Abstract
Background
Cardiac resynchronization therapy (CRT) reduces morbidity and mortality in heart failure (HF) patients with reduced left ventricular function and intraventricular conduction delay. However, individual outcomes vary significantly. This study sought to use a machine learning algorithm to develop a model to predict outcomes following CRT.
Methods and Results
Models were developed with machine learning algorithms to predict all-cause mortality or HF hospitalization at twelve months post CRT in the COMPANION trial. The best performing model was developed with the Random Forest algorithm. The ability of this model to predict all-cause mortality or HF hospitalization and all-cause mortality alone was compared to discrimination obtained using a combination of bundle branch block morphology and QRS duration. In the 595 CRT-D patients in the COMPANION trial, 105 deaths occurred (median follow-up 15.7 months). The survival difference across subgroups differentiated by bundle branch block morphology and QRS duration did not reach significance (p = 0.08). The Random Forest model produced quartiles of patients with an eight-fold difference in survival between those with the highest and lowest predicted probability for events (hazard ratio 7.96, p < 0.0001). The model also discriminated the risk of the composite endpoint of all-cause mortality or HF hospitalization better than subgroups based on bundle branch block morphology and QRS duration.
Conclusions
In the COMPANION trial, a machine learning algorithm produced a model that predicted clinical outcomes following CRT. Applied prior to device implant, this model may better differentiate outcomes over current clinical discriminators and improve shared decision-making with patients.
Keywords: cardiac resynchronization therapy, heart failure, shared decision-making, precision medicine, machine learning
Introduction
Multiple clinical trials have demonstrated the benefit of cardiac resynchronization therapy (CRT) on morbidity and mortality in heart failure (HF) patients with evidence of left ventricular dysfunction and intraventricular conduction delay.1–5 However, approximately 30% of patients, despite meeting criteria for implant, do not experience clinical benefit from CRT.6–9
Current ACC/AHA/HRS and ESC guidelines emphasize bundle branch block morphology and QRS duration for patient selection, with the strongest recommendations for CRT implant in patients with left bundle branch block (LBBB) and QRS duration greater than or equal to 150ms.9,10 Recent analyses from registries have demonstrated that bundle branch block morphology and QRS duration also predict long term outcomes following CRT; patients with LBBB and QRS of 150ms or greater experienced fewer deaths and re-admissions than other patients receiving CRT.11,12 However, clinical experience shows that not all patients with LBBB and wide QRS have a good outcome following CRT.13 Conversely, some patients with non-LBBB and QRS duration greater than 150ms or others with LBBB but QRS duration less than 150ms experience improvement.14–15 Additionally, device implantation carries risk with coronary sinus complications occurring in 2% of patients, the need for device failure requiring revision in as many as 5% of patients, and a six-month cumulative incidence of any complication as high as 10%.16–18
Therefore, estimating a patient's outcome following CRT is an important part of the shared decision-making process prior to implantation. In fact, the ACC/AHA/HRS guideline calls for shared decision-making with patients during the informed consent process.9 Unfortunately, the patient characteristics that are outlined in the ACC/AHA/HRS guidelines to estimate an individual patient's outcome, bundle branch block morphology and QRS duration, lack precision. The combination of these characteristics estimates a hazard ratio of only 1.5 between those patients with the best and worst mortality and less than 2 for heart failure admission.11
The Precision Medicine Initiative asks our profession to avoid oversimplification and to take individual variability into account to improve this shared decision-making process.19 A call to achieve precision cardiovascular care has been described.20 The tools outlined included improved analytic and bioinformatics methods to integrate data from the electronic health record to assist clinicians at the bedside. Machine learning is a computational discipline focused on building algorithms that model or recognize complex patterns or characteristics within large amounts of data. Machine learning algorithms have been applied within cardiology to understand the complex genetics of coronary artery disease, to improve prediction of 30-day readmission following coronary intervention, and to classify HF with preserved ejection fraction.21–23 Machine learning algorithms have also been used to generate models to predict echocardiographic response to CRT; however, to our knowledge, they have not been used to predict clinical outcomes like mortality and HF hospitalizations.24–26
We hypothesized that machine learning algorithms may produce a model that discriminates mortality and the composite endpoint of mortality or heart failure hospitalization following CRT implantation for individual patients better than the widely used clinical discriminators of bundle branch block morphology and QRS duration. We tested this hypothesis with data from the Comparison of Medical Therapy, Pacing and Defibrillation in Heart Failure (COMPANION) trial.
Methods
Study Population
The design and primary results of the COMPANION trial have been published elsewhere.1,27 The authors were given permission by the COMPANION executive committee to use the trial data. However, they do not control access to the data and cannot make the data, analytic methods, and study materials available to other researchers for purposes of reproducing the results or replicating the procedure. Briefly, COMPANION was a randomized, controlled multicenter trial that included 1,520 patients with advanced HF with left ventricular ejection fraction (LVEF) of 35% or less randomized in a 1:2:2 ratio to receive: optimal pharmacologic therapy (OPT) alone, or in combination with CRT-pacemaker (CRT-P) or CRT-defibrillator (CRT-D). All enrolled patients had New York Heart Association (NYHA) class III or IV symptoms.
Study Design
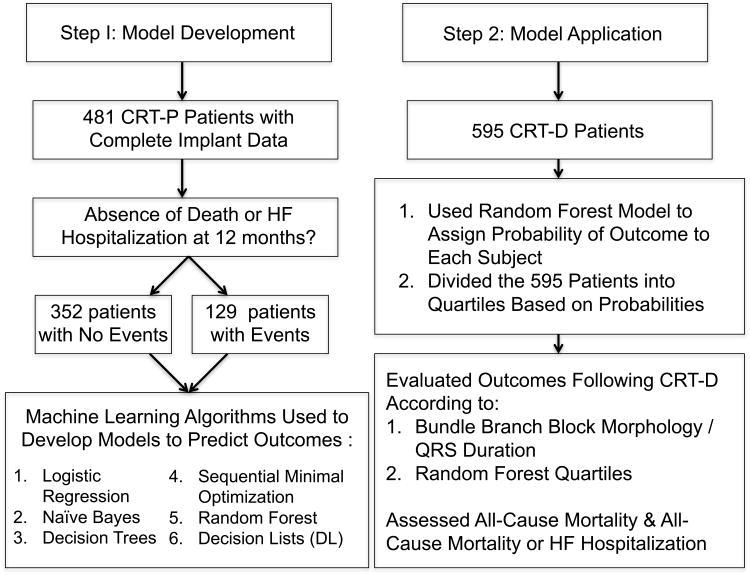
The present study included one cohort for model development and a second for validation (Figure 1). Machine learning models were developed using the CRT-P cohort of the COMPANION trial. Limiting model development to this cohort allowed for isolation of the benefits of CRT without the confounding effect of defibrillation. The best performing model was then validated in the CRT-D cohort.
Figure 1. Flow diagram of study.
Model development (left hand column) performed with 481 patients from the CRT-P cohort of the COMPANION trial for whom complete device implant data was available. Six machine learning algorithms were tested to predict the absence of death or HF (HF) hospitalization at 12 months. The best model was validated in the 595 CRT-D patients from the COMPANION trial (right hand column) by evaluating event driven outcomes.
Human Subjects approval
The COMPANION executive committee and the institutional review board of the University of Wisconsin-Madison approved the retrospective use of the de-identified data from the trial for the present study (IRB #2015-0657).
Model Development
Patient Selection
The CRT-P arm of COMPANION consisted of 617 patients; however, per the primary trial report, CRT implantation was successful in 539 patients.1 With the data available to the authors, successful implant could be confirmed in 481 patients; therefore, these patients were used for model development. This model development cohort was identified prior to merging pre-implant data with outcomes to avoid selection bias.
Machine Learning Algorithms
Model development included trials of several machine learning algorithms available in the Waikato Environment for Knowledge Analysis, an open-source unified workbench that allows access to state-of-the-art machine learning algorithms.28 Algorithms tested included Naïve Bayes classifier, sequential minimal optimization for training a support vector machine, decision lists, J48 decision tree, and the Random Forest algorithm. We also compared the performance of the machine learning algorithms to a standard multivariate logistic regression model. Forty-five pre-implant features (see Table 1) were used to characterize the patients.
Table 1. Forty-five Features Used in Final Model Development.
| Domain | Individual Features |
|---|---|
| Demographics | Gender, Age |
| Physical Characteristics | Body mass index, Heart rate, Systolic blood pressure, Diastolic blood pressure, Pulse pressure |
| Heart Failure | Etiology, Duration of heart failure, NHYA Class, Six minute walk distance, |
| LV Assessment | LV ejection fraction, Method of LV assessment, LV end-diastolic diameter |
| ECG | QRS duration, PR interval, QRS morphology |
| Comorbid Conditions | Intermittent atrial arrhythmia, Diabetes, Hepatic disease, Cerebrovascular disease, Peripheral vascular disease, Carotid artery disease, Pulmonary hypertension, Renal disease, Hypertension, Hyperlipidemia, Other, No comorbid conditions |
| Surgical Interventions | CABG, Valve replacement, angioplasty, Coronary stent, No cardiac surgeries |
| Medications Classes | ACE inhibitor, Angiotensin-receptor blocker, Anticoagulant, Lipid lowering, Anti-platelet, Beta-blocker, Calcium channel blocker, Digoxin, Nitrate, Aldosterone antagonist, Class III anti-arrhythmic |
NYHA = New York Heart Association, LV = Left ventricular, CABG = Coronary artery bypass grafting
Model Selection
The algorithms were used to create models to predict the composite end-point of the absence of death or HF hospitalization at 12 months post randomization in the 481 CRT-P patients. Ten-fold cross-validation was used to evaluate the predictive performance of each model by dividing the training data set into 10 mutually exclusive subsets, 9 of which were used for training and 1 for evaluation. This was repeated 10 times, thereby using 10 different, but overlapping training sets, and 10 unique testing sets.
Receiver operating characteristic curve analysis was used to evaluate the performance of each model. Comparisons between the area under the curve (AUC) for the receiver operating characteristic curve of each model were made using a paired t-test based on fold-by-fold AUC during the cross-validation. The classification performance at particular cutoff thresholds was also evaluated according to its sensitivity, specificity, positive predictive value, and negative predictive value.
The most informative model based on receiver operating characteristic curve AUC was the one produced using the Random Forest algorithm. This finding is consistent with other studies in clinical datasets in which the Random Forest out-performed other algorithms.29,30 The Random Forest algorithm is an ensemble of decision trees.31,32 A full discussion of this method is beyond the scope of this paper. Briefly, decision trees repeatedly dichotomize a dataset based on their determination of the most informative feature. At a given node (i.e., decision point), the algorithm finds the feature (e.g. QRS duration, QRS morphology, patient age, etc.) and threshold value that best partitions the cases into two subsets that differ in class distribution (did or did not experience and event in this example). The procedure continues recursively until each terminal node consists mostly of cases of one class. The resulting terminal nodes are each assigned the label that is the majority class of the cases in that node. In a Random Forest, decision trees are built from a training set constructed by sampling a number of cases with replacement at random from the data and sampling a number of features at random. This process is repeated to produce many decision trees (a forest) whose predicted outcomes are combined into a single value. Test cases are labelled by majority vote of the resulting trees.
Model Validation
As depicted in Figure 1, the best performing model was applied to the 595 CRT-D patients for validation. The CRT-D population was partitioned two ways: 1) according to a combination of bundle branch block morphology and QRS duration; “BBB / QRS 1” – LBBB and QRS duration of 150ms or greater, “BBB / QRS 2” – LBBB and QRS duration less than 150ms or non-LBBB and QRS duration of 150ms or greater, and “BBB / QRS 3” – non-LBBB with QRS duration less than 150ms; 2) according to quartiles based upon the probability of remaining event free as predicted by the Random Forest model; “Quartile 1” – highest probability of remaining event free to “Quartile 4” – lowest probability.
Event driven outcomes were then assessed for each subgroup. The outcomes assessed were (1) the composite of all-cause mortality or HF hospitalization and (2) all-cause mortality alone.
Comparison of Descriptive Statistics and Outcomes Across Subgroups
The differences in demographic, ECG, echocardiographic and clinical characteristics for each partition were compared using either χ2 or Fisher's exact tests for categorical variables and either t-test or ANOVA for continuous variables. All-cause mortality or HF hospitalizations and all-cause mortality alone were compared across subgroups for each method of partitioning with Kaplan-Meier analysis. Differences in events between subgroups were evaluated using the logrank test. Unadjusted Cox proportional hazards models were used to determine the independent association between subgroup and outcomes. Statistical analyses were performed using R version 3.2.2 (2015-08-04).
Results
Model Development
The characteristics of the patients used for model development are shown in Supplemental Table 1. One hundred twenty-nine patients (27%) experienced a death or HF hospitalization within twelve months of randomization. Patients who experienced this endpoint were significantly more likely to have the following characteristics: larger left ventricular end diastolic dimension, a shorter QRS duration, male sex, ischemic etiology for their cardiomyopathy, NYHA class IV, a shorter six-minute walk distance, not on a beta-blocker, history of renal disease, and a history of intermittent atrial arrhythmia.
The Random Forest algorithm with 550 trees produced a model with the best AUC (0.74, 95% CI: 0.72-0.76). The improvement in AUC for the Random Forest model was statistically significant compared to that of the other models (p < 0.001, see Figure 2 for comparison to multivariate logistic regression and sequential minimal optimization for training a support vector machine). The Random Forest model shown in Figure 2, operating at a threshold of 0.76 (patients with a model output of 0.76 or greater predicted to have no event), had a sensitivity of 52%, a negative predictive value of 38%, a specificity of 80%, and a positive predictive value of 88%.
Figure 2. Receiver operating characteristic curves for best models.
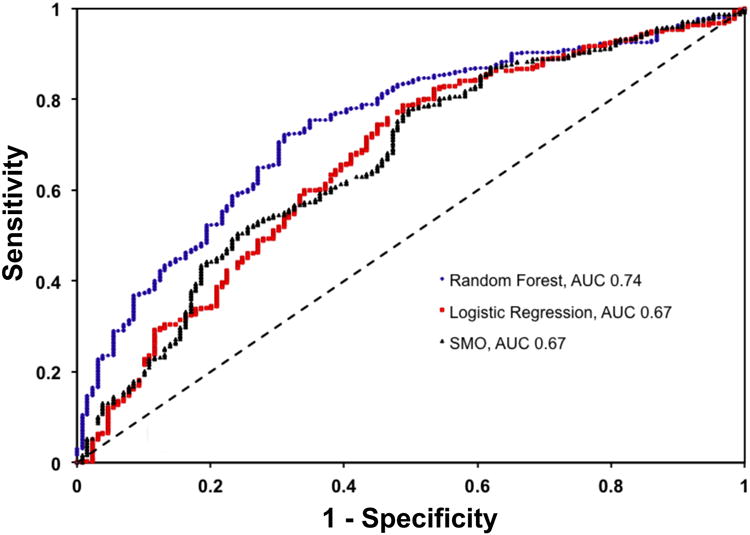
Using area under curve (AUC), the Random Forest model with 550 trees (blue, AUC = 0.74, 95% CI: 0.72-0.76) was superior to that produced by multivariate logistic regression (red, AUC = 0.67, 95% CI: 0.65-0.69) or sequential minimal optimization to train a support vector machine (SMO, black, AUC = 0.67, 95% CI: 0.65-0.68). The improvement in AUC for the Random Forest model was statistically significant compared to that of the multiple logistic regression or SMO model, p < 0.001 for both.
Validation of Model
The characteristics of the 595 patients in the CRT-D cohort as well as the subgroups defined by bundle branch block morphology / QRS duration and the Random Forest model are shown in Supplemental Table 2. While both schemes created variation between subgroups in some features, there were significant differences across Random Forest model quartiles for 33/45 features compared to only 12/45 for subgroups defined by bundle branch block morphology / QRS duration. The model performance in the CRT-D validation cohort was similar to that seen in the CRT-P cohort used for development. Operating at a threshold of 0.76 (patients with a model output of 0.76 or greater predicted to have no event) the model had a sensitivity of 51% in the CRT-D cohort (compared to 52% in training set), had a negative predictive value of 37% (compared to 38%), a specificity of 77% (compared to 80%), and a positive predictive value of 85% (compared to 88%).
In the CRT-D cohort of the COMPANION trial, there 214 events for the composite endpoint of death or HF hospitalization over a median follow-up of 15.7 months. The Kaplan-Meier analysis for this outcome for each method of partitioning is shown in Figure 3 (panels A and B) with hazard ratios (HR) for each subgroup (referenced to either LBBB and QRS ≥ 150ms for bundle branch block morphology / QRS duration or Quartile 1 for the Random Forest model) shown in panel C. There was a significant difference in the distribution of events across subgroups generated by bundle branch block morphology / QRS duration (panel A, logrank p value = 0.005); however, there was no significant difference in events between those patients with bundle branch block morphology / QRS duration subgroup 2 (BBB / QRS 2: non-LBBB and QRS ≥ 150ms or LBBB and QRS < 150ms,) and those with bundle branch block morphology / QRS duration subgroup 3 (BBB / QRS 3: non-LBBB and QRS < 150ms). The Random Forest model produced subgroups with a graded increase in events moving from Quartile 1 through Quartile 4 with patients in Quartile 4 having a 3-fold increase in events compared to those in Quartile 1 (HR 3.26, 95% confidence interval (CI): 2.13 to 5.00).
Figure 3. Survival free of all-cause mortality or HF hospitalization.
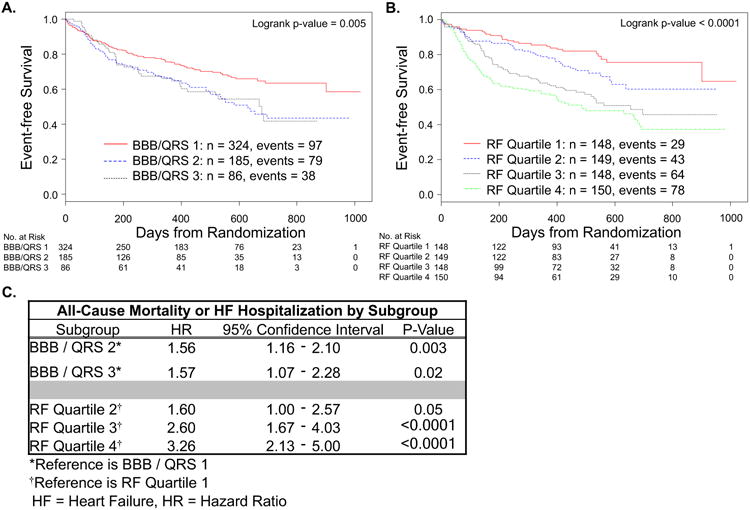
Kaplan-Meier curves for all-cause mortality or HF hospitalization partitioned by bundle branch block morphology / QRS duration: BBB / QRS 1 = LBBB and QRS ≥ 150 ms, BBB / QRS 2 = non-LBBB and QRS ≥ 150 ms or LBBB and QRS < 150 ms, and BBB / QRS 3 = non-LBBB and QRS < 150 ms (A) or Random Forest model sub-divided into quartiles with Quartile 1 expected to have the best outcomes and Quartile 4 the worst (B). Hazard ratio for each subgroup compared to reference for that partition (C).
The Kaplan-Meier analysis for all-cause mortality alone for both methods of partitioning is shown in Figure 4 (panels A and B) with HR for each subgroup (referenced to either LBBB and QRS ≥ 150ms or Quartile 1) shown in panel 4C. In the CRT-D cohort, there were 105 deaths over a median follow-up of 15.7 months. For the subgroups based on bundle branch block morphology / QRS duration, the difference in the survival distribution across subgroups did not reach significance (logrank p-value = 0.08). Compared to patients with LBBB and QRS ≥ 150ms, there was a significant decrease in survival for patients with non-LBBB and QRS ≥ 150ms or LBBB and QRS < 150ms (HR 1.58, 95% CI: 1.04 to 2.40) but only a trend toward decreased survival in patients with non-LBBB and QRS < 150ms (HR 1.42, 95% CI: 0.82 to 2.48). The results for subgroups created using the Random Forest model are shown in panel 5B. The difference in survival distribution across subgroups reached significance (p < 0.0001) with a nearly 8-fold increase in mortality in Quartile 4 compared to Quartile 1 (HR 7.96; 95% CI: 3.60 to 17.56).
Figure 4. Survival free of all-cause mortality.
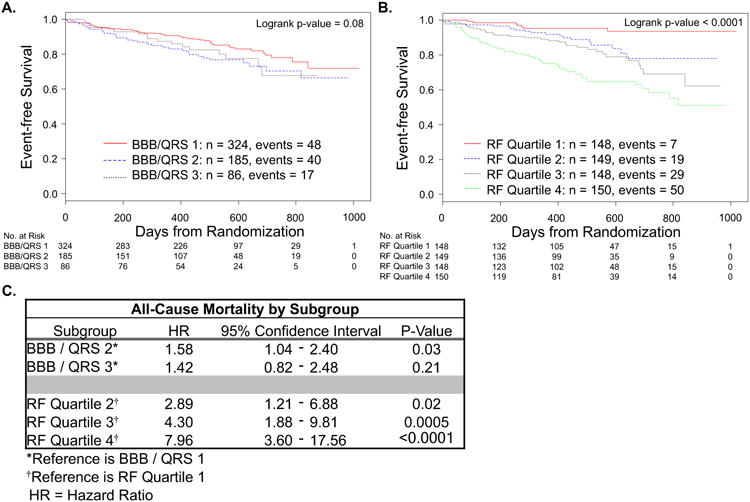
Kaplan-Meier curves for all-cause mortality partitioned by bundle branch block morphology / QRS duration: BBB / QRS 1 = LBBB and QRS ≥ 150 ms, BBB / QRS 2 = non-LBBB and QRS ≥ 150 ms or LBBB and QRS < 150 ms, and BBB / QRS 3 = non-LBBB and QRS < 150 ms (A) or Random Forest model sub-divided into quartiles with Quartile 1 expected to have the best outcomes and Quartile 4 the worst (B). Hazard ratio for each subgroup compared to reference for that partition (C).
The Random Forest model reclassified a significant number of patients compared to classification based on bundle branch block morphology / QRS duration (Figure 5). Over one-third of the patients in Random Forest Quartile 4 (52 patients) – the patients predicted to have the worst outcomes by this model - had a LBBB morphology and QRS duration ≥ 150ms, i.e. characteristics that would predict the best outcome using bundle branch block morphology and QRS duration alone. Conversely, over 20% of the patients in Random Forest Quartile 1 (32 patients) did not have LBBB morphology and QRS duration of ≥ 150ms. The 52 patients with LBBB morphology and QRS duration ≥ 150ms in Random Forest Quartile 4 experienced significantly more events compared to the 32 patients without LBBB and QRS duration ≥ 150ms in Random Forest Quartile 1 (HR 2.42, 95% CI: 1.09 to 5.34 for all-cause mortality or HF hospitalization; HR 6.62, 95% CI: 1.55 to 28.30 for all-cause mortality).
Figure 5. Reclassification from bundle branch block morphology / QRS duration to Random Forest quartile.
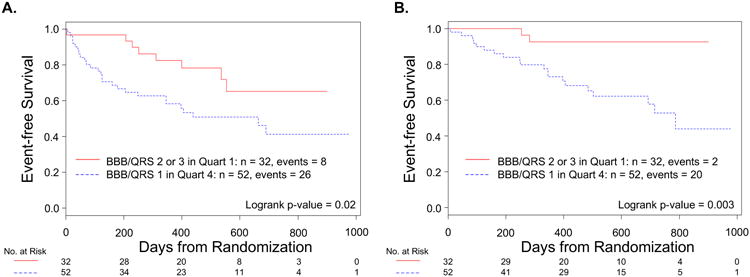
Fifty-two patients with LBBB and QRS duration ≥ 150 ms (BBB / QRS 1) were in Random Forest Quartile 4 and thirty-two patients without LBBB and QRS duration ≥ 150 ms (BBB / QRS 2 or 3) were in Quartile 1. Both all-cause mortality or HF hospitalizations (A) and all-cause mortality alone (B) were significantly different between these groups, favouring the patients with BBB / QRS 2 or 3 in Quartile 1.
Discussion
Using a Random Forest algorithm, we developed, validated and applied a model to predict CRT outcomes based on pre-implant characteristics in a retrospective analysis of the COMPANION trial. Compared to prediction methodology based on bundle branch block morphology and QRS duration, our data demonstrates that the Random Forest model more precisely predicted patient outcomes in the COMPANION trial. This improved prediction included reclassification of patients from a group expected to have a better outcome based on bundle branch block morphology and QRS duration to a more a precise group based on the Random Forest model.
Ideally, treatment recommendations for patients will include individualized outcome estimates. However, historically this has not been feasible and outcome estimates have been extrapolated from large clinical trial outcomes. While these are effective at predicting outcomes at the population level, there remains a significant gap in capability to predict an outcome for an individual patient. Machine learning is a powerful, computational method that could allow for improved description of phenotypes and development of decision support tools to predict clinical outcomes and better inform shared decision-making with patients.
For CRT, there is substantial variation in response and outcomes, and several studies have investigated predictors that contribute to this variation.33–39 Current ACC/AHA/HRS and ESC guidelines9,10 provide recommendations largely based on bundle branch block morphology and QRS duration. Recent analyses have shown that left bundle branch block morphology and QRS duration ≥ 150ms do predict all-cause mortality and HF hospitalizations at a population level following CRT.11,12 Here, when applied retrospectively to patients from the COMPANION trial, partitioning the CRT-D cohort according to bundle branch block morphology and QRS duration did discriminate outcomes. Patients without the combination of LBBB and QRS duration ≥ 150ms experienced a 1.5-fold increase in both endpoints evaluated: the composite endpoint of death or HF hospitalization and death alone, compared to those with LBBB and QRS duration ≥ 150ms. This result is very similar to previously published data regarding outcomes based on bundle branch block morphology and QRS duration.11
However, the Random Forest model provided greater differentiation across subgroups (Figures 3 and 4). For example, the 148 patients predicted to have the best outcome in the Random Forest model, i.e. those in Quartile 1, experienced 7 deaths during in-trial follow-up compared to 50 deaths in the 150 patients in Quartile 4 - a greater than 4-fold increase in events from Quartile 1 to Quartile 4. In contrast, using bundle branch block morphology and QRS duration, the difference in mortality across sub-groups was not as robust and did not reach statistical significance.
Additionally, the Random Forest model reclassified a significant number of patients from one subgroup based on bundle branch block morphology and QRS duration to a more precise group based on the Random Forest model. Thirty-two patients (22%) in Quartile 1 did not have a LBBB and QRS duration ≥ 150ms. Of these thirty-two patients, who would have been expected to have a worse outcome based solely on bundle branch block morphology and QRS duration, only 2 experienced death during follow-up. In contrast, fifty-two of the 150 patients (35%) in Quartile 4, did have LBBB and QRS duration ≥ 150ms and accounted for 40% of the deaths (20/50) in this quartile. This reclassification demonstrates how a model that incorporates a broad spectrum of clinical data (demographic data, ECG data, echocardiogram, patient history) can improve discussions with individual patients. Based on bundle branch block morphology and QRS duration, these 52 with LBBB and QRS duration ≥ 150ms patients would have been predicted to have a relatively good outcome prior to implant. However, when taken together with other available data, a different expectation could have been communicated to the patient prior to implant.
The Random Forest model and the reclassification observed with the model also provides an opportunity to understand what features (or variables) may contribute to the varied outcomes observed. One method to understand the feature importance in the data set is using its relative information gain.28 This can be quantified by how well a feature reduces “impurity” in the data set. In this regard, an ideal feature would split the data set perfectly according to the classification, i.e. in this case, the ideal feature would split the data set into those patients who experienced death or HF at twelve months post randomization and those who did not. For this data set (see Supplemental Figure 1), the five most important features based on information gain were 1) a history of renal disease, 2) the time from HF diagnosis, 3) NYHA Class, 4) QRS duration and 5) a history of intermittent atrial arrhythmia. The importance of these features in the Random Forest model becomes apparent when looking at the differences between Random Forest quartiles (Supplemental Table 2B). This table demonstrates a marked increase in the percentage of patients with a history of renal disease, a shorter duration of HF prior to implant, more patients with NYHA Class III symptoms, a longer QRS duration and fewer patients with a history of intermittent atrial arrhythmias when moving from RF Quartile 1 to RF Quartile 4.
Analyzing the patients reclassified with the Random Forest model offers another opportunity to assess features that should be considered when attempting to estimate outcomes for individual patients. Patients in the BBB / QRS subgroups 2 and 3 who were in RF quartile 1 (the 32 patients shown in Figure 5) were very similar to BBB / QRS subgroup 1 patients in RF quartile 1. There were statistically significant differences in only 4 features between these groups: QRS duration and percent of patients with LBBB as a consequence of the BBB / QRS partitioning, LVEF and the number of patients with a history of peripheral vascular disease (see Supplemental Table 3). However, patients in BBB / QRS subgroup 1 who were in RF quartile 4 (the 52 patients in Figure 5) had statistically significant differences from BBB / QRS subgroup 1 patients in RF quartile 1 in 26 different features. These included four of the five features identified based on information gain above (all except QRS duration as would be expected). Other features with statistical significant differences reaching p < 0.0001 included age, an ischemic etiology and other features associated with a history of ischemic heart disease, six-minute walk distance, PR interval, and whether or not patients were on a beta-blocker.
Interestingly, many of these features (QRS duration, NYHA Class, history of renal disease, ischemic disease and atrial arrhythmias, PR interval) have been described elsewhere as influencing CRT outcomes.14,35,40–43 Additionally, there has been a recent paper describing the influence of co-morbid conditions on CRT outcomes.13 However, to our knowledge, to date there has not been a model developed to bring together all of these features.
With the increasing use of electronic health records, the prospective application of models developed with machine learning algorithms is quickly becoming possible. The use of such models will represent a paradigm shift that will bring precision medicine closer to reality. An important element in shared decision-making with an individual patient is understanding the best available evidence for the risks and benefits of a therapy for that patient.44 For CRT, bundle branch block morphology and QRS duration are currently used to guide these discussions without incorporating other clinical characteristics and comorbid conditions into a decision-making tool. However, one can envision a model like the one developed with the Random Forest algorithm being applied via the electronic medical record to each patient presenting for a pre-implant discussion. The predicted probability of an event could be shared with the patient and would serve as a critical element to assist the implanter in developing guidance for that patient and to facilitate shared decision-making.
In many instances, traditional logistic regression analysis may be the best model to make this prediction. This is especially true if data sets are smaller and contain limited features. However, this is only one tool, and as data sets grow larger and additional features are added, other methods may improve prediction. If we expand our choices to include 10 or more additional leading machine learning algorithms, we increase our chances of finding a model that is suitable for the data in question. Therefore, machine learning algorithms may play an important role in precision cardiology.
Limitations
The current study utilized data from the COMPANION trial and included follow-up of patients for only 16 months. Therefore, it is not known whether a similar result would be obtained if follow-up was longer. The COMPANION trial also enrolled only patients with NYHA class III and IV symptoms. Therefore, the results of this study only extend to those with advanced HF. It is unknown whether these results are applicable to patients with NYHA I or II symptoms.
The models developed here rely on 45 features that are easily obtainable from the clinical history, ECG data or basic echocardiographic features. The features were consistently described in both the model development (CRT-P) and validation (CRT-D) cohorts as both cohorts were from the same clinical trial. Our finding that the Random Forest algorithm produced the best model for a data is consistent with other work using machine learning algorithms to predict clinical endpoints29,30; however, as has been pointed out previously with Random Forests, consistency in the reporting of features has limited their utility within cardiology to date.45 Moving forward, developing consistent standards for the definition of features will be necessary to fully utilize the strength of machine learning algorithms.
Finally, the models developed here utilize the data from one clinical trial. While the trial was multi-center, due to the retrospective nature of this study, potential unidentified confounders may exist. Therefore, the results of this study should be validated in additional study populations.
Conclusion
In this study, we used a machine learning algorithm to develop and validate a model to discriminate clinical outcomes with CRT using easily obtainable pre-implantation characteristics. Classification of patients using models like this may improve patient selection for CRT and enhance the shared decision-making process with patients prior to implant. As clinical data sets expand, application of machine learning algorithms will lead to further improvements in precision cardiovascular medicine.
Supplementary Material
What is Known?
Cardiac Resynchronization Therapy (CRT) provides a clear benefit to heart failure patients with reduced left ventricular function and intraventricular conduction delay.
Individual outcomes following CRT vary significantly.
What the Study Adds?
A Random Forest algorithm was used to develop a model that significantly improved the ability to discriminate outcomes following CRT.
The use of machine learning algorithms like the Random Forest may improve shared decision-making with patients and lead to improvements in precision cardiovascular medicine.
Acknowledgments
Sources of Funding: Support was provided by the Clinical and Translational Science Award (CTSA) program, through the NIH National Center for Advancing Translational Sciences (NCATS), grant UL1TR000427 (Dr. Page). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Disclosures: All authors report no conflicts of interest
References
- 1.Bristow MR, Saxon LA, Boehmer J, Krueger S, Kass DA, De Marco T, Carson P, DiCarlo L, DeMets D, White BG, DeVries DW, Feldman AM. Cardiac-resynchronization therapy with or without an implantable defibrillator in advanced chronic heart failure. N Engl J Med. 2004;350:2140–2150. doi: 10.1056/NEJMoa032423. [DOI] [PubMed] [Google Scholar]
- 2.Cleland JG, Daubert JC, Erdmann E, Freemantle N, Gras D, Kappenberger L, Tavazzi L. The effect of cardiac resynchronization on morbidity and mortality in heart failure. N Engl J Med. 2005;352:1539–1549. doi: 10.1056/NEJMoa050496. [DOI] [PubMed] [Google Scholar]
- 3.Moss AJ, Hall WJ, Cannom DS, Klein H, Brown MW, Daubert JP, Estes NM, III, Foster E, Greenberg H, Higgins SL, Pfeffer MA, Solomon SD, Wilber D, Zareba W. Cardiac-resynchronization therapy for the prevention of heart-failure events. N Engl J Med. 2009;361:1329–1338. doi: 10.1056/NEJMoa0906431. [DOI] [PubMed] [Google Scholar]
- 4.Tang AS, Wells GA, Talajic M, Arnold MO, Sheldon R, Connolly S, Hohnloser SH, Nichol G, Birnie DH, Sapp JL, Yee R, Healey JS, Rouleau JL. Cardiac-resynchronization therapy for mild-to-moderate heart failure. N Engl J Med. 2010;363:2385–2395. doi: 10.1056/NEJMoa1009540. [DOI] [PubMed] [Google Scholar]
- 5.Linde C, Abraham WT, Gold MR, St John Sutton M, Ghio S, Daubert C. Randomized trial of cardiac resynchronization in mildly symptomatic heart failure patients and in asymptomatic patients with left ventricular dysfunction and previous heart failure symptoms. J Am Coll Cardiol. 2008;52:1834–1843. doi: 10.1016/j.jacc.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Young JB, Abraham WT, Smith AL, Leon AR, Lieberman R, Wilkoff B, Canby RC, Schroeder JS, Liem LB, Hall S, Wheelan K. Combined cardiac resynchronization and implantable cardioversion defibrillation in advanced chronic heart failure: the MIRACLE ICD Trial. JAMA. 2003;289:2685–2694. doi: 10.1001/jama.289.20.2685. [DOI] [PubMed] [Google Scholar]
- 7.McAlister FA, Ezekowitz J, Hooton N, Vandermeer B, Spooner C, Dryden DM, Page RL, Hlatky MA, Rowe BH. Cardiac resynchronization therapy for patients with left ventricular systolic dysfunction: a systematic review. JAMA. 2007;297:2502–2514. doi: 10.1001/jama.297.22.2502. [DOI] [PubMed] [Google Scholar]
- 8.Chung ES, Leon AR, Tavazzi L, Sun JP, Nihoyannopoulos P, Merlino J, Abraham WT, Ghio S, Leclercq C, Bax JJ, Yu CM, Gorcsan J, St John Sutton M, De Sutter J, Murillo J. Results of the predictors of response to CRT (PROSPECT) Trial. Circulation. 2008;117:2608–2616. doi: 10.1161/CIRCULATIONAHA.107.743120. [DOI] [PubMed] [Google Scholar]
- 9.Tracy CM, Epstein AE, Darbar D, DiMarco JP, Dunbar SB, Estes NAM, Ferguson TB, Hammill SC, Karasik PE, Link MS, Marine JE, Schoenfeld MH, Shanker AJ, Silka MJ, Stevenson LW, Stevenson WG, Varosy PD, Anderson JL. 2012 ACCF/AHA/HRS focused update of the 2008 guidelines for device-based therapy of cardiac rhythm abnormalities: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. Circulation. 2012;126:1784–1800. doi: 10.1161/CIR.0b013e3182618569. [DOI] [PubMed] [Google Scholar]
- 10.Brignole M, Auricchio A, Baron-Esquivias GB, Bordachar P, Boriani G, Breithardt O, Cleland J, Deharo JC, Delgado V, Elliot PM, Gorenek B, Israel CW, Leclercq C, Linde C, Mont L, Padeletti L, Sutton R, Vardas PE. 2013 ESC Guidelines on cardiac pacing and cardiac resynchronization therapy: The Task Force on cardiac pacing and resynchronization therapy of the European Society of Cardiology (ESC). Developed in collaboration with the European Heart Rhythm Association (EHRA) Eur Heart J. 2013;34:2281–2329. doi: 10.1093/eurheartj/eht150. [DOI] [PubMed] [Google Scholar]
- 11.Peterson PN, Greiner MA, Qualls LG, Al-Khatib SM, Curtis JP, Fonarow GC, Hammill SC, Heidenreich PA, Hammill BG, Piccini JP, Hernandez AF, Curtis LH, Masoudi FA. QRS duration, bundle-branch block morphology, and outcomes among older patients with heart failure receiving cardiac resynchronization therapy. JAMA. 2013;310:617–626. doi: 10.1001/jama.2013.8641. [DOI] [PubMed] [Google Scholar]
- 12.Bilchick K, Kamath S, DiMarco J, Stukenborg G. Bundle-branch block morphology and other predictors of outcome after cardiac resynchronization therapy in medicare patients. Circulation. 2010;122:2022–2030. doi: 10.1161/CIRCULATIONAHA.110.956011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeitler EP, Friedman DJ, Daubert JP, Al-Khatib SM, Solomon SD, Biton Y, McNitt S, Zareba W, Moss AJ, Kutyifa V. Multiple Comorbidities and Response to Cardiac Resynchronization Therapy. J Am Coll Cardiol. 2017;69:2369–2379. doi: 10.1016/j.jacc.2017.03.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleland JG, Abraham WT, Linde C, Gold MR, Young JB, Claude Daubert J, Sherfesee L, Wells GA, Tang ASL. An individual patient meta-analysis of five randomized trials assessing the effects of cardiac resynchronization therapy on morbidity and mortality in patients with symptomatic heart failure. Eur Heart J. 2013;34:3547–3556. doi: 10.1093/eurheartj/eht290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varma N, Manne M, Nguyen D, He J, Niebauer M, Tchou P. Probability and magnitude of response to cardiac resynchronization therapy according to QRS duration and gender in nonischemic cardiomyopathy and LBBB. Heart Rhythm. 2014;11:1139–1147. doi: 10.1016/j.hrthm.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 16.van Rees JB, de Bie MK, Thijssen J, Borleffs CJW, Schalij MJ, van Erven L. Implantation-related complications of implantable cardioverter-defibrillators and cardiac resynchronization therapy devices. J Am Coll Cardiol. 2011;58:995–1000. doi: 10.1016/j.jacc.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Kirkfeldt RE, Johansen JB, Nohr EA, Jorgensen OD, Nielsen JC. Complications after cardiac implantable electronic device implantations: an analysis of a complete, nationwide cohort in Denmark. Eur Heart J. 2014;35:1186–1194. doi: 10.1093/eurheartj/eht511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta N, Kiley ML, Anthony F, Young C, Brar S, Kwaku K. Multi-center, community-based cardiac implantable electronic devices registry: population, device utilization, and outcomes. J Am Heart Assoc. 2016;5:e002798. doi: 10.1161/JAHA.115.002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah SH, Arnett D, Houser SR, Ginsburg GS, MacRae C, Mital S, Loscalzo J, Hall JL. Opportunities for the cardiovascular community in the precision medicine initiative. Circulation. 2016;133:226–231. doi: 10.1161/CIRCULATIONAHA.115.019475. [DOI] [PubMed] [Google Scholar]
- 21.Drenos F, Grossi E, Buscema M, Humphries SE. Networks in coronary heart disease genetics as a step towards systems epidemiology. PLOS ONE. 2015;10:e0125876. doi: 10.1371/journal.pone.0125876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasfy JH, Singal G, O'Brien C, Blumenthal DM, Kennedy KF, Strom JB, Spertus JA, Mauri L, Normand SLT, Yeh RW. Enhancing the prediction of 30-day readmission after percutaneous coronary intervention using data extracted by querying of the electronic health record. Circ Cardiovasc Qual Outcomes. 2015;8:477–485. doi: 10.1161/CIRCOUTCOMES.115.001855. [DOI] [PubMed] [Google Scholar]
- 23.Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang CC, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–279. doi: 10.1161/CIRCULATIONAHA.114.010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peressutti D, Bai W, Jackson T, Sohal M, Rinaldi A, Rueckert D, King A. Prospective identification of CRT super responders using a motion atlas and random projection ensemble learning. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Cham: Springer International Publishing; 2015. pp. 493–500. Available from: http://link.springer.com/10.1007/978-3-319-24574-4_59. [Google Scholar]
- 25.Huang H, Shen L, Zhang R, Makedon F, Hettleman B, Pearlman J. Cardiac motion analysis to improve pacing site selection in CRT. Acad Radiol. 2006;13:1124–1134. doi: 10.1016/j.acra.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz B, De Maria R, Gatsios D, Chrysanthakopoulou T, Landolina M, Gasparini M, Campolo J, Parolini M, Sanzo A, Galimberti P, Bianchi M, Lenders M, Brand E, Parodi O, Lunati M, Brand SM. Identification of genetic markers for treatment success in heart failure patients: insight from cardiac resynchronization therapy. Circ Cardiovasc Genet. 2014;7:760–770. doi: 10.1161/CIRCGENETICS.113.000384. [DOI] [PubMed] [Google Scholar]
- 27.Bristow MR, Feldman AM, Saxon LA. Heart failure management using implantable devices for ventricular resynchronization: comparison of medical therapy, pacing, and defibrillation in chronic heart failure (COMPANION) trial. J Card Fail. 2000;6:276–285. doi: 10.1054/jcaf.2000.9501. [DOI] [PubMed] [Google Scholar]
- 28.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. ACM SIGKDD Explor Newsl. 2009;11:10–18. [Google Scholar]
- 29.Weiss JC, Natarajan S, Peissig PL, McCarty CA, Page D. Machine learning for personalized medicine: predicting primary myocardial infarction from electronic health records. AI Magazine. 2012 Winter;:33–45. [PMC free article] [PubMed] [Google Scholar]
- 30.Churpek MM, Yuen TC, Winslow C, Meltzer DO, Kattan MW, Edelson DP. Multicenter Comparison of Machine Learning Methods and Conventional Regression for Predicting Clinical Deterioration on the Wards. Crit Care Med. 2016;44:368–374. doi: 10.1097/CCM.0000000000001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan J. C4.5: Programs for Machine Learning First. San Mateo, CA: Morgan Kaufmann Publishers Inc; 1993. [Google Scholar]
- 32.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 33.Cleland J, Freemantle N, Ghio S, Fruhwald F, Shankar A, Marijanowski M, Verboven Y, Tavazzi L. Predicting the long-term effects of cardiac resynchronization therapy on mortality from baseline variables and the early response. J Am Coll Cardiol. 2008;52:438–445. doi: 10.1016/j.jacc.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 34.Dupont M, Rickard J, Baranowski B, Varma N, Dresing T, Gabi A, Finucan M, Mullens W, Wilkoff BL, Tang WHW. Differential response to cardiac resynchronization therapy and clinical outcomes according to QRS morphology and QRS duration. J Am Coll Cardiol. 2012;60:592–598. doi: 10.1016/j.jacc.2012.03.059. [DOI] [PubMed] [Google Scholar]
- 35.Goldenberg I, Moss AJ, Hall WJ, Foster E, Goldberger JJ, Santucci P, Shinn T, Solomon S, Steinberg JS, Wilber D, et al. Predictors of response to cardiac resynchronization therapy in the Multicenter Automatic Defibrillator Implantation Trial with Cardiac Resynchronization Therapy (MADIT-CRT) Circulation. 2011;124:1527–1536. doi: 10.1161/CIRCULATIONAHA.110.014324. [DOI] [PubMed] [Google Scholar]
- 36.Lee AY, Moss AJ, Ruwald MH, Kutyifa V, McNitt S, Polonsky B, Zareba W, Ruwald AC. Temporal influence of heart failure hospitalizations prior to implantable cardioverter defibrillator or cardiac resynchronization therapy with defibrillator on subsequent outcome in mild heart failure patients (from MADIT-CRT) Am J Cardiol. 2015;115:1423–1427. doi: 10.1016/j.amjcard.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 37.Sipahi I, Chou JC, Hyden M, Rowland DY, Simon DI, Fang JC. Effect of QRS morphology on clinical event reduction with cardiac resynchronization therapy: meta-analysis of randomized controlled trials. Am Heart J. 2012;163:260–267. doi: 10.1016/j.ahj.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Bommel RJ, Borleffs CJW, Ypenburg C, Marsan NA, Delgado V, Bertini M, van der Wall EE, Schalij MJ, Bax JJ. Morbidity and mortality in heart failure patients treated with cardiac resynchronization therapy: influence of pre-implantation characteristics on long-term outcome. Eur Heart J. 2010;31:2783–2790. doi: 10.1093/eurheartj/ehq252. [DOI] [PubMed] [Google Scholar]
- 39.Zusterzeel R, Curtis JP, Caños DA, Sanders WE, Selzman KA, Piña IL, Spatz ES, Bao H, Ponirakis A, Varosy PD, Masoudi FA, Strauss DG. Sex-Specific Mortality Risk by QRS Morphology and Duration in Patients Receiving CRT. J Am Coll Cardiol. 2014;64:887–894. doi: 10.1016/j.jacc.2014.06.1162. [DOI] [PubMed] [Google Scholar]
- 40.Gold MR, Padhiar A, Mealing S, Sidhu MK, Tsintzos SI, Abraham WT. Long-Term Extrapolation of Clinical Benefits Among Patients With Mild Heart Failure Receiving Cardiac Resynchronization Therapy. JACC Heart Fail. 2015;3:691–700. doi: 10.1016/j.jchf.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Daimee UA, Moss AJ, Biton Y, Solomon SD, Klein HU, McNitt S, Polonsky B, Zareba W, Goldenberg I, Kutyifa V. Long-term outcomes with cardiac resynchronization therapy in patients with mild heart failure with moderate renal dysfunction. Circ Heart Fail. 2015;8:725–732. doi: 10.1161/CIRCHEARTFAILURE.115.002082. [DOI] [PubMed] [Google Scholar]
- 42.Hoppe UC. Effect of Cardiac Resynchronization on the Incidence of Atrial Fibrillation in Patients With Severe Heart Failure. Circulation. 2006;114:18–25. doi: 10.1161/CIRCULATIONAHA.106.614560. [DOI] [PubMed] [Google Scholar]
- 43.Friedman DJ, Bao H, Spatz ES, Curtis JP, Daubert JP, Al-Khatib SM. Association between a prolonged PR interval and outcomes of cardiac resynchronization therapy: a report from the National Cardiovascular Data Registry. Circulation. 2016;134:1617–1628. doi: 10.1161/CIRCULATIONAHA.116.022913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Legare F, Witteman HO. Shared Decision-making: Examining Key Elements And Barriers To Adoption Into Routine Clinical Practice. Health Aff (Millwood) 2013;32:276–284. doi: 10.1377/hlthaff.2012.1078. [DOI] [PubMed] [Google Scholar]
- 45.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.