Abstract
Emerging infectious diseases are reducing biodiversity on a global scale. Recently, the emergence of the chytrid fungus Batrachochytrium salamandrivorans resulted in rapid declines in populations of European fire salamanders. Here, we screened more than 5000 amphibians from across four continents and combined experimental assessment of pathogenicity with phylogenetic methods to estimate the threat that this infection poses to amphibian diversity. Results show that B. salamandrivorans is restricted to, but highly pathogenic for, salamanders and newts (Urodela). The pathogen likely originated and remained in coexistence with a clade of salamander hosts for millions of years in Asia. As a result of globalization and lack of biosecurity, it has recently been introduced into naïve European amphibian populations, where it is currently causing biodiversity loss.
Emerging infectious diseases play an important role in the ongoing sixth mass extinction (1). Fungi comprise a greater threat relative to other taxonomic classes of pathogens and have recently caused some of the most severe die-offs and extinctions among a wide range of organisms (2). The classical cause of amphibian chytridiomycosis (Batrachochytrium dendrobatidis) has resulted in extensive disease and declines in a wide variety of amphibian species across the three orders [i.e., frogs and toads (Anura), salamanders and newts (Urodela), and caecilians (Gymnophiona)] (2). Recently, a second highly pathogenic chytrid fungus (B. salamandrivorans) emerged as a novel form of amphibian chytridiomycosis and extirpated fire salamander populations in northern Europe (3, 4) in a region where B. dendrobatidis is in a state of stable coexistence with the amphibian communities (5).
To predict the potential impact of B. salamandrivorans on amphibian diversity more broadly, we first estimated its host range by experimentally exposing 35 species from the three amphibian orders (10 anurans, 24 urodelans, and 1 caecilian) to controlled doses of 5000 zoospores for 24 hours (3) (table S1). Except for five urodelan taxa for which wild-caught specimens were used, all other experimental animals were captive bred. With the exception of four urodelan taxa, all experimental animals derived from a single source population. After exposure, animals were monitored daily for clinical signs until at least 4 weeks after exposure. Infection loads were assessed weekly using quantitative polymerase chain reaction (qPCR) on skin swabs (6), and histopathology was performed on all specimens that died. Our results show that colonization by B. salamandrivorans was limited to Urodela, whereas none of the anuran and caecilian species became infected (Fig. 1, squares). Alarmingly, 41 out of 44 of the Western Palearctic salamanders (Salamandridae and Plethodontidae) rapidly died after infection with B. salamandrivorans. The propensity of B. salamandrivorans to infect these species was confirmed by its ability to successfully invade the skin of several urodelan, but none of the anuran, species. This was demonstrated with an immunohistochemical staining of the abdominal skin of amphibians after exposure to 10,000 zoospores for 24 hours (table S1 and fig. S1).
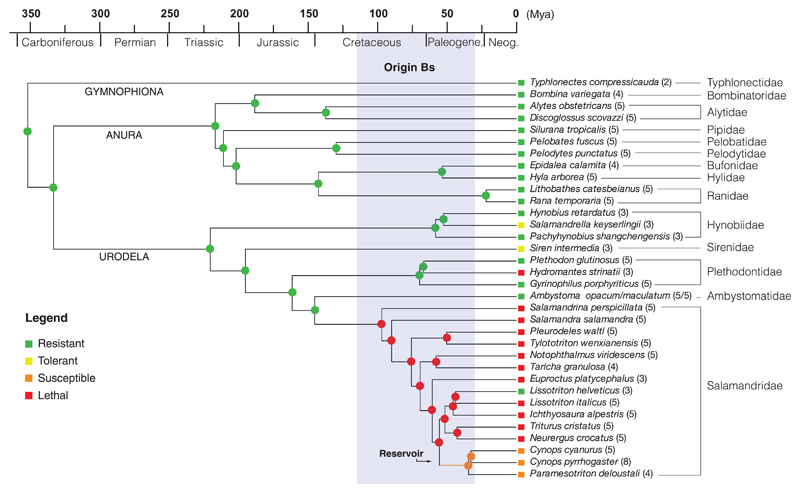
Fig. 1. Amphibian susceptibility to Batrachochytrium salamandrivorans (Bs) through time.
Molecular time scale (millions of years ago) for 34 species; rectangles indicate the species category based on the experimental infection tests. Resistant: no infection, no disease; tolerant: infection in the absence of disease; susceptible: infection resulting in clinical disease with possibility of subsequent recovery; lethal: infection resulting in lethal disease in all infected animals. Colored dots on nodes indicate the results of the maximum likelihood ancestral reconstructions (P > 0.95).The clade of susceptible Asian salamanders that originated in the early Paleogene is indicated in orange. The 95% highest posterior density for time of divergence between B. salamandrivorans and B. dendrobatidis is indicated in gray.
To estimate the current range of B. salamandrivorans infections, we used qPCR to screen 5391 wild amphibian individuals from four continents for the presence of B. salamandrivorans DNA in their skin (6) (tables S2 and S3). In accordance with the results of the experimentally determined host range, infections were detected only in urodeles. Furthermore, the detection of B. salamandrivorans DNA (all sequences were 100% identical with GenBank accession number KC762295) was limited to East Asia (Thailand, Vietnam, and Japan) in the absence of obvious disease, and Europe (Netherlands and Belgium) where it is associated with severe disease outbreaks [Netherlands, 2010 (3, 4), and Belgium, 2013 (Eupen, N50°37′23″; E6°05′19″) and 2014 (Robertville, N50°27′12″; E6°06′11″)]. These findings suggest long-term endemism in Asia and a recent incursion in Europe.
We used the results of our infection experiments as a proxy for classifying amphibians into four categories of response to B. salamandrivorans: resistant, tolerant, susceptible, and lethal (Fig. 1, squares). Although the limited number of source populations used does not allow estimation of within-species variation, responses to infection were highly consistent within a given population. Lethal responses were observed in specimens from both captive-bred (10 of 19 taxa) and wild (2 of 5 taxa) urodelans. Our infection experiments indicated three Asian salamanders (Cynops pyrrhogaster, Cynops cyanurus, and Paramesotriton deloustali) as potential reservoirs. Seven specimens of these species were capable of limiting clinical disease and either persisted with infection for up to at least 5 months with recurring episodes of clinical disease, or even totally cleared the infection (table S1 and fig. S2). The combined evidence of natural occurrence and experimental maintenance of B. salamandrivorans infections indicates that at least these three species may function as a reservoir in Asia.
To investigate whether these amphibian communities may have constituted a reservoir of infection in the past, we estimated when B. salamandrivorans diverged from B. dendrobatidis and used present-day patterns of susceptibility to reconstruct amphibian susceptibility through time. Our Bayesian estimates of divergence time with a broad prior calibration range resulted in a mean estimate of 67.3 million years ago (Ma) (fig. S3) and a 95% highest posterior density interval of 115.3 to 30.3 Ma, indicating that B. salamandrivorans diverged from B. dendrobatidis in the Late Cretaceous or early Paleogene (Fig. 1, gray bar). Maximum parsimony and maximum likelihood ancestral reconstructions (Fig. 1) of amphibian susceptibility suggest that the potential of being a reservoir evolved in the ancestors of modern Asian newts between 55 and 34 Ma in the Paleogene (Fig. 1, orange branch), shortly after the origin of their pathogen. These ancestors reached Asia after withdrawal of the Turgai Sea (7), suggesting that Asia has been a natural reservoir for B. salamandrivorans for the past 30 million years. Our detection of B. salamandrivorans in a >150-year-old museum sample of the Asian newt Cynops ensicauda (table S4, RMNH RENA 47344) is consistent with this reservoir hypothesis.
Given the discontinuity of the global distribution of B. salamandrivorans, introduction from Asia into Europe must have been human-mediated. Asian salamanders and newts are being traded internationally in large numbers annually (for instance, more than 2.3 million individuals of Cynops orientalis were imported into the United States from 2001 to 2009) (8). To assess the potential of B. salamandrivorans spread by captive amphibians, we tested 1765 skin samples from amphibians in pet shops in Europe, London Heathrow Airport, and an exporter in Hong Kong (tables S5 and S6) and 570 samples from other captive amphibians (tables S7 and S8) for B. salamandrivorans. We found three positive samples from captive individuals of the Asian newt species Tylototriton vietnamensis, two of which were imported to Europe in 2010. Furthermore, our transmission experiments showed that B. salamandrivorans can effectively be transmitted across multiple urodelan species (e.g., from Cynops pyrrhogaster to Salamandra salamandra, fig. S4) by direct contact, demonstrating the potential for pathogen spillover.
Our infection experiments show that B. salamandrivorans is lethal to at least some of the New World salamandrid species (genera Taricha and Notophthalmus). Although these combined genera contain only seven species, together they have a widespread distribution and are often very abundant. The outcome of exposure of three lineages of plethodontids (a family comprising 66% of global urodelan diversity) to B. salamandrivorans ranged from a lack of any detectable infection (Gyrinophilus), to transient skin invasion (Plethodon) and lethal infection (Hydromantes), making it likely that other species in this large family are vulnerable.
Our study demonstrates that the process of globalization with its associated human and animal traffic can rapidly erode ancient barriers to pathogen transmission, allowing the infection of hosts that have not had the opportunity to establish resistance. Thus, pathogens, such as those we describe here, have the potential to rapidly pose a threat of extinction.
Supplementary Material
Acknowledgments
We thank M. Schenkel and J. Beukema for providing samples and the National Museum of Natural History–Naturalis, Leiden, Netherlands, for providing museum specimens. We thank the many amphibian breeders (including S. Bogaerts, M. Sparreboom, H. Janssen, F. Maillet, A. Jamin, and S. Voitel) who provided offspring to conduct the infection experiments. Financial support was partly provided by the Dutch Ministry of Economic Affairs and by the UK Department for Environment, Food and Rural Affairs, project grant FC1195. M.B. is funded by a Dehousse grant from the Royal Zoological Society of Antwerp. P.V.R. is funded by Ghent University Special Research Fund (BOF13/PDO/130). F.P. and T.W.J.G. are funded by the Morris Animal Foundation (D12z0-002). M.C.F. and T.W.J.G. are funded by the UK Natural Environment Research Council (NERC). R.A.F. is supported by the Wellcome Trust. U.T. and B.R.S. are funded by the Vontobel Stiftung, Janggen-Pöhn Stiftung, Basler Stiftung für biologische Forschung, Stiftung Dr. Joachim De Giacomi, Zoo Zürich, Grün Stadt Zürich, European Union of Aquarium Curators, and Zürcher Tierschutz. J.B. is funded by Fundación General CSIC and Banco Santander. E.W. is funded by Economic and Social Research Council (ESRC)-NERC Interdisciplinary Ph.D. studentship. A.A.C. is supported by a Royal Society Wolfson research merit award. K.N. is funded by grants from the Ministry of Education, Science and Culture, Japan (nos. 20770066 and 23770084) and Japan Society for the Promotion of Science (JSPS) AA Core-to-Core program Type B. Asia-Africa Science Platforms. T.T.N. is funded by the JSPS RONPAKU program. F.B. is supported by European Research Council Starting Grant 204509 [project Tracing Antimicrobial Peptides and Pheromones in the Amphibian Skin (TAPAS)]. I.V.B is supported by a postdoctoral Fellowship from the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO). All data described in the paper are presented in the supplementary materials.
References
- 1.Wake DB, Vredenburg VT. Proc Natl Acad Sci USA. 2008;105(suppl. 1):11466–11473. doi: 10.1073/pnas.0801921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher MC, et al. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martel A, et al. Proc Natl Acad Sci USA. 2013;110:15325–15329. doi: 10.1073/pnas.1307356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spitzen-van der Sluijs A, et al. Amphib-reptil. 2013;34:233–239. [Google Scholar]
- 5.Spitzen-van der Sluijs A, et al. Conserv Biol. 2014;28:1302–1311. doi: 10.1111/cobi.12281. [DOI] [PubMed] [Google Scholar]
- 6.Blooi M, et al. J Clin Microbiol. 2013;51:4173–4177. doi: 10.1128/JCM.02313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang P, Papenfuss TJ, Wake MH, Qu L, Wake DB. Mol Phylogenet Evol. 2008;49:586–597. doi: 10.1016/j.ympev.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Herrel A, van der Meijden A. Herpetol J. 2014;24:103–110. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.