Abstract
The RNA-binding protein Sex-lethal (Sxl) is an important post-transcriptional regulator of sex determination and dosage compensation in female Drosophila. To prevent the assembly of the MSL dosage compensation complex in female flies, Sxl acts as a repressor of male-specific lethal-2 (msl-2) mRNA translation. It uses two distinct and mutually reinforcing blocks to translation that operate on the 5′ and 3′ untranslated regions (UTRs) of msl-2 mRNA, respectively. While 5′ UTR-mediated translational control involves an upstream open reading frame, 3′ UTR-mediated regulation strictly requires the co-repressor protein Upstream of N-ras (Unr), which is recruited to the transcript by Sxl. We have identified the protein Sister-of-Sex-lethal (Ssx) as a novel repressor of translation with Sxl-like activity. Both proteins have a comparable RNA-binding specificity and can associate with uracil-rich RNA regulatory elements present in msl-2 mRNA. Moreover, both repress translation when bound to the 5′ UTR of msl-2. However, Ssx is inactive in 3′ UTR-mediated regulation, as it cannot engage the co-repressor protein Unr. The difference in activity maps to the first RNA-recognition motif (RRM) of Ssx. Conversion of three amino acids within this domain into their Sxl counterpart results in a gain of function and repression via the 3′ UTR, allowing detailed insights into the evolutionary origin of the two proteins and into the molecular requirements of an important translation regulatory pathway.
Keywords: regulation of translation, Sex-lethal, male-specific lethal-2, protein evolution
INTRODUCTION
In Drosophila, sex determination is under the control of the switch gene Sex-lethal (Sxl) which encodes an RNA-binding protein that is expressed only in female flies. Sxl acts on multiple levels, post-transcriptionally controlling the synthesis of key factors involved in sex-specific traits to govern female development (Penalva and Sánchez 2003; Salz and Erickson 2010; Graindorge et al. 2011; Salz 2011; Venables et al. 2012; Moschall et al. 2017).
One of the best-characterized regulatory targets of Sxl is male-specific lethal 2 (msl-2) mRNA that encodes an essential component of the dosage compensation complex (DCC or MSL), which is involved in hyper-transcription of the single male X-chromosome (Prestel et al. 2010; Graindorge et al. 2011; Conrad and Akhtar 2012). Msl2 synthesis is required in male animals, but is deleterious in females (Kelley et al. 1995). To prevent Msl2 protein expression in females, Sxl binds to U-rich elements within the untranslated regions (UTRs) of msl-2 mRNA to control its splicing, export, and translation (Zhou et al. 1995; Bashaw and Baker 1997; Kelley et al. 1997; Gebauer et al. 1998; Merendino et al. 1999; Förch et al. 2000, 2001; Graindorge et al. 2013). Regulation on several levels and by multiple mechanisms ensures a robust and fail-safe repression of Msl2 protein production to effectively prevent DCC formation in female flies (for review, see Moschall et al. 2017).
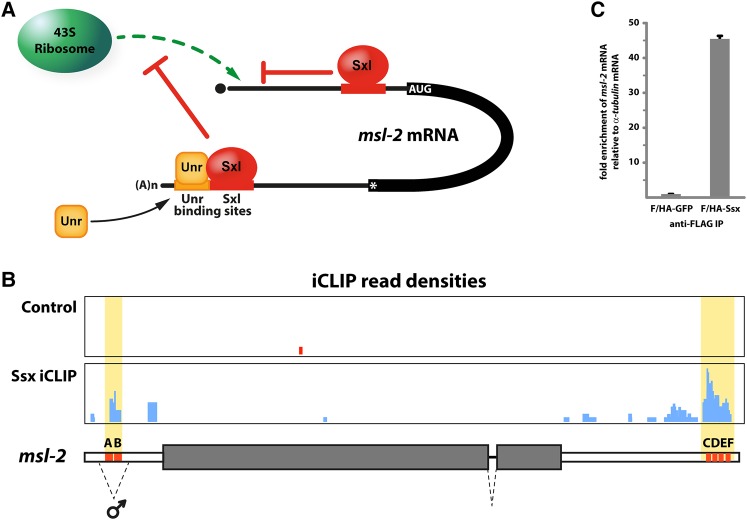
Sxl-mediated translational repression of msl-2 mRNA involves multiple RNA regulatory elements and at least two different regulatory mechanisms that interfere with translation initiation (Fig. 1A; Beckmann et al. 2005). Bound to regulatory sequences in the 3′ UTR of msl-2, Sxl recruits the co-repressor protein Upstream of N-ras (Unr) to adjacent RNA motifs. Once assembled, the repressor complex then interferes with recruitment of ribosomal preinitiation complexes to the 5′ end of the mRNA (Abaza et al. 2006; Duncan et al. 2006, 2009). Ribosomes that escape this first regulatory mechanism are then challenged by additional Sxl molecules bound to the 5′ UTR. Here, Sxl controls the activity of an upstream open reading frame (uORF) to block the progress of scanning ribosomal subunits to the msl-2 initiation codon, effectively blocking Msl2 protein production (Medenbach et al. 2011).
FIGURE 1.
Ssx associates with the UTRs of msl-2 mRNA. (A) Schematic representation of Sxl-mediated repression of msl-2 mRNA translation. Bound to 3′ UTR regulatory elements, Sxl recruits the co-repressor protein Unr to assemble a stable repressor complex (depicted at the bottom) that interferes with the recruitment of ribosomal initiation complexes to the 5′ end of the mRNA. Ribosomes that escape this regulatory pathway are then challenged by additional molecules of Sxl bound to the 5′ UTR of the msl-2 transcript (depicted at the top). Here, Sxl controls the activity of an upstream open reading frame to prevent scanning ribosomes from reaching the Msl2 coding sequence. (B) In the absence of Sxl, Ssx associates with known Sxl binding sites in the msl-2 transcript (areas highlighted in yellow, Sxl binding sites depicted as orange boxes and labeled A to F). The msl-2 gene locus is depicted schematically at the bottom. Protein-coding regions of the gene are shown as shaded boxes, UTRs as white boxes. Splicing events are depicted by dashed lines (the facultative intron in the 5′ UTR is marked ♂). Read densities from replicates of Ssx iCLIP experiments (blue) and control reactions (red) are shown at the top. (C) Ssx binds endogenous msl-2 mRNA. FLAG-HA-tagged GFP and Ssx were expressed in SL2 cells and immunoprecipitated by anti-FLAG antibodies. Bound RNAs were purified, reverse transcribed and subjected to quantitative real-time RT-PCR. Enrichment of msl-2 over α-tublin mRNA in Ssx IPs is plotted relative to the GFP control.
The Drosophila genome encodes another protein with high similarity to Sxl: Sister-of-Sex-lethal (Ssx). Sxl and Ssx are paralogs originating from a gene duplication event early in Drosophilid evolution (Traut et al. 2006; Cline et al. 2010). While Sxl has been well studied in past decades, the function of Ssx remains enigmatic. ssx knockout does not significantly affect viability or fecundity in either sex under standard laboratory conditions, even in combination with mutations in Sxl (Cline et al. 2010). Transposon insertion into the ssx locus however is immunocompromising and the mutant flies quickly succumb to Gram-positive bacterial infection, but not to infection with Gram-negative pathogens, suggesting a function of Ssx in immunity (Ayres et al. 2008).
Ssx and Sxl exhibit quite distinct N- and C-terminal regions; however, they share a highly similar central domain which in the case of Sxl is sufficient for translational repression of msl-2 mRNA (Grskovic et al. 2003; Moschall et al. 2017). Intriguingly, Sxl and Ssx have a similar RNA-binding specificity, and in the absence of Sxl, Ssx was found to bind to the U-rich RNA regulatory elements of msl-2 mRNA (Rogell et al. 2017); however, the functional consequence of this association remains unknown. Here we demonstrate that, like Sxl, Ssx acts as a repressor of translation when bound to the 5′ UTR of msl-2 mRNA. However, when bound to the 3′ UTR regulatory sequences, Ssx cannot recruit the co-repressor protein Unr and hence fails to regulate translation via this pathway. We biochemically map this difference in activity to three amino acids in the first RNA-recognition motif (RRM) of Ssx. Conversion of the critical amino acids into their Sxl counterparts results in a gain of function, recruitment of Unr, and translational repression.
RESULTS
Sister-of-Sex-lethal binds to U-rich elements in msl-2 mRNA
We performed individual-nucleotide resolution crosslinking-immunoprecipitation (iCLIP) experiments to determine the target mRNAs and binding specificity of Ssx. These analyses revealed that in cells with “male” characteristics, where Sxl is absent, Ssx associates with several U-rich RNA elements located in the 5′ and 3′ UTR of msl-2 mRNA (Fig. 1B), that play a key role in Sxl-dependent post-transcriptional regulation of msl-2 expression.
To confirm the interaction of Ssx with msl-2 mRNA, we expressed FLAG-HA-tagged, full-length Ssx or GFP as a control in cultured Drosophila SL2 cells, which have male characteristics and lack Sxl. After immunoprecipitation with antibodies against the tag, we analyzed bound RNAs by qRT-PCR. Relative to the GFP control, we could detect an almost 50-fold enrichment of msl-2 mRNA in Ssx immunoprecipitates, while a control RNA without a predicted Sxl/Ssx binding motif (α-tubulin) was not enriched (Fig. 1C). Recently, also specific RNP capture independently confirmed the association of Ssx with the UTRs of msl-2 mRNA (Rogell et al. 2017).
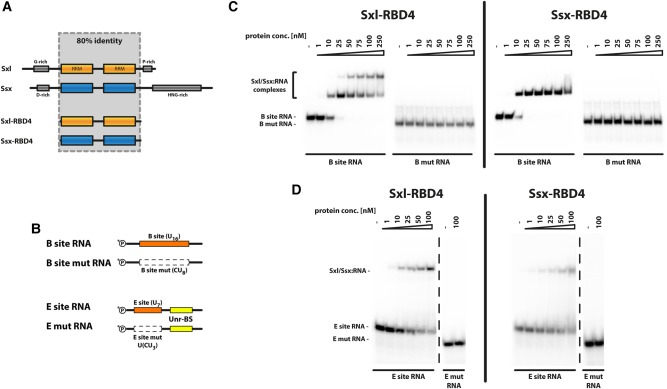
To further characterize the interaction of Ssx with msl-2 mRNA, we expressed and purified to homogeneity the central RNA-binding domains of Sxl (aa122–301, Sxl-RBD4, see Grskovic et al. 2003) and Ssx (aa93–269, Ssx-RBD4) (Fig. 2A; Supplemental Fig. 1A). We then assayed binding to a 5′ UTR fragment of msl-2 mRNA that contains a well characterized Sxl-binding site—a homopolymeric stretch of 16 U residues denoted B site (Fig. 2B). In electromobility shift assays (Fig. 2C), both recombinant proteins bind the B-site RNA with high affinity. Moreover, the interaction requires the homopolymeric U sequence as replacement of every second uracil by a cytosine (Fig. 2B) completely abolishes the interaction, demonstrating specificity (Fig. 2C). Using a different RNA fragment with a shorter Sxl-binding motif (E-site, U7) derived from the 3′ UTR of msl-2 mRNA yields comparable results (Fig. 2B,D).
FIGURE 2.
Ssx binds U-rich sequence motifs. (A) Schematic representation of Sxl and Ssx proteins and shortened versions thereof (RBD4). Eighty percent identity can be observed between the central domains of Sxl and Ssx (shaded area), which encompass two RNA-recognition motifs (RRMs, depicted in orange or blue, respectively). Low complexity regions in the N- and C-terminal regions are depicted as gray boxes. (B) Schematic representation of RNA oligonucleotides used for binding studies. Sxl binding sites (B site and E site) are depicted as orange boxes; the Unr-binding site is shown in yellow. As a specificity control, Sxl binding was abolished by mutation of every second U to C within the Sxl binding motif (white boxes with broken outline) yielding the mutant constructs. Radioactive labeling by 32P is indicated at the 5′ ends of the oligonucleotides. (C) Sxl and Ssx bind the msl-2-derived B site with comparable affinity. Electromobility shift assays using an RNA fragment derived from the 5′ UTR of msl-2 (B site) and its mutant derivative (as depicted in panel B). The radiolabeled RNA probes were incubated with the indicated amounts of the Sxl-RBD4 or Ssx-RBD4 proteins. RNP formation was analyzed by resolving the complexes by native PAGE. (D) Sxl and Ssx both associate with a U-rich fragment derived from the msl-2 3′ UTR. Electromobility shift assays as described for panel C, this time using E site and E mut RNAs (see panel B).
Differences between the two proteins can only be observed in the stoichiometry of the interaction on the longer RNA motif (Fig. 2C): At higher protein concentrations, two Sxl molecules associate with the RNA, resulting in a further shift in electrophoretic mobility. However, no such complex can be observed for Ssx.
Sister-of-Sex-lethal is a repressor of translation
Because Ssx can interact with the RNA regulatory elements in msl-2 mRNA, we wondered whether it can also—like Sxl—control its processing and/or translation. For Sxl it was demonstrated that the highly conserved RNA-binding domain is both necessary and sufficient for translational repression (Grskovic et al. 2003), but regulation of splicing additionally depends on the N-terminal, glycine-rich domain (Deshpande et al. 1999; Yanowitz et al. 1999). As conservation between the two proteins does not extend beyond the RNA-binding domain, we reasoned that Ssx instead of controlling splicing might rather be a regulator of translation.
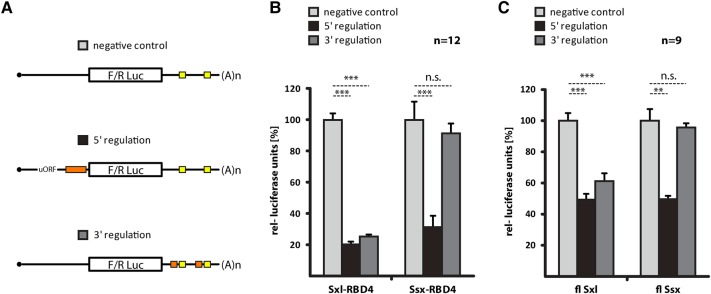
Previous work aiming to decipher the mechanism by which Sxl controls translation was enabled by an in vitro translation system based on Drosophila embryo extracts. Using this experimental setup, Sxl-dependent translational control can be faithfully recapitulated on luciferase reporter RNAs that bear fragments of the msl-2 UTRs (Gebauer et al. 1998, 2003; Grskovic et al. 2003; Beckmann et al. 2005; Duncan et al. 2006, 2009; Gebauer and Hentze 2007; Medenbach et al. 2011). To individually probe the two different translation regulatory mechanisms that operate on msl-2 mRNA, we used RNAs with mutations in either the 5′ or 3′ UTR Sxl binding sites (Fig. 3A; Beckmann et al. 2005; Medenbach et al. 2011). We then monitored translation in the absence or presence of recombinant Sxl-RBD4 or Ssx-RBD4 (Fig. 3B). As previously shown (Beckmann et al. 2005), Sxl can repress translation, acting via binding sites in either UTR (Fig. 3B, 5′ regulation and 3′ regulation), and mutation of all Sxl binding motifs abolishes regulation (negative control). Despite the fact that Sxl- and Ssx-RBD4 both bind the 3′ UTR regulatory sequences (Fig. 2D), Ssx only weakly represses a reporter that monitors 3′ UTR-mediated translational control (Fig. 3B). In contrast, repression via the 5′ UTR is strong and occurs at a level similar to Sxl-RBD4.
FIGURE 3.
Ssx represses translation via the 5′ UTR, but not the 3′ UTR of msl-2. (A) Schematic representation of the reporter RNAs used. A Firefly (in vitro assays) or Renilla (transfection into cultured cells) luciferase open reading frame is flanked by msl-2-derived UTRs or mutant derivatives thereof. The negative control (depicted at the top) lacks all Sxl-regulatory elements. To monitor 5′ UTR-dependent regulation, a construct with an upstream open reading frame (uORF, required for strong regulation) and a single 5′ UTR Sxl binding site (B site, orange box) is used (all Sxl binding sites in the 3′ UTR are mutated). Conversely, 3′ UTR-mediated regulation is monitored on a reporter that lacks Sxl binding sites in the 5′ UTR, but carries intact 3′ UTR Sxl- and Unr-binding sites (orange and yellow boxes). (B) In vitro translation assays using recombinant Sxl-RBD4 and Ssx-RBD4 (Fig. 2A; Supplemental Fig. 1A) to monitor 5′ and 3′ UTR-mediated translational repression of luciferase-based reporter RNAs relative to a nonregulated control (A). Firefly luciferase activity was determined in the absence or presence of the proteins indicated at the bottom and normalized to a cotranslated Renilla luciferase control RNA. Plotted are mean values and standard deviations of four experiments performed in biological triplicates. P-values: (***) P < 0.001, n.s., not significant. (C) Translational regulation assayed in cultured Drosophila SL-2 cells. Full-length, HA-tagged Sxl, Ssx, and mutant derivative proteins are expressed in cultured cells (for expression levels, see Supplemental Fig. 1B) and their impact on cotransfected msl-2-derived luciferase reporters is determined. As for panel B, regulation via 5′- or 3′ UTR Sxl binding sites is assessed individually using three different reporters (see panel A). All measurements are normalized to a nonregulated, cotransfected control construct. Mean values with standard deviation of three experiments performed in biological triplicates. P-values: (***) P < 0.001, (**) P < 0.01, n.s., not significant.
To confirm these findings in a more physiological setting, we transfected Drosophila SL2 cells with msl-2 reporter plasmids and monitored translational repression through 5′ or 3′ UTR regulatory elements (Medenbach et al. 2011). After expression of full-length Sxl or Ssx proteins, we measured reporter-derived luciferase activity relative to a nonregulated control (Fig. 3C). As previously demonstrated, Sxl can repress translation via both regulatory pathways (Medenbach et al. 2011). In contrast, Ssx was active when regulating via the 5′ UTR, but despite being expressed at levels slightly higher than Sxl (Supplemental Fig. 1B), we could not observe significant repression of the reporter with 3′ UTR binding sites.
RRM1 of Sxl and Ssx differ functionally
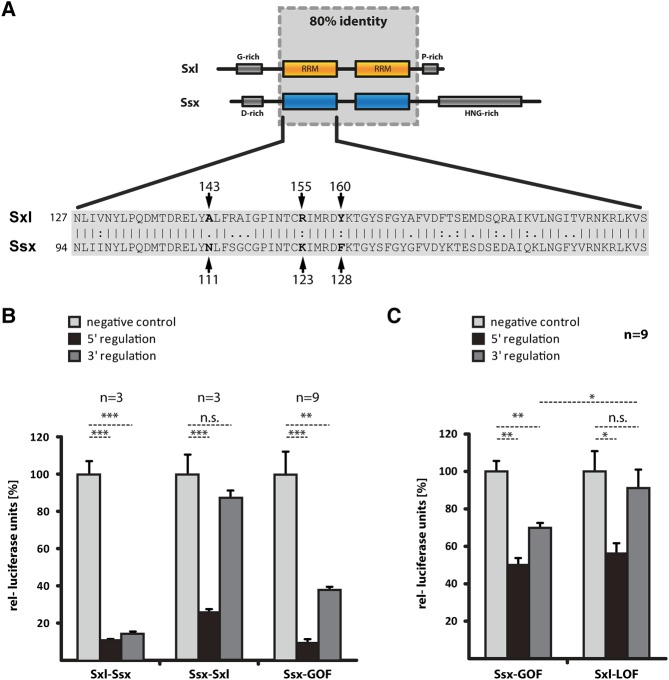
The data above show that Sxl and Ssx share many similarities—they both bind U-rich RNA sequences and repress msl-2 translation via the 5′ mechanism. However, they differ drastically in their ability to regulate translation via the 3′ UTR. To better understand this functional difference, we biochemically mapped the responsible protein region. We generated chimeric proteins with RRM1 of Sxl fused to RRM2 of Ssx (Sxl–Ssx) and vice versa (Ssx–Sxl). Both recombinant proteins are active in 5′ UTR-dependent translational regulation (Fig. 4B), demonstrating that the domain-swap does not interfere with protein folding. However, 3′ UTR-mediated regulation segregates with RRM1 of Sxl, indicating that the difference in activity is harbored in this domain.
FIGURE 4.
Mutation of Ssx RRM1 results in a gain of function and regulation via the msl-2 3′ UTR. (A) RRM1 of Sxl and Ssx differ in 18 amino acids. Schematic representation of Sxl and Ssx proteins (top) with a sequence alignment of RRM1 below. The critical amino acids A143, R155, and Y160 are highlighted (numbering according to Sxl, corresponding to Ssx positions N111, K123, and F128). (B) In vitro translation assays using chimeric proteins with RRM1 of Sxl fused to RRM2 of Ssx (Sxl–Ssx), or vice versa (Ssx–Sxl), and a Ssx mutant derivative with three mutations in RRM1 (N111A, K123R, and F128Y, denoted Ssx-GOF). Assays performed as described for Figure 3B. P-values: (***) P < 0.001, (**) P < 0.01, n.s., not significant. (C) Translational regulation assayed in cultured Drosophila SL-2 cells as described for Figure 3C, but using Ssx and Sxl mutant derivatives: Ssx-GOF (N111A, K123R, and F128Y) and Sxl-LOF (A143N, R155K, and Y160F). P-values: (**) P < 0.01, (*) P < 0.05, n.s., not significant
RRM1 of Sxl and Ssx are highly conserved and differ in only 18 amino acids (Fig. 4A). With the aim to further map the difference in activity, we introduced mutations in RRM1 of Ssx-RBD4, converting to their Sxl counterparts amino acids that differ between the two proteins. After purification of the mutant derivatives we scored for proper folding and RNA binding by monitoring repression of an msl-2 5′ UTR RNA reporter in translation extracts, followed by analysis of repression via the 3′ UTR (Supplemental Fig. 2). Combining three mutations that individually only showed minor effects on regulation via the 3′ UTR (N111A, K123R, and F128Y), we were able to generate an Ssx gain-of-function mutant protein (denoted Ssx-GOF) that represses translation of a 3′ UTR reporter with near Sxl-like activity (Fig. 4B). We validated these findings in the context of the full-length Ssx protein in tissue culture experiments (Fig. 4C). Moreover, we performed a converse experiment and replaced the identified critical amino acids in Sxl by their Ssx counterparts (A143N, R155K, and Y160F, protein denoted Sxl-LOF). As expected, this results in a loss of function and failure to repress translation of a 3′ UTR reporter (Fig. 4C).
Ssx fails to recruit Unr for repression
Sxl-dependent translational repression of msl-2 mRNA via the 3′ UTR critically requires the protein Unr. Despite being itself an RNA-binding protein that associates with many mRNAs in a sex-specific fashion, Unr binds to msl-2 mRNA in a Sxl-dependent manner (Abaza et al. 2006; Duncan et al. 2006; Abaza and Gebauer 2008). Hence, in male flies where Sxl is absent, Unr does not associate with msl-2 mRNA and synthesis of Msl2 protein is not repressed.
To understand if failure to repress translation stems from an inability of Ssx to recruit Unr to the RNA, we analyzed regulatory complex formation in vitro. Previously, it was demonstrated that the central RNA-binding domain of Sxl and the first of the five cold shock domains of Unr (UNR-CSD1) are sufficient for assembly of a highly stable complex on a short, msl-2-derived RNA fragment that harbors binding sites for the two proteins (Abaza et al. 2006; Duncan et al. 2006; Hennig et al. 2014). Furthermore, one of the Sxl critical residues analyzed here (Y160) was previously shown to be required for complex formation (Hennig et al. 2014).
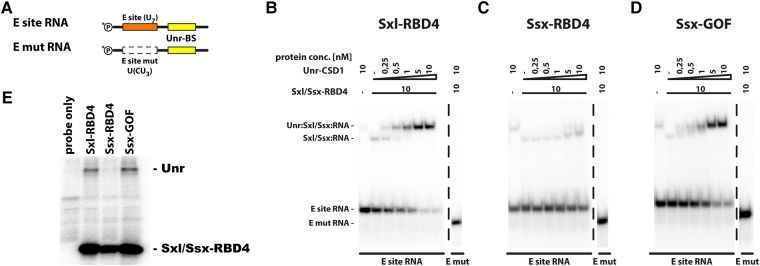
Sxl and Unr exhibit highly cooperative binding to the 3′ UTR of msl-2 mRNA. At a concentration of 10 nM, only weak binding of Sxl-RBD4 to an RNA fragment derived from the 3′ UTR of msl-2 RNA can be observed (E-site RNA, Fig. 2B,D). In contrast, upon additionally supplementing recombinant Unr-CSD1, the RNA can be almost quantitatively incorporated into a ternary complex that contains both recombinant proteins (Fig. 5B), while a mutant version of the RNA that lacks the Sxl binding site (E mut RNA, Fig. 5A) does not support complex formation (Fig. 5B).
FIGURE 5.
Ssx cannot recruit the co-repressor protein Unr to the 3′ UTR of msl-2. (A) Schematic representation of the RNA fragments used (as in Fig. 2B). (B–D) Analysis of Sxl/Ssx:Unr:RNA repressor complex formation. Assembly of a heterotrimeric complex on the msl-2-derived E site RNA (and its mutant derivative, Fig. 2B) is analyzed by EMSA. Recombinant Sxl-RBD4 (panel B), Ssx-RBD4 (panel C), or Ssx-GOF (panel D) are tested for their ability to recruit the recombinant cold shock domain 1 of Unr (Unr-CSD1) to the RNA. (E) Analysis of repressor complex formation in Drosophila embryo extracts by UV crosslinking. To allow repressor complex formation, an msl-2-derived, radiolabeled RNA fragment (similar to E-site RNA in panel A) was incubated in Drosophila embryo extract under translation conditions in the presence of recombinant proteins (as denoted above each lane, each GST-tagged). After UV-crosslinking and RNase digestion, proteins were purified via the GST tag, resolved by denaturing PAGE and visualized using a Phosphoimager. The identity of the proteins is indicated on the right.
In contrast to Sxl-RBD4, Ssx-RBD4 fails to form a ternary complex with Unr-CSD1, explaining lack of activity on 3′ UTR reporters (Fig. 5C).
Similar results are obtained using a previously described crosslinking-IP approach from Drosophila extracts (Grskovic et al. 2003; Abaza et al. 2006). A radiolabeled RNA fragment encompassing a stretch of the msl-2 3′ UTR can be efficiently photo-crosslinked to recombinant Sxl-RBD4 in extracts (Fig. 5E), whereas crosslinking of Ssx-RBD4 is somewhat reduced (see also Fig. 2D). Moreover, in the presence of recombinant Sxl-RBD4, endogenous Unr can be detected in complex with the RNA. However, this is not the case with Ssx-RBD4. The synergistic RNA binding of Sxl and Unr significantly contributes to stability of the resulting RNA-protein complex. Failure of Ssx to recruit Unr to the RNA likely results in formation of a Ssx:RNA complex with reduced stability which is reflected by a reduction in photo-crosslinking of Ssx-RBD4 to the RNA.
Notably, the Ssx-GOF protein efficiently forms a complex with recombinant Unr-CSD1 in vitro (Fig. 5D) and in native extracts with full-length, endogenous Unr (Fig. 5E), explaining the gain of function and its ability to regulate translation of 3′ UTR reporters.
DISCUSSION
In Drosophila melanogaster, the switch gene Sxl is the master regulator of female development. Functional Sxl protein is only produced in female animals, where it controls the expression of key factors involved in sex-specific traits, thus committing to female development.
However, sex determination in insects is evolving rapidly and the function of Sxl as master regulator of female development appears to be limited to Drosophilids (for review, see Sawanth et al. 2016). In many other Dipteran insects, the major sex-determining switch gene is not Sxl, but transformer (Traut et al. 2006; Salz 2011). This suggests that the function of Sxl in female development of D. melanogaster has been evolutionarily acquired rather recently (Cline et al. 2010). It has been proposed that the gene duplication event of the ancestral Sxl gene (which gave rise to the current ssx and Sxl genes in Drosophilids) was instrumental for the change in function. Having two copies of the ancestral Sxl gene presumably reduced selective pressure on one of them, allowing it to evolve more freely and to eventually adapt its current, Drosophila-specific role as a feminizing developmental switch gene (Traut et al. 2006; Cline et al. 2010).
This raises several questions: What was the function of the ancestral Sxl protein, before it became the master regulator of female development in D. melanogaster? Has the gene duplication event facilitated the loss of this ancestral, non-sex-specific function from Sxl (probably by subfunctionalization of Sxl and Ssx)? Or has Sxl, during its recent evolution, acquired novel activities that facilitated adaption to its novel role as master regulator of female development (neo-functionalization)?
Analyses of Sxl and Ssx protein sequences from a variety of insect species revealed that, upon adapting its function in the sex determination cascade in Drosophila, numerous adaptive changes occurred in Sxl, followed by purifying selection. In contrast, lessening of evolutionary pressure on its paralog ssx resulted in excessive positive selection. This suggests subfunctionalization of the two proteins rather than neo-functionalization (Mullon et al. 2012). However, alternative scenarios were also evoked with Sxl evolving a novel function in sex-determination and (i) ssx maintaining the ancestral function(s) (Traut et al. 2006), or (ii) Sxl and ssx subfunctionalizing the ancestral, non-sex-specific activities (Cline et al. 2010). Biochemical data in support of any of the hypotheses are so far missing.
Regardless of the mechanism, the current situation is that Drosophila melanogaster contains two paralogs that appear to have different functions as judged by their mutant phenotypes (sex-specific lethality for Sxl mutants vs. no apparent phenotype for ssx mutants in unchallenged animals). Here we report different activities of Sxl and Ssx in the repression of msl-2 mRNA translation. While both proteins repress translation when bound to the 5′ UTR of the RNA, Ssx fails to regulate via the 3′ UTR due to subtle differences in RRM1 that result in failure to recruit the co-repressor protein Unr. Even though Ssx represses msl-2 translation via the 5′ UTR, its presence in males is not deleterious because the 5′ UTR intron is removed from the majority of the transcripts (Zhou et al. 1995). Since the intron contains the Sxl/Ssx-binding sites, Ssx cannot exert regulation on transcripts lacking the 5′ UTR intron, suggesting that translational repression of msl-2 mRNA by Ssx serves fine-tuning of Msl2 protein levels in male flies.
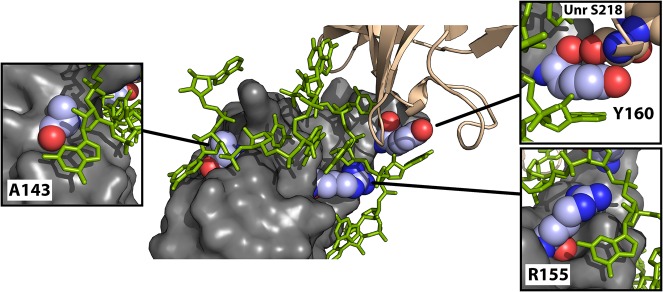
Three rather subtle amino acid substitutions in Ssx are sufficient to promote interaction with Unr and to generate a gain of function (N111A, K123R, and F128Y, Figs. 4, 5). As evident in the crystal structure of the co-repressor complex (Hennig et al. 2014), only Y160 (corresponding to F128 in Ssx) is directly at the interface between Unr and Sxl. This residue has previously been identified as being critical for regulation and is key for Sxl:Unr:RNA complex formation (Hennig et al. 2014). In a triple-zipper-like interaction, Y160 is sandwiched between an Adenosine base 3727 of the msl-2 mRNA and S215 of Unr (Fig. 6). The hydroxyl group of Y160 protrudes into a pocket formed by the peptide backbone of UNR (involving H213, F214, and S215) to further stabilize the interaction—a function that is not supported by F128 in Ssx. However, Y160 is not sufficient to provide complex formation, even together with conserved residues previously shown to establish essential contacts within the complex. These residues (D138, R139, Y142, Y164) are conserved in Ssx; however, the single F128Y mutation in Ssx does not provide significant GOF (Supplemental Fig. 2). Other residues in the vicinity of Y128, which we identify here as A111 and R123, critically contribute to stable complex formation.
FIGURE 6.
A143, R155, and Y160 play important roles in the stabilization of the Sxl:Unr:msl-2 repressor complex. Crystal structure of the Sxl:Unr:msl-2 co-repressor complex (Hennig et al. 2014, PDB: 4QQB). RRM1 of Sxl is shown in gray with the critical amino acids A143, R155, and Y160 highlighted in color and depicted in more detail in the individual panels. CSD1 of Unr is shown in salmon, the msl-2 mRNA in green.
In the absence of Unr, Sxl interactions with the RNA involve canonical contacts established by the β-sheet surface of the RRMs that are also typically observed for other RRM-containing proteins (Handa et al. 1999; Hennig et al. 2014; for review, see Moschall et al. 2017). Upon recruitment of Unr by Sxl, however, the RNA is chaperoned into a different path and becomes sandwiched between the two proteins to wrap around the RRM1 of Sxl in an almost 180° turn (see Fig. 6). This is facilitated by additional interactions of the RNA with the Sxl protein that involve residues of Sxl RRM1, which usually do not contact the RNA upon “canonical” binding (Hennig et al. 2014). Here R155 and A143 play an important role in facilitating interaction with the RNA, explaining why their substitution affects ternary Sxl:Unr:RNA complex formation: R155 interacts with the RNA backbone, positioning it in the proper orientation for wrapping around RRM1 of Sxl; A143 is small enough not to interfere with stacking of the adenosine base 3735 of msl-2 onto the backside helix of Sxl RRM1, allowing shape complementarity (Fig. 6). Our data suggest that the presence of N111 and K123 in Ssx (corresponding to A143 and R155 in Sxl) hinder complex formation with Unr by sterically interfering with packing of the RNA against the backside of Ssx RRM1.
The three critical amino acids (A143, R155, and Y160) are conserved over large evolutionary distances in Sxl orthologs from various insect species, but all members of the Ssx clade show substitutions in at least two of the residues (not shown). This suggests that other members of the Ssx protein clade most probably also lack the ability to recruit Unr.
Previously, it was reported that the Musca domestica Sxl ortholog (mSxl) is detected in both sexes and its expression in Drosophila has no feminizing effect (Meise et al. 1998). mSxl clearly resembles Sxl in the positions that we identified to be critical for regulation via the 3′ UTR (mSxl A94, R106, and Y111, corresponding to A143, R155, and Y160 in Sxl). It also resembles Sxl in other residues important for complex formation (Sxl residues D138, R139, Y142, R146) (Hennig et al. 2014). Surprisingly, however, similar to Ssx, mSXL represses translation of reporters with msl-2 mRNA 5′ UTR regulatory elements, but cannot do so on 3′ UTR reporters (Supplemental Fig. 3). Curiously, mSXL can associate with the regulatory elements in the 3′ UTR of msl-2 mRNA but fails to recruit the co-repressor Unr (Grskovic et al. 2003). Therefore, other unknown properties or residues of Sxl must contribute to recruitment of Unr. In sum, despite being more closely related to Sxl in sequence, mSxl appears to be a functional equivalent of Ssx when it comes to translational control of msl-2 mRNA. This suggests that either the two proteins mSxl and Ssx have lost their ability to recruit Unr, or, more likely, Sxl has recently evolved the ability to recruit Unr for regulation of msl-2 translation, arguing in favor of a neo-functionalization event.
MATERIALS AND METHODS
DNA constructs
For production of recombinant proteins in E.coli, the central RNA-binding domains of Ssx (aa93-269, Ssx-RBD4) and Sxl (aa122–301, Sxl-RBD4) were PCR-amplified and cloned into the pGEX6P3 vector using the BamHI and XhoI restriction sites. For transfection experiments in cultured insect cells, annealed oligonucleotides encoding a FLAG-3xHA sequence were introduced into a modified pCaSpeR-HS vector (Medenbach et al. 2011) using the EcoRI restriction site. Subsequently the full-length open reading frames of Sxl and Ssx were PCR amplified and cloned in-frame with the tag using the EcoRI and XbaI restriction sites. To generate Sxl and Ssx derivatives, site-directed PCR mutagenesis was performed, introducing the desired sequence changes with the primers.
Recombinant protein expression
Recombinant Ssx and Sxl proteins (and mutant derivatives thereof) were expressed by IPTG induction in E. coli (BL21Star [Invitrogen] transformed with the Rosetta 2 plasmid [Merck]) for 4 h at 23°C. Cells were pelleted and resuspended in buffer X (20 mM Tris/Cl pH 7.5, 1 M NaCl, 0.2 mM EDTA, 1 mM DTT, cOmplete Protease Inhibitor Cocktail [Roche]). After cell lysis, debris was removed by centrifugation for 20 min at 12kg and recombinant protein was purified by GSH-affinity chromatography using an ÄKTA FPLC system. Elution occurred in 100 mM HEPES/KOH pH 8.0, 50 mM glutathione, 50 mM KCl, 1 mM DTT. Fractions containing the protein were supplemented with 3C protease and dialyzed overnight against IEX buffer (20 mM HEPES/KOH pH 8.0, 50 mM KCl, 10% Glycerol, 0.2 mM EDTA, 0.01% NP-40) before performing ion exchange chromatography using a MonoS column. After salt elution, fractions containing the pure protein were dialyzed against Dignam buffer D (20 mM HEPES/KOH pH 8.0, 20% Glycerol, 0.2 mM EDTA, 0.01% NP-40, 1 mM DTT) and stored at −80°C. Unr CSD1 was expressed as previously described (Hennig et al. 2014).
RIP and qRT-PCR
Drosophila SL2 cells were propagated at 80% confluency in Express Five SFM supplemented with 10× Glutamax. Fugene HD (Promega) was used according to the manufacturer's instructions to transfect a Ssx-encoding plasmid followed by incubation for 48 h at 25°C. RNA-immunoprecipitation was essentially performed as previously reported (Hernández et al. 2013) with the following modifications: Transfected Drosophila cells were harvested, washed in PBS, and lysed in a buffer containing 50 mM Tris/Cl pH 8.0, 150 mM NaCl, 2 mM MgCl2, 1% NP-40, and 1× cOmplete Protease Inhibitor Cocktail (Roche). The lysate was cleared by centrifugation, and binding to anti-FLAG magnetic beads (Life Technologies) occurred for 3 h at 4°C rotating head over tail. Beads were washed repeatedly with a buffer containing 50 mM Tris/Cl pH 8.0, 150 mM NaCl, 2 mM MgCl2, and 0.1% NP-40. After treatment with proteinase K (30 min at 50°C), RNAs were purified by organic extraction and, after DNase I treatment, subjected to RT-qPCR. Reverse transcription was performed using random hexamer primers and SuperscriptIII (ThermoFisher) according to the manufacturer's instructions, followed by qPCR using the following primers: α tubulin GCTTCCTCATCTTCCACTCG and AATCAGACGGTTCAGGTTGG; msl-2 ACTGGGGAAGGGAACCGAAGCC and CTTCTGCCCCCATAAGCCTAGTGCCG. Amplification efficiencies were calculated based on serial dilutions of the sample and melting curve analysis was performed to ensure purity of the product.
Electromobility shift assays and repressive complex formation
To allow ribonucleoprotein complex formation for EMSA experiments, 10 fmol of 32P-labeled RNA were incubated for 30 min at 4°C with the indicated amounts of protein in a reaction containing 10 mM Tris/Cl pH 7.4, 50 mM KCl, 1 mM EDTA, 1 mM DTT, and 0.2 µg/µL yeast tRNA. Subsequently, complexes were resolved by 8% native PAGE (acrylamide/bisacrylamide 37.5:1) for 2 h at 4°C and 10 V/cm. Detection occurred using the Personal Molecular Imager System (Bio-Rad).
Repressive complex formation in Drosophila embryo extracts was performed essentially as described before (Grskovic et al. 2003; Abaza et al. 2006), except that the complexes were precipitated with Glutathione Sepharose.
Translation assays
In vitro translation assays and tissue culture-based reporter assays were performed as previously described (Medenbach et al. 2011). In brief, msl-2-based, capped and polyadenylated firefly luciferase reporter RNAs were generated by in vitro transcription in the presence of a 3′-O-Me-m7(5′)Gppp(5′)G (“anti-reverse”) cap analog (NEB). 130 fmol of the purified firefly luciferase reporter RNA and 23 fmol of a Renilla luciferase control RNA were then subjected to translation in a reaction containing 24 mM HEPES/KOH (pH 7.4), 100 mM KOAc, 0.5 mM Mg(OAc)2, 60 mM amino acids, 20 mM creatine phosphate, 800 ng creatine kinase, and 40% Drosophila embryo extract (Gebauer et al. 1999). Translation occurred either in the absence or presence of 1 µM of the indicated recombinant Sxl or Ssx proteins. After incubation for 90 min at 25°C, luciferase activities were assayed with the Dual Luciferase Assay System (Promega) in a microplate luminometer (Berthold) and normalized to control reactions supplemented with buffer instead of Ssx or Sxl protein.
For translation experiments in cultured cells, Drosophila SL-2 cells were transfected at a confluency of ∼50% using FugeneHD according to the manufacturer's instructions with a ratio of 1:3 DNA to FugeneHD. Per well of a 48-well plate, equal amounts (45 ng) of a msl-2 mRNA based Renilla luciferase reporter plasmid and a firefly luciferase-encoding control plasmid were transfected along with 75 ng of a Sxl expression plasmid (or derivatives thereof) or an empty vector control. After 48 h the cells were harvested, washed, and lysed in 1× passive lysis buffer (Promega). After clearing the extracts by centrifugation, Renilla and firefly luciferase activity was determined using the Dual Luciferase Assay System (Promega) in a microplate luminometer (Berthold).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the German Research Foundation (grant ME4238/1-1 and SFB 960/2 to J.M.); the Bavarian State Ministry for Education, Science and the Arts (Bavarian Research Network for Molecular Biosystems, BioSysNet, to J.M.); the German Federal Ministry of Education and Research (BMBF, grant 01ZX1401D to J.M.); and the Spanish Ministry of Economy and Competitiveness (MINECO) and the European Regional Development Fund (ERDF) (grants BFU2012-37135, BFU2015-68741, and CSD2009-00080 to F.G.). We acknowledge support of the MINECO “Centro de Excelencia Severo Ochoa 2013-2017,” SEV-2012-0208. We thank O. Rossbach, G. Lehmann, N. Eichner, and G. Meister for help with the CLIP assay, and B. Rogell and MW. Hentze for sharing of unpublished data.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063776.117.
REFERENCES
- Abaza I, Gebauer F. 2008. Functional domains of Drosophila UNR in translational control. RNA 14: 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaza I, Coll O, Patalano S, Gebauer F. 2006. Drosophila UNR is required for translational repression of male-specific lethal 2 mRNA during regulation of X-chromosome dosage compensation. Genes Dev 20: 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Freitag N, Schneider DS. 2008. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics 178: 1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw GJ, Baker BS. 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789–798. [DOI] [PubMed] [Google Scholar]
- Beckmann K, Grskovic M, Gebauer F, Hentze MW. 2005. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila. Cell 122: 529–540. [DOI] [PubMed] [Google Scholar]
- Cline TW, Dorsett M, Sun S, Harrison MM, Dines J, Sefton L, Megna L. 2010. Evolution of the Drosophila feminizing switch gene Sex-lethal. Genetics 186: 1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. 2012. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet 13: 123–134. [DOI] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Schedl PD. 1999. The N-terminal domain of Sxl protein disrupts Sxl autoregulation in females and promotes female-specific splicing of tra in males. Development 126: 2841–2853. [DOI] [PubMed] [Google Scholar]
- Duncan K, Grskovic M, Strein C, Beckmann K, Niggeweg R, Abaza I, Gebauer F, Wilm M, Hentze MW. 2006. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: translational repression for dosage compensation. Genes Dev 20: 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan KE, Strein C, Hentze MW. 2009. The SXL-UNR corepressor complex uses a PABP-mediated mechanism to inhibit ribosome recruitment to msl-2 mRNA. Mol Cell 36: 571–582. [DOI] [PubMed] [Google Scholar]
- Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, Anderson P, Valcárcel J. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 6: 1089–1098. [DOI] [PubMed] [Google Scholar]
- Förch P, Merendino L, Martínez C, Valcárcel J. 2001. Modulation of msl-2 5′ splice site recognition by Sex-lethal. RNA 7: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. 2007. Studying translational control in Drosophila cell-free systems. Methods Enzymol 429: 23–33. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Merendino L, Hentze MW, Valcárcel J. 1998. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA 4: 142–150. [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Corona DF, Preiss T, Becker PB, Hentze MW. 1999. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J 18: 6146–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Grskovic M, Hentze MW. 2003. Drosophila sex-lethal inhibits the stable association of the 40S ribosomal subunit with msl-2 mRNA. Mol Cell 11: 1397–1404. [DOI] [PubMed] [Google Scholar]
- Graindorge A, Militti C, Gebauer F. 2011. Posttranscriptional control of X-chromosome dosage compensation. Wiley Interdiscip Rev RNA 2: 534–545. [DOI] [PubMed] [Google Scholar]
- Graindorge A, Carré C, Gebauer F. 2013. Sex-lethal promotes nuclear retention of msl2 mRNA via interactions with the STAR protein HOW. Genes Dev 27: 1421–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grskovic M, Hentze MW, Gebauer F. 2003. A co-repressor assembly nucleated by Sex-lethal in the 3′UTR mediates translational control of Drosophila msl-2 mRNA. EMBO J 22: 5571–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. 1999. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398: 579–585. [DOI] [PubMed] [Google Scholar]
- Hennig J, Militti C, Popowicz GM, Wang I, Sonntag M, Geerlof A, Gabel F, Gebauer F, Sattler M. 2014. Structural basis for the assembly of the Sxl-Unr translation regulatory complex. Nature 515: 287–290. [DOI] [PubMed] [Google Scholar]
- Hernández G, Miron M, Han H, Liu N, Magescas J, Tettweiler G, Frank F, Siddiqui N, Sonenberg N, Lasko P. 2013. Mextli is a novel eukaryotic translation initiation factor 4E-binding protein that promotes translation in Drosophila melanogaster. Mol Cell Biol 33: 2854–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI. 1995. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81: 867–877. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Wang J, Bell L, Kuroda MI. 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195–199. [DOI] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW. 2011. Translational control via protein-regulated upstream open reading frames. Cell 145: 902–913. [DOI] [PubMed] [Google Scholar]
- Meise M, Hilfiker-Kleiner D, Dübendorfer A, Brunner C, Nöthiger R, Bopp D. 1998. Sex-lethal, the master sex-determining gene in Drosophila, is not sex-specifically regulated in Musca domestica. Development 125: 1487–1494. [DOI] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martínez C, Valcárcel J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Moschall R, Gaik M, Medenbach J. 2017. Promiscuity in post-transcriptional control of gene expression: Drosophila Sex-lethal and its regulatory partnerships. FEBS Lett 591: 1471–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullon C, Pomiankowski A, Reuter M. 2012. Molecular evolution of Drosophila Sex-lethal and related sex determining genes. BMC Evol Biol 12: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva LO, Sánchez L. 2003. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol Mol Biol Rev 67: 343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestel M, Feller C, Becker PB. 2010. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol 11: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogell B, Fischer B, Rettel M, Krijgsveld J, Castello A, Hentze MW. 2017. Specific RNP capture with antisense LNA/DNA mixmers. RNA 23: 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz HK. 2011. Sex determination in insects: a binary decision based on alternative splicing. Curr Opin Genet Dev 21: 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz HK, Erickson JW. 2010. Sex determination in Drosophila: the view from the top. Fly 4: 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawanth SK, Gopinath G, Sambrani N, Arunkumar KP. 2016. The autoregulatory loop: a common mechanism of regulation of key sex determining genes in insects. J Biosci 41: 283–294. [DOI] [PubMed] [Google Scholar]
- Traut W, Niimi T, Ikeo K, Sahara K. 2006. Phylogeny of the sex-determining gene Sex-lethal in insects. Genome 49: 254–262. [DOI] [PubMed] [Google Scholar]
- Venables JP, Tazi J, Juge F. 2012. Regulated functional alternative splicing in Drosophila. Nucleic Acids Res 40: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanowitz JL, Deshpande G, Calhoun G, Schedl PD. 1999. An N-terminal truncation uncouples the sex-transforming and dosage compensation functions of sex-lethal. Mol Cell Biol 19: 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Yang Y, Scott MJ, Pannuti A, Fehr KC, Eisen A, Koonin EV, Fouts DL, Wrightsman R, Manning JE, et al. 1995. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J 14: 2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.