Graphical abstract
A cellular protease-mediated graphene-based nanosystem is developed for co-delivery of a membrane-associated cytokine (TRAIL) and an intracellular-acting small-molecule drug (DOX). The nanocarrier realizes the intramembrane enzyme-mediated extracellular release of TRAIL and endocytotic acidity-responsive intracellular release of DOX, which enables them to target to their distinct sites of action. This formulation starts a new generation of 2D nanomaterials with programmed-release therapeutics capability for combination cancer treatment.
Keywords: furin, graphene oxide, cytokine, drug delivery, nanomedicine
Combination cancer therapy has attracted considerable attentions due to its advantages of enhancing therapeutic efficacy, decreasing possibility of drug resistance and reducing side effects over monotherapy.[1] Co-encapsulation of multiple anticancer agents in one single nanocarrier can optimize their pharmacokinetic profiles and biodistribution behaviors in a spatiotemporal co-delivery fashion, resulting in a more efficient synergistic antitumor activity, compared to the conventional “cocktail”-based drug mixtures.[2] For example, irinotecan/floxuridine co-loaded liposome has proved to be more effective than the cocktails of individual free and liposomal drugs in both animal tumor models and patients with advanced solid tumors concordantly.[3]
Co-delivery systems incorporating therapeutic protein/peptides with small-molecule drugs or macromolecular nuclear acids has also been validated as a promising strategy for enhancement of cancer treatment.[4] Different from small-molecule drugs and nuclear acids that mostly function within the cells, the action sites of proteins are mainly classified to be localized on the cell membrane and inside the cells, respectively. For instance, tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), a typical membrane-associated cytokine, acts on the death receptor on the cell membrane for induction of apoptosis,[5] while caspase 3 takes effect in the cytoplasm to activate the caspase-mediated apoptosis.[6] However, a co-delivery system capable of differentiating the extracellular and intracellular targets still remains elusive. In our previous work, we have developed a gel-liposome-based system for sequential and site-specific co-delivery of TRAIL and doxorubicin (DOX), a small-molecule anticancer drug. This formulation was limited by relatively low drug-loading capacity and undesirable leakage of physically-encapsulated protein.[4d]
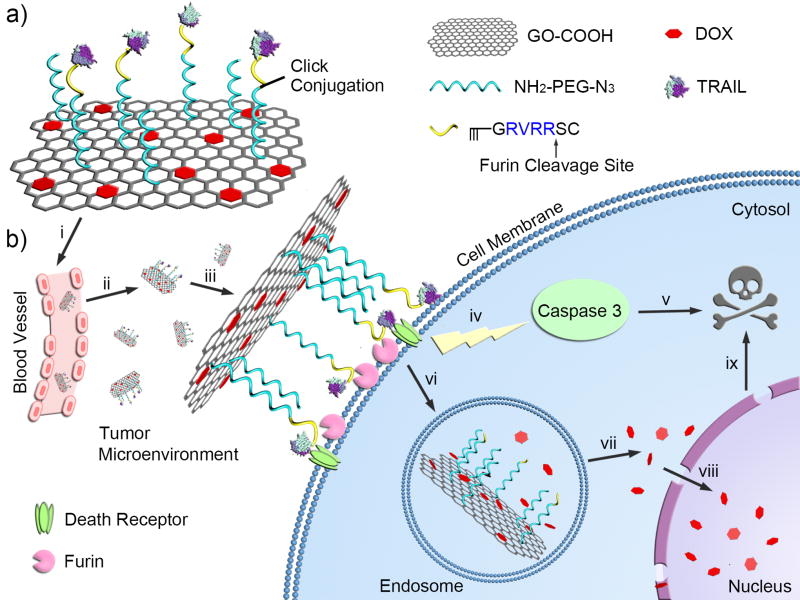
We herein report a new graphene-based co-delivery nanosystem of a cell-membrane-targeted anticancer protein and a chemotherapeutic agent for combination cancer treatment (Figure 1). This designed nanocarrier is composed of graphene oxide (GO), polyethylene glycol (PEG) linker and a furin-cleavable peptide, which is achieved by step-by-step site-specific conjugations (Figure 1a). The GO nanosheet is applied to bear DOX due to a strong supramolecular π-π stacking interaction between GO and DOX.[7] A heterobifunctional PEG linker containing amino and azide terminal groups (NH2-PEG-N3) was first utilized for connection between the carboxylated GO (GO-COOH) nanosheet and the furin-cleavable peptide, which also provides a good dispersity and stability of GO nanosheet due to the high hydrophilicity of the long PEG chain. The alkynylated peptide sequence in a typical form of RX(K/R)RY (R: arginine; K: lysine; X: any amino acid; Y: the cleavage site) is linked to the PEG chain via a click chemistry reaction, which can be specifically recognized and cleaved by furin, a protease highly expressed on the cell membrane and Golgi complex of many cancer cells.[8] Finally, tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), that acts on the death receptor on the cell membrane to activate apoptosis-mediated cell death, is conjugated to the sulfhydryl (cysteine) groups of the peptide using an amine-to-sulfhydryl crosslinker. Of note, TRAIL itself can also serve as “targeting ligand” for the whole delivery system.[9]
Figure 1.
Schematic design of the cellular protease-mediated graphene-based co-delivery system. a) Main components of TRAIL/DOX-fGO, consisting of DOX-loaded GO, PEG linker and TRAIL-conjugated furin-cleavable peptide. b) Site-specific delivery of TRAIL to cell membrane and DOX to nuclei for enhanced synergistic cancer treatment. i: intravenous administration of GO; ii: accumulation of GO at the tumor site through passive and active targeting effects; iii: TRAIL binding on the death receptor and degradation of peptide linker by furin on the cell membrane; iv: activation of caspase-mediated apoptosis; v: induction of cell death; vi: endocytosis of GO by the tumor cells; vii: acid-promoted DOX release in endosome; viii: accumulation of released DOX into nucleus; ix: induction of DNA damage-mediated apoptosis and cytotoxicity.
The GO-based nanocarrier preferentially accumulates at the tumor site by means of the enhanced permeability and retention (EPR) effect after intravenous administration, and covered on the tumor cells along with the binding of TRAIL on the death receptor.[5c, 10] The plasma membrane-located furin digests the peptide linker, which results in the extracellular release of TRAIL and prevents its internalization together with the GO into the cancer cells for enhanced apoptosis induction. The GO nanosheet carrying DOX is then internalized into the cells.[11] In the endosomes, the acidic endocytotic vesicles, the release of DOX is promoted as expected, resulting from the increased solubility of protonated DOX in the acidic environment. The intracellularly released DOX specifically accumulates into the nuclei to produce DNA damage-mediated cytotoxicity,[12] which combines with the apoptosis-inducing effect of TRAIL for an optimal synergistic antitumor activity (Figure 1b).
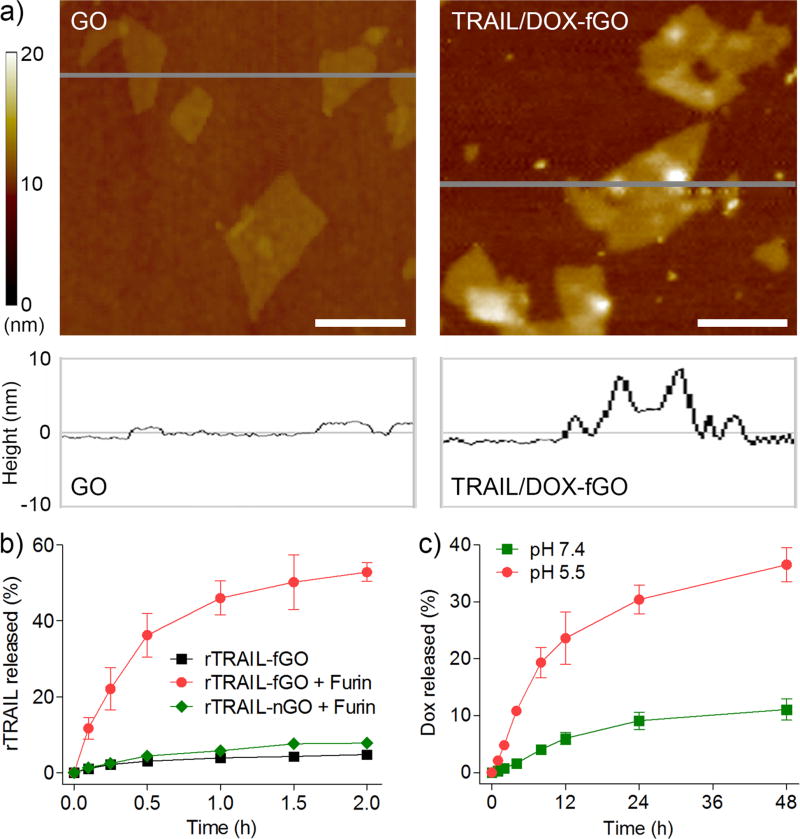
To substantiate our design, we first synthesized TRAIL/DOX-fGO using a sequential and site-specific conjugation technique (Figure S1a in the Supporting Information). A nano-sized GO sheet was obtained after ultrasonication and purification, which had an average size of 152 nm determined by the dynamic light scattering measurement[13] (Figure S2a). The atomic force microscope (AFM) imaging showed a typical two-dimensional (2D) appearance of GO with the thickness of 2 nm (Figure 2a). The GO nanosheet was then carboxylated and conjugated with NH2-PEG-N3 via carbodiimide catalyzed amide formation.[14] PEGylation was confirmed by the infrared spectroscopy (Figure S3). Upon the click chemistry-mediated linkage between the furin-cleavable peptide and the PEG chain, TRAIL was conjugated to the sulfhydryl group of cysteine in the peptide using an amino- and sulfhydryl-directed heterobifunctional crosslinker (Sulfo-SMCC) to acquire TRAIL-conjugated furin-cleavable GO (TRAIL-fGO). After incubation of TRAIL-fGO with triethylamine (TEA)-treated DOX, the DOX-loaded and TRAIL-conjugated furin-cleavable GO (TRAIL/DOX-fGO) was obtained by centrifugal filtration to remove the excessive TEA and unbound DOX (Figure S4a), which had an average size of 194 nm (Figure S2b) and a DOX-loading capacity up to 43.8%, far exceeding that of many lipid- and polymer-based nanocarriers. TRAIL/DOX-fGO with the long PEG chain (molecular weight: 5000 Da) showed higher stability than both GO and TRAIL/DOX-fGO with the short PEG chain (molecular weight: 350 Da) in the culture medium containing 10% fetal bovine serum (Figure S4b, c). The AFM image showed that TRAIL/DOX-fGO had a lateral width of less than 200 nm and a topological height of over 8 nm (Figure 2a), which was significantly higher than that of the PEG-GO conjugate as about 5 nm (Figure S5), suggesting the conjugation of the peptide-linked TRAIL on the PEG-modified GO nanosheet. Additionally, the TRAIL conjugation was assessed using the protein displacement method (Figure S6). No significant change in the fluorescence intensity of rTRAIL was found in the filtrate after centrifugal filtration following the incubation of rTRAIL-fGO with different concentrations of bovine serum albumin for 1 h, which further indicated that TRAIL was conjugated on the surface of GO.
Figure 2.
a) AFM images of GO and TRAIL/DOX-fGO. Scale bars indicate 200 nm. b) In vitro release profiles of rTRAIL from rTRAIL-fGO or rTRAIL-nGO in the absence and presence of furin. c) In vitro release profiles of DOX from rTRAIL/DOX-fGO at pH 7.4 and 5.5.
To evaluate the furin-mediated release of TRAIL, we determined the in vitro release profile of rhodamine-labeled TRAIL (rTRAIL) from rTRAIL-fGO in the presence and absence of furin using a centrifuge filter tube (Figure 2b). Only 4.8% of rTRAIL was detected within 2 h in the absence of furin. In sharp contrast, 11.7% of rTRAIL was released from rTRAIL-fGO in the first 6 min after incubation with furin, and nearly 52.9% was released within 2 h. Furthermore, we have utilized rTRAIL-conjugated GO without furin-cleavable substrate (rTRAIL-nGO) as a control (Figure S1b). After 24 h of incubation with furin, the cumulative amount of rTRAIL released from rTRAIL-nGO was determined to be only 7.8%. It was suggested that the specific peptide linker between TRAIL and GO could be efficiently cleaved by furin and therefore led to the release of TRAIL. The circular dichroism spectrum of the released TRAIL was consistent with that of the native TRAIL (Figure S7), indicating that this furin-directed peptide degradation had no impact on the protein secondary structure of TRAIL. In addition, the release behavior of DOX from TRAIL/DOX-fGO was tested using a dialysis tube (Figure 2c). TRAIL/DOX-fGO displayed extremely higher DOX release rate at pH 5.5 than pH 7.4, implying that when TRAIL/DOX-fGO was endocytosed into the endosome, the acidic environment allowed the promoted release of DOX inside rather than outside the cells.
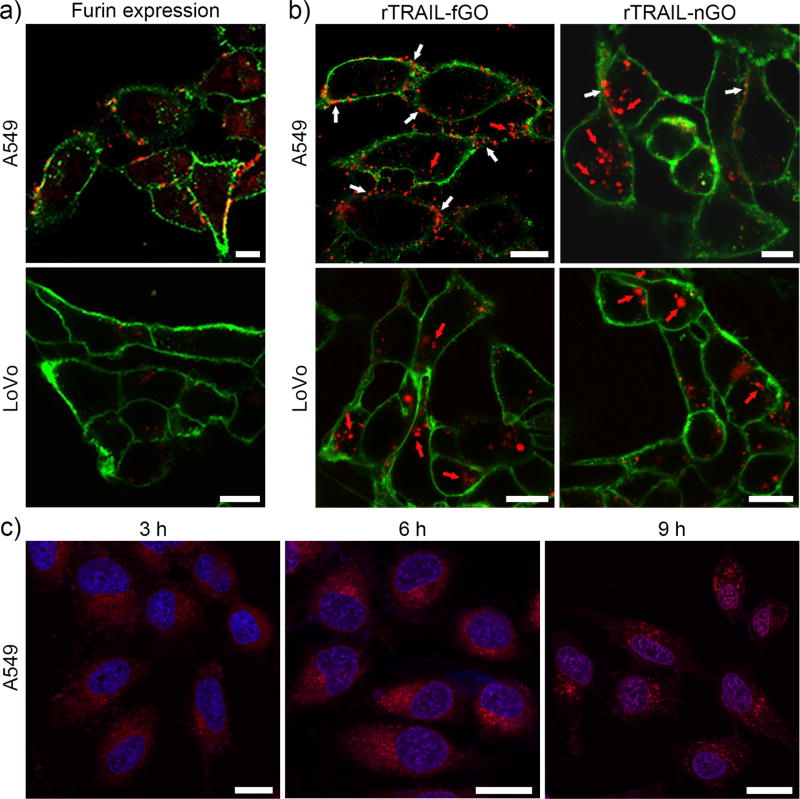
Next, the human lung adenocarcinoma epithelial (A549) cells that express sufficient furin[15] were applied as a cell model to investigate the site-specific delivery of TRAIL and DOX by TRAIL/DOX-fGO. The furin-deficient LoVo cells, the human colon adenocarcinoma cells, were taken as a control.[16] We first determined the level of furin expression on the cancer cells using the immunofluorescence staining (Figure 3a and Figure S8). As expected, endoprotease furin (red) was apparently detected to be localized both on the cell membranes (green) and within the cytosol of A549 cells, whereas few were found in LoVo cells. To validate that the membrane-localized furin was able to degrade the peptide, thus causing the released TRAIL bind onto the plasma membrane, A549 and LoVo cells were incubated with rTRAIL-fGO or rTRAIL-nGO for 2 h, followed by the confocal laser scanning microscope (CLSM) observation (Figure 3b and Figure S9). An evident difference in the distribution of rTRAIL delivered by rTRAIL-fGO and rTRAIL-nGO was observed in A549 cells. A large number of rTRAIL (red) bound to the cell membrane (green) after incubation with rTRAIL-fGO, while most of rTRAIL was detected within the cells that were treated with rTRAIL-nGO, which indicated that the conjugated TRAIL on nGO without the furin-cleavable site was inevitably internalized with GO into the cell, but the released TRAIL from TRAIL-fGO mediated by the cell surface furin could efficiently bind onto the cell membrane for enhanced apoptosis-inducing effect. On the contrary, for LoVo cells, rTRAIL of both formulations was mainly monitored inside the cells, further confirming the furin-mediated extracellular release and membrane binding of TRAIL.
Figure 3.
a) CLSM images of A549 and LoVo cells after immunofluorescence staining with the furin antibody and the rhodamine-labeled secondary antibody. The cell membranes were stained with AF488-WGA. Red: furin; Green: cell membrane. Scale bars indicate 10 µm. b) CLSM images of A549 and LoVo cells after incubation with rTRAIL-fGO for 2 h. The cell membranes were stained with AF488-WGA. Red: rTRAIL; Green: cell membrane. Scale bars indicate 10 µm. c) CLSM images of A549 cells after incubation with TRAIL/DOX-fGO for different time. The nuclei were stained with Hoechst 33342. Red: DOX; Blue: nuclei. Scale bars indicate 20 µm.
To assess the endocytotic capability of TRAIL/DOX-fGO, we compared the cellular uptake of TRAIL/DOX-fGO on A549 cells at 37 °C and 4 °C (Figure S10). The uptake of TRAIL/DOX-fGO was significantly inhibited at 4 °C, suggesting that the GO nanocarrier permeated into the cells through a typical internalization pathway. The intracellular distribution of DOX in A549 cells was evaluated using CLSM (Figure 3c and Figure S11). After the cells were incubated for 3 h, the DOX signal (red) evenly distributed within the cells, suggesting that TRAIL/DOX-fGO was taken up by the cells. As time increased to 6 h, DOX was released from the GO nanosheet and accumulated into the nuclei (blue). After 9 h of incubation, most of DOX showed nucleus targeting as visualized by the magenta fluorescence for subsequent DNA damage-induced apoptosis.
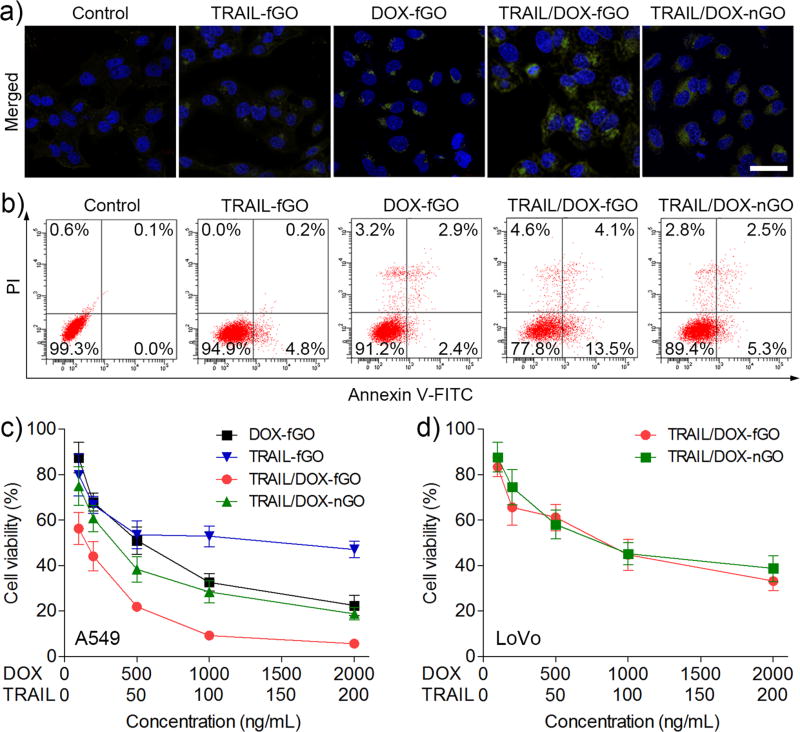
The synergistic apoptosis-inducing activities of TRAIL and DOX delivered site-specifically by TRAIL/DOX-fGO were evaluated toward A549 cells using the FITC (fluorescein isothiocyanate) active caspase 3 apoptosis kit and the Annexin-FITC apoptosis detection kit, respectively. Caspase 3, known as an effector for death signal, has a central effect on the execution phase of cell apoptosis. As shown in Figure 4a, the fluorescence signal (green) was hardly detected within the cells after treated with TRAIL-fGO; this is because A549 cells were TRAIL resistant and did not show a clear evidence of apoptosis even at a high concentration of TRAIL.[17] Chemotherapeutic agents including DOX can sensitize TRAIL-resistant A549 cells to TRAIL-mediated apoptosis.[18] It has been validated that the uptake of anticancer drugs remarkably increased the expression of death receptors in a variety of cancer cells for enhanced apoptotic effects.[19] The combination treatment of TRAIL and DOX using TRAIL/DOX-nGO on A549 cells resulted in a stronger green fluorescence signal from the active caspase 3 in the cytosol. Of note, TRAIL/DOX-fGO compared to TRAIL/DOX-nGO presented a significantly greater apoptosis-inducing capability, arising from the apoptosis-enhancement by TRAIL that was targetedly delivered on the cell membrane by fGO, combined with the apoptosis-sensitization by DOX. Furthermore, the quantitative determination results obtained from the Annexin V-FITC/PI (propidium iodide) detection assay verified the optimal synergistic apoptotic efficacy of TRAIL/DOX-fGO (Figure 4b). TRAIL/DOX-fGO showed the highest total apoptotic ratio of 17.6% (a sum of the early apoptotic ratio of 13.5% and the late apoptotic ratio of 4.1%) and the lowest viability of 77.8% compared to other formulations.
Figure 4.
a) CLSM images of A549 cells stained with FITC-labeled active caspase 3 antibody after treated with different formulations. The nuclei were stained with Hoechst 33342. Green: active caspase 3; Blue: nuclei. Scale bars are 40 µm. b) Flow cytometric analysis of A549 cell apoptosis induced by different formulations. The dot plots represent the typical viable and apoptotic cell populations (%) obtained using Annexin V-FITC/PI staining. c) In vitro cytotoxicity of different formulations against A549 cells for 48 h. d) In vitro cytotoxicity of TRAIL/DOX-fGO and TRAIL/DOX-nGO against LoVo cells for 48 h.
The in vitro cytotoxicity of TRAIL/DOX-fGO against A549 cells were examined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Figure 4c). The half-maximal inhibitory concentration (IC50) of TRAIL-fGO on A549 cells was 119 ng/mL, suggesting the TRAIL-insensitivity of A549 cells, while IC50 of DOX-fGO was 509 ng/mL. In comparison, TRAIL/DOX-nGO showed increased cytotoxicity with decreased IC50 of 33 ng/mL (TRAIL concentration) and 329 ng/mL (DOX concentration). More significantly, IC50 of TRAIL/DOX-fGO was determined to be 14 ng/mL (TRAIL concentration) and 140 ng/mL (DOX concentration), which increased cytotoxicities of TRAIL-fGO, DOX-fGO and TRAIL/DOX-fGO to 8.5-, 3.6- and 2.4-fold, respectively. We next compared the cytotoxicities of TRAIL/DOX-fGO and TRAIL/DOX-nGO against the furin-deficient LoVo cells (Figure 4d). TRAIL/DOX-fGO showed a comparable cytotoxicity with IC50 of 759 ng/mL (DOX concentration) to 884 ng/mL of TRAIL/DOX-nGO, due to the similar transportation property of TRAIL/DOX-fGO and TRAIL/DOX-nGO toward LoVo cells, which further evidenced that the important role of furin-mediated membrane-targeted delivery of TRAIL by fGO on enhanced synergistic anticancer activity with DOX. In addition, the blank fGO without TRAIL and DOX had negligible cytotoxicities on both A549 and LoVo cells at all the studied concentrations (Figure S12). Taken together, the specific delivery of TRAIL and DOX to their distinct sites of action supports the improvement on their combination cytotoxicity.
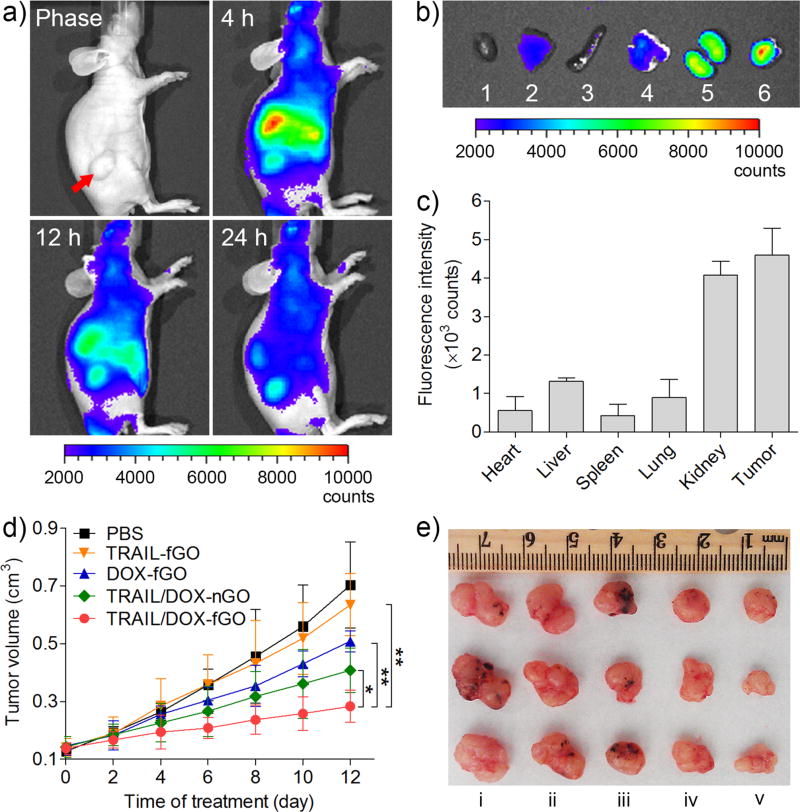
To explore the tumor-targeting capability of the GO nanocarrier, the in vivo biodistribution of Cy5.5-labeled TRAIL-conjugated fGO (Cy5.5-TRAIL-fGO) after intravenous administration into the A549 tumor-bearing nude mice was monitored using a non-invasive optical imaging technique. As displayed in Figure 5a, after administration for 4 h, a strong fluorescence signal of Cy5.5-TRAIL-fGO was obviously detected at the tumor site. As time extended, Cy5.5-TRAIL-fGO showed a higher Cy5.5 signal in the tumor than in the normal tissues after administration for 24 h, suggesting a remarkable tumor targetability of the GO nanosheet. At 24 h post administration, the mice were sacrificed. The tumor and normal organs were harvested, followed by the ex vivo imaging. The strongest fluorescence signal was observed at the tumor compared to other normal organs (Figure 5b), which was in agreement with the quantitative region-of-interest (ROI) analysis (Figure 5c). The fluorescence intensity of Cy5.5 at the tumor tissue was 8.14-, 3.5-, 10.7-, 5.1- and 1.1-fold that at heart, liver, spleen, lung and kidney, respectively. Collectively, it was confirmed that TRAIL/DOX-fGO allowed the high accumulation of drugs at the tumor site via a combination of passive and active targeting effects.
Figure 5.
a) In vivo fluorescence imaging of the A549 tumor-bearing nude mice at 4, 12, 24 h after intravenous administration of Cy5.5-TRAIL-fGO (30 nmol/kg Cy5.5). Red arrow indicates the tumor site. b) Ex vivo fluorescence imaging of the tumor and normal organs collected from the mice at 24 h after administration. 1: heart; 2: liver; 3: spleen; 4: lung; 5: kidney; 6: tumor. c) ROI analysis of fluorescent signals of the tumor and normal organs. d) The A549 tumor growth curves of the mice after treatment with different formulations (0.16 mg/kg TRAIL, 2 mg/kg DOX). *P < 0.05, **P < 0.01. e) The representative images of the A549 xenograft tumors collected from the mice after treatment with different formulations at Day 12. i: PBS; ii: TRAIL-fGO; iii: DOX-fGO; iv: TRAIL/DOX-nGO; v: TRAIL/DOX-fGO.
The in vivo antitumor efficacy of TRAIL/DOX-fGO was investigated on the A549 tumor-bearing nude mice. As shown in Figure 5d, the growth of tumor was significantly inhibited after the mice were successively treated with different DOX formulations including DOX-fGO, TRAIL/DOX-nGO and TRAIL/DOX-fGO, except TRAIL-fGO due to the A549 tumor resistant to TRAIL, compared to phosphate buffered saline (PBS) as a negative control. TRAIL/DOX-nGO showed a stronger tumor inhibition capability than DOX-fGO, indicating that the synergistic antitumor activity by a combination of TRAIL and DOX. It is notable that TRAIL/DOX-fGO brought about the strongest effect on suppressing the tumor growth, which substantiated that site-specific delivery of TRAIL and DOX by TRAIL/DOX-fGO enabled the reinforcement on the synergistic therapeutic efficacy. The images and weights of tumors harvested from the mice after treatment further indicated the most significant tumor size shrinking caused by TRAIL/DOX-fGO (Figure 5e and Figure S13a). No noticeable change in the body weight of the mice was monitored during the treatment of TRAIL/DOX-fGO (Figure S13b). We further applied the hematoxylin and eosin (HE) and the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining to evaluate the antitumor efficiency of TRAIL/DOX-fGO, respectively (Figure S14a). The images of HE-stained tumor tissue presented that a massive remission of tumor cells occurred after the mice were treated with TRAIL/DOX-fGO. The fluorescence images obtained from the in situ TUNEL staining showed that the highest level of the AlexaFluor 488-stained apoptotic DNA fragmentation (green) in the tumor tissue of the mice receiving TRAIL/DOX-fGO, suggesting that the pronounced inhibition on tumor growth was attributed mainly to the elevated apoptosis induced by TRAIL/DOX-fGO. Additionally, the histologic images of other organs, such as heart, liver, spleen, lung, kidney, collected from the mice treated with TRAIL/DOX-fGO exhibited no obvious pathological abnormalities in all the studied normal organs compared to those treated with PBS (Figure S14b).
In summary, we have developed a sequentially-functionalized GO nanostructure as a new cellular protease-mediated programmed co-delivery system integrating membrane-related proteins and intracellular-functioning small-molecule drugs. Such GO nanocarrier was able to efficiently release its cargoes, TRAIL and DOX to their distinct sites of action separately, in a site-specific manner. We believe that it will start a new generation of 2D nanomaterials with programmed-release therapeutics capability for combination cancer treatment with enhanced efficacy.
Supplementary Material
Acknowledgments
This work was supported by NC TraCS (grant 550KR51307), NIH’s Clinical and Translational Science Awards (CTSA, 1UL1TR001111) at UNC-CH, the NC State Faculty Research and Professional Development Award, the start-up package from the Joint BME Department of UNC-CH and NCSU to Z.G., and the High-Level Talent Program for Young Researchers and the start-up package from China Pharmaceutical University to R.M..
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr Tianyue Jiang, State Key Laboratory of Natural Medicines and Jiangsu Key Laboratory of Drug Discovery for Metabolic Diseases, Center of Drug Discovery, China Pharmaceutical University, Nanjing 210009, China; Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Wujin Sun, Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Qiuwen Zhu, State Key Laboratory of Natural Medicines and Jiangsu Key Laboratory of Drug Discovery for Metabolic Diseases, Center of Drug Discovery, China Pharmaceutical University, Nanjing 210009, China.
Nancy A. Burns, Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, NC 27695, USA
Dr Saad A. Khan, Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, NC 27695, USA.
Dr Ran Mo, State Key Laboratory of Natural Medicines and Jiangsu Key Laboratory of Drug Discovery for Metabolic Diseases, Center of Drug Discovery, China Pharmaceutical University, Nanjing 210009, China; Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Dr Zhen Gu, Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
References
- 1.a) Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]; b) Al-Lazikani B, Banerji U, Workman P. Nat. Biotechnol. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]; c) Chow EK, Ho D. Sci. Transl. Med. 2013;5:216rv4. doi: 10.1126/scitranslmed.3005872. [DOI] [PubMed] [Google Scholar]; d) Naahidi S, Jafari M, Edalat F, Raymond K, Khademhosseini A, Chen P. J. Controlled Release. 2013;166:182–194. doi: 10.1016/j.jconrel.2012.12.013. [DOI] [PubMed] [Google Scholar]; e) Sun W, Jiang T, Lu Y, Reiff M, Mo R, Gu Z. J. Am. Chem. Soc. 2014;136:14722–14725. doi: 10.1021/ja5088024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Kolishetti N, Dhar S, Valencia PM, Lin LQ, Karnik R, Lippard SJ, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17939–17944. doi: 10.1073/pnas.1011368107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen Z, Penet MF, Nimmagadda S, Li C, Banerjee SR, Winnard PT, Jr, Artemov D, Glunde K, Pomper MG, Bhujwalla ZM. ACS Nano. 2012;6:7752–7762. doi: 10.1021/nn301725w. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen Y, Gao Y, Chen H, Zeng D, Li Y, Zheng Y, Li F, Ji X, Wang X, Chen F. Adv. Funct. Mater. 2012;22:1586–1597. [Google Scholar]; d) Huang S, Shao K, Liu Y, Kuang Y, Li J, An S, Guo Y, Ma H, Jiang C. ACS Nano. 2013;7:2860–2871. doi: 10.1021/nn400548g. [DOI] [PubMed] [Google Scholar]
- 3.a) Mayer LD, Harasym TO, Tardi PG, Harasym NL, Shew CR, Johnstone SA, Ramsay EC, Bally MB, Janoff AS. Mol. Cancer Ther. 2006;5:1854–1863. doi: 10.1158/1535-7163.MCT-06-0118. [DOI] [PubMed] [Google Scholar]; b) Batist G, Gelmon KA, Chi KN, Miller WH, Jr, Chia SK, Mayer LD, Swenson CE, Janoff AS, Louie AC. Clin. Cancer Res. 2009;15:692–700. doi: 10.1158/1078-0432.CCR-08-0515. [DOI] [PubMed] [Google Scholar]
- 4.a) Ko YT, Falcao C, Torchilin VP. Mol. Pharmaceutics. 2009;6:971–977. doi: 10.1021/mp900006h. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bae S, Ma K, Kim TH, Lee ES, Oh KT, Park ES, Lee KC, Youn YS. Biomaterials. 2012;33:1536–1546. doi: 10.1016/j.biomaterials.2011.10.050. [DOI] [PubMed] [Google Scholar]; c) Kim I, Byeon HJ, Kim TH, Lee ES, Oh KT, Shin BS, Lee KC, Youn YS. Biomaterials. 2013;34:6444–6453. doi: 10.1016/j.biomaterials.2013.05.018. [DOI] [PubMed] [Google Scholar]; d) Jiang T, Mo R, Bellotti A, Zhou J, Gu Z. Adv. Funct. Mater. 2014;24:2295–2304. [Google Scholar]; e) Hu Q, Katti PS, Gu Z. Nanoscale. 2014;6:12273–12286. doi: 10.1039/c4nr04249b. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Lu Y, Sun W, Gu Z. J. Controlled Release. 2014;194:1–19. doi: 10.1016/j.jconrel.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Mo R, Jiang T, Gu Z. Nanomedicine. 2014;9:1117–1120. doi: 10.2217/nnm.14.62. [DOI] [PubMed] [Google Scholar]; h) Sun W, Lu Y, Gu Z. Part. Part. Syst. Charact. 2014 doi: 10.1002/ppsc.201400140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Dranoff G. Nat. Rev. Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]; b) Johnstone RW, Frew AJ, Smyth MJ. Nat. Rev. Cancer. 2008;8:782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]; c) Mocellin S, Rossi CR, Pilati P, Nitti D. Cytokine Growth Factor Rev. 2005;16:35–53. doi: 10.1016/j.cytogfr.2004.11.001. [DOI] [PubMed] [Google Scholar]; d) Mitchell MJ, Wayne E, Rana K, Schaffer CB, King MR. Proc. Natl. Acad. Sci. U. S. A. 2014;111:930–935. doi: 10.1073/pnas.1316312111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Lakhani SA, Masud A, Kuida K, Porter GA, Jr, Booth CJ, Mehal WZ, Inayat I, Flavell RA. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lowe SW, Cepero E, Evan G. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 7.a) Yang K, Feng L, Shi X, Liu Z. Chem. Soc. Rev. 2013;42:530–547. doi: 10.1039/c2cs35342c. [DOI] [PubMed] [Google Scholar]; b) Feng L, Li K, Shi X, Gao M, Liu J, Liu Z. Adv. Healthcare Mater. 2014;3:1261–1271. doi: 10.1002/adhm.201300549. [DOI] [PubMed] [Google Scholar]; c) Wang F, Liu B, Ip ACF, Liu J. Adv. Mater. 2013;25:4087–4092. doi: 10.1002/adma.201301183. [DOI] [PubMed] [Google Scholar]; d) Chen Y, Xu P, Shu Z, Wu M, Wang L, Zhang S, Zheng Y, Chen H, Wang J, Li Y, Shi J. Adv. Funct. Mater. 2014;24:4386–4396. [Google Scholar]
- 8.a) Thomas G. Nat. Rev. Mol. Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Molloy SS, Thomas L, VanSlyke JK, Stenberg PE, Thomas G. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Mallet WG, Maxfield FR. J. Cell Biol. 1999;146:345–360. doi: 10.1083/jcb.146.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Biswas A, Joo K, Liu J, Zhao M, Fan G, Wang P, Gu Z, Tang Y. ACS Nano. 2011;5:1385–1394. doi: 10.1021/nn1031005. [DOI] [PubMed] [Google Scholar]
- 9.a) Pan LQ, Wang HB, Xie ZM, Li ZH, Tang XJ, Xu YC, Zhang C, Naranmandura H, Chen SQ. Adv. Mater. 2013;25:4718–4722. doi: 10.1002/adma.201301385. [DOI] [PubMed] [Google Scholar]; b) Kohlhaas SL, Craxton A, Sun X-M, Pinkoski MJ, Cohen GM. J. Biol. Chem. 2007;282:12831–12841. doi: 10.1074/jbc.M700438200. [DOI] [PubMed] [Google Scholar]
- 10.a) Pennarun B, Meijer A, de Vries EG, Kleibeuker JH, Kruyt F, de Jong S. Biochim. Biophys. Acta. 2010;1805:123–140. doi: 10.1016/j.bbcan.2009.11.004. [DOI] [PubMed] [Google Scholar]; b) Pan G, O'Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 11.a) Sun X, Liu Z, Welsher K, Robinson JT, Goodwin A, Zaric S, Dai H. Nano Res. 2008;1:203–212. doi: 10.1007/s12274-008-8021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang L, Xia J, Zhao Q, Liu L, Zhang Z. Small. 2010;6:537–544. doi: 10.1002/smll.200901680. [DOI] [PubMed] [Google Scholar]; c) Yang K, Feng L, Hong H, Cai W, Liu Z. Nat. Protoc. 2013;8:2392–2403. doi: 10.1038/nprot.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 13.a) Tang L, Wang Y, Liu Y, Li J. ACS Nano. 2011;5:3817–3822. doi: 10.1021/nn200147n. [DOI] [PubMed] [Google Scholar]; b) Hong BJ, Compton OC, An Z, Eryazici I, Nguyen ST. ACS Nano. 2012;6:63–73. doi: 10.1021/nn202355p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Liu Z, Robinson JT, Sun X, Dai H. J. Am. Chem. Soc. 2008;130:10876–10877. doi: 10.1021/ja803688x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang H, Zhang Q, Chu X, Chen T, Ge J, Yu R. Angew. Chem. Int. Ed. 2011;50:7065–7069. doi: 10.1002/anie.201101351. [DOI] [PubMed] [Google Scholar]
- 15.a) Bassi DE, Fu J, Lopez de Cicco R, Klein-Szanto AJ. Mol. Carcinogen. 2005;44:151–161. doi: 10.1002/mc.20134. [DOI] [PubMed] [Google Scholar]; b) Ma YC, Fan WJ, Rao SM, Gao L, Bei ZY, Xu ST. Cancer Cell Int. 2014;14:43. doi: 10.1186/1475-2867-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Takahashi S, Kasai K, Hatsuzawa K, Kitamura N, Misumi Y, Ikehara Y, Murakami K, Nakayama K. Biochem. Biophys. Res. Commun. 1993;195:1019–1026. doi: 10.1006/bbrc.1993.2146. [DOI] [PubMed] [Google Scholar]; b) Ohnishi Y, Shioda T, Nakayama K, Iwata S, Gotoh B, Hamaguchi M, Nagai Y. J. Virol. 1994;68:4075–4079. doi: 10.1128/jvi.68.6.4075-4079.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cao CY, Shen YY, Wang JD, Li L, Liang GL. Sci. Rep. 2013;3:1024. doi: 10.1038/srep01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Zhuang H, Jiang W, Cheng W, Qian K, Dong W, Cao L, Huang Q, Li S, Dou F, Chiu JF. Lung Cancer. 2010;68:27–38. doi: 10.1016/j.lungcan.2009.05.014. [DOI] [PubMed] [Google Scholar]; b) Seol JY, Park KH, Hwang CI, Park WY, Yoo CG, Kim YW, Han SK, Shim C-T, Lee YS. Cancer Gene Ther. 2003;10:540–548. doi: 10.1038/sj.cgt.7700597. [DOI] [PubMed] [Google Scholar]
- 18.Song JH, Tse MC, Bellail A, Phuphanich S, Khuri F, Kneteman NM, Hao C. Cancer Res. 2007;67:6946–6955. doi: 10.1158/0008-5472.CAN-06-3896. [DOI] [PubMed] [Google Scholar]
- 19.a) Min YJ, Lee JH, Choi SJ, Chi HS, Lee JS, Kim WK, Lee KH. Leuk. Res. 2004;28:359–365. doi: 10.1016/j.leukres.2003.08.015. [DOI] [PubMed] [Google Scholar]; b) Gibson SB, Oyer R, Spalding AC, Anderson SM, Johnson GL. Mol. Cell Biol. 2000;20:205–212. doi: 10.1128/mcb.20.1.205-212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.