) The FLEXAMP memory cassette used for immortalization of embryonic INPs into the adult brain; modified from Bertet et al., 2014. (B-P) Central complex neuropil regions from flies containing FLEXAMP memory cassette reared at different temperature regimes to permanently label neurons born within all development (29°C positive control), no stage of development (18°C negative control) or specifically during late embryogenesis (29°C pulse) stained for GFP (green) and NC82 (magenta). (B-F) Positive controls reared at 29°C from embryo to adult with over 500 (n=4) of immortalized neurons innervating the protocerebral bridge (PB), the fan-shaped body (FB), the ellipsoid body (EB) and the noduli (NO). (G-K) Negative control adult brains of flies reared at 18°C from embryo to adult showing 10±5 (n=5) neurons from the adult 9D11-gal4 pattern innervating only the dorsal region of the FB. (L-R) Experimental adult brains from flies reared for 6 h pulse at 29°C at late embryonic stages, then reared at 18°C until adult (see Materials and Methods); there are 64±4 (n=12) neurons that innervate the PB, FB, EB but not the NO. (P′-R) Experimental adult brains with differences in innervation pattern within the EB (n=12). (P′) Single z plane from the anterior region shown in P with innervation of two wedges within the EB (yellow arrowheads) seen in 12/12 brains. (P″) Single z plane from posterior region shown in P showing widefield neuron innervation within the EB seen in 9/12 brains. (Q) EB with innervation of two wedges, lacking the widefield innervation (n=1). (R) EB with innervation of one wedge, lacking the widefield innervation (n=1). Yellow arrowheads in Q,R indicate wedge labeling in ellipsoid body. Scale bars: 20 μm. n>5 for positive control (29°C constitutive) and negative control (18°C constitutive); n>10 for experimental (29°C pulse).
We found robust labeling of >500 neurons in the positive control brains raised at 29°C, including many cell bodies innervating the protocerebral bridge, fan-shaped body, ellipsoid body and noduli (Fig. 7B-F). The negative control (18°C permanently) showed labeling of only the approximately ten neurons that project to the dorsal part of the fan-shaped body (Fig. 7G-K), which is similar to the adult pattern of R9D11 (FlyLight). We suspect the ‘leaky’ expression at 18°C might reflect the inefficiency of Gal80 repression in these adult neurons. Importantly, FLEXAMP immortalization of embryonic INP progeny showed labeling of additional neurons (64±4) that project to three central complex regions: the protocerebral bridge, a large portion of the fan-shaped body and the ellipsoid body, but notably not the noduli (Fig. 7L-P). Within the ellipsoid body, we observed variation in labeling. Most brains contained one or two wedge neurons (Fig. 7P′, arrows) and widefield neuron innervation throughout the posterior region of the ellipsoid body (Fig. 7P″; n=12). Interestingly, a few brains contained only the wedge neurons, suggesting that the widefield neuron innervation might be an early-born neuron within the lineages (see Discussion) (n=3/12; Fig. 7Q,R). Additionally, FLEXAMP immortalization of embryonic INP progeny identified neurons innervating the central complex accessory neuropils lateral accessory lobe and the gall, which were never labeled in the 18°C negative control (Fig. S4). We conclude that embryonic INPs generate progeny that persist into the adult brain, and innervate three neuropils of the central complex.
DISCUSSION
It has been difficult to link embryonic neuroblasts to their larval counterparts in the brain and thoracic segments owing to the period of quiescence at the embryo-larval transition, and owing to dramatic morphological changes of the CNS that occur at late embryogenesis. Recent work has revealed the embryonic origin of some larval neuroblasts: the four mushroom body neuroblasts in the central brain and about 20 neuroblasts in thoracic segments (Kunz et al., 2012; Lacin and Truman, 2016). Here, we use molecular markers and clonal analysis to identify all eight known type II neuroblasts in each brain lobe and show they all form during embryogenesis, perhaps the last-born central brain neuroblasts. We were unable to identify each neuroblast individually, however, owing to their tight clustering, movements of the brain lobes, and the lack of markers for specific type II neuroblasts.
The single previously reported embryonic type II neuroblast formed from PntP1+ neuroectodermal cells with apical constrictions called a placode (Hwang and Rulifson, 2011). We have not investigated this neuroectodermal origin of type II neuroblasts in much detail, but we also observe multiple type II neuroblasts developing from PntP1+ neuroectoderm (data not shown). In the future, it would be interesting to determine whether all type II neuroblasts arise from PntP1+ neuroectoderm or from neuroectodermal placodes. Interestingly, one distinguishing molecular attribute of type II neuroblasts is PntP1, which is not detected in type I neuroblasts (Zhu et al., 2011; Xie et al., 2016). Thus, a candidate for distinguishing type I/type II neuroblast identity is EGF signaling, which can be detected in the three head placodes (de Velasco et al., 2007; Hwang and Rulifson, 2011) and is required for PntP1 expression (Gabay et al., 1996). Clearly, there are more PntP1+ neuroectodermal cells than there are type II neuroblasts, and expression of an EGF negative regulator such as Argos (Rebay, 2002) might be necessary to divert some of these neuroectodermal cells away from type II neuroblast specification. The earliest steps of type II neuroblast formation represent an interesting spatial patterning question for future studies.
Now that we have identified the embryonic type II neuroblasts, it is worth considering whether there are differences between embryonic and larval type II neuroblasts or their INP progeny. To date, molecular markers do not reveal any differences between embryonic and larval type II neuroblasts, with the exception that embryonic neuroblasts transiently express the temporal transcription factor Pdm (see below). Interestingly, type I embryonic neuroblasts require Cas to close the Pdm expression window (Grosskortenhaus et al., 2006; Tran and Doe, 2008), whereas we find that cas mutants do not exhibit extension of the Pdm expression window in the earliest-born type II neuroblast or de novo expression of Pdm in the later-forming neuroblasts (Fig. S1C,D). Are there differences between embryonic and larval INPs? Larval INPs mature over a period of 6 h and then divide four to six times with a cell cycle of about 1 h (Bello et al., 2008). In contrast, embryonic INPs might have a more rapid maturation because we see Elav+ neurons within 9D11+ INP lineages by stage 14, just 3 h after the first type II neuroblast forms. We found that INPs undergo quiescence at the embryo-larval transition, as shown by the pools of INPs at stage 16 that do not stain for the mitotic marker pH3. The fate of these quiescent INPs – whether they resume proliferation, differentiate or die – remains to be determined.
Neuroblasts in the embryonic ventral nerve cord use the temporal transcription factor cascade Hb→Krüppel→Pdm→Cas→Grh to generate neural diversity (Brody and Odenwald, 2002; Kohwi et al., 2013; Allan and Thor, 2015; Kang and Reichert, 2015; Doe, 2017). Here, we show that the type II neuroblasts are among the last neuroblasts to form in the embryonic brain, and that they sequentially express only the late temporal transcription factors Pdm (in the earliest-forming neuroblast) followed by Cas and Grh (in all eight type II neuroblasts). It is unknown why most type II neuroblasts skip the early Hb→Krüppel→Pdm temporal transcription factors; perhaps it is due to their late time of formation, although several earlier-forming thoracic neuroblasts also skip Hb (NB3-3), Hb→Krüppel (NB5-5), or Hb→Krüppel→Pdm (NB6-1) (Cui and Doe, 1992; Tsuji et al., 2008; Benito-Sipos et al., 2010). This is another interesting spatial patterning question for the future. Furthermore, misexpression of the early factors (Hb and Krüppel) would be unlikely to affect the progeny produced by type II NBs during embryogenesis, as the competence window for Hb (i.e. the stage at which neuroblasts are responsive to Hb expression) closes with the loss of Dan/Danr expression in all neuroblasts at stage 12 (Kohwi et al., 2013). Thus, most embryonic type II neuroblasts form after closing of the Hb competence window and would probably be unresponsive.
Type I neuroblasts show persistent expression of the temporal transcription factors within neurons born during each window of expression (i.e. a Hb+ neuroblast divides to produce a Hb+ GMC which makes Hb+ neurons). In contrast, we find that type II lineages do not show persistent Cas or Grh expression in INPs born during each expression window, but do contain some Cas+ neurons (data not shown). Both Cas and Grh transcription factors can be seen in INPs immediately adjacent to the parental neuroblast, consistent with transient perdurance from the parental neuroblast, but they are typically lacking in INPs more distant (data not shown). The function of Pdm, Cas and Grh in embryonic type II neuroblasts awaits identification of specific markers for neural progeny born during each expression window.
During larval neurogenesis, virtually all INPs sequentially express the temporal transcription factors Dichaete→Grh→Ey (Bayraktar and Doe, 2013). In contrast, embryonic INPs express only Dichaete. These data, together with the short time frame of embryogenesis, suggest that INP quiescence occurs during the Dichaete window, preventing expression of the later Grh→Ey cascade. Interestingly, INPs in the posterior cluster (presumptive DL1 and DL2 type II neuroblast progeny) completely lack Dichaete; this is similar to the DL1 and DL2 larval lineages, which also do not express Dichaete (Fig. S2). It is possible that DL1/DL2 neuroblasts make INPs that generate identical progeny (and thus do not require an INP temporal cascade), or perhaps these two neuroblasts use a novel temporal cascade in both embryonic and larval stages.
Larval type II neuroblasts produce many intrinsic neurons of the adult central complex (Bayraktar and Doe, 2013; Ito et al., 2013; Yu et al., 2013). Here, we show that embryonic INPs also produce neurons that contribute to the adult central complex. Our data show ∼54 neurons (64 minus 10 due to ‘leaky’ expression) born from embryonic-born INPs survive to adulthood and innervate the central complex. It is likely that this is an underestimate, however, because (1) 9D11-gal4 expression is lacking from a few INPs in the embryonic brain and (2) the time to achieve sufficient FLP protein levels to achieve immortalization could miss the earliest born neurons. The variation in immortalization of the widefield ellipsoid body neuron might represent a neuron born early in the type II lineages, thus unlabeled in a subset of embryos. Additionally, some embryonic-born neurons might perform important functions in the larval/pupal stages but die prior to eclosion.
Further studies will be required to understand the function of neurons born from embryonic type II lineages. It remains to be experimentally determined whether some or all embryonic progeny of type II neuroblasts (1) remain functionally immature in both the larval and adult brain, but serve as pioneer neurons to guide larval-born neurons to establish the central complex, (2) remain functionally immature in the larval brain, but differentiate and function in the adult central complex, or (3) differentiate and perform a function in both the larval and adult CNS. It will be informative to ablate embryonic-born neurons selectively and determine the effect on the assembly of the larval or adult central complex, and their role in generating larval and adult behavior.
MATERIALS AND METHODS
Fly stocks
Male and female Drosophila melanogaster were used. The chromosomes and insertion sites of transgenes (if known) are shown next to genotypes. Unless indicated, lines were obtained from Bloomington Stock Center (FlyBase IDs shown). Enhancer gal4 lines, mutants and reporters were: cas24/TM3, P[GAL4]pnt14-94 (III) (gift of Y. N. Jan, UCSF), R9D11-gal4 (III, attP2), R9D11-CD4-tdTomato (III, attP2), 10XUAS-IVS-mCD8::GFP [III, su(Hw)attP2; referred to as UAS-GFP], hs-FLPG5;;MCFO (I and III; FBst0064086). For the FLEXAMP experiment, y,w,UAS-FLP; tubGAL80ts/CyO; R9D11-gal4/TM3 and 13Xlex-Aop2-myr::GFP; tubGAL80ts/CyO; P[nSyb(FRT.stop)LexA.p65] were used.
Immunofluorescence staining
Primary antibodies were: rat anti-Dpn (1:50, Abcam, 11D1BC7), guinea pig anti-Dpn (1:1000, Jim Skeath, Washington University, USA), chicken anti-GFP (1:1000, Aves Laboratories, GFP-1020), guinea pig anti-Dichaete (1:500, John Nambu, University of Massachusetts, USA), rabbit anti-Ey (1:2500, Uwe Walldorf, Saarland University, Germany), rabbit anti-phospho-histone H3 (ser10) (1:20,000, Millipore, 06-570), rabbit anti-PntP1 (1:1000, Jim Skeath), rat anti-Grh (1:1000, Stefan Thor, Linkoping University, Sweden), rabbit anti-DsRed (1:1000, Clontech Laboratories, 632496), rabbit anti-Ase (1:1000, Cheng-Yu Lee, University of Michigan, USA), mouse anti-Hunchback (1:500, Abcam, F18-1G10.2), guinea pig anti-Krüppel (1:500, Doe Lab, immunogen information available on request), rat anti-Pdm2 (1:1000, Abcam, 6G6AC11), guinea pig anti-Asense (1:1000, Hongyan Wang, Duke-NUS Medical School, Singapore), rabbit anti-Cas (1:1000, Ward Odenwald, NIH, distributed by the Doe lab), mouse anti-NC82 (1:200, Developmental Studies Hybridoma Bank). Secondary antibodies were from Molecular Probes or Jackson ImmunoResearch and were used at 1:400.
Embryos were blocked overnight in 0.3% PBST (1× PBS with 0.3% Triton X-100) with 5% normal goat serum and 5% donkey serum (PDGS) (Vector Laboratories), followed by incubation in primary antibody overnight at 4°C. Next, embryos underwent four washes, 15 min each, in PDGS, followed by a 2 h secondary antibody incubation at 25°C. Embryos were then either dehydrated with ethanol and mounted in dibutyl phytalate in xylene (DPX) according to Janelia protocol (Wolff et al., 2015) or were cleared with a glycerol series: 25% for 10 min, 50% for 10 min, 90% for 10 min then into 90% glycerol with 4% n-propyl gallate overnight before imaging.
Larval brains were dissected in PBS, fixed in 4% formaldehyde in PBST for 25 min, rinsed for 30 min in PBST, and blocked in PDGS overnight at 4°C. Staining was carried out as above for embryos, but after the secondary antibody incubation brains were mounted in Vectashield (Vector Laboratories).
Adult brains were fixed in 2% formaldehyde in PBST, rinsed, and blocked in PDGS with 0.5% Triton X-100. Brains were incubated in primary antibodies for 4 days at 4°C, then in secondary antibodies for 2 days at 4°C. Brains were mounted in DPX according to Janelia protocol.
Clones
For type II clones, P[GAL4]pnt14-94 (III)×hs FLPG5;;MCFO (I and III; FBst0064086) embryos were collected for 4 h at 25°C, aged 4 h and heat shocked at 37°C for 12 min, then left to develop until desired stages.
FLEXAMP immortalization of embryonic INPs
The FLEXAMP experiment used 1- to 3-day-old adult females from crossing y,w,UAS-FLP; tubGAL80ts/CyO; R9D11-gal4/TM3 to 13Xlex- Aop2-myr::GFP; tubGAL80ts/CyO; P[nSyb(FRT.stop)LexA.p65] to label embryonic INPs permanently (Bertet et al., 2014). Negative controls were raised continuously at 18°C to maintain Gal80 repression; positive controls were raised continuously at 29°C inactivate Gal80 and allow 9D11-gal4 expression. To ‘immortalize’ embryonic INPs and their progeny, we exposed embryos aged 5-6 h to 29°C for 10 h to allow R9D11-gal4 expression and then shifted all unhatched embryos to 18°C to block R9D11-gal4 expression during larval, pupal and adult stages.
Cell proliferation analysis
Number of proliferating INPs was calculated by dividing the number of pH3-positive by the total number of INPs within each cluster of neuroblasts at different stages. Each circle represents one cluster of INPs. Error bars represent s.e.m.
Imaging
Images were captured with a ZeissLSM700 or ZeissLSM710 confocal microscope with a z-resolution of 1.0 μm, and processed in the open source software FIJI (http://fiji.sc) and Photoshop CS5 (Adobe). Figures were made in Illustrator CS5 (Adobe). Three-dimensional brain reconstructions in Figs 3 and 6 were generated using Imaris (Bitplane).
Supplementary Material
Acknowledgements
We thank Luis Sullivan, Emily Sales, Laurina Manning and Sen-Lin Lai for fly stocks, technical assistance and helpful discussions; Sen-Lin Lai and Volker Hartenstein for comments on the manuscript; Jim Skeath, John Nambu, Uwe Walldorf, Stefan Thor, Cheng-Yu Lee, Claude Desplan, Hongyan Wang, Y. N. Jan and Ward Odenwald for reagents; and Fernando Diaz-Benjumea and Volker Hartenstein for sharing unpublished work. We acknowledge the Bloomington Drosophila Stock Center (NIH P40OD018537) and the Developmental Studies Hybridoma Bank (DSHB).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.T.W., C.Q.D.; Methodology: K.T.W., C.Q.D.; Software: K.T.W., C.Q.D.; Validation: K.T.W., C.Q.D.; Formal analysis: K.T.W.; Investigation: K.T.W.; Resources: K.T.W., C.Q.D.; Data curation: K.T.W., C.Q.D.; Writing - original draft: K.T.W., C.Q.D.; Writing - review & editing: K.T.W., C.Q.D.; Visualization: K.T.W., C.Q.D.; Supervision: C.Q.D.; Project administration: C.Q.D.; Funding acquisition: K.T.W., C.Q.D.
Funding
This work was funded by the National Institutes of Health (NIH) (HD27056 to C.Q.D.), a Genetics Training Grant from the NIH (T32 GM007413 to K.T.W.) and the Howard Hughes Medical Institute, where C.Q.D. is an Investigator. Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.157826.supplemental
References
- Allan D. W. and Thor S. (2015). Transcriptional selectors, masters, and combinatorial codes: regulatory principles of neural subtype specification. Wiley Interdiscip. Rev. Dev. Biol. 4, 505-528. 10.1002/wdev.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsio J. M., Tarchini B., Cayouette M. and Livesey F. J. (2013). Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc. Natl. Acad. Sci. USA 110, E716-E725. 10.1073/pnas.1215707110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M., Karlsson D., Salmani B. Y., Bivik C., MacDonald R. B., Gunnar E. and Thor S. (2014). Global programmed switch in neural daughter cell proliferation mode triggered by a temporal gene cascade. Dev. Cell 30, 192-208. 10.1016/j.devcel.2014.06.021 [DOI] [PubMed] [Google Scholar]
- Bayraktar O. A. and Doe C. Q. (2013). Combinatorial temporal patterning in progenitors expands neural diversity. Nature 498, 449-455. 10.1038/nature12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar O. A., Boone J. Q., Drummond M. L. and Doe C. Q. (2010). Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 5, 26 10.1186/1749-8104-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B. C., Izergina N., Caussinus E. and Reichert H. (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5 10.1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J. A., Pollack A. J. and Ritzmann R. E. (2010). Neural activity in the central complex of the insect brain is linked to locomotor changes. Curr. Biol. 20, 921-926. 10.1016/j.cub.2010.03.054 [DOI] [PubMed] [Google Scholar]
- Benito-Sipos J., Estacio-Gomez A., Moris-Sanz M., Baumgardt M., Thor S. and Diaz-Benjumea F. J. (2010). A genetic cascade involving klumpfuss, nab and castor specifies the abdominal leucokinergic neurons in the Drosophila CNS. Development 137, 3327-3336. 10.1242/dev.052233 [DOI] [PubMed] [Google Scholar]
- Berger C., Urban J. and Technau G. M. (2001). Stage-specific inductive signals in the Drosophila neuroectoderm control the temporal sequence of neuroblast specification. Development 128, 3243-3251. [DOI] [PubMed] [Google Scholar]
- Bertet C., Li X., Erclik T., Cavey M., Wells B. and Desplan C. (2014). Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell 158, 1173-1186. 10.1016/j.cell.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. Q. and Doe C. Q. (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68, 1185-1195. 10.1002/dneu.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G. and Knoblich J. A. (2008). The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535-546. 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyan G. S. and Reichert H. (2011). Mechanisms for complexity in the brain: generating the insect central complex. Trends Neurosci. 34, 247-257. 10.1016/j.tins.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Brody T. and Odenwald W. F. (2000). Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev. Biol. 226, 34-44. 10.1006/dbio.2000.9829 [DOI] [PubMed] [Google Scholar]
- Brody T. and Odenwald W. F. (2002). Cellular diversity in the developing nervous system: a temporal view from Drosophila. Development 129, 3763-3770. [DOI] [PubMed] [Google Scholar]
- Cenci C. and Gould A. P. (2005). Drosophila Grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development 132, 3835-3845. 10.1242/dev.01932 [DOI] [PubMed] [Google Scholar]
- Chu-LaGraff Q. and Doe C. Q. (1993). Neuroblast specification and formation regulated by wingless in the Drosophila CNS. Science 261, 1594-1597. 10.1126/science.8372355 [DOI] [PubMed] [Google Scholar]
- Cui X. and Doe C. Q. (1992). ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development 116, 943-952. [DOI] [PubMed] [Google Scholar]
- Datta S. (1995). Control of proliferation activation in quiescent neuroblasts of the Drosophila central nervous system. Development 121, 1173-1182. [DOI] [PubMed] [Google Scholar]
- de Velasco B., Erclik T., Shy D., Sclafani J., Lipshitz H., McInnes R. and Hartenstein V. (2007). Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev. Biol. 302, 309-323. 10.1016/j.ydbio.2006.09.035 [DOI] [PubMed] [Google Scholar]
- Doe C. Q. (2008). Neural stem cells: balancing self-renewal with differentiation. Development 135, 1575-1587. 10.1242/dev.014977 [DOI] [PubMed] [Google Scholar]
- Doe C. Q. (2017). Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219-240. 10.1146/annurev-cellbio-111315-125210 [DOI] [PubMed] [Google Scholar]
- Egger B., Chell J. M. and Brand A. H. (2008). Insights into neural stem cell biology from flies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 39-56. 10.1098/rstb.2006.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estacio-Gómez A. and Díaz-Benjumea F. J. (2014). Roles of Hox genes in the patterning of the central nervous system of Drosophila. Fly (Austin) 8, 26-32. 10.4161/fly.27424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay L., Scholz H., Golembo M., Klaes A., Shilo B. Z. and Klambt C. (1996). EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development 122, 3355-3362. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R., Robinson K. J. and Doe C. Q. (2006). Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 20, 2618-2627. 10.1101/gad.1445306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero P., Estacio-Gómez A., Moris-Sanz M., Alvarez-Rivero J. and Diaz-Benjumea F. J. (2014). Origin and specification of the brain leucokinergic neurons of Drosophila: similarities to and differences from abdominal leucokinergic neurons. Dev. Dyn. 243, 402-414. 10.1002/dvdy.24083 [DOI] [PubMed] [Google Scholar]
- Hwang H. J. and Rulifson E. (2011). Serial specification of diverse neuroblast identities from a neurogenic placode by Notch and Egfr signaling. Development 138, 2883-2893. 10.1242/dev.055681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T., Pearson B., Holbrook S. and Doe C. Q. (2001). Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511-521. 10.1016/S0092-8674(01)00465-2 [DOI] [PubMed] [Google Scholar]
- Ito M., Masuda N., Shinomiya K., Endo K. and Ito K. (2013). Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Curr. Biol. 23, 644-655. 10.1016/j.cub.2013.03.015 [DOI] [PubMed] [Google Scholar]
- Izergina N., Balmer J., Bello B. and Reichert H. (2009). Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 4, 44 10.1186/1749-8104-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M. I., Okabe M. and Hiromi Y. (2005). seven-up Controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Dev. Cell 8, 203-213. 10.1016/j.devcel.2004.12.014 [DOI] [PubMed] [Google Scholar]
- Kang K. H. and Reichert H. (2015). Control of neural stem cell self-renewal and differentiation in Drosophila. Cell Tissue Res. 359, 33-45. 10.1007/s00441-014-1914-9 [DOI] [PubMed] [Google Scholar]
- Karcavich R. and Doe C. Q. (2005). Drosophila neuroblast 7-3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. J. Comp. Neurol. 481, 240-251. 10.1002/cne.20371 [DOI] [PubMed] [Google Scholar]
- Kohwi M. and Doe C. Q. (2013). Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823-838. 10.1038/nrn3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M., Lupton J. R., Lai S.-L., Miller M. R. and Doe C. Q. (2013). Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152, 97-108. 10.1016/j.cell.2012.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz T., Kraft K. F., Technau G. M. and Urbach R. (2012). Origin of Drosophila mushroom body neuroblasts and generation of divergent embryonic lineages. Development 139, 2510-2522. 10.1242/dev.077883 [DOI] [PubMed] [Google Scholar]
- Lacin H. and Truman J. W. (2016). Lineage mapping identifies molecular and architectural similarities between the larval and adult Drosophila central nervous system. Elife 5, e13399 10.7554/eLife.13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E. C., Dry K. E., Alaimo D. R., Rudd K. T., Cillo A. R., Clenshaw M. E., Negre N., White K. P. and Truman J. W. (2012). Ultrabithorax confers spatial identity in a context-specific manner in the Drosophila postembryonic ventral nervous system. Neural Dev. 7, 31 10.1186/1749-8104-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.-R., Raabe T. and Heisenberg M. (1999). Central complex substructures are required for the maintenance of locomotor activity in Drosophila melanogaster. J. Comp. Physiol. A 185, 277-288. 10.1007/s003590050387 [DOI] [PubMed] [Google Scholar]
- Maurange C. and Gould A. P. (2005). Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila. Trends Neurosci. 28, 30-36. 10.1016/j.tins.2004.10.009 [DOI] [PubMed] [Google Scholar]
- Maurange C., Cheng L. and Gould A. P. (2008). Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133, 891-902. 10.1016/j.cell.2008.03.034 [DOI] [PubMed] [Google Scholar]
- McDonald J. A., Holbrook S., Isshiki T., Weiss J., Doe C. Q. and Mellerick D. M. (1998). Dorsoventral patterning in the Drosophila central nervous system: the vnd homeobox gene specifies ventral column identity. Genes Dev. 12, 3603-3612. 10.1101/gad.12.22.3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler U., Vogler G. and Urban J. (2006). Timing of identity: spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero. Development 133, 429-437. 10.1242/dev.02229 [DOI] [PubMed] [Google Scholar]
- Moris-Sanz M., Estacio-Gomez A., Alvarez-Rivero J. and Diaz-Benjumea F. J. (2014). Specification of neuronal subtypes by different levels of Hunchback. Development 141, 4366-4374. 10.1242/dev.113381 [DOI] [PubMed] [Google Scholar]
- Moris-Sanz M., Estacio-Gomez A., Sanchez-Herrero E. and Diaz-Benjumea F. J. (2015). The study of the Bithorax-complex genes in patterning CCAP neurons reveals a temporal control of neuronal differentiation by Abd-B. Biol. Open 4, 1132-1142. 10.1242/bio.012872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Pfeiffer B. D. and Rubin G. M. (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112, E2967-E2976. 10.1073/pnas.1506763112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny T., Eiselt R. and Urban J. (2002). Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development 129, 1027-1036. [DOI] [PubMed] [Google Scholar]
- Ofstad T. A., Zuker C. S. and Reiser M. B. (2011). Visual place learning in Drosophila melanogaster. Nature 474, 204-207. 10.1038/nature10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhou Y., Guo C., Gong H., Gong Z. and Liu L. (2009). Differential roles of the fan-shaped body and the ellipsoid body in Drosophila visual pattern memory. Learn. Mem. 16, 289-295. 10.1101/lm.1331809 [DOI] [PubMed] [Google Scholar]
- Poeck B., Triphan T., Neuser K. and Strauss R. (2008). Locomotor control by the central complex in Drosophila-An analysis of the tay bridge mutant. Dev. Neurobiol. 68, 1046-1058. 10.1002/dneu.20643 [DOI] [PubMed] [Google Scholar]
- Prokop A. and Technau G. M. (1994). Early tagma-specific commitment of Drosophila CNS progenitor NB1-1. Development 120, 2567-2578. [DOI] [PubMed] [Google Scholar]
- Rebay I. (2002). Keeping the receptor tyrosine kinase signaling pathway in check: lessons from Drosophila. Dev. Biol. 251, 1-17. 10.1006/dbio.2002.0806 [DOI] [PubMed] [Google Scholar]
- Reichert H. (2011). Drosophila neural stem cells: cell cycle control of self-renewal, differentiation, and termination in brain development. Results Probl. Cell Differ. 53, 529-546. 10.1007/978-3-642-19065-0_21 [DOI] [PubMed] [Google Scholar]
- Renn S. C. P., Armstrong J. D., Yang M., Wang Z., An X., Kaiser K. and Taghert P. H. (1999). Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J. Neurobiol. 41, 189-207. [DOI] [PubMed] [Google Scholar]
- Riebli N., Viktorin G. and Reichert H. (2013). Early-born neurons in type II neuroblast lineages establish a larval primordium and integrate into adult circuitry during central complex development in Drosophila. Neural Dev. 8, 6 10.1186/1749-8104-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J. D. and Jayaraman V. (2011). Studying sensorimotor processing with physiology in behaving Drosophila. Int. Rev. Neurobiol. 99, 169-189. 10.1016/B978-0-12-387003-2.00007-0 [DOI] [PubMed] [Google Scholar]
- Seelig J. D. and Jayaraman V. (2013). Feature detection and orientation tuning in the Drosophila central complex. Nature 503, 262-266. 10.1038/nature12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J. D. and Jayaraman V. (2015). Neural dynamics for landmark orientation and angular path integration. Nature 521, 186-191. 10.1038/nature14446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath J. B. and Thor S. (2003). Genetic control of Drosophila nerve cord development. Curr. Opin. Neurobiol. 13, 8-15. 10.1016/S0959-4388(03)00007-2 [DOI] [PubMed] [Google Scholar]
- Skeath J. B., Zhang Y., Holmgren R., Carroll S. B. and Doe C. Q. (1995). Specification of neuroblast identity in the Drosophila embryonic central nervous system by gooseberry-distal. Nature 376, 427-430. 10.1038/376427a0 [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R., Cheng L. Y. and Gould A. P. (2010). Regulating neural proliferation in the Drosophila CNS. Curr. Opin. Neurobiol. 20, 50-57. 10.1016/j.conb.2009.12.005 [DOI] [PubMed] [Google Scholar]
- Strauss R. (2002). The central complex and the genetic dissection of locomotor behaviour. Curr. Opin. Neurobiol. 12, 633-638. 10.1016/S0959-4388(02)00385-9 [DOI] [PubMed] [Google Scholar]
- Syed M. H., Mark B. and Doe C. Q. (2017). Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. Elife 6, e26287 10.7554/eLife.26287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K. D. and Doe C. Q. (2008). Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development 135, 3491-3499. 10.1242/dev.024349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman J. W. and Bate M. (1988). Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 125, 145-157. 10.1016/0012-1606(88)90067-X [DOI] [PubMed] [Google Scholar]
- Tsuji T., Hasegawa E. and Isshiki T. (2008). Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development 135, 3859-3869. 10.1242/dev.025189 [DOI] [PubMed] [Google Scholar]
- Ulvklo C., MacDonald R., Bivik C., Baumgardt M., Karlsson D. and Thor S. (2012). Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression. Development 139, 678-689. 10.1242/dev.074500 [DOI] [PubMed] [Google Scholar]
- Urbach R. and Technau G. M. (2003). Early steps in building the insect brain: neuroblast formation and segmental patterning in the developing brain of different insect species. Arthropod. Struct. Dev. 32, 103-123. 10.1016/S1467-8039(03)00042-2 [DOI] [PubMed] [Google Scholar]
- Urban J. and Mettler U. (2006). Connecting temporal identity to mitosis: the regulation of Hunchback in Drosophila neuroblast lineages. Cell Cycle 5, 950-952. 10.4161/cc.5.9.2727 [DOI] [PubMed] [Google Scholar]
- Wang Z., Pan Y., Li W., Jiang H., Chatzimanolis L., Chang J., Gong Z. and Liu L. (2008). Visual pattern memory requires foraging function in the central complex of Drosophila. Learn. Mem. 15, 133-142. 10.1101/lm.873008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. B., Von Ohlen T., Mellerick D. M., Dressler G., Doe C. Q. and Scott M. P. (1998). Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev. 12, 3591-3602. 10.1101/gad.12.22.3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessnitzer J. and Webb B. (2006). Multimodal sensory integration in insects--towards insect brain control architectures. Bioinspir. Biomim. 1, 63-75. 10.1088/1748-3182/1/3/001 [DOI] [PubMed] [Google Scholar]
- Wolff T., Iyer N. A. and Rubin G. M. (2015). Neuroarchitecture and neuroanatomy of the Drosophila central complex: a GAL4-based dissection of protocerebral bridge neurons and circuits. J. Comp. Neurol. 523, 997-1037. 10.1002/cne.23705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Li X., Deng X., Hou Y., O'Hara K., Urso A., Peng Y., Chen L. and Zhu S. (2016). The Ets protein pointed prevents both premature differentiation and dedifferentiation of Drosophila intermediate neural progenitors. Development 143, 3109-3118. 10.1242/dev.137281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.-H., Awasaki T., Schroeder M. D., Long F., Yang J. S., He Y., Ding P., Kao J. C., Wu G. Y., Peng H. et al. (2013). Clonal development and organization of the adult Drosophila central brain. Curr. Biol. 23, 633-643. 10.1016/j.cub.2013.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Barshow S., Wildonger J., Jan L. Y. and Jan Y.-N. (2011). Ets transcription factor Pointed promotes the generation of intermediate neural progenitors in Drosophila larval brains. Proc. Natl. Acad. Sci. USA 108, 20615-20620. 10.1073/pnas.1118595109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.