Abstract
Purpose
We previously completed a comprehensive profile of the mouse lens transcriptome. Here, we investigate the proteome of the mouse lens through mass spectrometry–based protein sequencing at the same embryonic and postnatal time points.
Methods
We extracted mouse lenses at embryonic day 15 (E15) and 18 (E18) and postnatal day 0 (P0), 3 (P3), 6 (P6), and 9 (P9). The lenses from each time point were preserved in three distinct pools to serve as biological replicates for each developmental stage. The total cellular protein was extracted from the lens, digested with trypsin, and labeled with isobaric tandem mass tags (TMT) for three independent TMT experiments.
Results
A total of 5404 proteins were identified in the mouse ocular lens in at least one TMT set, 4244 in two, and 3155 were present in all three TMT sets. The majority of the proteins exhibited steady expression at all six developmental time points; nevertheless, we identified 39 proteins that exhibited an 8-fold differential (higher or lower) expression during the developmental time course compared to their respective levels at E15. The lens proteome is composed of diverse proteins that have distinct biological properties and functional characteristics, including proteins associated with cataractogenesis and autophagy.
Conclusions
We have established a comprehensive profile of the developing murine lens proteome. This repository will be helpful in identifying critical components of lens development and processes essential for the maintenance of its transparency.
Keywords: proteome, lens proteins, mass spectrometry
The murine lens serves as an excellent system to investigate intricate details of development and differentiation. During embryonic development, the lens arises from the head ectoderm that thickens to form the lens placode.1,2 The lens placode invaginates alongside the optic vesicle, forming the lens pit that separates from the ectoderm to form the lens vesicle, which further divides into two single-cell layers.1,2 The anterior layer differentiates into the lens epithelium while cells of the posterior layer differentiate into primary fiber cells.1,2 During the late embryonic and early postnatal stages, the lens grows rapidly as lens epithelial cells proliferate and elongate, continuously differentiating into secondary fiber cells, adding to the fiber cell mass.1,2
The ocular lens is an avascular transparent tissue, and a deficiency in the processes responsible for the maintenance of lens transparency results in cataractogenesis. Cataracts, cloudiness or opacity of the lens, are the leading cause of blindness worldwide and account for one-third of the cases of blindness in children.3,4 Inherited cataracts comprise a significant fraction of the global burden of cataractogenesis.5
Previously, two-dimensional gel electrophoresis (2-DGE)-based investigations have been performed to characterize the lens proteome, which mostly covered the posttranslational modifications and differential expression pattern of crystallin proteins in the human and mouse lens.6–11 Although ideal for constructing an initial proteome map, these investigations failed to characterize the less abundant noncrystallin proteins in the lens. Recent advancements in mass spectrometry–based proteome sequencing methodologies have made it possible to characterize the entire proteome of the human or mouse ocular lens with unprecedented speed and accuracy.12–14
Three independent investigations used the mass spectrometry–based approach to identify 200, 951, and 506 proteins from the plasma membranes of mouse, human, and bovine lens fiber cells, respectively.15–17 Shang and colleagues18 reported 2052 proteins in the wild-type mouse lens at postnatal day 0 (P0). Importantly, none of these studies examined the lens proteome at more than one developmental time point.
Here, we present a comprehensive mouse lens proteome achieved through mass spectrometry–based in-depth proteome profiling at multiple developmental stages including two embryonic and four postnatal time points. Our proteome profiling yielded a total of 5404 proteins present in the developing mouse ocular lens. Of these, 39 proteins exhibited an 8-fold differential (higher or lower) expression during the developmental time course compared with their respective expression at embryonic day 15 (E15).
Materials and Methods
Sample Collection
The use of mice in this study was approved by the Johns Hopkins Animal Care and Use Committee (ACUC), and all experiments were performed in accordance with a protocol approved by the Johns Hopkins ACUC. This study was carried out in compliance with ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Lenses were obtained at six different developmental stages, including two embryonic (E15 and E18) and four postnatal stages (P0, P3, P6, and P9). The ocular lenses were extracted from mouse eyes as described previously.19 Briefly, mice were first anesthetized with isoflurane and subsequently euthanized through cervical dislocation, and the lenses were isolated using forceps under a microscope. Three biological replicates, each consisting of lenses from 23 embryos for E15, 10 embryos for E18, and eight pups for P0, P3, P6, and P9, were used for the respective developmental stages in each of the three tandem mass tag (TMT) runs. Upon extraction, all lenses were frozen immediately at −80°C until further processing for total protein extraction.
Protein Extraction, Digestion, and Labeling
The mouse lenses were lysed by vortexing for 2 minutes with glass beads (0.5, 1, and 3 mm in diameter) in 8 M urea and 50 mM triethylammonium bicarbonate (TEABC). Protein lysates were centrifuged at 16,000g at 4°C to exclude cell debris (pelleting at the bottom), and protein concentration was estimated using a bicinchoninic acid (BCA) assay. A total of 100 μg each sample was reduced with 10 mM dithiothreitol at room temperature for 1 hour and alkylated with 30 mM iodoacetamide for 20 minutes in the dark. The protein samples were digested overnight at 37°C using sequencing-grade trypsin (1:50) followed by desalting and labeling with 10-plex TMT reagents according to the manufacturer's instructions (Thermo Fisher Scientific, Rockford, IL, USA), and the first six channels (126, 127N, 127C, 128N, 128C, and 129N) were used for labeling. The labeling reaction was performed for 1 hour at room temperature, followed by quenching with 100 mM Tris-HCl (pH 8.0). The digested and labeled peptides from all six time points were pooled and desalted with C18 Sep-Pak (Waters Corporation, Milford, MA, USA).
The peptides were fractionated by basic pH reversed-phase liquid chromatography (bRPLC) into 96 fractions, followed by concatenation into 24 fractions by combining every 24th fraction. Agilent (Santa Clara, CA, USA) 1260 offline LC system was used for bRPLC fractionation, which includes a binary pump, variable wavelength detector, an autosampler, and an automatic fraction collector. Lyophilized samples were reconstituted in solvent A (10 mM triethylammonium bicarbonate, pH 8.5) and loaded onto XBridge C18, 5-μm 250 × 4.6-mm column (Waters Corporation). Peptides were resolved using a gradient of 3% to 50% solvent B (10 mM triethylammonium bicarbonate in 90% acetonitrile, pH 8.5) at a flow rate of 1 mL/min over 50 minutes, collecting 96 fractions. Subsequently, the fractions were concatenated into 24 fractions followed by vacuum drying using SpeedVac (Thermo Fisher Scientific, San Jose, CA, USA). The dried peptides were suspended in 15 μL of 0.1% formic acid, and all 15 μL was injected.
Mass Spectrometry–Based Proteome Sequencing
The fractionated peptides were analyzed on an Orbitrap Fusion Lumos Tribrid Mass Spectrometer coupled with the EASY-nLC 1200 nano-flow liquid chromatography system (Thermo Fisher Scientific). The peptides from each fraction were reconstituted in 15 μL of 0.1% formic acid, and all the 15 μL was loaded on an Acclaim PepMap100 Nano-Trap Column (100 μm × 2 cm, Thermo Fisher Scientific) packed with 5-μm C18 particles at a flow rate of 5 μL/min. Peptides were resolved at 250 nL/min flow rate using a linear gradient of 10% to 35% solvent B (0.1% formic acid in 95% acetonitrile) over 95 minutes on an EASY-Spray column (50 cm × 75 μm ID, Thermo Fisher Scientific) packed with 2-μm C18 particles, which was fitted with an EASY-Spray ion source that was operated at a voltage of 2.0 kV.
Mass spectrometry (MS) analysis was carried out in a data-dependent manner with a full scan in the mass-to-charge ratio (m/z) range of 350 to 1550 in the “Top Speed” setting, 3 seconds per cycle. MS1, MS2, and MS3 were acquired for the peptide fragmentation ions in MS2 level and the reporter ions in MS3 level. MS1 scans were measured at a resolution of 120,000 at an m/z of 200. MS2 scan was acquired by fragmenting precursor ions using the higher-energy collisional dissociation (HCD) method and detected at a mass resolution of 30,000 at an m/z of 200. MS3 scan was acquired by isolating the top five fragment ions monitored in the MS2 scan and fragmenting the top five selected ions using the HCD method and detected at a mass resolution of 30,000 at an m/z of 200. Automatic gain control for MS1 was set to 1 million ions and for MS2 and MS3 was set to 0.05 million ions. A maximum ion injection time was set to 50 milliseconds (ms) for MS1 and 100 ms for MS2 and MS3. MS1 were acquired in profile mode, and MS2 and MS3 were acquired in centroid mode. Higher-energy collisional dissociation was set to 35 for MS2 and 65 for MS3. Dynamic exclusion was set to 30 seconds, and singly charged ions were rejected. Internal calibration was carried out using the lock mass option (m/z 445.1200025) from ambient air.
Data Analysis
The Proteome Discoverer (v 2.1; Thermo Fisher Scientific) suite was used for quantitation and identification. During MS/MS preprocessing, the top 10 peaks in each window of 100 m/z were selected for database search. The tandem mass spectrometry data were then searched using SEQUEST algorithms against a mouse UniProt (Swiss-Prot only) database (released June 2017; http://www.uniprot.org/peptidesearch/) with common contaminant proteins. The search parameters used were as follows: (a) trypsin as a proteolytic enzyme (with up to two missed cleavages); (b) peptide mass error tolerance of 10 ppm; (c) fragment mass error tolerance of 0.02 Da; and (d) carbamidomethylation of cysteine (+57.02146 Da) and TMT tags (+229.162932 Da) on lysine residues and peptide N-termini as a fixed modification and oxidation of methionine (+15.99492 Da) as a variable modification. The minimum peptide length was set to seven amino acids, and proteins identified by only one peptide were filtered out. Peptides and proteins were filtered at a 1% false-discovery rate (FDR) at the peptide spectrum match (PSM) level using percolator node and at the protein level using protein FDR validator node, respectively.
The protein quantification was performed with following parameters and methods. The most confident centroid option was used for the integration mode while the reporter ion tolerance was set to 20 ppm. The MS order was set to MS3, and the activation type was set to HCD. Unique and razor peptides both were used for peptide quantification while protein groups were considered for peptide uniqueness. Reporter ion abundance was computed based on intensity, and the missing intensity values were replaced with the minimum value. The quantification value corrections for isobaric tags, average reporter signal-to-noise threshold, and data normalization were disabled while the co-isolation threshold was set to 30%.
Protein grouping was performed with the strict parsimony principle to generate the final protein groups. All proteins sharing the same set or subset of identified peptides were grouped while protein groups with no unique peptides were filtered out. The Proteome Discoverer iterated through all spectra and selected PSMs with the highest number of unambiguous and unique peptides.
All three TMT experiments were processed in a single Proteome Discoverer analysis using all three TMT sets as replicates. The Proteome Discoverer summed all the reporter ion intensities of PSMs for the corresponding proteins in each of the TMT runs, separately.
Subsequent to protein identification and quantification, the protein table was exported to Perseus 1.5.2.6 software for normalization of the reporter ion intensities.20 Proteins with reporter ion intensity higher than 2500 for at least one of the six quantification channels were retained. The reporter ion intensities of E18, P0, P3, P6, and P9 were divided by the reporter ion intensity value of E15 for each TMT set separately. Finally, each column was divided by the median value of the corresponding column to remove systemic deviation.
The abundance values of reporter ion intensities from each TMT set (from Proteome Discoverer platform) were imported into Partek Genomics Suite v6.6 (Partek, Inc., St. Louis, MO, USA) for principal component analysis (PCA) and creating plots from these expression values. The normalized reporter ion intensities (normalized against E15 in each TMT set, independently) were imported into Spotfire DecisionSite with Functional Genomics v9.1.2 (TIBCO Spotfire, Boston, MA, USA) for time course analysis.
Partek Genomics Suite was used to investigate the differential expression of proteins at different developmental time points. The normalized reporter ion intensities (normalized against E15 in each TMT set, independently) were examined for the standard deviation (SD) to investigate the differential expression between time points in each of the five comparisons, that is, E18 vs. E15, P0 vs. E15, P3 vs. E15, P6 vs. E15, and P9 vs. E15. The P values were estimated by a 2-tailed one-sample t-test statistical procedure, assuming a hypothesized mean of 0 change. The normalized dataset ratios were converted to log2 scale (becoming the conventional “log ratios” or “log2 fold changes”) for statistical and graphic representation.
To investigate differential expression of proteins between embryonic and postnatal time points, we quantile normalized reporter ion intensities from each TMT set, independently. Briefly, reporter ion intensities from each TMT set (from Proteome Discoverer platform) were imported into Partek Genomics Suite v6.6. The reporter ion intensities of proteins that were reliably quantitated in all three TMT sets were first transformed in log2 and subsequently, each TMT set was independently quantile normalized. The geometric means of E15 and E18 samples were compared to the geometric means of P0, P3, P6, and P9 samples to determine relative embryonic versus postnatal fold changes of each protein and its statistical significance (P value). These P values were determined by a 2-way analysis of variance, an extension of the Student's t-test statistic, including set and assuming a hypothesized mean of 0 change between classes. Note that quantile normalization was used only to investigate the expression differential between grouped embryonic and postnatal time points.
Gene Ontology Functional Enrichment Analysis
A functional annotation analysis of differentially expressed mouse lens proteins at multiple developmental time points was performed using Visual Annotation Display (VLAD; ver. 1.6.0), a Web-based tool from the Mouse Genome Informatics (MGI).21 The VLAD performs the statistical analysis to test the enrichment of gene ontology (GO) terms based on their annotations to gene function.21 A complete set of mouse genes was used as a reference annotation dataset, and ontological terms annotated with the evidence code ND (no biological data) were excluded from the enrichment analysis. The statistically significant enriched terms were sorted based on their corrected P value (≤0.01) calculated using multiple testing and positive FDR for each term.
Results
We performed mass spectrometry–based proteome sequencing of the mouse ocular lens at six time points including E15, E18, P0, P3, P6, and P9. Three biological replicates were used for each time point, divided into three sets, and each set was sequenced and analyzed independently. Proteome sequencing generated 207,459 total PSMs yielding a total of 56,972 peptides in all three sets. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository,22 with the dataset identifier PXD006381.
We identified 4630, 4426, and 3747 proteins in TMT set 1, set 2, and set 3, respectively (Supplementary Table S1). Of these, 5404 proteins were present in at least one TMT set while 4244 and 3155 proteins were present in at least two and all three TMT sets, respectively (Supplementary Table S1). In addition to the above-mentioned proteins, we identified 1344 proteins in our mass spectrometry–based proteome of the mouse ocular lens; however, these 1344 proteins were not reliably quantitated (Supplementary Table S1).
The mouse genome (GENCODE Ver. M14; GRCm38.p5) includes 21,948 protein-coding genes. We identified 5404 proteins representing nearly 25% of the protein-coding genes in the developing mouse lens. The lens proteome exhibits a wide spectrum of expression levels with maximum and minimum reporter ion intensities of 1,457,114,681 and 506, respectively (Supplementary Table S1). We identified high expression levels of crystallin proteins, lens-specific intermediate filament proteins, and heat shock proteins in the mouse lens proteome (Supplementary Table S1). In contrast, molecules with binding and catalytic activities are among the proteins expressed at low levels in the mouse lens proteome (Supplementary Table S1).
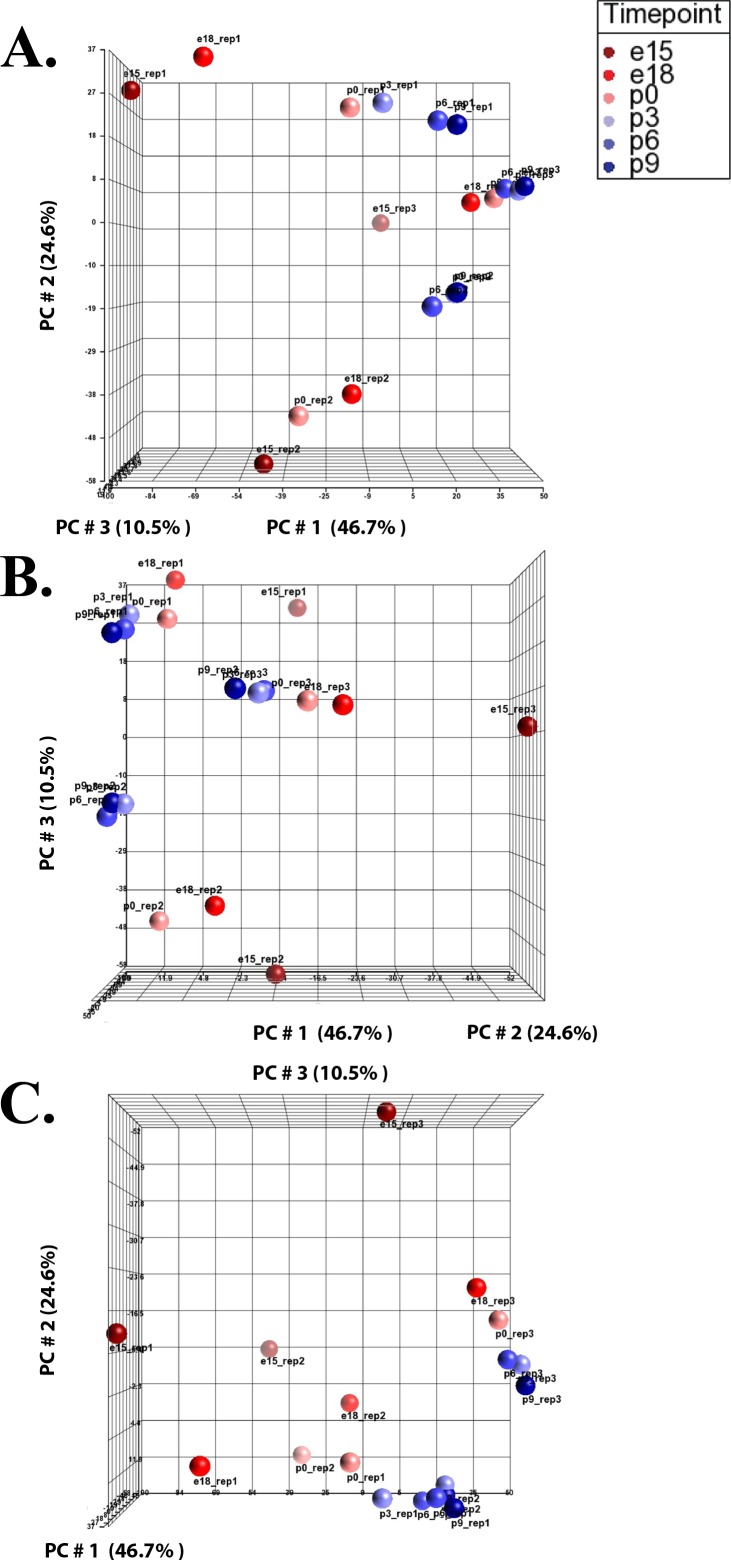
Principal component analysis (PCA) can provide an overall understanding of expression differentials and illustrates how samples differ at different time points. Here, we used reporter ion intensities of each TMT dataset exported independently from the Proteome Discoverer and employed PCA to provide a global proteome expression profile in the developing lens (Fig. 1). The PCA plot analysis revealed that there is indeed a time course running from E15 to P9 in all three independent TMT datasets, reflected in the chronological order of these time points. The time course for set 1 and set 2 are clearly visible in Figure 1A while the time course for set 3 is better visible from different angles as shown in Figures 1B and 1C. The analysis of PCA plots revealed three cluster formations (each of six developmental time points for the three TMT sets) indicating a batch effect in three independent TMT experiments. The batch effect is further illustrated by uneven distribution of reporter ion intensities in the TMT datasets shown in the box plots and histogram analyses (Supplementary Figs. S1, S2).
Figure 1.
Principal component analysis (PCA) illustrating direct comparisons among developmental time points. Partek Genomics Suite was used to create PCA plots using reporter ion intensities of each TMT dataset, independently. (A–C) Illustration of the PCA plot at different angles with x-, y-, and z-axes depicting the three largest components of variation in expression, expressed in percentage of the total, PC#1, PC#2, and PC#3, respectively.
Next, we performed a time course analysis to investigate changes in protein expression in the developing lens, and to intuitively track the trajectory of each protein over time. The earliest developmental time point, E15, was set as the reference point, and the remaining five time points were separately compared to E15 in each TMT set (Supplementary Tables S2–S4), thus yielding individual protein fold changes between the E15 starting point and increasingly greater development. The expression profile plots show that a considerable proportion of the proteins exhibit a change in their expression levels, either up or down, from their levels at E15, and, further, that the degree of change increases as time passes (Supplementary Fig. S3). We identified 39 proteins that exhibited an 8-fold expression differential compared with translational levels at E15 (Supplementary Table S5). Of these, we identified 32 proteins that showed an increase and 7 proteins that showed a decrease in expression compared with expression at E15 (Supplementary Table S5). Upon increasing the stringency by raising the expression differential to 20-fold, we identified a total of 14 proteins, including 11 upregulated and 3 downregulated in the mouse lens proteome (Supplementary Table S5).
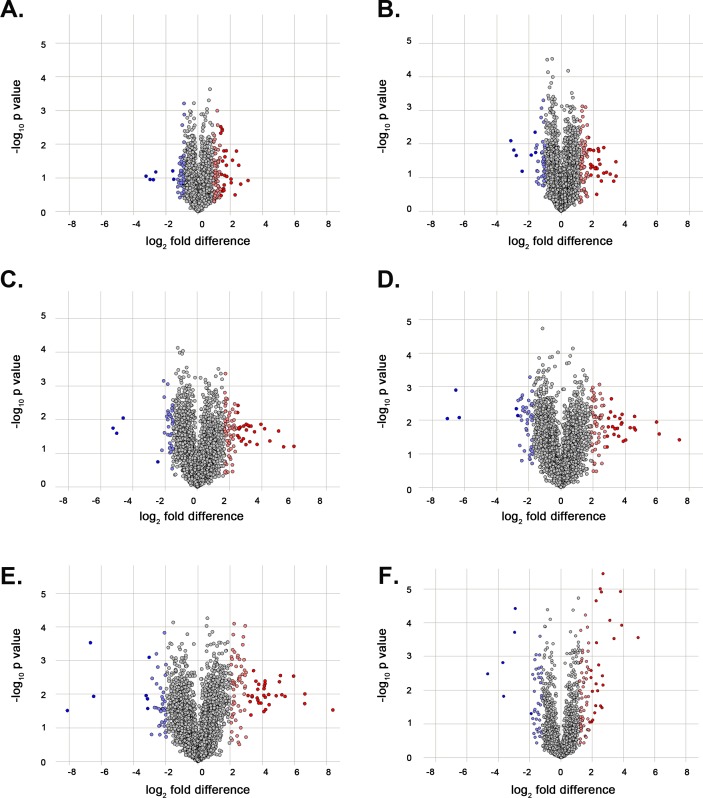
We compared the expression at E15 with the five later developmental time points, that is, E18, P0, P3, P6, and P9, individually (Fig. 2A–E). We identified a set of crystallin proteins, lens-specific intermediate filament proteins, and heat shock proteins that exhibit an upregulation trend starting at E18 and consistent at later developmental time points (i.e., P0, P3, P6, and P9), compared to their respective expression levels at E15 (Supplementary Tables S6–S10). All crystallin proteins identified in the mouse lens proteome displayed the highest differential expression at P9 (Supplementary Table S10). We additionally examined the protein differential expression between embryonic and postnatal time points (Fig. 2F) identifying crystallin proteins, lens-specific intermediate filament proteins, and heat shock proteins, among others, upregulated in the postnatal mouse lens (Supplementary Table S11).
Figure 2.
Volcano plots illustrating the differentially expressed proteins in the mouse lens at different developmental time points. The proteins identified in all three TMT datasets (i.e., 3155) were examined to classify significantly upregulated and downregulated proteins, compared with their respective expression levels at embryonic day 15 (E15). Volcano plots showing differentially expressed proteins at (A) embryonic day 18 (E18), (B) postnatal day 0 (P0), (C) postnatal day 3 (P3), (D) postnatal day 6 (P6), and (E) postnatal day 9 (P9) when compared with the earliest time point E15, and (F) the differential expression of proteins between embryonic time points (E15 and E18) and postnatal time points (P0, P3, P6, and P9). The x-axis corresponds to the log2 fold change value, and the y-axis displays the −log10 P value. Proteins that are significantly upregulated (≥ +2 standard deviations) are highlighted in red and light red, and those with significant downregulation (≥ −2 standard deviations) are highlighted in blue and light blue.
The GO-based functional enrichment of the differentially expressed proteins was investigated in the mouse lens at different developmental time points. The gene ontologies of the mouse lens proteome were characterized based on the molecular function, biological process, and cellular component. The analysis revealed the enrichment of molecular function GO term (GO: 0005212) associated with structural constituents of the eye lens in the mouse lens proteome (Supplementary Tables S12–S16). In addition, the analysis identified the enrichment of six biological process GO terms associated with lens development in camera-type eye (GO: 0002088), camera-type eye development (GO: 0043010), eye development (GO: 0001654), sensory organ development (GO: 0007423), lens fiber cell differentiation (GO: 0070306), and lens fiber cell development (GO: 0070307) in the mouse lens proteome (Supplementary Tables S12–S16).
Finally, we sought to establish a correlation between the next-generation–based transcriptomics and mass spectrometry–based proteomics datasets. We previously published a mouse lens transcriptome and identified 14,465 genes across the same six developmental time points.19 A comparative analysis identified 5404 species overlapping in the transcriptome and protein datasets. We further selected six genes (CryαA, CryαB, CryβA1, and CryγC, Hsf4, and Pax6) and compared their expressions in both the transcriptome and the proteome datasets. As shown in Supplementary Figure S4, the expression of all four crystallin mRNA and their respective proteins are directly proportional, increasing at postnatal time points (Supplementary Fig. S4A–D). Interestingly, Hsf4 exhibited an expression profile similar to CryαA, CryαB, CryβA1, and CryγC (Supplementary Fig. S4E). In contrast, a relatively higher expression of Pax6 (and the respective protein) was observed at early time points that gradually decreased with age, resulting in lower expression at postnatal time points consistent with the role of Pax6 (Supplementary Fig. S4F).
Discussion
Here, we report a comprehensive proteome profile of embryonic and postnatal mouse lens using mass spectrometry–based protein sequencing. The mouse lenses were extracted at two embryonic and four postnatal time points, maintained as three biological replicates, and examined in three independent TMT experiments. The analysis of raw data revealed 5404 reliably quantitated proteins in the mouse lens. Our data comprise a comprehensive repository of the lens proteome profile at six developmental time points.
In total, we identified 5404 proteins representing nearly 25% of the protein-coding genes in the developing mouse lens. While we present the largest catalogue of the lens proteome reported to date, caution must be exercised when exploring this repository. It is important to understand that absence of protein(s) in our repository may not necessarily mean lack of expression in the lens proteome but rather lack of identification due to the limitations of the current methodologies.
To quantify proteins in biological samples, isobaric tagging (or labeling) is among the most commonly used methods that permit multiplexing of different samples; however, batch effects are common in mass spectrometry in combination with TMT peptide labeling.23–25 The time points from the three TMT sets did not cluster together in the PCA plot, suggesting a batch effect in the TMT experiments (Fig. 1). Likewise, the uneven distribution of reporter ion intensities in box plots and histogram analyses were further suggestive of a batch effect (Supplementary Figs. S1, S2). Maes and colleagues24 reported that the variability induced by TMT labeling is a small fraction of the total variation in mass spectrometry–based proteome sequencing with TMT peptide labeling. As the TMT-labeling efficiency is similar in all three TMT sets, we reason that TMT labeling does not contribute significantly to the total batch effect, and although the major contributing factor(s) responsible for the variability among the three TMT sets remains elusive, the mean normalized expression of the three TMT sets best represents the physiological expression at any developmental time point.
We identified proteins with catalytic activities, autophagy-associated proteins, cytoskeleton-associated proteins, heat shock proteins, mitochondrial-associated proteins, proteasome pathway–associated proteins, translation regulating proteins, ubiquitination process-associated proteins, proteins with receptor activities, and proteins with transporter activities in the mouse lens proteome (Supplementary Table S1). We identified 300-plus transcription factors in the developing mouse lens proteome. In contrast to crystallin proteins, expression of transcription factor proteins is more abundant at embryonic time points and gradually decreases as the lens ages (Supplementary Table S1). Homeobox (SIX3, SIX6, and PROX1) and paired box (PAX6) transcription factors were among the most abundantly expressed proteins in the mouse lens proteome (Supplementary Table S1).
We identified 19 different crystallin proteins originating from 46,150 PSMs, representing 22.2% of the total number of PSMs identified in the developing mouse lens proteome. These included CRYαA, CRYαB, CRYβA1, CRYβA2, CRYβA4, CRYβB1, CRYβB2, CRYβB3, CRYγA, CRYγB, CRYγC, CRYγD, CRYγE, CRYγF, CRYγN, CRYγS, CRYM, CRYZ, and CRYZL1. All crystallin proteins identified in the mouse lens proteome displayed the highest expression at P9 (Supplementary Table S1). We identified a 2- to 4-fold increase in expression for crystallin proteins at E18 compared to their respective expression at E15 (Supplementary Table S6). Interestingly, CRYγF exhibited an 8-fold upregulation at E18 compared to E15 (Supplementary Table S6). The upward trend in expression of crystallin proteins continued at P0, P3, P6, and P9 compared to E15, with a significant increase in expression of CRYβB2, CRYγF, and CRYγS (Supplementary Tables S7–S10).
The hallmark characteristic of the lens is a terminal differentiation of epithelial cells to fiber cells through a synchronized gene expression pattern. Analysis of proteome data identified a set of proteins (LGSN, BFSP1, BFSP2, and DNASE2β) with significantly higher expression in the postnatal mouse lens that was consistent with transcriptome data.19 In a recent study, Harding and colleagues26 identified expression of lgsn in differentiating secondary lens fiber cells, and knockdown of lgsn in zebrafish results in a smaller lens with defective cortex due to impaired secondary fiber cell morphogenesis.26 Likewise, Bfsp1, Bfsp2, and Dnase2β are dominantly expressed in the fiber cells of developing and adult lenses and have been implicated in the autosomal recessive and dominant forms of cataractogenesis.27,28 Moreover, secreted frizzled-related proteins (Sfrp) play an important role in lens development with strong expression in lens fiber cells.29,30 We identified two secreted frizzled-related proteins, SFRP1 and SFRP2, in the developing mouse lens proteome. The expression levels of SFRP2 decreased as lens development progressed while SFRP1 maintained a steady expression pattern, consistent with the transcriptome data.19
Cataracts are the leading cause of blindness worldwide and a major cause of childhood blindness in the Third World countries. The Cat-Map database includes genes associated with congenital, age-related cataracts and ocular and nonocular syndromes with cataractogenesis.5 We identified 29 proteins associated with autosomal dominant congenital cataracts and 20 proteins associated with autosomal recessive congenital cataracts (Supplementary Table S17). We also identified 21 proteins associated with age-related cataracts and 88 proteins associated with cataractogenesis with other ocular and nonocular anomalies in the mouse lens proteome.
We previously reported multiple loss-of-function mutations in FYCO1, an autophagy-associated gene, responsible for autosomal recessive congenital cataracts.31 Moreover, we recently reported that haploinsufficiency of DNAJB1, an autophagy-associated cochaperone, results in Peters anomaly.32 We searched the proteins in the mouse lens proteome and identified 404 autophagy-associated proteins (Supplementary Table S18). Interestingly, proteins involved in autophagy exhibited a relatively higher expression in postnatal lens consistent with the notion that autophagy plays a critical role in lens fiber cell differentiation.
In conclusion, we present a comprehensive repository of the lens proteome for six developmental time points using quantitative mass spectrometry–based protein sequencing. Investigating the proteomic landscape in a model tissue that closely mimics the human ocular lens will serve as a resource for the lens research community and will add to our understanding of the proteome's role in the maintenance of lens transparency and disease manifestation.
Supplementary Material
Acknowledgments
Supported by National Eye Institute Grant R01EY022714 (SAR; Bethesda, MD, USA).
Disclosure: S.Y. Khan, None; M. Ali, None; F. Kabir, None; S. Renuse, None; C.H. Na, None; C.C. Talbot Jr, None; S.F. Hackett, None; S.A. Riazuddin, None
References
- 1. McAvoy JW, Chamberlain CG, de Iongh RU, Hales AM, Lovicu FJ. . Lens development. Eye (Lond). 1999; 13 pt 3b: 425– 437. [DOI] [PubMed] [Google Scholar]
- 2. Lovicu FJ, McAvoy JW. . Growth factor regulation of lens development. Dev Biol. 2005; 280: 1– 14. [DOI] [PubMed] [Google Scholar]
- 3. Robinson GC, Jan JE, Kinnis C. . Congenital ocular blindness in children, 1945 to 1984. Am J Dis Child. 1987; 141: 1321– 1324. [DOI] [PubMed] [Google Scholar]
- 4. Hejtmancik JF, Smaoui N. . Molecular genetics of cataract. Dev Ophthalmol. 2003; 37: 67– 82. [DOI] [PubMed] [Google Scholar]
- 5. Shiels A, Bennett TM, Hejtmancik JF. . Cat-Map: putting cataract on the map. Mol Vis. 2010; 16: 2007– 2015. [PMC free article] [PubMed] [Google Scholar]
- 6. Datiles MB, Schumer DJ, Zigler JS Jr, Russell P, Anderson L, Garland D. . Two-dimensional gel electrophoretic analysis of human lens proteins. Curr Eye Res. 1992; 11: 669– 677. [DOI] [PubMed] [Google Scholar]
- 7. Jungblut PR, Otto A, Favor J,et al. Identification of mouse crystallins in 2D protein patterns by sequencing and mass spectrometry. Application to cataract mutants. FEBS Lett. 1998; 435: 131– 137. [DOI] [PubMed] [Google Scholar]
- 8. Lampi KJ, Ma Z, Hanson SR,et al. Age-related changes in human lens crystallins identified by two-dimensional electrophoresis and mass spectrometry. Exp Eye Res. 1998; 67: 31– 43. [DOI] [PubMed] [Google Scholar]
- 9. Lampi KJ, Shih M, Ueda Y, Shearer TR, David LL. . Lens proteomics: analysis of rat crystallin sequences and two-dimensional electrophoresis map. Invest Ophthalmol Vis Sci. 2002; 43: 216– 224. [PubMed] [Google Scholar]
- 10. Ma Z, Hanson SR, Lampi KJ, David LL, Smith DL, Smith JB. . Age-related changes in human lens crystallins identified by HPLC and mass spectrometry. Exp Eye Res. 1998; 67: 21– 30. [DOI] [PubMed] [Google Scholar]
- 11. Ueda Y, Duncan MK, David LL. . Lens proteomics: the accumulation of crystallin modifications in the mouse lens with age. Invest Ophthalmol Vis Sci. 2002; 43: 205– 215. [PubMed] [Google Scholar]
- 12. Elsobky S, Crane AM, Margolis M, Carreon TA, Bhattacharya SK. . Review of application of mass spectrometry for analyses of anterior eye proteome. World J Biol Chem. 2014; 5: 106– 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kyselova Z. . Mass spectrometry-based proteomics approaches applied in cataract research. Mass Spectrom Rev. 2011; 30: 1173– 1184. [DOI] [PubMed] [Google Scholar]
- 14. Semba RD, Enghild JJ, Venkatraman V, Dyrlund TF, Van Eyk JE. . The Human Eye Proteome Project: perspectives on an emerging proteome. Proteomics. 2013; 13: 2500– 2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bassnett S, Wilmarth PA, David LL. . The membrane proteome of the mouse lens fiber cell. Mol Vis. 2009; 15: 2448– 2463. [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Z, Han J, David LL, Schey KL. . Proteomics and phosphoproteomics analysis of human lens fiber cell membranes. Invest Ophthalmol Vis Sci. 2013; 54: 1135– 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z, Schey KL. . Proteomic analysis of lipid raft-like detergent-resistant membranes of lens fiber cells. Invest Ophthalmol Vis Sci. 2015; 56: 8349– 8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shang F, Wilmarth PA, Chang ML,et al. Newborn mouse lens proteome and its alteration by lysine 6 mutant ubiquitin. J Proteome Res. 2014; 13: 1177– 1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khan SY, Hackett SF, Lee MC, Pourmand N, Talbot CC Jr, Riazuddin SA. . Transcriptome profiling of developing murine lens through RNA sequencing. Invest Ophthalmol Vis Sci. 2015; 56: 4919– 4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tyanova S, Temu T, Sinitcyn P,et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016; 13: 731– 740. [DOI] [PubMed] [Google Scholar]
- 21. Richardson JE, Bult CJ. . Visual annotation display (VLAD): a tool for finding functional themes in lists of genes. Mamm Genome. 2015; 26: 567– 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vizcaino JA, Csordas A, Del-Toro N,et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44: 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gregori J, Villarreal L, Mendez O, Sanchez A, Baselga J, Villanueva J. . Batch effects correction improves the sensitivity of significance tests in spectral counting-based comparative discovery proteomics. J Proteomics. 2012; 75: 3938– 3951. [DOI] [PubMed] [Google Scholar]
- 24. Maes E, Valkenborg D, Baggerman G,et al. Determination of variation parameters as a crucial step in designing TMT-based clinical proteomics experiments. PLoS One. 2015; 10: e0120115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuligowski J, Perez-Guaita D, Lliso I,et al. Detection of batch effects in liquid chromatography-mass spectrometry metabolomic data using guided principal component analysis. Talanta. 2014; 130: 442– 448. [DOI] [PubMed] [Google Scholar]
- 26. Harding RL, Howley S, Baker LJ,et al. Lengsin expression and function during zebrafish lens formation. Exp Eye Res. 2008; 86: 807– 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song S, Landsbury A, Dahm R, Liu Y, Zhang Q, Quinlan RA. . Functions of the intermediate filament cytoskeleton in the eye lens. J Clin Invest. 2009; 119: 1837– 1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishimoto S, Kawane K, Watanabe-Fukunaga R,et al. Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature. 2003; 424: 1071– 1074. [DOI] [PubMed] [Google Scholar]
- 29. Sugiyama Y, Shelley EJ, Wen L,et al. Sfrp1 and Sfrp2 are not involved in Wnt/beta-catenin signal silencing during lens induction but are required for maintenance of Wnt/beta-catenin signaling in lens epithelial cells. Dev Biol. 2013; 384: 181– 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wawersik S, Purcell P, Rauchman M, Dudley AT, Robertson EJ, Maas R. . BMP7 acts in murine lens placode development. Dev Biol. 1999; 207: 176– 188. [DOI] [PubMed] [Google Scholar]
- 31. Chen J, Ma Z, Jiao X,et al. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am J Hum Genet. 2011; 88: 827– 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khan SY, Vasanth S, Kabir F,et al. FOXE3 contributes to Peters anomaly through transcriptional regulation of an autophagy-associated protein termed DNAJB1. Nat Commun. 2016; 7: 10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.