Abstract
The Fragile X-related disorders are a group of 3 clinical conditions resulting from the instability of a CGG-repeat tract at the 5’ end of the FMR1 transcript. Fragile X-associated tremor/ataxia syndrome (FXTAS) and Fragile X-associated primary ovarian insufficiency (FXPOI) are disorders seen in carriers of FMR1 alleles with 55–200 repeats. Female carriers of these premutation (PM) alleles are also at risk of having a child who has an FMR1 allele with >200 repeats. Most of these full mutation (FM) alleles are epigenetically silenced resulting in a deficit of the FMR1 gene product, FMRP. This results in Fragile X Syndrome (FXS), the most common heritable cause of intellectual disability and autism. The diagnosis and study of these disorders is challenging in part because the detection of alleles with large repeat numbers has, until recently, been either time-consuming or unreliable. This problem is compounded by the mosaicism for repeat length and/or DNA methylation that is frequently seen in PM and FM carriers. Furthermore, since AGG interruptions in the repeat tract affect the risk that a FM allele will be maternally transmitted, the ability to accurately detect these interruptions in female PM carriers is an additional challenge that must be met. This review will discuss some of the pros and cons of some recently described assays for these disorders, including those that detect FMRP levels directly, as well as emerging technologies that promise to improve the diagnosis of these conditions and to be useful in both basic and translational research settings.
Keywords: FMR1-related disorders (FMR1 disorders), FX-associated tremor and ataxia syndrome (FXTAS), FX-associated primary ovarian insufficiency (FXPOI), Fragile X syndrome (FXS)
Background
The Fragile X-related disorders are a group of clinical conditions that include Fragile X syndrome (FXS; OMIM 300624), the most common heritable cause of intellectual disability and a frequent cause of autism, as well as Fragile X-associated tremor/ataxia syndrome (FXTAS; OMIM 300623) and Fragile X-associated primary ovarian insufficiency (FXPOI; OMIM 300624). These disorders all result from mutations in FMR1 (OMIM *309550), a gene on the long arm of the X chromosome. FXTAS, FXPOI and almost all cases of FXS result from expansion of an unstable CGG-repeat tract at the 5’ end of the FMR1 transcript.
The American College of Medical Genetics divides FMR1 alleles into 4 classes based on CGG-repeat number, normal alleles with 6–44 repeats, intermediate (IM) or gray zone alleles with 45–54 repeats, premutation (PM) alleles with 55–200 repeats and full mutations (FM) with more than 200 repeats (Monaghan et al. 2013). This 200 repeat threshold is based on rough estimates of the number of repeats seen in the alleles of FXS carriers analyzed by Southern blot. However, recent data using higher resolution techniques suggest that the threshold may be somewhat higher (Brykczynska et al. 2016; Zhou et al. 2016). A recent screen of 14,207 newborns in the US showed a prevalence of IM alleles of 1 in 66 females and 1 in 112 males, with PM alleles found in 1 in 209 females and 1 in 430 males (Tassone et al. 2012), while the US prevalence of FM alleles as been estimated to be 1 in 5000 males and 1 in 2500–8000 females (Coffee et al. 2009). However, the prevalence of IM, PM and FM alleles varies significantly between different populations. For example, the prevalence of PM alleles ranges from a low of 1 in ~1,674 females in Japan to a high of 1 in 100 in Columbia and Israel (reviewed in (Hagerman and Hagerman 2016)). These differences are likely due to the effect of founder mutations (Eichler et al. 1994).
Inheritance of a PM allele confers risk of FXTAS and FXPOI with the risk being related to the number of repeats in the PM allele (Murray 2000; Sherman 2000; Tassone et al. 2007a). Pathology in these individuals arises from some deleterious consequence of expressing high levels of FMR1 mRNA with large numbers of CGG repeats (Oh et al. 2015; Sellier et al. 2013; Sellier et al. 2010; Todd et al. 2010). Recent evidence suggests that even carriers of IM alleles may be at risk of these disorders (reviewed in (Loesch and Hagerman 2012)). In addition to the risk of FXTAS and FXPOI, female PM carriers are also at risk of transmitting a FM allele to their children. This risk is directly related to the number of maternal CGG-repeats but is reduced by the presence of one or more AGG interruptions in the repeat tract (Eichler et al. 1994; Latham et al. 2014; Nolin et al. 2015; Nolin et al. 2013; Yrigollen et al. 2012; Yrigollen et al. 2014). In contrast to PM alleles, most FM alleles are epigenetically silenced (Coffee et al. 2002; Coffee et al. 1999; Kumari and Usdin 2010; Oberle et al. 1991; Pieretti et al. 1991; Sutcliffe et al. 1992), and thus make little of the FMR1 gene product, FMRP, a protein important for learning and memory. It is this FMRP deficit that is responsible for FXS.
In real life the CGG-repeat is prone to expansion and contraction both in the germ line and in somatic cells. Thus many individuals are mosaic for the repeat size, having some combination of normal, IM, PM and FM alleles. To complicate matters, methylation mosaicism is also seen. In fact, it is estimated that as many as 40% of FM carriers exhibit some sort of size and/or methylation mosaicism (Nolin et al. 1994). Occasionally, for reasons that are still the subject of debate (Zhou et al. 2016), some individuals escape silencing altogether. These individuals may make large amounts of the deleterious FMR1 transcript (Basuta et al. 2015; Hagerman et al. 1994; Loesch et al. 2012; Tabolacci et al. 2008a; Wohrle et al. 1998). However, since large alleles are not efficiently translated (Feng et al. 1995), these individuals may also have an FMRP deficit. They thus can have the symptoms of both FXTAS and FXS (Basuta et al. 2011; Basuta et al. 2015). PM carriers may also have an FMRP deficit resulting either from low levels of methylation that are sometimes seen in such individuals (Pretto et al. 2014) or from inefficient translation of large PM alleles. There is thus significant clinical overlap seen in PM and FM carriers. This has led to the suggestion that the term Fragile X spectrum disorder (FXSD) would better reflect the continuity of clinical involvement seen in individuals with PM or FM alleles (Lozano et al. 2014).
Prior to the identification of the gene responsible for these disorders, the only genetic test for FXS was chromosome-based. FXS cells show a folate-sensitive fragile site, FRAXA, on the X chromosome at Xq27.3, coincident with what we now know is the FMR1 gene. This site can be visualized in metaphase spreads of cells grown in specific media (Sutherland and Ashforth 1979). However, the assay for chromosome fragility is time-consuming and not always reliable (Jenkins et al. 1988; Rocchi et al. 1985). This test is also not informative for IM and PM alleles, provides no information as to the repeat number or methylation status of FM alleles. This test is no longer considered an acceptable diagnostic method even for FXS (Monaghan et al. 2013).
We now know that some combination of the following factors need to be ascertained for appropriate diagnosis and clinical management of these disorders: the repeat number and extent of repeat size mosaicism, the AGG interruption pattern and the extent of methylation mosaicism. In females, knowledge of the activation ratio is also useful since skewed X chromosome inactivation (XCI) would impact disease severity. In some situations, direct measures of the amount of FMRP may be preferred. The ability to accurately determine all of these factors is important in the research setting as well, in part because factors like repeat number and methylation status can vary tremendously with growth in culture (Zhou et al. 2016). Until recently, our ability to accurately determine all these factors has been limited. This review will discuss some of the recent advances in molecular techniques that have improved our ability to do so.
DNA-based assays
Southern blotting
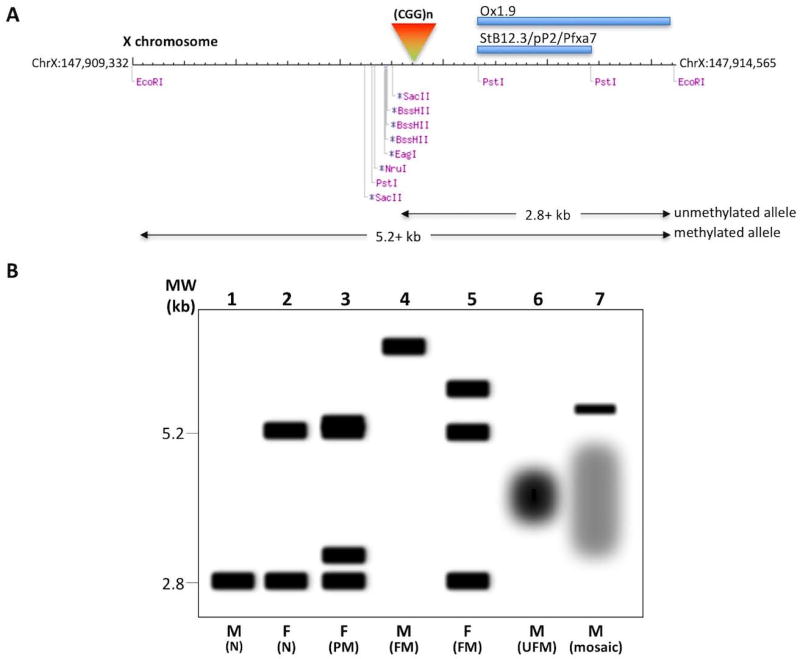
The first DNA-based assays for these disorders made use of Southern blotting strategies in which genomic DNA was digested with a restriction enzyme like EcoRI that cuts the FMR1 locus whether the gene was methylated or not, and a methylation-sensitive enzyme that only cuts unmethylated alleles, like BssHII EagI, NruI, or SacII (Rousseau et al. 1991). Samples are then resolved by agarose gel electrophoresis. These gels are best run without ethidium bromide to avoid aberrant fragment mobilities that can be mistaken for size mosaicism (Nolin et al. 2008). After transfer to a suitable membrane, the blot is exposed to a radioactive or, more recently, a digoxigenin-labeled probe derived from the 5’ end of the FMR1 gene (Gold et al. 2000). Commonly used probes include Stb12.3, aka pP2 or Pfxa7, and Ox1.9 that hybridize to a region downstream of both the repeats and the recognition sites of the methylation-sensitive enzymes. As illustrated in Fig. 1A., the methylation-sensitive enzymes all cut within ~200 bp of one another and thus cutting at any one of these sites produces a band of ~2.8 kb in normal males when EcoRI is used as the methylation-insensitive enzyme (Fig. 1B, lane 1). Since FMR1 is on the X chromosome, the same digestion of DNA from normal females produces two bands, one of 2.8 kb corresponding to the allele on the active X chromosome and a second of 5.2 kb, corresponding to the allele on the inactive X as illustrated in Fig. 1B, lane 2. In this assay, unmethylated PM alleles (Fig. 1B, lane 3) and most unmethylated FM alleles would be smaller than 5.2 kb, while methylated PM or FM alleles would be larger than 5.2 kb (Fig. 1B, lanes 3–5, 7). For example, an unmethylated FM with 400 repeats would be present on a fragment of 3.5 kb, while a methylated allele with the same repeat number would be present on a fragment that was ~ 6.1 kb in size. In practice, things may be more complex. For example, large unmethylated alleles tend to be unstable, resulting in smears rather than discrete bands as illustrated in Fig. 1B, lanes 6 and 7. The net effect of the size and methylation mosaicism is that the effective concentration of each allele in the population is reduced. This makes detection of these alleles much more challenging than for a typical single copy gene.
Fig. 1. Southern blotting assay for the FMR1-related disorders.
A) Diagrammatic representation of the 5.2 kb EcoRI fragment containing the 5’ end of the FMR1 gene. The coordinates are from the GRCh38/hg38 build of the human genome. The location of various commonly used methylation-sensitive restriction enzyme recognition sites, as well as the location of the CGG repeat tract and the 2 sets of commonly used probes are shown. The predicted fragment sizes that would be obtained from an unmethylated and a methylated allele is illustrated below. B) A graphic representation of the expected fragment sizes that would be seen in male and female carriers of normal, PM and FM alleles including carriers of unmethylated FM (UFM) alleles and a male mosaic for both a methylated FM allele and a heterogenous mixture of unmethylated PM and FM alleles. In practice samples can be more heterogeneous and the signal to noise ratio lower than depicted here.
There are other disadvantages to this technique, including the fact that it is time consuming and requires micrograms of high molecular weight genomic DNA. The resolution of agarose gels is also limited, precluding the precise determination of repeat number. As with the other FMR1 assays described below, it is also important to bear in mind the existence of rare gene variants that could lead to a false positive or false negative result. For example, mutations within the repeat have been reported that result in a novel EagI site and thus mimic a deletion (Cecconi et al. 2008; Tabolacci et al. 2008b; Tarleton et al. 2002), while a G-to-A mutation in the upstream EcoRI site has been reported that mimics a FM allele (Liang et al. 2008). In addition, deletions that involve the region downstream of the repeats may lead to an underestimation of the true number of repeats. In principle, deletions that involve the loss of the recognition sites of the methylation-sensitive enzyme used may result in the false diagnosis of a FM allele even if the deletion removed most of the repeats. Furthermore, deletions that remove the transcription start site or the translation start site may result in alleles with small repeat numbers that nonetheless would result in symptoms of FXS (Meijer et al. 1994).
However, on the positive side, in addition to providing the repeat size and the extent of any repeat size heterogeneity, this assay does provide important information as to the extent of methylation of different alleles including any evidence of skewed XCI.
Amplification-based assays
Assays for repeat size
The use of amplification-based assays overcomes some of the drawbacks of Southern blotting in that the assay takes hours, rather than days, and requires nanograms, rather than micrograms, of DNA. However, the CGG-repeat is difficult to amplify by the Polymerase Chain Reaction (PCR), likely because of the stable secondary structures that they form (Fry and Loeb 1994; Mitas et al. 1995; Usdin and Woodford 1995; Yu et al. 1997). Over the years there have been many efforts to improve the PCR yield, including the use of osmolytes such as betaine and dimethylsulfoxide and dGTP analogs such as 7-deaza-2’-GTP that reduce the stability of secondary structures. A variety of DNA polymerases able to amplify other G+C-rich templates were also tested. Nonetheless, most of these early assays were unable to amplify FM alleles reliably (Brown et al. 1996; Chong et al. 1994; Pergolizzi et al. 1992; Saluto et al. 2005; Tassone et al. 2008). However, recent assays have been described that do enable large FM alleles to be detected reliably even in females and mosaic males (Chen et al. 2010; Filipovic-Sadic et al. 2010; Hayward et al. 2016; Orpana et al. 2012). These assays are all capable of amplifying even very large full mutation alleles with as many as 940 repeats. The assay described by Filipovic-Sadic et al uses a commercially available product whose reagents are proprietory, so it is unclear what makes this assay work, although it was reported that more than 60 primer pairs and 1000s of buffer/additive combinations had to be tested before arriving at the optimal combination (Filipovic-Sadic et al. 2010). The other assays make use of buffers that lack K+ and include high concentrations of betaine, at least for very large alleles (Hayward et al. 2016; Orpana et al. 2012). Potassium ions stabilize the structures formed by the CGG-repeats (Usdin and Woodford 1995). Omission of this cation may thus improve PCR yield by minimizing the stability of these structures. Betaine may improve the PCR yield by further destabilizing these structures (Rees et al. 1993). One of these assays uses DyNAzyme EXT DNA polymerase, a mixture of DyNAzyme II DNA polymerase and a proof-reading polymerase, which unfortunately is no longer commercially available (Orpana et al. 2012). In addition to this specific polymerase mix, the assay uses multiple heat pulse extension steps during amplification. The second assay uses a predigestion step with HindIII, followed directly without further purification by PCR using Phusion® DNA polymerase (Hayward et al. 2016). This assay does not require heat pulses, perhaps because the predigestion step reduces the template size and improves the efficiency of early rounds of PCR. The lack of heat pulse steps simplifies the PCR set up and significantly reduces the PCR cycling time required. However, both assays are able to amplify large alleles and do so relatively inexpensively, thus reducing the barrier to the use of these assays for large scale population screens and for routine research use.
The PCR products generated in these assays can be resolved on agarose gels and detected using standard DNA stains like ethidium bromide or Sybr® Green. Because the PCR product is smaller than the fragments typically produced using Southern blotting strategies, the estimation of repeat number is also likely to be more accurate. However, for more precise determinations of the repeat number in PM and small FM alleles, capillary electrophoresis is currently the method of choice. For this purpose, one of the PCR primers is typically fluorescently labeled.
One important point to bear in mind is that, in capillary electrophoresis the top strand of the repeat-containing region migrates faster than the bottom strand and neither strand migrates in a manner consistent with its molecular weight. This is likely due to the formation of different unusual DNA structures by the G-rich and C-rich strands (Fry and Loeb 1994; Mitas et al. 1995; Usdin and Woodford 1995; Yu et al. 1997). Thus for accurate repeat size determinations a set of FMR1 reference standards with different repeat numbers is necessary. These are typically PCR products generated from patient cell lines or from individual plasmid clones generated from these lines that have been amplified with the same primer pairs (Hayward et al. 2016). Because of the limited resolving power of capillary electrophoresis, the repeat numbers of larger FM alleles are only approximations.
Furthermore, despite improvements to the PCR protocols it is always important to be aware of the potential for allele dropout (ADO). ADO can be potentially be an issue in females with large FM alleles and in males who are mosaic since larger alleles can be less efficiently amplified than smaller ones. In addition, it is necessary to be aware of factors that may produce false positive or negative reactions. For example, 0.3% of X chromosomes in the Finnish population have a ~49 bp tandem duplication in the region immediately upstream of the repeat. A primer located upstream of this region will produce a PCR product consistent with an allele with an additional 16 repeats, while a primer within the duplicated region would produce an extra PCR product (Mononen et al. 2007). A number of deletions that include the region flanking the repeat have also been reported (reviewed in (Coffee et al. 2008; Hammond et al. 1997)). This can potentially result in the failure to amplify the allele if one of the primer binding sites has been lost. This may lead to the incorrect assumption that a woman with this sort of mutation is homozygous for a normal allele. Since a common deletion hotspot has been described 53–75 bp proximal to the repeat (de Graaff et al. 1995), it is preferable to use a forward primer that is upstream of this region, but even more distally located primers may fail to amplify larger deletions.
Assays for methylation status
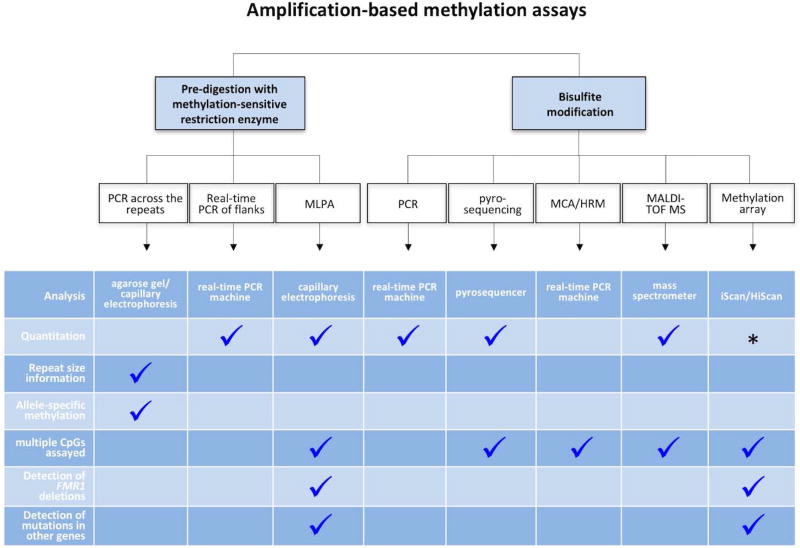
In addition to Southern blotting assays, a number of different assays have been used in recent years to monitor the methylation status of the FMR1 gene (Fig. 2). For example, a number of PCR based strategies have been described that incorporate a predigestion step with a methylation-sensitive enzyme that has one or more recognition sites within the amplicon being analyzed. Since the accuracy of these assays depends on the digestion being complete, an appropriate digestion control should always be included.
Fig. 2. Schematic representation of the various amplification-based assays for the extent of methylation in the FMR1-related disorders.
The asterisk indicates an assay that is partially quantitative in being able to distinguish between normal males and males with and without methylation mosaicism. However, it is not sensitive enough for use in females. Allele-specific methylation refers to the ability of the assay to identify which allele is methylated, knowledge that is relevant in females where skewed XCI may modify disease severity. The MLPA assay can detect abnormal FMR2 methylation and some contractions. In principle, the probe set could be modified to include those for other genetic disorders. The methylation array assay can detect abnormal methylation genome-wide and can thus also detect imprinting disorders.
Repeat PCR assays able to amplify large FM alleles have been adapted to provide information about the methylation status by including a predigestion step with HpaII (Chen et al. 2011; Grasso et al. 2014; Hayward et al. 2016).
Comparison of digested and mock-digested samples allows the determination of both the total number of repeats and the extent of allele methylation (Chen et al. 2011; Grasso et al. 2014; Hayward et al. 2016). The use of primers with different fluorescent labels allows both the digested and undigested samples to be run in the same capillary if CE is being used for detection (Chen et al. 2011). This not only reduces costs but eliminates any variability between samples that is related to electrophoresis. The ratio of the peak height in the digested sample to the peak height in the undigested sample is an indicator of the extent of methylation of any particular allele. A small synthetic DNA fragment containing the primer binding sites and at least one recognition site for the methylation sensitive enzyme can be added to the genomic DNA as a control for digestion (Grasso et al. 2014; Hayward et al. 2016).
These assays are informative for both repeat size and methylation and are thus particularly useful in cases of skewed XCI. However, because of differences in the efficiency with which different sized alleles are amplified and the presence of multiple allele sizes in some individuals, accurate quantitation of the overall extent of methylation can sometimes be difficult.
Assays that monitor the methylation state of one or more CpG residues in the flanking region are much easier to quantify. For example, quantitative real time (qRT)-PCR can be used to analyze a region flanking the repeat using DNA after pre-digestion with a methylation-sensitive enzyme such as HpaII (Hayward et al. 2016). Comparison of the yield of this PCR product with the yield from a mock digested sample allows the % methylation to be determined. This can be done relatively inexpensively using Sybr® dyes to quantify the PCR product. While these sorts of assays can only interrogate a limited number of CpG-residues, they can show a good correlation with the extent of methylation determined by assays that examine multiple CpG residues when CpGs that are consistently methylated in FM carriers are interrogated (Hayward et al. 2016).
A methylation-specific multiplex ligation-dependent probe amplification assay (MS-MLPA) has also been described for FXS (Nygren et al. 2008). In this assay multiple oligonucleotide probe pairs are hybridized to a denatured DNA sample, with one primer in each probe pair covering a recognition site for the methylation-sensitive restriction endonuclease HhaI. After simultaneous treatment with ligase-65, a heat-stable ligase, and HhaI, MLPA probes that are hybridized to unmethylated sites are digested and therefore not amplified in a subsequent PCR. The ratio of the peak heights produced with and without HhaI, allow the methylation status of the analyzed sample to be determined. The original protocol involved a mixture of 45 probes. Eleven of these probes were specific for FMR1, 5 of which are methylation-sensitive. The probe set also includes a Y chromosome-specific probe and methylation-specific probes from regions elsewhere in the genome that act as digestion controls. Importantly, since the FMR1-specific probes are distributed across the entire gene, including 7 probes in exon 1, this assay could potentially provide information about deletions that may be missed in other assays. Another advantage of this type of assay is that multiple targets can be tested simultaneously. For example, the original probe set also contained 14 probes specific for FMR2, another X-linked gene whose methylation is also associated with intellectual disability. However, it has been reported that the test may be less robust for FXS in some females because of contributions of the normal, inactive X to the peak ratios (Gatta et al. 2013). It is also unable to reliably quantify low levels of methylation (Gatta et al. 2013).
Bisulfite modification of genomic DNA can also be used to determine the methylation status. Bisulfite converts unmethylated cytosines into uracils. PCR then generates a template in which the uracils have been replaced with thymines. The net result are templates that have both an altered sequence and a lower G+C content. The PCR product can be cloned and sequenced thus giving information about the extent of amplification of individual CpG residues on individual alleles (Pietrobono et al. 2002; Stoger et al. 1997). However, this is a relatively labor intensive process that is not feasible for high throughput applications or a diagnostic setting. Assays more suitable for these settings include PCR-based assays with primers that distinguish between methylated and unmethylated alleles. One issue with such assays is the potential for differential amplification of methylated and unmethylated alleles because of differences in their G+C content. However, the Q-MSP assay, an assay that uses primers with mixed bases that amplify both the methylated and unmethylated alleles (Coffee et al. 2009), followed by detection of these alleles using specific Taqman™ probes and real-time PCR, has been successfully used to screen the dried blood spots of 36,124 newborn males. Impressively, this was done using initial pools consisting of samples from 44 different blood spots.
The average methylation of individual CpGs can also be assessed using pyrosequencing, a reiterative sequencing-by-synthesis approach that measures the real-time incorporation of templated bases, allowing the extent of methylation to be calculated from the ratio of C (formerly methylated cytosine) to T (formerly unmethylated cytosine) at each CpG residue present in the original sequence. Thus, unlike the sequencing of cloned bisulfite-treated DNA, pyrosequencing provides information about the average methylation per site rather than the methylation status of individual DNA molecules. However, this assay has been shown to be linear over a wide range of methylation levels and is sensitive enough to detect as little as 5% difference in methylation between samples (Dejeux et al. 2009; Zhou et al. 2016).
Other assays using bisulfite modification followed by PCR and melt curve analysis (MCA)/high resolution melting (HRM) have also been described (Dahl et al. 2007; Elias et al. 2011; Inaba et al. 2014; Rajan-Babu et al. 2015). These assays exploit the fact that bisulfite conversion of unmethylated alleles results in templates that have a lower G+C content than methylated alleles, and thus a lower melting temperature. Bisulfite-modified and PCR amplified products can also be processed by in vitro transcription, followed by base-specific cleavage and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) (Godler et al. 2010). This assay is based on the fact that every unmethylated CpG in the original DNA results in an RNA fragment that is 16Da lighter than its methylated equivalent due to the replacement of a G with A in the transcript.
More recently, an Illumina DNA methylation microarray assay has been used to identify FXS males (Schenkel et al. 2016) using probes that query the methylation status of 10 CpGs within the FMR1 CpG island. This approach relies on isothermal amplification of bisulfite modified DNA and hybridization to BeadChips containing locus- and methylation-specific 50mers. After binding to the template DNA, a single-base extension beyond the queried C residue with different fluorescently labeled nucleotides, allows the ratio of extension from an unmethylated CpG residue to the corresponding methylated residue to be determined by imaging of the BeadChip using a iScan or HiScan laser scanner. While this assay was not able to distinguish methylation differences between normal females and females with a FM allele, the fact that the DNA methylation chip contains probes for multiple genomic regions, makes it possible to simultaneously test for abnormal methylation characteristic of other disorders including Prader-Willi Syndrome and Sotos Syndrome, 2 disorders frequently considered in the differential diagnosis of FXS (Schenkel et al. 2016). The downside to many of these assays is that they require significant sample processing and/or specialized equipment and may thus not be viable for high throughput screens.
It is also important to remember that methylated FMR1 alleles are seen not only in males with FXS but also in those with Klinefelter syndrome (KS; XXY), whose prevalence in 1:500 males makes it significantly more common than FXS. Thus for all assays that monitor the methylation status of the flanking sequence of the FMR1 gene rather than the repeat itself, an additional assay capable of revealing the presence of 2 X chromosomes in males may be warranted. This would be particularly important in population screenings or other scenarios where the existence of a PM or FM allele has not been directly tested (Coffee et al. 2009).
Assays for AGG-interruptions
PCR-based repeat assays can also be modified to detect AGG-interruptions. One such assay takes advantage of the fact that the restriction enzyme EciI has a recognition site, GGCGGA (Hayward et al. 2016; Tassone et al. 2007b). As such this enzyme will cut repeat tracts containing AGG interruptions. Post-PCR digestion of amplicons with this enzyme produces a series of bands on agarose gels or peaks on CE corresponding to cleavage in the vicinity of each AGG. This produces a profile that is relatively simple to interpret and produces relatively few samples for which a definitive answer cannot be obtained. However, this assay has some drawbacks including the fact that the EciI enzyme is not very stable (https://www.neb.com/products/r0590-ecii). This can result in incomplete digests that may be difficult to interpret.
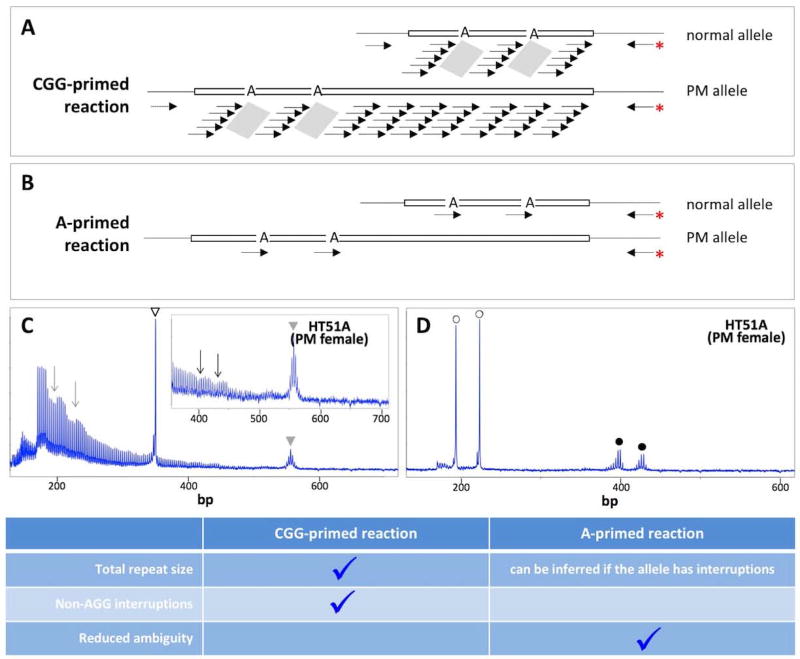
A second class of PCR-based assays for the detection of AGG interruptions involves a triplet-primed (TP) PCR reaction. First generation TP-assays make use of a hybrid PCR primer that had a number of CCG (Tassone et al. 2008) or CGG-repeats (Chen et al. 2010) at its 3’ end. The 5’ end of the primer consisted of bases that were not homologous to the FMR1 gene to increase the optimal annealing temperature of the primer. This primer is used together with FMR1-specific primers from both upstream and downstream of the repeat. Amplification with the hybrid primer and the appropriate gene-specific primer results in a PCR profile that in the case of uninterrupted alleles resembles a slope declining uniformly from a maximum at the beginning of the repeat tract to a minimum at the end (Chen et al. 2010). In the case of interrupted repeats, the slope is punctuated by sharp dips. These dips correspond to the region of the repeat tract containing the AGG interruptions that do not bind the triplet-containing primer, as illustrated in Fig. 3A. The gene-specific primers also generate a PCR product that includes all the repeats. Thus these assays can be used to determine both repeat size and interruption status simultaneously. In principle, these assays would also be able to detect those rare non-AGG interruptions that are occasionally seen in human populations (Cecconi et al. 2008; Tabolacci et al. 2008b; Tarleton et al. 2002), since they too prevent binding of the repeat-containing primer (Hayward and Usdin, unpublished observations).
Fig. 3. Triplet-primed assays for the detection of AGG interruptions.
A–B) Potential primer binding sites in the first generation (CGG-primed) and second generation (A-primed) reactions. The grey parallelograms in panel A illustrate the places on the template that the primer will not bind. The red asterisk indicates the fluorescently labeled primer. C) The output of the CGG-primed reaction on DNA from a female PM carrier. The open arrowhead in panel C marks the PCR product resulting from the flanking PCR primers on the normal allele, while the grey arrowhead indicates the PCR product resulting from the use of the flanking primers on the PM allele. The arrows indicate the PCR products resulting from the use of the triplet containing primer and the fluorescently labeled reverse primer. The inset is a magnification of the 400–700 bp region of the electrophoretogram. D) The output of the A-primed reaction on the same PM carrier shown in panel C. The open circles indicate the PCR products corresponding to priming at each of the 2 AGG interruptions in the normal allele while the filled circles indicate the PCR products corresponding to priming at each of the 2 AGG interruptions in the PMl allele. This figure was adapted from our previously published work for which we retain the copyright (Hayward and Usdin In Press).
However, both these TP-assays have significant disadvantages. As can be seen in Fig. 3C, when the repeat-containing primer contains CGG-repeats, the dips in the PCR profile can be difficult to discern on larger alleles since these alleles amplify relatively poorly and the dips may be obscured because of PCR artifacts that introduce heterogeneity into the PCR profile. In contrast, reactions using the CCG-repeat containing primer allow the dips to be seen more readily since the PCR products are smaller and less prone to PCR artifacts. This assay is useful for males since they have just a single FMR1 allele. However, in females interpretation is complicated by the presence of 2 FMR1 alleles because the dips resulting from interruptions on either allele would co-localize on the electrophoretogram. This makes it difficult to know whether interruptions are on the expanded allele or the normal one. Thus, only where both alleles have the same interruption pattern, can an unambiguous answer can be deduced for the expanded allele.
To obviate these problems, we have recently developed 2nd generation TP-assays, ones that involve a hybrid primer containing either GGA or CCT at its 3’ end and a second primer from the flanking region (Hayward and Usdin In Press). Rather than priming wherever the interruption isn’t, as in the first generation assays, the triplet-containing primer only primes at AGG interruptions (Fig. 3B). Thus, these assays show a single peak for each interruption making this assay much easier to interpret and much less likely to produce an ambiguous outcome (Fig. 3D). The GGA-primed assay is best done in combination with a specially designed internal control for successful amplification of long alleles, thus allowing false negatives to be easily identified. The downside to these assays is that since they make use of only one flanking primer, they do not give information about the overall length of the repeat tract. However, for alleles with interruptions the most likely size of the allele can be inferred from the size of the largest AGG-primed product.
Single-molecule sequencing assays
Single-molecule, real-time (SMRT) sequencing is an emerging technology based on the real-time detection of a base-specific fluorophore that is released as the base is incorporated into the nascent DNA strand by a DNA polymerase/template complex located at the bottom of a zero-mode waveguide (ZMW) nanowell. This approach does not suffer from the diminishing resolution inherent in Sanger sequencing and allows read-lengths of up to 15 kb. SMRT sequencing has been shown to work on PCR products generated from an allele with ~750 repeats (Loomis et al. 2013) or containing AGG interruptions (Ardui et al. 2017; Loomis et al. 2013). However, these assays require extensive sample preparation and significant data processing and thus SMRT sequencing is currently not viable for routine diagnostic applications. However, since modified bases alter the rate of base incorporation during sequencing thus affecting the inter-pulse duration (IPD), technological improvements that permit direct sequencing of genomic DNA may make it possible to provide nucleotide level information about the repeat tract itself, including DNA modifications like methylation and hydroxymethylation. Thus, this technology may be useful for addressing important research questions.
FMRP protein assays
The severity of FXS symptoms is inversely related to FMRP levels and is thus an important predictor of clinical involvement in FXS. However, protein-based assays are not currently used for routine diagnostic purposes. This is due in part because of their limited use in females and in the identification of premutation carriers who have little, if any, reduction in FMRP. However, such assays may nonetheless be useful in a variety of other situations and a recent proof of principle demonstration suggests that some of these assays may be suitable for future new born screening programs (LaFauci et al. 2013).
Cell-based assays
Early assays for FMRP in cells included antibody staining in hair follicles, blood smears and chorionic villi (de Graaff et al. 1996; Lambiris et al. 1999; Rife et al. 2004; Smeets et al. 1995; Willemsen et al. 1999; Willemsen et al. 1997; Willemsen et al. 2003). However, these assays are difficult to quantify and results were often reported simply as the % of cells expressing FMRP (Garcia Arocena et al. 2000). This reduced the diagnostic power in FM females and mosaic males and made it more challenging to detect any FMRP deficit in PM carriers.
However, recent improvements in automated digital imaging and image processing have allowed a more quantitative assessment of protein levels in cells. Such a high-content screening assay for FMRP has been used to carry out a phenotypic screen of a library of 50,000 compounds (Kaufmann et al. 2015). Multiparametric data analysis coupled to machine learning made it possible to identify several new compounds that weakly induced FMRP production in FM cells and to examine the heterogeneous response of cells in the population to these compounds (Kaufmann et al. 2015). In principle, such assays could be applied to quantifying FMRP directly from patient cells.
Cell extract-based assays
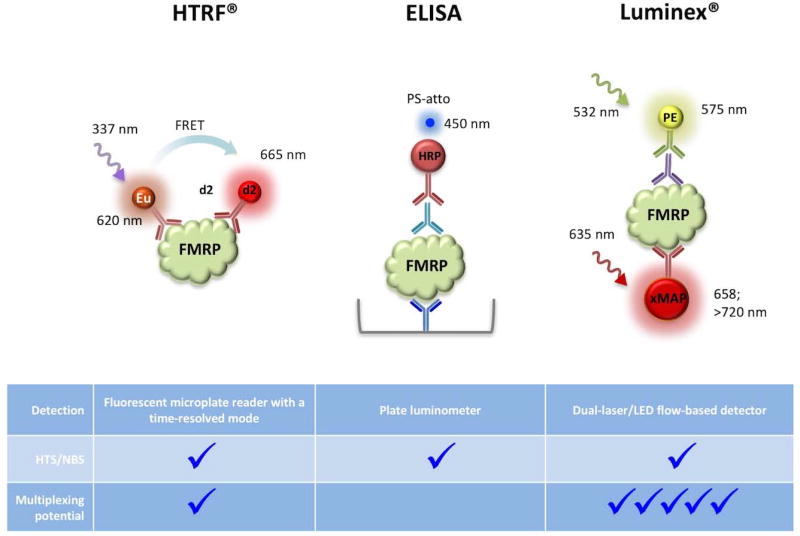
Early assays on cell lysates included western blotting (Devys et al. 1993; Lessard et al. 2012) and slot blot assays (Kenneson et al. 2001) using a single anti-FMRP antibody. For example, one such western blot assay using cell extracts made from 5×106 platelets was able to readily distinguish both males and females with FXS from unaffected individuals. (Lessard et al. 2012). However, more recent assays have been described that are more amenable to population screens and high throughput drug screening since they can be readily miniaturized and many steps can be automated. They also rely on the binding of 2 FMRP-specific antibodies, potentially making them more specific. These assays include those based on Homogeneous Time-Resolved Fluorescence (HTRF®) (Kumari et al. 2015; Schutzius et al. 2013), a chemiluminescent enzyme-linked immunosorbent based assay (ELISA) (Iwahashi et al. 2009) and qFMRP, a Luminex®-based assay (LaFauci et al. 2013). The principles of these assays are illustrated in Fig. 4.
Fig. 4. Ilustration of the principle of recently described dual-FMRP antibody assays for FMRP.
The colors of the fluorescent/luminscent labels indicates the part of the electromagnetic spectrum in which the label emits light. The HTRF®-based FMRP assays involve a FMRP antibody coupled to either Europium cryptate (Kumari et al. 2015) as illustrated here, or Lumi4-Tb cryptate (Schutzius et al. 2013), and a second antibody tagged with an energy acceptor such as d2. Irradiation of the sample at ~337 nm excites the cryptate moiety and if the two antibodies bind to epitopes on FMRP that are within ~70–90 Å, FRET occurs. This results in the excitation of the second fluorophore and emission at 665 nm. The ELISA assay described for FMRP uses a chicken anti-FMRP antibody shown in blue to coat the well of the microplate and capture FMRP. The second FMRP antibody, shown in aqua, is a mouse monoclonal antibody used to detect FMRP via its interaction with a third antibody, shown in red, an anti-mouse IgG antibody conjugated to horseradish peroxidase (HRP). After addition of the HRP substrate, PS-atto, the samples are detected at 450 nm. The Luminex®-based assay for FMRP uses a mouse monoclonal FMRP antibody, shown in red, that is coupled to xMAP beads or microspheres to capture the FMRP. The second FMRP antibody, shown in purple, is a rabbit polyclonal Ab. Detection is carried out at 575 nm using a goat-anti-rabbit phycoerythrin-conjugated IgG antibody, shown in green. Note that while both the HTRF® and the Luminex®-based assays can in principle be multiplexed, the multiplexing potential of the Luminex®-based assay is much higher than that of HTRF® assays as denoted by the larger number of checkmarks.
HTRF® combines standard fluorescence resonance energy transfer (FRET) technology with antibodies labeled with long-lived fluorophores that emit in the near infra-red (IR). This assay combines the benefits of a lower level of shortlived background fluorescence resulting from the time-resolved measurement, with the reduced likelihood of autofluorescence by the medium in the near IR range. In principle, the requirement that the two antibodies be within the distance required for FRET to occur increases the specificity of the assay. In addition, because the assay does not involve detection of either antibody directly, the antibodies are not limited to those derived from different animal species as in the other 2 antibody assays. HTRF® assays were readily able to distinguish between carriers of normal and FM alleles using small numbers of cells (Kumari et al. 2015; Schutzius et al. 2013), to detect low levels of FMRP produced in FM cells after treatment with 1 µM of the DNA methyltransferase inhibitor 5-aza-2’-deoxycytidine (AZA) (Kumari et al. 2015) and to identify compounds able to modestly reactivate the FMR1 gene in a HTS assay (Kumari et al. 2015). However, with the antibody pairs used in both published HTRF assays, the ratio of signal to noise was relatively low making it difficult to detect low levels of FMRP. Antibodies with higher affinity and avidity would likely address this problem.
The FMRP ELISA assay makes use of a chicken anti-FMRP antibody to coat the wells of a 96 well plate to capture FMRP from a sample solution and a mouse anti-FMRP antibody as the second FMRP antibody (Iwahashi et al. 2009). An anti-mouse IgG antibody coupled to horseradish peroxidase is used to detect the second FMRP antibody. PS-atto is used as the chemiluminescent substrate having the advantages of a broad dynamic range, low picogram to femtogram sensitivity and sustained luminescence. This assay allowed a 12% decline in the amount of FMRP to be seen in the peripheral lymphocytes of 23 PM males (Iwahashi et al. 2009).
In the Luminex®-based qFMRP assay, the sample is mixed with xMAP® Microplex microspheres that are pre-coated with a mouse monoclonal antibody to FMRP (LaFauci et al. 2013). A rabbit polyclonal antibody to FMRP is added to create an antibody-antigen sandwich. A goat-anti-rabbit phycoerythrin (PE)-conjugated antibody is then added to the mixture and the amount of PE measured by irradiation with a green laser or LED at 532 nm. This assay has some advantages over ELISA including requiring much smaller amounts of both reagents and sample. The good performance characteristics of this assay are likely due, at least in part, to the use of antibodies with high affinity and avidity that had been developed by LaFauci et al. (LaFauci et al. 2013).
The qFMRP assay is the most sensitive FMRP assay described thus far. It has a limit of detection of 0.5 pmol/L and a signal to noise ratio of 7.8-fold and 42-fold for mosaic and non-mosaic FM males respectively (LaFauci et al. 2016). The assay correctly identified the 17 FM dried blood spots (DBS) in a sample of 103 male DBS (LaFauci et al. 2013). This accomplishment is particularly impressive since some of the DBS had been stored for up to 4 years. This assay has also been used to screen 2000 randomly-selected DBS from a newborn screening program (Adayev et al. 2014). Of the 14 samples that had FMRP levels lower than 2 standard deviations below the mean, 13 had normal alleles, while the remaining sample was from a female carrying a large PM allele that was on the active X in 90% of her leukocytes.
While this has not yet been explored in the context of FMRP detection, Luminex® technology permits multiplexing because antibodies to different proteins can be coupled to xMAP® microspheres that are dyed with different combinations of red and infrared fluorophores. This allows up to 500 different proteins to be detected simultaneously in the same well. As the sample passes through the dual Laser/LEDs of the flow-based detection instrument used in Luminex® assays, an initial irradiation at 635 nm excites the dye on the microsphere resulting in a characteristic emission at 658 nm and above depending on the specific combinations of color-coded beads. This classifies the microsphere thus identifying the protein being detected. A second irradiation with the 532 nm laser/LED then allows the magnitude of the PE-derived signal associated with that microsphere, and thus the amount of the protein that is present in the mixture, to be determined. Thus, the qFMRP assay could perhaps be expanded to include other cellular proteins for use as controls for differences in sample loading and degradation. It may also be possible to use this sort of assay to screen simultaneously for multiple disorders that result from abnormal protein levels or from the production of abnormal proteins for which there are antibodies available.
Conclusions
There are now a variety of assays that can be used to accurately determine the repeat size, the AGG interruption pattern, the methylation status of the FMR1 repeat tract and FMRP levels. This has increased the accuracy and turnaround time for diagnostic testing and facilitated newborn screening pilot studies. It also provides useful, and in some cases quite affordable, tools for basic research and drug discovery. One research area in which this should be particularly valuable is in tissue cultures studies where the variability in repeat size and thus potentially methylation status is a potentially confounding factor, that up until now has not always been evaluated properly. The ultimate choice of which assay or assays to use in different circumstances varies, with some assays offering advantages in terms of scalability and cost, whilst other more specialized techniques may provide advantages that are more valuable in small scale patient studies.
While current assays allow us to examine the FMR1 repeat and the levels of FMRP much more robustly than previously possible, it is important to remember that there are a small fraction of FXS cases that are not associated with a repeat expansion. For those cases presenting with strong FXS-like symptoms but no evidence of repeat expansion or an FMRP deficit, other sorts of assays like HRM of the FMR1 coding sequence (Handt et al. 2014), array-based sequencing (Collins et al. 2010b), massively parallel sequencing (Collins et al. 2010a) or sequencing of target custom capture libraries (Quartier et al. 2017) may be required.
Acknowledgments
Grant Sponsor: Intramural program of the NIDDK, NIH (DK057808)
Footnotes
Conflict of Interest: The authors have no conflict of interest
References
- Adayev T, LaFauci G, Dobkin C, Caggana M, Wiley V, Field M, Wotton T, Kascsak R, Nolin SL, Glicksman A, Hosmer N, Brown WT. Fragile X protein in newborn dried blood spots. BMC Med Genet. 2014;15:119. doi: 10.1186/s12881-014-0119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardui S, Race V, Zablotskaya A, Hestand MS, Van Esch H, Devriendt K, Matthijs G, Vermeesch JR. Detecting AGG Interruptions in Male and Female FMR1 Premutation Carriers by Single-Molecule Sequencing. Hum Mutat. 2017;38:324–331. doi: 10.1002/humu.23150. [DOI] [PubMed] [Google Scholar]
- Basuta K, Narcisa V, Chavez A, Kumar M, Gane L, Hagerman R, Tassone F. Clinical phenotypes of a juvenile sibling pair carrying the fragile X premutation. Am J Med Genet A. 2011;155A:519–25. doi: 10.1002/ajmg.a.33446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basuta K, Schneider A, Gane L, Polussa J, Woodruff B, Pretto D, Hagerman R, Tassone F. High functioning male with fragile X syndrome and fragile X-associated tremor/ataxia syndrome. Am J Med Genet A. 2015;167A:2154–61. doi: 10.1002/ajmg.a.37125. [DOI] [PubMed] [Google Scholar]
- Brown WT, Nolin S, Houck G, Jr, Ding X, Glicksman A, Li SY, Stark-Houck S, Brophy P, Duncan C, Dobkin C, Jenkins E. Prenatal diagnosis and carrier screening for fragile X by PCR. Am J Med Genet. 1996;64:191–5. doi: 10.1002/(SICI)1096-8628(19960712)64:1<191::AID-AJMG34>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Pecho-Vrieseling E, Thiemeyer A, Klein J, Fruh I, Doll T, Manneville C, Fuchs S, Iazeolla M, Beibel M, Roma G, Naumann U, Kelley N, Oakeley EJ, Mueller M, Gomez-Mancilla B, Buhler M, Tabolacci E, Chiurazzi P, Neri G, Bouwmeester T, Di Giorgio FP, Fodor BD. CGG Repeat-Induced FMR1 Silencing Depends on the Expansion Size in Human iPSCs and Neurons Carrying Unmethylated Full Mutations. Stem Cell Reports. 2016;7:1059–1071. doi: 10.1016/j.stemcr.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi M, Forzano F, Rinaldi R, Cappellacci S, Grammatico P, Faravelli F, Dagna Bricarelli F, Di Maria E, Grasso M. A single nucleotide variant in the FMR1 CGG repeat results in a "Pseudodeletion" and is not associated with the fragile X syndrome phenotype. J Mol Diagn. 2008;10:272–5. doi: 10.2353/jmoldx.2008.070163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hadd A, Sah S, Filipovic-Sadic S, Krosting J, Sekinger E, Pan R, Hagerman PJ, Stenzel TT, Tassone F, Latham GJ. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J Mol Diagn. 2010;12:589–600. doi: 10.2353/jmoldx.2010.090227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hadd A, Sah S, Houghton JF, Filipovic-Sadic S, Zhang W, Hagerman PJ, Tassone F, Latham GJ. High-resolution methylation polymerase chain reaction for fragile X analysis: evidence for novel FMR1 methylation patterns undetected in Southern blot analyses. Genet Med. 2011;13:528–538. doi: 10.1097/GIM.0b013e31820a780f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SS, Eichler EE, Nelson DL, Hughes MR. Robust amplification and ethidium-visible detection of the fragile X syndrome CGG repeat using Pfu polymerase. Am J Med Genet. 1994;51:522–6. doi: 10.1002/ajmg.1320510447. [DOI] [PubMed] [Google Scholar]
- Coffee B, Ikeda M, Budimirovic DB, Hjelm LN, Kaufmann WE, Warren ST. Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis: a case report and review of the literature. Am J Med Genet A. 2008;146A:1358–67. doi: 10.1002/ajmg.a.32261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Keith K, Albizua I, Malone T, Mowrey J, Sherman SL, Warren ST. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet. 2009;85:503–14. doi: 10.1016/j.ajhg.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am J Hum Genet. 2002;71:923–32. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- Collins SC, Bray SM, Suhl JA, Cutler DJ, Coffee B, Zwick ME, Warren ST. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet A. 2010a;152A:2512–20. doi: 10.1002/ajmg.a.33626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SC, Coffee B, Benke PJ, Berry-Kravis E, Gilbert F, Oostra B, Halley D, Zwick ME, Cutler DJ, Warren ST. Array-based FMR1 sequencing and deletion analysis in patients with a fragile X syndrome-like phenotype. PLoS One. 2010b;5:e9476. doi: 10.1371/journal.pone.0009476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl C, Gronskov K, Larsen LA, Guldberg P, Brondum-Nielsen K. A homogeneous assay for analysis of FMR1 promoter methylation in patients with fragile X syndrome. Clin Chem. 2007;53:790–3. doi: 10.1373/clinchem.2006.080762. [DOI] [PubMed] [Google Scholar]
- de Graaff E, de Vries BB, Willemsen R, van Hemel JO, Mohkamsing S, Oostra BA, van den Ouweland AM. The fragile X phenotype in a mosaic male with a deletion showing expression of the FMR1 protein in 28% of the cells. Am J Med Genet. 1996;64:302–8. doi: 10.1002/(SICI)1096-8628(19960809)64:2<302::AID-AJMG14>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- de Graaff E, Rouillard P, Willems PJ, Smits AP, Rousseau F, Oostra BA. Hotspot for deletions in the CGG repeat region of FMR1 in fragile X patients. Hum Mol Genet. 1995;4:45–9. doi: 10.1093/hmg/4.1.45. [DOI] [PubMed] [Google Scholar]
- Dejeux E, El abdalaoui H, Gut IG, Tost J. Identification and quantification of differentially methylated loci by the pyrosequencing technology. Methods Mol Biol. 2009;507:189–205. doi: 10.1007/978-1-59745-522-0_15. [DOI] [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–40. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Holden JJ, Popovich BW, Reiss AL, Snow K, Thibodeau SN, Richards CS, Ward PA, Nelson DL. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Elias MH, Ankathil R, Salmi AR, Sudhikaran W, Limprasert P, Zilfalil BA. A new method for FMR1 gene methylation screening by multiplex methylation-specific real-time polymerase chain reaction. Genet Test Mol Biomarkers. 2011;15:387–93. doi: 10.1089/gtmb.2010.0191. [DOI] [PubMed] [Google Scholar]
- Feng Y, Zhang F, Lokey LK, Chastain JL, Lakkis L, Eberhart D, Warren ST. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–4. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- Filipovic-Sadic S, Sah S, Chen L, Krosting J, Sekinger E, Zhang W, Hagerman PJ, Stenzel TT, Hadd AG, Latham GJ, Tassone F. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin Chem. 2010;56:399–408. doi: 10.1373/clinchem.2009.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc Natl Acad Sci U S A. 1994;91:4950–4. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Arocena D, de Diego Y, Oostra BA, Willemsen R, Mirta Rodriguez M. A fragile X case with an amplification/deletion mosaic pattern. Hum Genet. 2000;106:366–9. doi: 10.1007/s004390051052. [DOI] [PubMed] [Google Scholar]
- Gatta V, Gennaro E, Franchi S, Cecconi M, Antonucci I, Tommasi M, Palka G, Coviello D, Stuppia L, Grasso M. MS-MLPA analysis for FMR1 gene: evaluation in a routine diagnostic setting. BMC Med Genet. 2013;14:79. doi: 10.1186/1471-2350-14-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godler DE, Slater HR, Amor D, Loesch DZ. Methylation analysis of fragile X-related epigenetic elements may provide a suitable newborn screening test for fragile X syndrome. Genet Med. 2010;12:595. doi: 10.1097/GIM.0b013e3181f07088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Radu D, Balanko A, Chiang CS. Diagnosis of Fragile X syndrome by Southern blot hybridization using a chemiluminescent probe: a laboratory protocol. Mol Diagn. 2000;5:169–78. doi: 10.1054/modi.2000.9404. [DOI] [PubMed] [Google Scholar]
- Grasso M, Boon EM, Filipovic-Sadic S, van Bunderen PA, Gennaro E, Cao R, Latham GJ, Hadd AG, Coviello DA. A novel methylation PCR that offers standardized determination of FMR1 methylation and CGG repeat length without southern blot analysis. J Mol Diagn. 2014;16:23–31. doi: 10.1016/j.jmoldx.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman P. Fragile X-associated tremor/ataxia syndrome - features, mechanisms and management. Nat Rev Neurol. 2016;12:403–12. doi: 10.1038/nrneurol.2016.82. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Hull CE, Safanda JF, Carpenter I, Staley LW, O'Connor RA, Seydel C, Mazzocco MM, Snow K, Thibodeau SN, et al. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. Am J Med Genet. 1994;51:298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- Hammond LS, Macias MM, Tarleton JC, Shashidhar Pai G. Fragile X syndrome and deletions in FMR1: new case and review of the literature. Am J Med Genet. 1997;72:430–4. [PubMed] [Google Scholar]
- Handt M, Epplen A, Hoffjan S, Mese K, Epplen JT, Dekomien G. Point mutation frequency in the FMR1 gene as revealed by fragile X syndrome screening. Mol Cell Probes. 2014;28:279–83. doi: 10.1016/j.mcp.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Hayward BE, Usdin K. Improved assays for AGG interruptions in Fragile X premutation carriers. J Mol Diag. doi: 10.1016/j.jmoldx.2017.06.008. (In Press) doi: http://dx.doi.org/10.1016/j.jmoldx.2017.06.008. [DOI] [PMC free article] [PubMed]
- Hayward BE, Zhou Y, Kumari D, Usdin K. A Set of Assays for the Comprehensive Analysis of FMR1 Alleles in the Fragile X-Related Disorders. J Mol Diagn. 2016;18:762–74. doi: 10.1016/j.jmoldx.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba Y, Schwartz CE, Bui QM, Li X, Skinner C, Field M, Wotton T, Hagerman RJ, Francis D, Amor DJ, Hopper JL, Loesch DZ, Bretherton L, Slater HR, Godler DE. Early detection of fragile X syndrome: applications of a novel approach for improved quantitative methylation analysis in venous blood and newborn blood spots. Clin Chem. 2014;60:963–73. doi: 10.1373/clinchem.2013.217331. [DOI] [PubMed] [Google Scholar]
- Iwahashi C, Tassone F, Hagerman RJ, Yasui D, Parrott G, Nguyen D, Mayeur G, Hagerman PJ. A quantitative ELISA assay for the fragile x mental retardation 1 protein. J Mol Diagn. 2009;11:281–9. doi: 10.2353/jmoldx.2009.080118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins EC, Brown WT, Krawczun MS, Duncan CJ, Lele KP, Cantu ES, Schonberg S, Golbus MS, Sekhon GS, Stark S, et al. Recent experience in prenatal fra(X) detection. Am J Med Genet. 1988;30:329–36. doi: 10.1002/ajmg.1320300133. [DOI] [PubMed] [Google Scholar]
- Kaufmann M, Schuffenhauer A, Fruh I, Klein J, Thiemeyer A, Rigo P, Gomez-Mancilla B, Heidinger-Millot V, Bouwmeester T, Schopfer U, Mueller M, Fodor BD, Cobos-Correa A. High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen. 2015;20:1101–11. doi: 10.1177/1087057115588287. [DOI] [PubMed] [Google Scholar]
- Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet. 2001;10:1449–54. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- Kumari D, Swaroop M, Southall N, Huang W, Zheng W, Usdin K. High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated From Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. Stem Cells Transl Med. 2015;4:800–8. doi: 10.5966/sctm.2014-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari D, Usdin K. The distribution of repressive histone modifications on silenced FMR1 alleles provides clues to the mechanism of gene silencing in fragile X syndrome. Hum Mol Genet. 2010;19:4634–42. doi: 10.1093/hmg/ddq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFauci G, Adayev T, Kascsak R, Brown WT. Detection and Quantification of the Fragile X Mental Retardation Protein 1 (FMRP) Genes (Basel) 2016:7. doi: 10.3390/genes7120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFauci G, Adayev T, Kascsak R, Kascsak R, Nolin S, Mehta P, Brown WT, Dobkin C. Fragile X screening by quantification of FMRP in dried blood spots by a Luminex immunoassay. J Mol Diagn. 2013;15:508–17. doi: 10.1016/j.jmoldx.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Lambiris N, Peters H, Bollmann R, Leschik G, Leisti J, Salonen R, Cobet G, Oostra BA, Willemsen R. Rapid FMR1-protein analysis of fetal blood: an enhancement of prenatal diagnostics. Hum Genet. 1999;105:258–60. doi: 10.1007/s004390051098. [DOI] [PubMed] [Google Scholar]
- Latham GJ, Coppinger J, Hadd AG, Nolin SL. The role of AGG interruptions in fragile X repeat expansions: a twenty-year perspective. Front Genet. 2014;5:244. doi: 10.3389/fgene.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard M, Chouiali A, Drouin R, Sebire G, Corbin F. Quantitative measurement of FMRP in blood platelets as a new screening test for fragile X syndrome. Clin Genet. 2012;82:472–7. doi: 10.1111/j.1399-0004.2011.01798.x. [DOI] [PubMed] [Google Scholar]
- Liang S, Bass HN, Gao H, Astbury C, Jamehdor MR, Qu Y. A pseudo-full mutation identified in fragile X assay reveals a novel base change abolishing an EcoRI restriction site. J Mol Diagn. 2008;10:469–74. doi: 10.2353/jmoldx.2008.080059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesch D, Hagerman R. Unstable mutations in the FMR1 gene and the phenotypes. Adv Exp Med Biol. 2012;769:78–114. doi: 10.1007/978-1-4614-5434-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesch DZ, Sherwell S, Kinsella G, Tassone F, Taylor A, Amor D, Sung S, Evans A. Fragile X-associated tremor/ataxia phenotype in a male carrier of unmethylated full mutation in the FMR1 gene. Clin Genet. 2012;82:88–92. doi: 10.1111/j.1399-0004.2011.01675.x. [DOI] [PubMed] [Google Scholar]
- Loomis EW, Eid JS, Peluso P, Yin J, Hickey L, Rank D, McCalmon S, Hagerman RJ, Tassone F, Hagerman PJ. Sequencing the unsequenceable: expanded CGG-repeat alleles of the fragile X gene. Genome Res. 2013;23:121–8. doi: 10.1101/gr.141705.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Rosero CA, Hagerman RJ. Fragile X spectrum disorders. Intractable Rare Dis Res. 2014;3:134–46. doi: 10.5582/irdr.2014.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer H, de Graaff E, Merckx DM, Jongbloed RJ, de Die-Smulders CE, Engelen JJ, Fryns JP, Curfs PM, Oostra BA. A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum Mol Genet. 1994;3:615–20. doi: 10.1093/hmg/3.4.615. [DOI] [PubMed] [Google Scholar]
- Mitas M, Yu A, Dill J, Haworth IS. The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn. Ganti base pairs. Biochemistry. 1995;34:12803–11. doi: 10.1021/bi00039a041. [DOI] [PubMed] [Google Scholar]
- Monaghan KG, Lyon E, Spector EB, erican College of Medical G, Genomics ACMG Standards and Guidelines for fragile X testing: a revision to the disease-specific supplements to the Standards and Guidelines for Clinical Genetics Laboratories of the American College of Medical Genetics and Genomics. Genet Med. 2013;15:575–86. doi: 10.1038/gim.2013.61. [DOI] [PubMed] [Google Scholar]
- Mononen T, von Koskull H, Airaksinen RL, Juvonen V. A novel duplication in the FMR1 gene: implications for molecular analysis in fragile X syndrome and repeat instability. Clin Genet. 2007;72:528–31. doi: 10.1111/j.1399-0004.2007.00903.x. [DOI] [PubMed] [Google Scholar]
- Murray A. Premature ovarian failure and the FMR1 gene. Semin Reprod Med. 2000;18:59–66. doi: 10.1055/s-2000-13476. [DOI] [PubMed] [Google Scholar]
- Nolin SL, Ding XH, Houck GE, Brown WT, Dobkin C. Fragile X full mutation alleles composed of few alleles: implications for CGG repeat expansion. Am J Med Genet A. 2008;146A:60–5. doi: 10.1002/ajmg.a.32087. [DOI] [PubMed] [Google Scholar]
- Nolin SL, Glicksman A, Ersalesi N, Dobkin C, Brown WT, Cao R, Blatt E, Sah S, Latham GJ, Hadd AG. Fragile X full mutation expansions are inhibited by one or more AGG interruptions in premutation carriers. Genet Med. 2015;17:358–64. doi: 10.1038/gim.2014.106. [DOI] [PubMed] [Google Scholar]
- Nolin SL, Glicksman A, Houck GE, Jr, Brown WT, Dobkin CS. Mosaicism in fragile X affected males. Am J Med Genet. 1994;51:509–12. doi: 10.1002/ajmg.1320510444. [DOI] [PubMed] [Google Scholar]
- Nolin SL, Sah S, Glicksman A, Sherman SL, Allen E, Berry-Kravis E, Tassone F, Yrigollen C, Cronister A, Jodah M, Ersalesi N, Dobkin C, Brown WT, Shroff R, Latham GJ, Hadd AG. Fragile X AGG analysis provides new risk predictions for 45–69 repeat alleles. Am J Med Genet A. 2013;161A:771–8. doi: 10.1002/ajmg.a.35833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren AO, Lens SI, Carvalho R. Methylation-specific multiplex ligation-dependent probe amplification enables a rapid and reliable distinction between male FMR1 premutation and full-mutation alleles. J Mol Diagn. 2008;10:496–501. doi: 10.2353/jmoldx.2008.080053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–102. doi: 10.1126/science.252.5009.1097. 252/5009/1097 [pii] 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Oh SY, He F, Krans A, Frazer M, Taylor JP, Paulson HL, Todd PK. RAN translation at CGG repeats induces ubiquitin proteasome system impairment in models of fragile X-associated tremor ataxia syndrome. Hum Mol Genet. 2015;24:4317–26. doi: 10.1093/hmg/ddv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orpana AK, Ho TH, Stenman J. Multiple heat pulses during PCR extension enabling amplification of GC-rich sequences and reducing amplification bias. Anal Chem. 2012;84:2081–7. doi: 10.1021/ac300040j. [DOI] [PubMed] [Google Scholar]
- Pergolizzi RG, Erster SH, Goonewardena P, Brown WT. Detection of full fragile X mutation. Lancet. 1992;339:271–2. doi: 10.1016/0140-6736(92)91334-5. [DOI] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–22. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Pietrobono R, Pomponi MG, Tabolacci E, Oostra B, Chiurazzi P, Neri G. Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic Acids Res. 2002;30:3278–85. doi: 10.1093/nar/gkf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretto DI, Mendoza-Morales G, Lo J, Cao R, Hadd A, Latham GJ, Durbin-Johnson B, Hagerman R, Tassone F. CGG allele size somatic mosaicism and methylation in FMR1 premutation alleles. J Med Genet. 2014;51:309–18. doi: 10.1136/jmedgenet-2013-102021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quartier A, Poquet H, Gilbert-Dussardier B, Rossi M, Casteleyn AS, Portes VD, Feger C, Nourisson E, Kuentz P, Redin C, Thevenon J, Mosca-Boidron AL, Callier P, Muller J, Lesca G, Huet F, Geoffroy V, El Chehadeh S, Jung M, Trojak B, Le Gras S, Lehalle D, Jost B, Maury S, Masurel A, Edery P, Thauvin-Robinet C, Gerard B, Mandel JL, Faivre L, Piton A. Intragenic FMR1 disease-causing variants: a significant mutational mechanism leading to Fragile-X syndrome. Eur J Hum Genet. 2017;25:423–431. doi: 10.1038/ejhg.2016.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan-Babu IS, Law HY, Yoon CS, Lee CG, Chong SS. Simplified strategy for rapid first-line screening of fragile X syndrome: closed-tube triplet-primed PCR and amplicon melt peak analysis. Expert Rev Mol Med. 2015;17:e7. doi: 10.1017/erm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees WA, Yager TD, Korte J, von Hippel PH. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry. 1993;32:137–44. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- Rife M, Nadal A, Mila M, Willemsen R. Immunohistochemical FMRP studies in a full mutated female fetus. Am J Med Genet A. 2004;124A:129–32. doi: 10.1002/ajmg.a.20342. [DOI] [PubMed] [Google Scholar]
- Rocchi M, Pecile V, Archidiacono N, Monni G, Dumez Y, Filippi G. Prenatal diagnosis of the fragile-X in male monozygotic twins: discordant expression of the fragile site in amniocytes. Prenat Diagn. 1985;5:229–31. doi: 10.1002/pd.1970050311. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Heitz D, Biancalana V, Blumenfeld S, Kretz C, Boue J, Tommerup N, Van Der Hagen C, DeLozier-Blanchet C, Croquette MF, et al. Direct diagnosis by DNA analysis of the fragile X syndrome of mental retardation. N Engl J Med. 1991;325:1673–81. doi: 10.1056/NEJM199112123252401. [DOI] [PubMed] [Google Scholar]
- Saluto A, Brussino A, Tassone F, Arduino C, Cagnoli C, Pappi P, Hagerman P, Migone N, Brusco A. An enhanced polymerase chain reaction assay to detect pre- and full mutation alleles of the fragile X mental retardation 1 gene. J Mol Diagn. 2005;7:605–12. doi: 10.1016/S1525-1578(10)60594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel LC, Schwartz C, Skinner C, Rodenhiser DI, Ainsworth PJ, Pare G, Sadikovic B. Clinical Validation of Fragile X Syndrome Screening by DNA Methylation Array. J Mol Diagn. 2016;18:834–841. doi: 10.1016/j.jmoldx.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Schutzius G, Bleckmann D, Kapps-Fouthier S, di Giorgio F, Gerhartz B, Weiss A. A quantitative homogeneous assay for fragile X mental retardation 1 protein. J Neurodev Disord. 2013;5:8. doi: 10.1186/1866-1955-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, Tassone F, Willemsen R, Disney MD, Hagerman PJ, Todd PK, Charlet-Berguerand N. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–80. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–61. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman SL. Premature ovarian failure in the fragile X syndrome. Am J Med Genet. 2000;97:189–94. doi: 10.1002/1096-8628(200023)97:3<189::AID-AJMG1036>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Smeets HJ, Smits AP, Verheij CE, Theelen JP, Willemsen R, van de Burgt I, Hoogeveen AT, Oosterwijk JC, Oostra BA. Normal phenotype in two brothers with a full FMR1 mutation. Hum Mol Genet. 1995;4:2103–8. doi: 10.1093/hmg/4.11.2103. [DOI] [PubMed] [Google Scholar]
- Stoger R, Kajimura TM, Brown WT, Laird CD. Epigenetic variation illustrated by DNA methylation patterns of the fragile-X gene FMR1. Hum Mol Genet. 1997;6:1791–801. doi: 10.1093/hmg/6.11.1791. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Sutherland GR, Ashforth PL. X-linked mental retardation with macro-orchidism and the fragile site at Xq 27 or 28. Hum Genet. 1979;48:117–20. doi: 10.1007/BF00273283. [DOI] [PubMed] [Google Scholar]
- Tabolacci E, Moscato U, Zalfa F, Bagni C, Chiurazzi P, Neri G. Epigenetic analysis reveals a euchromatic configuration in the FMR1 unmethylated full mutations. Eur J Hum Genet. 2008a;16:1487–98. doi: 10.1038/ejhg.2008.130. [DOI] [PubMed] [Google Scholar]
- Tabolacci E, Pomponi MG, Pietrobono R, Chiurazzi P, Neri G. A unique case of reversion to normal size of a maternal premutation FMR1 allele in a normal boy. Eur J Hum Genet. 2008b;16:209–14. doi: 10.1038/sj.ejhg.5201949. [DOI] [PubMed] [Google Scholar]
- Tarleton J, Kenneson A, Taylor AK, Crandall K, Fletcher R, Casey R, Hart PS, Hatton D, Fisch G, Warren ST. A single base alteration in the CGG repeat region of FMR1: possible effects on gene expression and phenotype. J Med Genet. 2002;39:196–200. doi: 10.1136/jmg.39.3.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Adams J, Berry-Kravis EM, Cohen SS, Brusco A, Leehey MA, Li L, Hagerman RJ, Hagerman PJ. CGG repeat length correlates with age of onset of motor signs of the fragile X-associated tremor/ataxia syndrome (FXTAS) Am J Med Genet B Neuropsychiatr Genet. 2007a;144B:566–9. doi: 10.1002/ajmg.b.30482. [DOI] [PubMed] [Google Scholar]
- Tassone F, Beilina A, Carosi C, Albertosi S, Bagni C, Li L, Glover K, Bentley D, Hagerman PJ. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. 2007b;13:555–62. doi: 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Iong KP, Tong TH, Lo J, Gane LW, Berry-Kravis E, Nguyen D, Mu LY, Laffin J, Bailey DB, Hagerman RJ. FMR1 CGG allele size and prevalence ascertained through newborn screening in the United States. Genome Med. 2012;4:100. doi: 10.1186/gm401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Pan R, Amiri K, Taylor AK, Hagerman PJ. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 2008;10:43–9. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, Pandey UB, Di Prospero NA, Min KT, Taylor JP, Paulson HL. Histone deacetylases suppress CGG repeat-induced neurodegeneration via transcriptional silencing in models of fragile X tremor ataxia syndrome. PLoS Genet. 2010;6:e1001240. doi: 10.1371/journal.pgen.1001240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–9. doi: 10.1093/nar/23.20.4202. doi: 5j0289 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Anar B, De Diego Otero Y, de Vries BB, Hilhorst-Hofstee Y, Smits A, van Looveren E, Willems PJ, Galjaard H, Oostra BA. Noninvasive test for fragile X syndrome, using hair root analysis. Am J Hum Genet. 1999;65:98–103. doi: 10.1086/302462. doi: S0002-9297(07)63732-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Smits A, Mohkamsing S, van Beerendonk H, de Haan A, de Vries B, van den Ouweland A, Sistermans E, Galjaard H, Oostra BA. Rapid antibody test for diagnosing fragile X syndrome: a validation of the technique. Hum Genet. 1997;99:308–11. doi: 10.1007/s004390050363. [DOI] [PubMed] [Google Scholar]
- Willemsen R, Smits A, Severijnen LA, Jansen M, Jacobs A, De Bruyn E, Oostra B. Predictive testing for cognitive functioning in female carriers of the fragile X syndrome using hair root analysis. J Med Genet. 2003;40:377–9. doi: 10.1136/jmg.40.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohrle D, Salat U, Glaser D, Mucke J, Meisel-Stosiek M, Schindler D, Vogel W, Steinbach P. Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J Med Genet. 1998;35:103–11. doi: 10.1136/jmg.35.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yrigollen CM, Durbin-Johnson B, Gane L, Nelson DL, Hagerman R, Hagerman PJ, Tassone F. AGG interruptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genet Med. 2012;14:729–36. doi: 10.1038/gim.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yrigollen CM, Martorell L, Durbin-Johnson B, Naudo M, Genoves J, Murgia A, Polli R, Zhou L, Barbouth D, Rupchock A, Finucane B, Latham GJ, Hadd A, Berry-Kravis E, Tassone F. AGG interruptions and maternal age affect FMR1 CGG repeat allele stability during transmission. J Neurodev Disord. 2014;6:24. doi: 10.1186/1866-1955-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Barron MD, Romero RM, Christy M, Gold B, Dai J, Gray DM, Haworth IS, Mitas M. At physiological pH, d(CCG)15 forms a hairpin containing protonated cytosines and a distorted helix. Biochemistry. 1997;36:3687–99. doi: 10.1021/bi9625410. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Kumari D, Sciascia N, Usdin K. CGG-repeat dynamics and FMR1 gene silencing in fragile X syndrome stem cells and stem cell-derived neurons. Mol Autism. 2016;7:42. doi: 10.1186/s13229-016-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]