Here, Estaras et al. researched how the Hippo effector YAP represses hESC differentiation and demonstrate that YAP binds to the WNT3 gene enhancer and prevents the gene from being induced by Activin in proliferating hESCs. Their findings indicate that YAP maintains hESC pluripotency by preventing WNT3 expression in response to Activin, thereby blocking a direct route to embryonic cardiac mesoderm formation.
Keywords: hESCs, YAP, stem cell differentiation, cardiac mesoderm development
Abstract
Activin/SMAD signaling in human embryonic stem cells (hESCs) ensures NANOG expression and stem cell pluripotency. In the presence of Wnt ligand, the Activin/SMAD transcription network switches to cooperate with Wnt/β-catenin and induce mesendodermal (ME) differentiation genes. We show here that the Hippo effector YAP binds to the WNT3 gene enhancer and prevents the gene from being induced by Activin in proliferating hESCs. ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data show that YAP impairs SMAD recruitment and the accumulation of P-TEFb-associated RNA polymerase II (RNAPII) C-terminal domain (CTD)-Ser7 phosphorylation at the WNT3 gene. CRISPR/CAS9 knockout of YAP in hESCs enables Activin to induce Wnt3 expression and stabilize β-catenin, which then synergizes with Activin-induced SMADs to activate a subset of ME genes that is required to form cardiac mesoderm. Interestingly, exposure of YAP−/− hESCs to Activin induces cardiac mesoderm markers (BAF60c and HAND1) without activating Wnt-dependent cardiac inhibitor genes (CDX2 and MSX1). Moreover, canonical Wnt target genes are up-regulated only modestly, if at all, under these conditions. Consequently, YAP-null hESCs exposed to Activin differentiate precisely into beating cardiomyocytes without further treatment. We conclude that YAP maintains hESC pluripotency by preventing WNT3 expression in response to Activin, thereby blocking a direct route to embryonic cardiac mesoderm formation.
In human embryonic stem cells (hESCs) and the developing blastocyst, Wnt and Activin signals cooperate to induce key mesendodermal (ME) differentiation genes that are needed to form the primitive streak (PS), mesoderm, and endoderm layers (Nostro et al. 2008; Wang and Chen 2016). In self-renewing hESCs, Activin signaling maintains pluripotency by activating NANOG expression (Sakaki-Yumoto et al. 2013). Prior to gastrulation and formation of the PS, Activin induces the expression of the WNT3 ligand, which activates β-catenin and downstream ME differentiation genes, including MIXL1, T, and EOMES. Once activated, the combination of Wnt and Activin signaling sustains the expression of these genes through a positive feedback loop (Arkell et al. 2013; Robertson 2014). Much is known about the transcription networks that control the expression of proliferation genes in hESCs. In contrast, much remains to be learned about how the Activin and Wnt signaling pathways selectively induce ME genes in early differentiation and what mechanisms prevent the inadvertent expression of these genes in self-renewing stem cells.
The coordinate up-regulation of ME differentiation genes by Wnt and Activin also provides an important paradigm for understanding the mechanistic basis of transcriptional synergy (Sumi et al. 2008; Loh et al. 2014; Naujok et al. 2014). Wnt signaling transiently stabilizes the β-catenin coactivator, which associates with nuclear LEF-1/TCF DNA-binding proteins to activate downstream target genes (Clevers and Nusse 2012). In hESCs, binding of β-catenin:LEF-1 complexes to distal ME gene enhancers induces cohesin-dependent enhancer–promoter looping and transcription initiation (Estarás et al. 2015). Expression of the TGF-β ligands Activin and Nodal mobilizes the SMAD2,3 DNA-binding proteins, which bind to ME gene promoters (Hansen et al. 2015; Luo 2017) in a p53-dependent manner (Wang et al. 2017) and further enhance transcription. Transcription at Wnt-activated ME genes is accompanied by the assembly of the RNA polymerase II (RNAPII) pause–release complex at the promoter, which establishes a requirement for the P-TEFb elongation factor (CycT1:CDK9) (Jonkers and Lis 2015). Subsequent recruitment of the SMAD2,3 proteins, which bind tightly to P-TEFb (Alarcón et al. 2009), dramatically increases P-TEFb recruitment, transcription elongation, and RNAPII C-terminal domain (CTD)-Ser7 phosphorylation (Ser7P) levels within the gene body (Czudnochowski et al. 2012) to support optimal ME gene expression in vivo (Estarás et al. 2015).
In hESCs, activation of the ME genes is directly counteracted by the Hippo effector YAP. YAP controls cell and organ size during development through various effects on cell proliferation and apoptosis (Lian et al. 2010; Barry and Camargo 2013; Hansen et al. 2015; Yu et al. 2015; Meng et al. 2016). In the nucleus, YAP/TAZ proteins associate with TEAD1–4 DNA-binding proteins and can regulate P-TEFb levels at target genes (Liu-Chittenden et al. 2012; Galli et al. 2015). Although YAP/TAZ proteins most often function as coactivators, they can also repress transcription (Barry et al. 2013; Azzolin et al. 2014; Kim et al. 2015), as is the case at ME genes. YAP repression involves recruitment of the NuRD complex, which prevents the displacement of promoter-proximal nucleosomes at the ME genes (Beyer et al. 2013). ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data further show that YAP dramatically reduces P-TEFb occupancy at ME genes and blocks the accumulation of RNAPII CTD-Ser7P in the transcribed region (Estarás et al. 2015). Interestingly, YAP does not regulate canonical Wnt or Activin target genes but instead functions as a ME gene-specific corepressor.
In this study, we examine the role of YAP in proliferating hESCs and uncover a previously unsuspected role as a selective inhibitor of hESC differentiation to cardiac mesoderm. We found that YAP binds to an intragenic enhancer of the WNT3 gene and prevents its premature activation by Activin in self-renewing hESCs. In the absence of YAP, Activin-induced SMAD2,3 proteins up-regulate WNT3 expression and specifically induce key ME genes. Moreover, YAP knockout cells exposed to Activin alone for a short period were found to differentiate specifically to cardiac mesoderm. This process did not stimulate most canonical Wnt target genes or activate the Wnt-responsive cardiac developmental inhibitors CDX2 or MSX1. As a consequence, a short exposure of YAP knockout cells to Activin was sufficient to trigger the full cardiomyocyte differentiation program. Taken together, our data show that YAP masks a previously unknown direct route toward cardiac mesoderm formation in hESCs. These findings may be exploited in the future to facilitate cardiac differentiation procedures for use in regenerative stem cell therapies (Atmanli and Domian 2017; Duelen and Sampaolesi 2017).
Results
YAP prevents hESC differentiation in response to Activin signaling
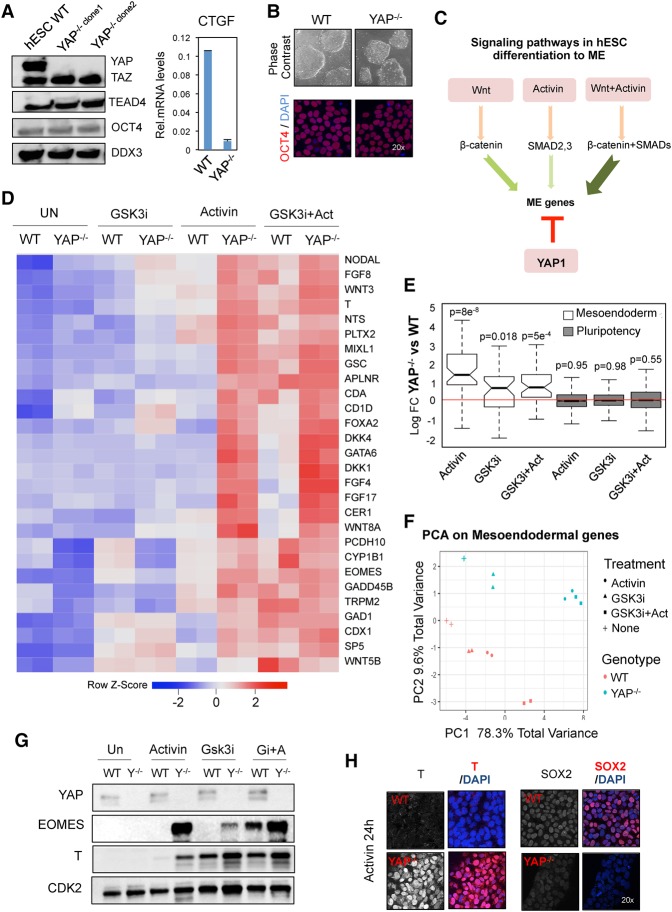
To better understand how YAP regulates hESC differentiation, YAP knockout hESCs were developed using the clustered regularly interspaced short palindromic repeats CRISPR/CAS9 gene-editing approach (Ran et al. 2013). Immunoblot analysis confirmed the loss of YAP expression in two independently derived clones (Fig. 1A, left panel). Importantly, ablation of YAP had no effect on the expression of the closely related protein TAZ and did not affect levels of the TEAD4 DNA-binding proteins. As expected, expression of the well-characterized YAP target gene CTGF was greatly reduced in YAP knockout cells (Fig. 1A, right panel; Supplemental Fig. S1A). The YAP knockout hESCs displayed a higher rate of apoptosis but nevertheless readily formed colonies that express high levels of pluripotency markers, such as OCT4 (Fig. 1A,B), indicating that YAP is not critical for hESC self-renewal. Moreover, the observation that CTGF expression is dramatically impaired in YAP-ablated cells despite normal expression of TAZ indicates that the two paralogs do not have identical functions in hESCs.
Figure 1.
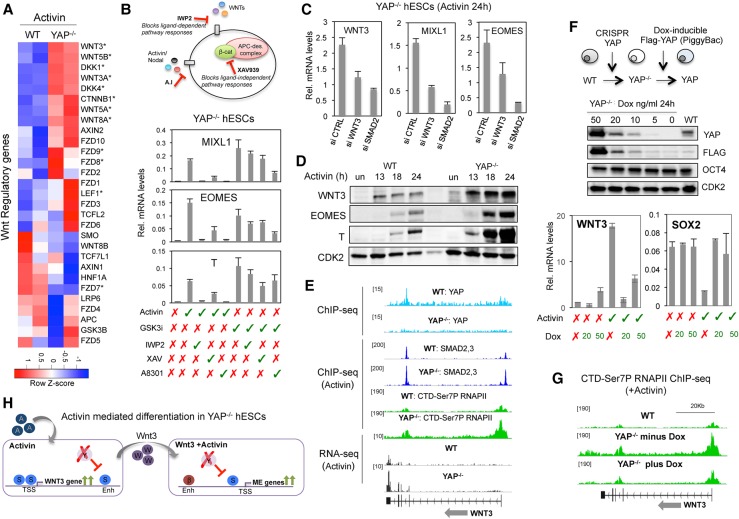
YAP selectively prevents hESC differentiation to mesendoderm in response to Activin. (A) Isolation of YAP knockout hESC clones. (Left) Immunoblot of two different YAP knockout clones. (Right) Quantitative PCR (qPCR) analysis of CTGF mRNA levels in wild-type and YAP knockout hESCs. Mean (SD). n = 3. (B) Phase contrast microscopy captures show colonies of wild-type and YAP knockout hESCs. OCT4 protein levels were analyzed by immunostaining. (C) Protocol for Activin- and Wnt-dependent hESC differentiation to ME cells. Activin and Wnt pathways synergize to induce optimal levels of ME gene transcription. YAP selectively blocks ME gene expression through an unknown mechanism. (D) Heat map display of RNA sequencing (RNA-seq) data from wild-type and YAP knockout hESCs treated with either 50 ng/mL Activin, GSK3 inhibitor (GSK3i; low concentration; 6 µM ChIR99021 [ChIR]), or Activin+GSK3i for 24 h. The heat map ranks the genes most strongly affected by Activin in YAP knockout cells. (E) Box plot showing the log fold change expression of pluripotency and ME differentiation genes in wild-type or YAP knockout hESCs after the indicated treatments. The adjusted P-value for multiple testing correction is shown above each box. (F) ME gene expression profiles (52 genes) projected onto the first two principal components. hESC lines and treatments are designated by colors and symbols. (G) Representative immunoblot of the indicated proteins in wild-type and YAP knockout cells treated for 24 h as indicated above each lane. n ≥ 3. (H) Immunostaining of wild-type and YAP knockout cells following 24 h of exposure to Activin.
As indicated in Figure 1C, Activin synergizes with Wnt ligands to induce ME developmental genes, whereas YAP selectively represses these genes. To identify additional targets for YAP in proliferating hESCs, we used RNA sequencing (RNA-seq) (Fig. 1D) to analyze the transcriptome of control and YAP knockout hESCs treated with either Activin, a GSK3 inhibitor (GSK3i) that activates Wnt signaling, or the combination of both molecules (GSK3i+Activin). The GSK3i ChIR99021 (ChIR) was used for this experiment because Wnt cytokines are not stable over the 24-h incubation period. Of note, a low concentration of GSK3i (6 µM ChIR) was used for the RNA-seq experiment to elevate ME gene expression to levels comparable with that observed with Activin alone in wild-type hESCs (Supplemental Fig. S1B). Interestingly, the ME developmental genes (MIXL1, EOMES, and T) were much more strongly induced by Activin alone in YAP knockout hESCs than in wild-type hESCs, as visualized in the heat map display of the RNA-seq data set (Fig. 1D,E; Supplemental Fig. S1C; Supplemental Table S1). Indeed, Activin was as effective as the combination of Wnt+Activin for inducing ME differentiation genes in YAP−/− cells (Fig. 1D–F). On the other hand, loss of YAP had little or no effect on ME gene activation in response to the Wnt/β-catenin inducer GSK3i/ChIR on its own (Fig. 1D,E; Supplemental Fig. S1C; Supplemental Table S1). Immunoblot and immunofluorescence analysis confirmed the dramatic increase of EOMES and T protein expression as well as the loss of SOX2 in YAP knockout cells exposed to Activin (Fig. 1G,H). Importantly, knockdown of YAP in H9 hESCs or the EC-11 iPSCs (induced pluripotent stem cells) also induced ME gene expression in response to Activin signaling (Supplemental Fig. S1D). We conclude that YAP acts to safeguard proliferating hESCs by protecting against the aberrant activation of ME differentiation genes in the presence of Activin.
β-Catenin is required for Activin-induced differentiation of YAP knockout hESCs
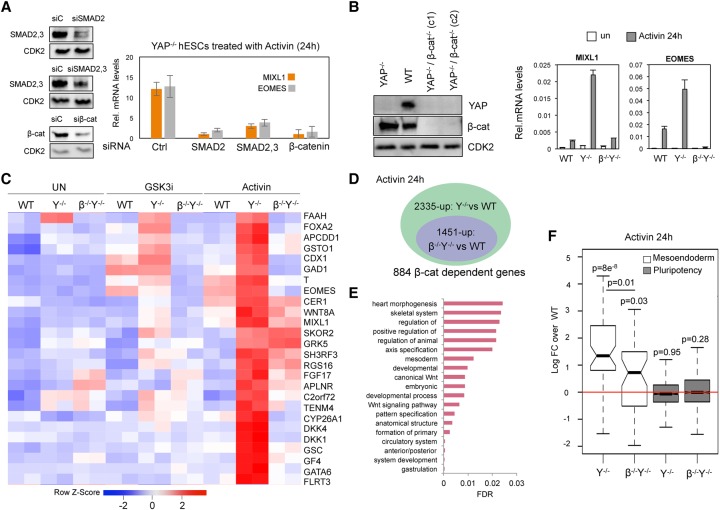
The surprising observation that Activin signaling was sufficient to induce ME genes in YAP knockout hESCs raised the question of whether the activation of these genes remains dependent on β-catenin despite the absence of exogenous Wnt ligands. Unexpectedly, we found that transient knockdown of β-catenin in YAP knockout cells blocks the induction of ME genes by Activin (Fig. 2A), as did depletion of the SMAD2,3 proteins. To further test whether functional β-catenin is present in Activin-treated YAP knockout cells, luciferase reporter vectors containing composite SMAD- and β-catenin-binding sites from the MIXL1 gene were transfected in wild-type and YAP knockout cells. As shown in Supplemental Fig. S2A, Activin alone was sufficient to induce the MIXL1 reporter gene in YAP knockout cells but not in wild-type hESCs. Taken together, these data suggest that β-catenin is critical for the activation of ME genes in Activin-treated YAP knockout hESCs.
Figure 2.
β-Catenin is required for Activin-induced differentiation of YAP knockout hESCs. (A) The efficiency of siRNA-mediated knockdown of the indicated proteins was analyzed by immunoblot. CDK2 was included as a loading control. The graph at the right shows the mRNA levels of the EOMES and MIXL1 genes in Activin-treated YAP knockout hESCs transfected with the indicated siRNAs. Mean (SD). n = 3. (B) Immunoblot analysis of the YAP and β-catenin double-knockout hESC lines. Analysis by qRT–PCR reveals that EOMES and MIXL1 are not induced by Activin in the YAP:β-catenin double-knockout (β−/−Y−/−) cells. Mean (SD). n = 3. (C) Heat map presentation of RNA-seq data ranking the genes most strongly affected by Activin treatment in YAP knockout cells. (D) The diagram depicts the number of genes that are induced by Activin signaling in YAP knockout cells but no longer induced in YAP/β-catenin knockout cells. (E) Gene ontology analysis of the genes down-regulated in the YAP:β-catenin knockout cells compared with YAP knockout cells. (F) The box plot shows the log fold change of YAP knockout and YAP:β-catenin knockout cell lines compared with wild-type hESCs for the set of genes involved in pluripotency and ME differentiation. The adjusted P-value for multiple testing correction is shown above each box.
To further test this conclusion, the endogenous β-catenin gene was removed from the YAP knockout hESCs using CRISPR technology (Fig. 2B). Interestingly, the double-knockout cell line remained pluripotent, although these cells grew slower than either YAP knockout or wild-type cells (data not shown). Analysis of MIXL1 and EOMES gene transcripts in these cells by quantitative PCR (qPCR) confirmed that they failed to respond to Activin in the absence of β-catenin (Fig. 2B; Supplemental Fig. S2B,C). To identify additional gene targets, RNA-seq experiments were carried out to compare the transcriptomes of the YAP knockout and YAP:β-catenin double-knockout cell lines treated with either GSK3i or Activin. A heat map display of the results revealed that expression of many key ME differentiation genes was severely impaired in the double-knockout cell line (Fig. 2C). Most notably, this approach identified 884 Activin-induced genes in the YAP knockout cells that require β-catenin (Fig. 2D). The majority of these β-catenin-regulated genes was associated with mesoderm and heart development (Fig. 2E). In contrast, the expression of canonical Activin-regulated genes, such as SMAD6 and LEFTY1, or defined pluripotency genes, such as OCT4, did not require β-catenin (Fig. 2F; Supplemental Table S1). Taken together, these data indicate that Activin selectively induces ME differentiation genes in YAP knockout cells, acting through an unknown mechanism to mobilize nuclear β-catenin.
Activin induces genome-wide recruitment of β-catenin to chromatin in YAP knockout cells
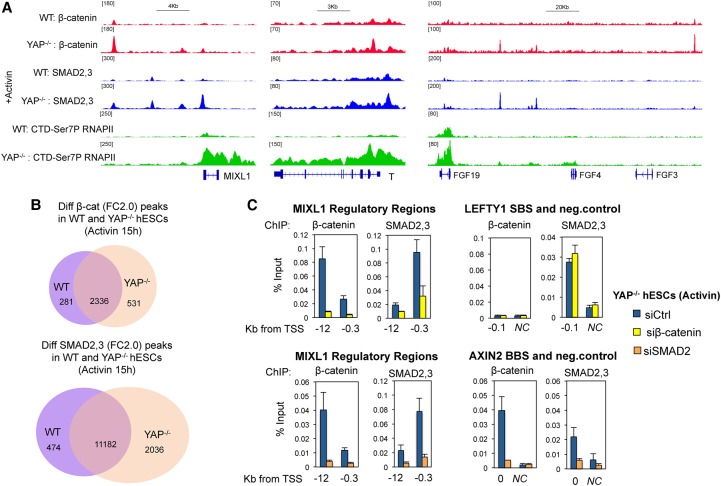
Next, we evaluated the binding of β-catenin to ME gene enhancers in wild-type and YAP knockout cells treated with Activin. The cells were treated with Activin for 15 h, and the number and distribution of β-catenin peaks were analyzed by ChIP-seq. In parallel, we also carried out ChIP-seq experiments to examine the binding profiles of SMAD2,3 proteins and the accumulation of the elongation-competent RNAPII CTD-Ser7P in gene bodies as a readout of transcriptional activity. Figure 3A and Supplemental Figure S3A show that β-catenin was indeed recruited to ME genes, such as MIXL1, T, and FGF, in Activin-treated YAP knockout cells. Bioinformatics analysis identified 531 new β-catenin peaks in YAP knockout cells compared with wild-type cells. Interestingly, the genome-wide recruitment of SMAD2,3 proteins was also greater in the YAP knockout cells treated with Activin (2036 new peaks) (Fig. 3B).
Figure 3.
Activin induces binding of β-catenin genome-wide in YAP knockout hESCs. (A) Wild-type and YAP knockout hESCs were treated with Activin for 15 h prior to ChIP-seq analysis of SMAD2,3, β-catenin, and elongation-competent RNAPII CTD-Ser7P at target genes genome-wide. Captures show the distribution of immunoprecipitated proteins at the selected ME genes. (B) Venn diagrams depicting the number of SMAD and β-catenin peaks in Activin-treated wild-type and YAP knockout cells. Note that 531 new β-catenin peaks appear in YAP knockout cells, which are absent in wild-type hESCs. (C) ChIP-qPCR in YAP knockout cells transfected with siRNAs against SMAD2 or β-catenin, as indicated. (SBS) SMAD-binding site; (BBS) β-catenin-binding site; (NC) negative control. Mean (SD) n = 3.
Previous studies have shown that β-catenin is a limiting factor for induction of ME differentiation, and Wnt ligands act upstream of SMAD signaling in hESCs (Estarás et al. 2015; Wang et al. 2017). Supporting this idea, our ChIP-seq analysis indicates that 41% of the new β-catenin peaks were accompanied by new SMAD2,3 peaks (±50 kb), whereas only 8.8% of the new SMAD peaks were located near β-catenin peaks (Supplemental Fig. S3B). To assess whether SMAD occupancy is affected by β-catenin, we transfected a siRNA against β-catenin in YAP knockout cells and assessed binding of SMAD2,3 to specific ME genes and the canonical Activin target genes LEFTY and OCT4. Interestingly, reduced levels of β-catenin impaired SMAD2,3 recruitment to ME genes, including MIXL1 and EOMES, but did not affect SMAD2,3 binding to the LEFTY or OCT4 gene promoters (Fig. 3C; Supplemental Fig. S3C). Of note, β-catenin knockdown affected the binding of SMAD proteins to ME gene regulatory regions that are distal to β-catenin-binding sites. For example, loss of the β-catenin peak at the −12-kb MIXL1 enhancer affected SMAD binding at the −0.3-kb enhancer of the same gene. Therefore, in YAP knockout cells, Activin induces β-catenin, which further stabilizes binding of SMAD2,3 proteins to ME genes in part as a downstream consequence of β-catenin-induced enhancer–gene looping and transcription initiation (Estarás et al. 2015).
To evaluate whether SMAD proteins also affect the binding of β-catenin, we transfected siRNAs against SMAD2 in YAP knockout cells treated with Activin and analyzed β-catenin occupancy by ChIP-qPCR. Surprisingly, siRNAs directed against SMAD proteins reduced β-catenin binding to not only the ME differentiation genes but also canonical Wnt target genes such as LEF-1 and AXIN2, where SMAD is not bound (Fig. 3C; Supplemental Fig. S3C). Rather than recruiting β-catenin to chromatin, these data suggest that SMADs function at an upstream step to activate nuclear β-catenin in YAP knockout cells.
YAP represses the WNT3 and NODAL enhancers in hESCs
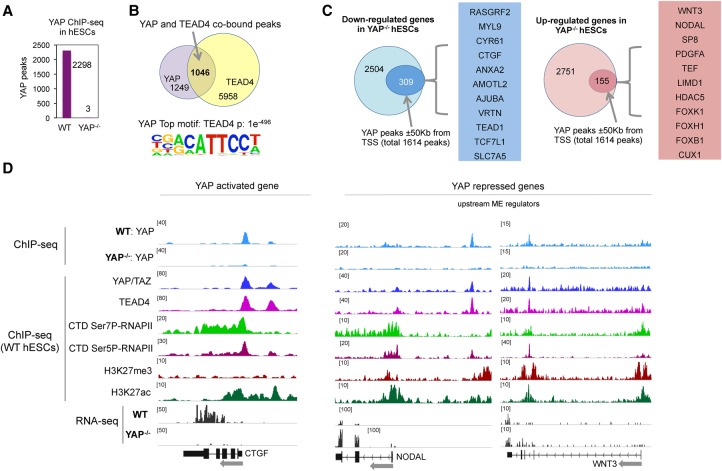
To identify the genes directly regulated by YAP in hESCs, ChIP-seq experiments were carried out to analyze YAP- and TEAD4-binding sites genome-wide. Ablation of YAP led to a loss of >99.9% of the identified peaks, confirming the specificity of the antibody (Fig. 4A; Supplemental Table S2). Bioinformatics analysis revealed that the TEAD4 element was the top DNA motif bound by YAP, and 1046 elements were cobound by YAP and TEAD4 in hESCs (Fig. 4B; Supplemental Fig. S4A). The majority of YAP peaks localized to intergenic and intronic regions (>80%), with only 11% of the peaks mapping to gene promoters (Supplemental Fig. S4B). Among these, 842 YAP peaks overlapped with the active enhancer mark H3K27ac, whereas 130 peaks were associated with repressive H3K27me3 (Supplemental Fig. S4C). Overall, the majority of YAP peaks maps to gene enhancer regions in hESCs.
Figure 4.
YAP binds to enhancers of developmental genes in hESCs. (A) Graph showing the number of YAP ChIP-seq peaks in wild-type and YAP knockout hESCs. (B) Venn diagram of overlapping of TEAD4 and YAP peaks. The bottom panel shows the top motif bound by YAP in hESCs. (C) Circle diagrams showing the top regulated genes ([blue] down-regulated; [red] up-regulated) that are differentially expressed in YAP knockout versus wild-type hESCs, based on the RNA-seq data. Genes that have a YAP peak within 50 kb from transcriptional start site and are transcriptionally regulated are indicated with small circles. A representative selection of these genes is shown in the columns. (D) Genome browser captures of ChIP-seq and RNA-seq experiments. The immunoprecipitated proteins are indicated at the left. The examples shown are from the YAP-activated CTGF gene and the directly repressed NODAL and WNT3 genes.
Analysis of the transcriptional profile of YAP knockout cells revealed that 2504 genes are down-regulated, whereas 2751 genes are up-regulated (false discovery rate [FDR] > 0.05) relative to wild-type hESCs (Fig. 4C,D; Supplemental Fig. S4D; Supplemental Table S3). Included among the list of genes dependent on YAP were several well-known YAP targets, including CTGF and CYR61. In contrast, only 155 genes appeared to be directly repressed by YAP in hESCs. Importantly, the list of repressed genes includes NODAL and WNT3, which regulate the formation of the PS in developing embryos.
Analysis of the ChIP-seq data revealed that YAP binds to a −14-kb enhancer in the NODAL gene. This site includes other signatures of gene enhancers, including an enhancer-specific form of RNAPII that contains high levels of CTD-Ser5P and low levels of CTD-Ser7P (Estarás et al. 2015) as well as the repressive histone H3K27me3 mark. Loss of YAP strongly induced NODAL transcription (Fig. 4D), indicating that YAP directly represses NODAL in proliferating hESCs. Moreover, increased NODAL expression led to enhanced binding of SMAD2,3 proteins genome-wide in YAP knockout hESCs (Fig. 3B). Similarly, we noted a YAP peak in an intragenic enhancer of the WNT3 gene (+45 kb) and found that loss of YAP modestly induced WNT3 gene transcription (Fig. 4D). Thus, YAP repression of WNT3 and NODAL prevents their ectopic activation in self-renewing hESCs, and, in the absence of YAP, hESCs become primed to differentiate toward ME cell fates.
WNT3 activation is essential for Activin-induced differentiation in YAP knockout cells
How does YAP regulate the Wnt/β-catenin pathway? Figure 5A shows a heat map display of Wnt regulatory genes ranked from the RNA-seq analysis of wild-type and YAP knockout cells after 24 h of exposure to Activin. Included among the up-regulated genes are those encoding the Wnt ligands WNT3 and WNT5A as well as the Wnt receptors FZD8 and FZD9. To confirm the importance of this pathway in the YAP knockout hESCs, we next tested two different small-molecule Wnt inhibitors. Addition of either the Wnt palmitoylation inhibitor IWP-2 or the Axin2-stabilizing drug XAV939 was sufficient to block activation of the EOMES, MIXL1, and T ME genes in Activin-treated YAP knockout cells (Fig. 5B). In contrast, the inhibitors had no effect on transcription of the Activin target gene LEFTY1 (Supplemental Fig. S5A). Among the various Wnt pathway proteins up-regulated in YAP knockout cells (Fig. 5A), we considered WNT3 to be particularly relevant because it is essential for formation of the PS and gastrulation. In support of this possibility, transfection of YAP knockout cells with siRNAs directed against WNT3 was sufficient to block the Activin-induced expression of the MIXL1 and EOMES genes (Fig. 5C). Moreover, a time course analysis of WNT3, EOMES, and T protein expression in Activin-treated cells revealed that expression of the Wnt ligand precedes the induction of ME differentiation genes (Fig. 5D). Furthermore, WNT3 mRNA levels were also induced by Activin in H9 and EC-11 iPSC lines transfected with siRNA against YAP (Supplemental Fig. S5B). Taken together, these data strongly suggest that derepression of the WNT3 gene is responsible for the induction of Wnt+Activin codependent ME genes in YAP knockout cells (Fig. 5H).
Figure 5.
YAP repression of WNT3 prevents premature differentiation in response to Activin. (A) Heat map showing expression levels of Wnt regulatory genes in wild-type and YAP knockout cells following treatment with Activin. (B) Diagram showing the mechanism of action of the Wnt inhibitors IWP2 and XAV and the Activin inhibitor A8301. The graphs show qPCR analysis of the indicated genes in YAP knockout hESCs treated with the specific compounds, as listed below the graphs. Mean (SD). n = 3. (C) YAP knockout hESCs were transfected with siRNAs against Wnt3 or SMAD2, and expression levels of the WNT3, EOMES, and MIXL1 genes were analyzed by qPCR. Mean (SD). n = 2. (D) Immunoblot showing expression levels of WNT3, EOMES, and T proteins after Activin treatment for the indicated times in wild-type and YAP knockout cells. CDK2 was used a loading control. (E) Genome browser captures of ChIP-seq and RNA-seq experiments at the WNT3 gene. Cell lines and immunoprecipitated proteins are indicated above each lane. (F) YAP knockout cells were transfected with the transposon-based vector PiggyBAC containing an intact Flag-YAP cDNA under a doxycycline promoter. The immunoblot shows YAP levels in cells exposed to different levels of doxycycline. The graphs illustrate the inverse correlation of WNT3 and YAP mRNAs in hESCs. Mean (SD). n = 2. (G) Genome browser capture of RNAPII CTD-Ser7P ChIP-seq in Activin-treated wild-type and PiggyYAP cell lines before and after doxycycline treatment. (H) A schematic depiction of the results of the figure. (S) SMAD2,3; (β) β-catenin; (Y) YAP; (enh) enhancer; (TSS) transcriptional start site.
YAP prevents SMAD2,3 binding to the WNT3 promoter and counteracts transcription elongation
We next tested whether YAP blocks Activin/SMAD binding to the WNT3 gene. Consistent with this possibility, the loss of YAP enhanced SMAD2,3 binding at the WNT3 gene promoter and increased levels of elongating RNAPII CTD-Ser7P within the transcribed region (Fig. 5E) concomitantly with enhanced WNT3 gene transcription (Fig. 5A,E). Taken together, these data indicate that YAP represses transcription in part through blocking the binding of SMAD2,3 proteins to the promoter, which prevents P-TEFb/CDK9 recruitment and accumulation of RNAPII CTD-Ser7P at the gene.
To test whether YAP can directly control these steps, it was important to determine whether reintroduced wild-type YAP protein could rescue the defect observed in YAP knockout cells. For this purpose, the YAP knockout cells were stably transfected with the transposon-based vector PiggyBAC containing a Flag-YAP cDNA construct under the control of a doxycycline-inducible promoter. This system allowed us to control YAP levels in the same hESC line from zero (−doxycycline) to overexpressed (50 ng/mL doxycycline) levels (Fig. 5F). Analysis of WNT3 mRNA levels by qPCR revealed that re-expression of YAP into the YAP knockout cells severely impaired the induction of the WNT3 gene by Activin (Fig. 5F). As expected, this correlated with greatly reduced RNAPII CTD-Ser7P in the transcribed region of the WNT3 gene (Fig. 5G). The expression of other ME differentiation genes, such as MIXL1, T, and EOMES, was also compromised by re-expression of YAP (Supplemental Fig. S5C,D).
Activin-treated YAP knockout cells induce the cardiac mesoderm program
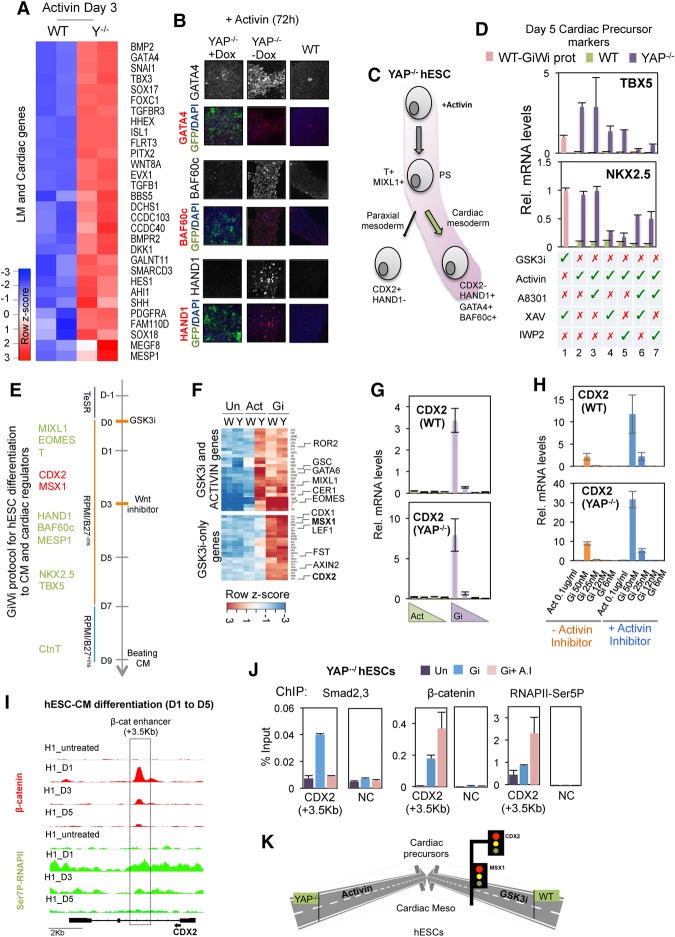
Upon formation of the PS, ME progenitors segregate into endoderm and mesoderm derivatives. Among the mesoderm derivatives, the lateral mesoderm gives rise to cardiac mesoderm, which in turn generates cardiomyocytes and other heart constituents (Supplemental Fig. S6A). To address the details of this pathway further, we carried out an RNA-seq analysis of wild-type and YAP knockout cells after 3 d of Activin treatment. Interestingly, many genes involved in lateral mesoderm and early cardiac development were up-regulated (including BAF60C, HAND1, EVX1, GRRP1, MESP1, SNAI1, GATA4, and ISL1) in the YAP knockout cells but not in wild-type hESCs (Fig. 6A; Supplemental Fig. S6B; Supplemental Table S1), suggesting that the Activin-treated YAP knockout cells had successfully differentiated to cardiac mesoderm. This possibility was confirmed by immunostaining and qPCR analysis of several master cardiac mesoderm marker proteins, including BAF60c, GATA4/6, MESP1, and HAND1, in Activin-treated YAP knockout cells (Fig. 6B,C; Supplemental Fig. S6C,D).
Figure 6.
Activin selectively promotes differentiation to cardiac mesoderm in YAP knockout hESCs. (A) Heat map plot showing the lateral mesoderm (LM) and early cardiac genes up-regulated in YAP knockout versus wild-type hESCs at day 3 following an initial 24 h of exposure to Activin. (B) Analysis of HAND1, GATA4, and BAF60c proteins by immunofluorescence in wild-type and PiggyYAP hESCs before and after doxycycline treatment. (C) Schematic showing the developmental pathway induced by Activin-treated YAP knockout cells. (D) Graphs showing the expression of cardiac precursor markers NKX2.5 and TBX5 in response to different treatments, as indicated below the graph; Activin or GSK3i treatments were added from day 0 to day 1, and XAV, IWP2, or A83-01 inhibitors were added from day 1 to day 2. Cells were collected at day 5 for analysis. For example, lanes 2 and 3 indicate 24 h of Activin treatment followed by A83-01 exposure for an additional 24 h (lane 3) or without further treatment (lane 2). (Lane 1) For comparison, wild-type hESCs were treated following the GiWi protocol, as indicated by the pink bar. Mean (SD). n = 3. (E) Scheme showing the GiWi protocol for hESC differentiation to cardiomyocytes (CM) (Lian et al. 2013). The main cardiac regulators that express sequentially are listed at the left. Proteins required for cardiomyocyte development are shown in green, and cardiac inhibitors are shown in red. (F) RNA-seq was performed in wild-type and YAP knockout cells treated with the specified cytokines for 30 h. Note that the GSK3i concentration used was equivalent to that used for the cardiomyocyte induction protocol (50 nM XV). The heat map shows mRNA levels of genes induced by Activin and GSK3i in wild-type and YAP knockout cells (top) and genes induced by only GSK3i treatment in wild-type and YAP knockout cells (bottom). (G) qPCR analysis shows mRNA levels of CDX2 in wild-type and YAP knockout hESCs after different concentrations of GSK3i (50–5 nM XV) and Activin (100–5 ng/mL) treatment for 24 h. The graph shows the average of two representative experiments of at least four independent replicates. Mean (SEM). n = 2. (H) qPCR analysis shows mRNA levels of CDX2 in wild-type and YAP knockout hESCs after treatment with GSK3i or Activin in the presence or absence of 1 µM Activin inhibitor A8301. The graphs show the average of two representative experiments of at least four independent replicates. Mean (SEM). n = 2. (I) Genome browser captures show β-catenin and CTD-Ser7P-RNAPII distribution on CDX2 gene in hESCs at day 0 and at days 1, 3, and 5 after initial differentiation following the GiWi protocol (see also Fig. 6E). (J) ChIP-qPCR analysis of β-catenin, Smad2,3, and RNAPII-Ser5P binding to the CDX2 enhancer (+3.5 kb) after treatment with GSK3i in the presence or absence of Activin inhibitor in YAP knockout hESCs. (NC) Negative control region. Mean (SEM). n = 2. (K) A schematic depiction to summarize the results of the figure.
Current protocols for differentiating hESCs to cardiomyocytes involve initial exposure to Wnt ligands and later treatment with small-molecule Wnt inhibitors (GiWi protocol) (Fig. 6E; Lian et al. 2013). Following this procedure, cardiac mesoderm cells progress toward cardiac precursors concomitant with the induction of cardiac lineage-specific markers TBX5 and NKX2.5 (Fig. 6E; Supplemental Fig. S6A). Consequently, we monitored the levels of these markers to track the differentiation state of Activin-treated YAP knockout cells (Fig. 6D; Supplemental Fig. S6E). In particular, we analyzed the cells treated with either Activin alone at day 0 for 24 h (Fig. 6D, lane 2), Activin plus Wnt inhibitor added at day 3 (IWP2 or XAV) (Fig. 6D, lanes 4,5), Activin inhibitor (A8301) (Fig. 6D, lane 3), or a combination of both types of inhibitors (Fig. 6D, lane 6,7). For comparison, we also analyzed gene expression in wild-type cells treated with the GiWi protocol at day 5 (Fig. 6D; Supplemental Fig. S6E, pink bars). Surprisingly, we found that the Activin-alone condition efficiently generated cardiac precursor cells in YAP knockout cells. Moreover, differentiation of the YAP knockout cells was not affected by the addition of an Activin inhibitor and was blocked upon exposure to Wnt inhibitors (Fig. 6D; Supplemental Fig. S6E). We conclude that a short exposure to Activin is both necessary and sufficient for YAP knockout hESC differentiation to cardiac precursor cells.
Cardiac inhibitors MSX1 and CDX2 are not induced in Activin-treated YAP knockout cells
The addition of Wnt inhibitors to the cardiomyocyte differentiation protocol in hESCs is required to block the expression of a subset of Wnt target genes that interferes with the formation of cardiac precursor cells, including CDX2 and MSX1 (Fig. 6E; Rao et al. 2016). Accordingly, RNA-seq analysis revealed that GSK3i treatment of wild-type and YAP knockout cells at a concentration used to induce cardiomyocyte differentiation (GSK3i: 50 nM XV) activated both ME genes and CDX2 and MSX1 (Fig. 6F,G; Supplemental Fig. S6F,G; Supplemental Table S4). As reported previously (Rao et al. 2016), the Wnt inhibitors rapidly repressed the CDX2 and MSX1 genes (Supplemental Fig. 6F, left panel, cf. days 3 and 5). In contrast, Activin signaling efficiently induced ME differentiation genes in YAP knockout cells but did not activate CDX2 or MSX1 (Fig. 6F,G; Supplemental Fig. S6F,G). Our RNA-seq analysis revealed that, similar to MSX1 and CDX2, Activin does not induce other Wnt target genes, such as LEF1 and AXIN2, in YAP−/− hESCs (Fig. 6F, bottom panel). Overall, these data indicate that although ME genes are efficiently induced by Activin in YAP knockout cells (Fig. 6F, top panel), there is a set of Wnt target genes not induced by Activin in YAP-null cells, suggesting that different mechanisms operate in the regulation of Wnt target genes.
Activin-induced Smad2,3 counteract β-catenin activity on the CDX2 gene
We explored further the regulation of CDX2 and found that CDX2 expression is higher in Wnt signaling cells treated with an Activin inhibitor, indicating that Activin represses CDX2 expression (Fig. 6H). We next analyzed the levels of β-catenin and Ser7P-RNAPII at the CDX2 gene in wild-type hESCs at days 1, 3, and 5 following the cardiomyocyte differentiation protocol (GiWi protocol) (Fig. 6I). Our ChIP-seq analysis revealed that β-catenin binds to an intragenic enhancer in the CDX2 gene in a manner that correlates with high levels of Ser7P-RNAPII and gene activation (Fig. 6I), similar to the pattern observed on MIXL1 gene (Supplemental Fig. S6I). Therefore, CDX2 is a direct target of Wnt/β-catenin signaling in hESCs. To test whether Activin interferes with β-catenin activity on the CDX2 enhancer, we performed a ChIP-qPCR experiment in wild-type and YAP knockout cells treated with GSK3i in the presence and absence of the Activin inhibitor. Of note, SMAD2,3 proteins were recruited to the CDX2 enhancer in response to GSK3i treatment and not in the presence of the Activin inhibitor (Fig. 6J; Supplemental Fig. S6J). Reduced SMAD2,3 binding was accompanied by increased binding of β-catenin and RNAPII-CTD-Ser5P levels at the CDX2 enhancer and enhanced transcription (Fig. 6H,J; Supplemental Fig. S6J). Therefore, SMAD2,3 binding to the CDX2 gene blocks the ability of β-catenin to activate the CDX2 gene. We conclude that low levels of β-catenin robustly synergize with Activin to up-regulate transcription at ME target genes but are insufficient to activate transcription of LEF1, AXIN, or other canonical Wnt target genes. Moreover, genes like CDX2, which are induced by Wnt but repressed by Activin, are not up-regulated in the Activin-treated YAP−/− hESCs (Supplemental Fig. 6K). A direct consequence is that, unlike Wnt-treated cells, Activin-treated YAP knockout hESCs can specifically develop into cardiac mesoderm (HAND1+:CDX2−) cells that progress into cardiac precursors (NKX2.5+/TBX5+ cells) following 24 h of Activin exposure (Fig. 6K).
Activin-treated YAP knockout hESCs progress toward cardiomyocytes
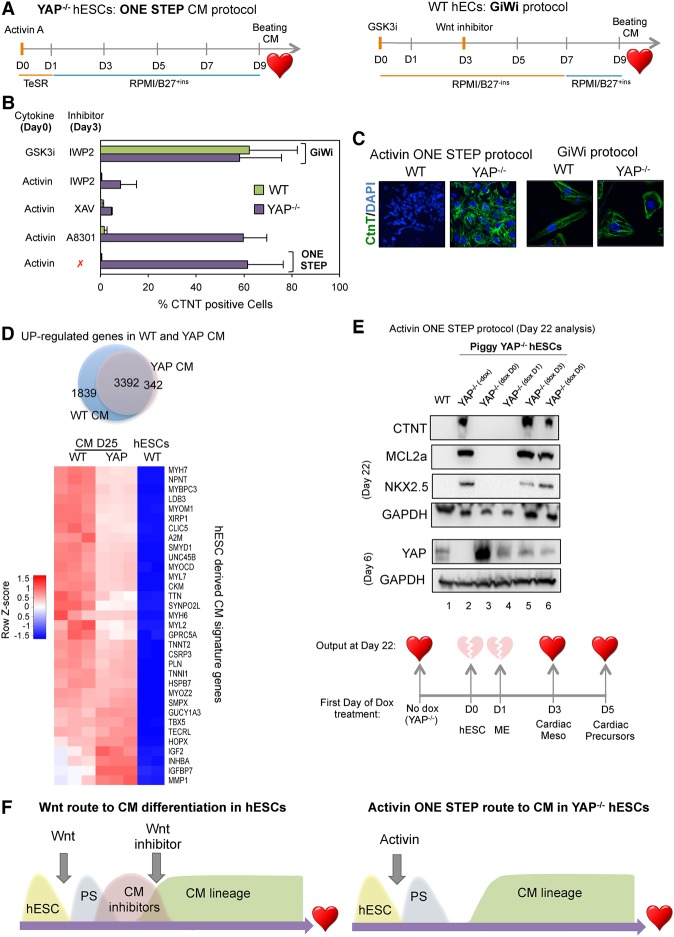
The observation that Activin-induced YAP knockout cells become cardiac precursors without inducing cardiac inhibitor genes raises the possibility that these cells may differentiate to cardiac cells in the absence of exogenous Wnt inhibitors (Fig. 6). Consequently, we next examined the potential of these cells to differentiate into beating cardiomyocytes. To address this question, Activin-treated YAP knockout hESCs were further incubated in RPMI/B27 culture medium without additional treatment (Fig. 7A), henceforth designated the one-step cardiomyocyte differentiation protocol. Importantly, spontaneously beating cardiac cells were detected by day 9 (Supplemental Video S1). By day 20, FACS and immunostaining analysis of the cardiac troponin marker CTNT revealed an average yield of 60% cardiomyocytes following this single-step protocol (Fig. 7B,C; Supplemental Fig. S7A). This efficiency was similar to the yield that we observed with wild-type hESCs following the GiWi protocol (Fig. 7B). Importantly, YAP-depleted H9 hESCs and EC-11 iPSCs also expressed cardiac precursor markers after a short Activin exposure (Supplemental Fig. S7B). Consistent with the previous molecular analysis, addition of Activin or Wnt inhibitors did not improve the efficiency of cardiomyocyte generation in YAP−/− cells, although these inhibitors are essential for cardiac differentiation in wild-type hESCs (Supplemental Fig. S7C). In agreement with these results, RNA-seq analysis confirmed that GiWi-induced wild-type cardiomyocytes and Activin-induced YAP knockout cardiomyocytes share a common cardiomyocyte gene signature (Fig. 7D; Supplemental Fig. S7D; Supplemental Table S5). Importantly, we found that YAP knockout cardiomyocytes can be maintained in culture for >50 d. Taken together, these results show that YAP blocks a direct route toward cardiomyocyte generation but may otherwise be dispensable for the survival of these cells.
Figure 7.
Human cardiomyocyte differentiation using a one-step protocol. (A) The schematic outline of the one-step protocol used to differentiate YAP knockout hESCs into beating cardiomyocytes in comparison with the GiWi protocol. (B) Wild-type and YAP knockout cells were differentiated using the conditions indicated at the left of the graph. Around day 22 following the initial treatment, cells were stained with the cardiac marker CTNT and subjected to FACS analysis. Mean (SD). n ≥ 3. (C) Immunostaining of wild-type- and YAP knockout-derived cardiomyocytes using the protocol indicated below the images. (D) The Venn diagram shows up-regulated genes in wild-type and YAP knockout cardiomyocytes compared with hESCs. (Below) The heat map shows a list of up-regulated cardiac markers in wild-type and YAP−/− cardiomyocytes. (E) Immunoblot analysis of cardiac markers CTNT, NKX2.5, and MCL2a in wild-type and PiggyYAP cells at day 22 after initial differentiation according to the Activin one-step protocol. Doxycycline was added at different time points (not added, day 0, day 1, day 3, or day 5). Note that the reintroduction of YAP at day 0 and day 1 impaired Activin-mediated differentiation, whereas expression of YAP in later stages (day 3 and day 5) did not affect differentiation, as indicated in the bottom illustration. (F) A schematic diagram summarizing the results of the figure. (CM) Cardiomyocyte.
YAP does not block intermediate stages of cardiomyogenesis
These data indicate that Activin signaling is sufficient for full differentiation of YAP knockout hESCs to cardiomyocytes. To address whether YAP also affects intermediate stages of cardiac differentiation, we used the PiggyYAP engineered cells to fine-tune the expression of YAP at different stages of differentiation (Fig. 7E). In this experiment, the differentiation efficiency of YAP knockout cells (−doxycycline) (Fig. 7E, lane 2) was compared with that of cells expressing wild-type YAP at either day 0 (Fig. 7E, lane 3), day 1 (Fig. 7E, lane 4), day 3 (Fig. 7E, lane 5), or day 5 (Fig. 7E, lane 6) of differentiation. Immunoblot analysis of the cardiomyocyte markers CTNT, NKX2.5, and MCL2.A revealed that YAP knockout cells (Fig. 7E, lane 2) and cells that express YAP after day 3 or day 5 were able to differentiate efficiently toward cardiomyocytes. In contrast, wild-type hESCs or hESCs that express YAP prior to day 3 did not differentiate in response to Activin. Therefore, YAP blocks a very early step of hESC differentiation but does not affect the maturation of cardiac mesoderm or the generation of cardiac precursors (Fig. 7E). Most importantly, these data show that early YAP deletion enables Activin to promote hESC differentiation toward the cardiomyocyte lineage (Fig. 7F).
Discussion
The transcriptional mechanisms that control the transition from pluripotency to differentiation for stem cells are not fully defined. Here we uncover a previously unknown role for the Hippo pathway regulator YAP in preventing hESC differentiation along the cardiac mesoderm lineage. Differentiation of hESCs to the mesendoderm is controlled by the Wnt and Activin signaling pathways, which synergistically activate key ME developmental genes in the first step toward formation of differentiated hepatocytes, cardiomyocytes, and pancreatic precursor cells. YAP has been shown to regulate organ size through contact inhibition of cell proliferation and control of stem cell fate (Barry and Camargo 2013; Yu et al. 2015; Meng et al. 2016) and has many roles in cardiac differentiation, proliferation, regeneration, and repair (Cai et al. 2016; Zhou et al. 2016). We reported previously that YAP inhibits hESC differentiation to the mesoderm through a selective role in repressing the expression of Wnt–Activin-coregulated ME genes (Estarás et al. 2015). To better understand the role of YAP in hESCs, we compared the differentiation potential of wild-type and YAP-null hESCs. Unexpectedly, we discovered that hESCs lacking YAP differentiate to beating cardiomyocytes in a single step following short exposure to Activin. Further analysis revealed that deletion of YAP increases the level of endogenous Wnt ligands in hESCs to overcome the block to differentiation along the ME cell lineage. RNA-seq analysis of the Activin-induced transcriptome revealed that YAP knockout cells specifically induce a subset of ME genes required for hESCs to develop into cardiac mesoderm, including the BAF60c, HAND1, and GATA4 markers. Importantly, unlike wild-type hESCs treated with exogenous Wnt ligand, the Activin-induced cardiac mesoderm cells fail to express cardiac inhibitors CDX2 and MSX1 and can therefore progress directly to cardiomyocytes.
Our previous study also revealed that YAP blocks recruitment of the P-TEFb transcription elongation complex to ME genes in Wnt+Activin signaling cells, which prevents the accumulation of RNAPII-CTD-Ser7P at these genes (Estarás et al. 2015). ChIP studies further established that YAP is recruited to the EOMES and MIXL1 enhancers in response to Wnt+Activin signaling, where it interferes with the cooperation between β-catenin and SMAD complexes that is normally required for efficient transcription elongation. In addition to a direct role in repression, YAP1 can regulate β-catenin ubiquitylation and stability within the APC destruction complex (Azzolin et al. 2014) both in the cytoplasm and potentially also at Wnt/LEF-1 enhancers (Choi et al. 2013). Here, we show that YAP has an additional repressive function in self-renewing hESCs, where it binds to enhancer elements and blocks expression of the WNT3 and NODAL genes. Our ChIP-seq data show that YAP binds to an intragenic enhancer of the WNT3 gene in a region containing high levels of histone H3K27ac and an enhancer-specific form of RNAPII-CTD-Ser5Phi:Ser7Plow (Estarás et al. 2015). Knockout of YAP enables SMAD DNA-binding complexes to access the WNT3 gene promoter and activate transcription in response to Activin. The Activin-induced SMAD2,3 proteins bound at promoters synergize with Wnt3-induced nuclear β-catenin at the enhancer to induce ME differentiation genes. Taken together, our findings indicate that YAP reinforces the block to ME differentiation at several steps in proliferating hESCs. Thus, YAP bound to the WNT3 and NODAL genes blocks SMAD binding to prevent aberrant gene activation in response to Activin. YAP also associates with SMAD complexes at ME gene promoters to block P-TEFb elongation and recruit the NURD complex to establish promoter-proximal nucleosomes that reinforce transcription repression (Beyer et al. 2013). Under conditions of prodifferentiation Wnt+Activin signaling, YAP bound to β-catenin:LEF-1 complexes at ME gene enhancers can dampen transcription levels by interfering with P-TEFb recruitment and enforcing suboptimal transcription elongation of these genes. The loss of YAP releases each of these separate constraints and enables full-blown transcription of ME genes in Activin-treated hESCs.
Studies using knockout mouse models have shown that formation of the PS requires specification of the posterior epiblast by WNT3/β-catenin and Nodal/Activin transforming growth factor-β (TGFβ)-related factors (Arkell et al. 2013; Robertson 2014). In vivo expression of WNT3 is activated by Nodal, which in turns reinforces a feedback loop to maintain NODAL expression in the epiblast. However, the mechanism through which Wnt3 ligand expression is initially established is not well understood. Activation of the WNT3 gene promoter by p53 was shown recently to initiate Wnt+Activin cooperation in mESCs lacking LIF (Wang et al. 2017). Our findings also show that YAP directly represses WNT3 gene expression in hESCs and prevents Activin/SMADs from prematurely inducing Wnt ligand expression and mobilizing β-catenin. Thus, in the absence of YAP, Activin/SMADs may cooperate with p53 to activate the WNT3 gene, as was observed at ME target genes (Wang et al. 2017). Interestingly, the YAP/TAZ genes were shown recently to be activated by the Wnt3 and Wnt5 ligands through an alternative Wnt signaling pathway that operates independently of β-catenin to inhibit Wnt signaling (Park et al. 2015). Taken together, the observations raise the possibility that the WNT3 gene may be subject to regulation through a negative feedback loop.
In YAP knockout cells, WNT3 is induced and β-catenin is recruited genome-wide to >500 new sites in response to Activin. Interestingly, we also observed >2000 new SMAD2,3 peaks in YAP knockout cells, likely resulting from the strongly increased NODAL expression. Nevertheless, ∼20% of the new SMAD-bound genes and 30% of β-catenin-bound genes are associated with increased transcription. Importantly, many Activin targets, such as LEFTY1 and BAMBI, were not induced in YAP-null cells. We speculate that increased SMAD2,3 binding to canonical Activin genes may not further increase transcription at these active genes. Of note, Activin-induced Wnt3 ligand expression was insufficient to activate canonical Wnt target genes, such as AXIN2 and LEF-1, which presumably require higher levels of endogenous Wnt signaling. Consistent with this scenario, Activin signaling strongly enriches levels of elongation RNAPII-CTD-Ser7P in the coding region of ME genes in YAP knockout cells, whereas this elongation-associated form of RNAPII does not accumulate at Wnt target genes. Consequently, the modest increase in β-catenin in YAP−/− cells synergizes strongly with Activin to selectively up-regulate ME gene expression and preferentially predisposes stem cells to differentiate along the cardiomyocyte pathway.
One of the goals of regenerative stem cell therapy for cardiomyopathies is to overcome the limited potential of transplanted cardiac progenitor cells to differentiate as needed to repair damage created by oxidative or mechanical stress (Atmanli and Domian 2017; Duelen and Sampaolesi 2017). We found that the YAP knockout hESCs express the lateral mesoderm markers BAF60c, HAND1, and GATA4 following a short exposure to exogenous Activin. In the developing embryo and in hESCs, the specification of cardiogenic mesoderm is regulated by a combination of Wnt inhibition and active BMP signaling. Subsequent inhibition of Wnt signaling is needed to shut down the expression of two cardiac inhibitor genes: CDX2 and MSX1 (Rao et al. 2016). Here we show that, unlike ME genes, CDX2 and MSX1 are up-regulated by Wnt signals but inhibited by Activin, and therefore these genes are not induced in YAP knockout cells. Consequently, YAP knockout cells differentiate into beating cardiomyocytes without the need for either Wnt ligands or Wnt inhibitors. In this respect, it is interesting that the DAN family protein Cerberus (Cer1), a potent inhibitor of NODAL and BMP signaling, has also been shown to block Wnt ligand expression during cardiac precursor specification and enable BMPs to activate cardiac precursors, such as NKX2.5 and MESP1 (Ruiz-Villaba et al. 2016). Our data reveal that YAP knockout cells express much higher levels of Cer1 and other Wnt inhibitors such as DKK1, DKK4, and APCDD1 (Supplemental Table S1) following Activin treatment compared with wild-type cells. Therefore, the increased expression of these factors may also contribute to the low level of endogenous Wnt target gene expression and allow efficient differentiation of YAP knockout cells to cardiomyocytes in the absence of Wnt inhibitors.
In summary, our findings show that YAP is a master transcriptional regulator that determines the biological outcome of Activin signaling in hESCs by regulating Wnt activity levels. The combination of endogenous Wnt signaling and high levels of Activin in YAP knockout cells has uncovered a direct route toward cardiogenesis in hESCs. These findings highlight new mechanisms that control ME genes in early development and suggest new approaches for preparing cardiomyocytes from pluripotent cells for stem cell therapies.
Materials and methods
hESC culture
H1 and H9 hESCs and EC-11 iPSCs were cultured in mTeSR1 medium on Matrigel-coated tissue culture plates. For expansion, colony fragments were split in a 1:3 ratio following dispase treatment. The medium was replaced daily. For differentiation experiments, colonies were disaggregated into single cells using accutase in the presence of Rock inhibitors.
Genetic manipulation of hESCs
Knockout cell lines were generating using the CRISPR/CAS9 vector pX458 (Addgene). The single-guide RNA sequences cloned are in Supplemental Table 5. Positive knockout clones were identified by sequencing the gene of interest and confirmed by analyzing the protein levels. The doxycycline-inducible YAP cell line was generated using a PiggyBac transposon system. The Flag-YAP cDNA was cloned into the KA0717 vector (pPB-hCMV1-cDNA-IRESVenus) and then cotransfected into YAP knockout cells together with transactivator and transposase-encoding vectors pCAG-PBase (Austin Smith laboratory) (Guo et al. 2009) and KA0637 pPBCAG-rtTAM2-IN. After G418 selection, two clones were isolated and used for experiments. The plasmids were kindly provided by Dr. Kenjiro Adachi (Max Planck Institute for Molecular Biomedicine).
A single-step protocol for formation of beating cardiomyocytes and metabolic selection
Wild-type and Yap knockout H1 hESCs were cultured on Matrigel-coated plates until 80%–90% confluence. At day 0, YAP knockout cells were treated with 100 ng/mL Activin for 24 h in TeSR medium. Next, the medium was aspirated, replenished with RPMI/B-27, and replaced every 2 d. The spontaneous beating phenotype should occur from day 9 onward. From day 20 to day 25, the culture was fed with RPMI (no glucose) supplemented with 4 mM sodium L-lactate solution to allow metabolic selection of cardiomyocytes. From day 25 onward, the cardiomyocytes were fed with RPMI/B-27 medium and replaced every 3 d. The purified cardiac cells were used for RNA-seq experiments in Figure 7 (see also the Supplemental Material).
High-throughput sequencing and bioinformatic analysis
High-throughput sequencing using Illumina HiSeq 2500 or 4000 devices was carried out at the Next-Generation Sequencing Core or with Dr. Joe Ecker (The Salk Institute). ChIP-seq and RNA-seq experimental procedures and bioinformatic analyses are detailed in the Supplemental Material.
Accession numbers
Processed and filtered data are in the Supplemental Tables. Full data sets of these and additional experiments have been submitted to the NCBI Gene Expression Omnibus database in a MIAME-compliant format under superseries accession number GSE99202. The H3K27me3, H3K27ac, and RNAPII CTD-Ser5P ChIP-seq experiments can be accessed through GSE64758.
Supplementary Material
Acknowledgments
Support for this project was provided through the Salk Bioinformatics Facility and Stem Cell Cores. Funding for the RNA-seq and ChIP-seq data reported here was provided by the California Institute for Regenerative Medicine (GC1R-06673-B). C.E. was supported by a fellowship from the SalkExCellerators Program. This study was funded by National Institutes of Health/National Cancer Institute grant RO1CA125535 (K.A.J.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.307512.117.
References
- Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. 2009. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkell RM, Fossat N, Tam PP. 2013. Wnt signaling in mouse gastrulation and anterior development: new players in the pathway and signal output. Curr Opin Genet Dev 23: 454–460. [DOI] [PubMed] [Google Scholar]
- Atmanli A, Domian IJ. 2017. Recreating the cardiac microenvironment in pluripotent stem cell models of human physiology and disease. Trends Cell Biol 27: 352–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, et al. 2014. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 158: 157–170. [DOI] [PubMed] [Google Scholar]
- Barry ER, Camargo FD. 2013. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol 25: 247–253. [DOI] [PubMed] [Google Scholar]
- Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, et al. 2013. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature 493: 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL. 2013. Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep 5: 1611–1624. [DOI] [PubMed] [Google Scholar]
- Cai W-F, Wang L, Liu G-S, Zhu P, Paul C, Yigang W. 2016. Manipulating the Hippo-YAP signal cascade in stem cells for heart regeneration. Ann Palliat Med 5: 125–134. [DOI] [PubMed] [Google Scholar]
- Choi SH, Estarás C, Moresco JJ, Yates JR III, Jones KA. 2013. α-Catenin interacts with APC to regulate β-catenin proteolysis and transcriptional repression of Wnt target genes. Genes Dev 27: 2473–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. 2012. Wnt/β-catenin signaling and disease. Cell 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Czudnochowski N, Bosken CA, Geyer M. 2012. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun 3: 842. [DOI] [PubMed] [Google Scholar]
- Duelen R, Sampaolesi M. 2017. Stem cell technology in cardiac regeneration: a pluripotent stem cell promise. EbioMedicine 16: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estarás C, Benner C, Jones KA. 2015. SMADs and YAP compete to control elongation of β-catenin:LEF-1-recruited RNAPII during hESC differentiation. Mol Cell 58: 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, et al. 2015. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol Cell 60: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. 2009. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136: 1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CG, Moroishi T, Guan KL. 2015. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol 25: 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Lis JT. 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kim T, Johnson RL, Lim DS. 2015. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep 11: 270–282. [DOI] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. 2010. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 24: 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. 2013. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 8: 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. 2012. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 26: 1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CY, Nichane M, et al. 2014. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell 14: 237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K. 2017. Signaling cross-talk between TGF-β/SMAD and other signaling pathways. Cold Spring Harb Perspect Biol 9: a022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan KL. 2016. Mechanisms of Hippo pathway regulation. Genes Dev 30: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujok O, Diekmann U, Lenzen S. 2014. The generation of definitive endoderm from human embryonic stem cells is initially independent from Activin A but requires canonical Wnt-signaling. Stem Cell Rev 10: 480–493. [DOI] [PubMed] [Google Scholar]
- Nostro MC, Cheng X, Keller GM, Gadue P. 2008. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2: 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Kim YC, Yu B, Moroishi T, Mo J-S, Plouffe SW, Meng Z, Lin KC. 2015. Alternative WNT signaling activates YAP/TAZ. Cell 162: 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR–Cas9 system. Nat Protoc 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J, Pfeiffer MJ, Frank S, Adachi K, Piccini I, Quaranta R, Araúzo-Bravo M, Schwarz J, Schade D, Leidel S, et al. 2016. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell 18: 341–353. [DOI] [PubMed] [Google Scholar]
- Robertson EJ. 2014. Dose-dependent Nodal/SMAD signals pattern the early mouse embryo. Semin Cell Dev Biol 32: 73–79. [DOI] [PubMed] [Google Scholar]
- Ruiz-Villaba A, Hoppler S, van den Hoff MJB. 2016. Wnt signaling in the heart fields: variations on a common theme. Dev Dyn 245: 249–306. [DOI] [PubMed] [Google Scholar]
- Sakaki-Yumoto M, Liu J, Ramalho-Santos M, Yosida N, Derynck R. 2013. Smad2 is essential for maintenance of the human and mouse primed pluripotent stem cell state. J Biol Chem 288: 18546–18560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. 2008. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/β-catenin, Activin/Nodal and BMP signaling. Development 135: 2969–2979. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen YG. 2016. Signaling control of differentiation of embryonic stem cells toward mesendoderm. J Mol Biol 428: 1409–1422. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zou Y, Nowotschin S, Kim SY, Li QV, Soh C-L, Su J, Zhang C, Shu W, Xi Q, et al. 2017. The p53 family coordinates Wnt and Nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell 20: 70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. 2015. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163: 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li L, Zhao B, Guan KL. 2016. The Hippo pathway in heart development, regeneration and diseases. Circ Res 116: 303311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.