Abstract
Experiments measuring DNA extension in nanochannels are at odds with even the most basic predictions of current scaling arguments for the conformations of confined semiflexible polymers such as DNA. We show that a theory based on a weakly self-avoiding, one-dimensional “telegraph” process collapses experimental data and simulation results onto a single master curve throughout the experimentally relevant region of parameter space and explains the mechanisms at play.
As the carrier of genetic information, DNA plays a key role in biology. At the same time, recent advancements in fluorescence microscopy allow DNA to serve as a model polymer for investigating fundamental questions in polymer physics [1,2]. Nowhere is this dual importance more apparent than in the problem of DNA confinement in a nanochannel [3–5]. When the radius of gyration of the DNA molecule is larger than the channel width, it extends along the channel. This stretching lies at the heart of genome mapping in nanochannels [6]. Here the stretched DNA molecules, usually greater than 150 kilobase pairs in length, contain fluorescent markers that reveal sequence-specific information with kilobase pair resolution. This new method serves as a complement to next-generation de novo DNA sequencing [6–8].
Polymer confinement has been investigated for four decades, starting with the scaling arguments of Daoud and de Gennes [9]. Yet there is to date no microscopic theory explaining the experimental data of recent genome-mapping experiments [6–8,10] in narrow nanochannels. The difficulty is that the channels are too wide to apply scaling arguments derived for strong confinement [11,12], yet too narrow for the scaling arguments and theory [11,13–16] in wider channels to hold.
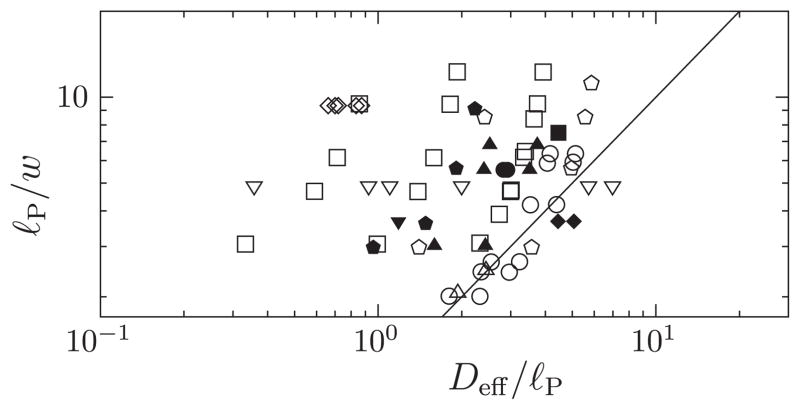
The challenge in developing a theory for the extension of nanoconfined DNA arises from its semiflexible nature. Semiflexible polymers are characterized by three length scales: the contour length L, the persistence length ℓP quantifying the stiffness of the chain, and the effective width w that appears in the Onsager excluded volume [17]. For polyelectrolytes such as DNA, both the persistence length [18–21] and the effective polymer width [22] depend on electrostatic interactions. Recent experiments are often conducted in high ionic-strength buffers. In this case ℓP is approximately 50 nm [23] while w is around 5 nm [24], and thus w ≪ ℓP. This inequality emphasizes the intrinsic difficulty of describing DNA in a wide range of situations. DNA is considerably stiffer than typical synthetic polymers, yet the number of persistence lengths L/ℓP in genomic DNA samples is large. Any theory for the conformational statistics of channel-confined DNA must account for both the local stiffness of the polymer and excluded-volume interactions. This is a formidable challenge. Matters are further complicated by the fact that most recent genome-mapping experiments are performed in nanochannels that are about 50 nm wide [6–8,10]. Essentially all experiments involving DNA (Fig. 1) take place in channel sizes D of the order of ℓP and do not satisfy the criterion required for the scaling arguments of Ref. [9] to apply.
FIG. 1.
Parameters for experiments on nanoconfined DNA: inverted triangle [25], square [26], filled square [27], circle [28], filled circle [29], triangle [30], filled triangle [31], inverted filled triangle [32], diamond [10], filled diamond [33], pentagon [34], and filled pentagon [35]. For experiments using funnels [29,31,33] only maximum and minimum channel widths are indicated. The methods for selecting the data sets, and for computing the “effective channel width” Deff, ℓP, and w from the experimental parameters, are described in the Supplemental Material [36]. Solid line shows .
There is nomicroscopic theory for the extension of confined DNA for , despite numerous attempts [28,38,39]. Scaling arguments [13,14,16] following Refs. [9,11] yield the most useful description. If D ≫ ℓP, they suggest that the extension X scales as X ~ D−2/3. The problem is that the theory fails when D ≈ ℓP, and as a result it proves to be a poor description of most recent DNA experiments in nanochannels. The earliest experiments [25], for instance, reported a much larger exponent X ~ D−0.85, and subsequent studies [32,33,40] continue to report exponents exceeding the theoretical prediction.
We take a different approach in this Letter. We show that the DNA-confinement problem for w ≪ ℓP and maps to the simple one-dimensional telegraph process in Fig. 2, describing the correlated walk of a particle moving with velocity v0 along the channel axis. The velocity changes sign at rate r, creating hairpin configurations in the particle path. The process lasts from t = 0 to t = T. When the particle revisits a position it has previously visited, it incurs a penalty ε. We show that this model collapses experimental and simulation data for the extension throughout the experimentally relevant parameter range onto a universal master curve as a function of a new scaling variable α that measures the combined effects of confinement, local stiffness, and self-avoidance.
FIG. 2.
Illustration of the telegraph process along the channel axis (x axis). The walk is one dimensional, but for clarity it is expanded vertically, to show the changes in direction that create hairpin configurations of the confined DNA molecule.
We start by considering narrow channels, D ≪ ℓP, and later extend the arguments to channel widths up to . Since we want to compute the extension of the DNA molecule along the channel axis, it suffices to consider the projection of the three-dimensional DNA configurations x(s) to the channel axis x. Here s is the contour-length coordinate; it corresponds to time t in the telegraph process. We decompose the probability P[x(s)] of observing the projected conformation x(s) as
| (1) |
The functional Pideal[x(s)] is the probability of observing the conformation x(s) in an ensemble of ideal confined polymers, disregarding self-avoidance. The functional 𝒜[x(s)] captures the effect of self-avoidance. It equals the fraction of three-dimensional polymer configurations corresponding to x(s) that contains no segments that collide with any other polymer segment.
When D < ℓP, the local conformation statistics are determined by Odijk’s theory for narrow channels [41], while the global statistics are dominated by a random sequence of direction changes (hairpins) [42]. Neglecting self-avoidance, the separation between neighboring hairpin bends is exponentially distributed [42]. On length scales much larger than the deflection length [41] λ ≡ (ℓPD2)1/3, the central-limit theorem assures that local alignment fluctuations are negligible [11]. These two facts imply that the ideal problem maps onto the one-dimensional telegraph process in Fig. 2. The correlation function of vx(s), the channel-axis component of the tangent vector of the ideal polymer, decays exponentially [36]:
| (2) |
The telegraph velocity has similar correlations:
| (3) |
Comparing Eqs. (2) and (3) we see that the contour parameter s maps to the time t in the telegraph process, whereupon the polymer-contour length L maps to the total time T in the telegraph model. The parameter a quantifies the tendency of the tangent vectors to align with the channel [31]. The parameter g is the global persistence length [42], characterizing the typical distance between hairpin turns. These parameters map to those of the telegraph process as follows: a = v0 and g = (2r)−1. We measured how a and g depend on the physical parameters of the full three-dimensional problem from simulations of confined ideal polymers. It turns out that it suffices to determine just two curves [Figs. 3(a) and 3(b)], since a and g/ℓP depend on D/ℓP only.
FIG. 3.
Results from ideal-polymer simulations [36] showing how a = v0, g = 1/(2r), ε, and α depend on D/ℓP.
Now consider the effect of self-avoidance. In general it is very difficult to derive an expression for 𝒜[x(s)]. But for a weakly self-avoiding polymer, the problem simplifies considerably when the channel is so narrow that interactions between the polymer and the channel wall cause the three-dimensional configurations to lose correlations. We show in the Supplemental Material [36] that
| (4) |
if w ≪ ℓP. Here ℒ(x)dx is the total amount of contour in the interval [x; x þ dx] [43]. The parameter ε penalizes overlaps. It is determined by the probability that two short polymer segments overlapping in one dimension collide in three dimensions [36]:
| (5) |
The average is over the conformations of the confined ideal polymer, and y and z are the transverse channel coordinates of a short polymer segment of length ℓ. Primed coordinates belong to a second, independent segment, and vex is the excluded volume. The excluded volume depends on the segment orientation. If ℓ ≫ w, we have vex = 2wℓ2 sin θ, where θ is the angle between the two segments [17]. Figure 3(c) shows how ε depends on D/ℓP, obtained by evaluating the average in Eq. (5) from three-dimensional simulations of confined ideal polymers [36]. A single curve is sufficient to determine how ε depends on the physical parameters, because εD2/w is a function of D/ℓP only (see Supplemental Material [36]).
In the telegraph model self-avoidance is incorporated in the same way. Here, ℒ has units of (time)/(position). Equation (4) then shows that ε has units of (position)/(time)2. Since r has units of (time)−1, and v0 of (position)/(time), the only dimensionless combination of ε, r, and v0 is
| (6) |
In the limit of large T, only α can have physical significance. Invoking our mapping between telegraph model and polymer problem, we conclude that α is given by
| (7) |
This parameter measures the expected number of overlaps between the two strands of a hairpin of length g.
Equation (7) has two important consequences. First, Eq. (7) allows us to generalize the mapping to all channel widths up to . To show this, consider first the ideal part. Strictly speaking, the simple picture outlined above breaks down when D ~ ℓP because the typical hairpin length g becomes of the same order as ℓP. But consider how α changes as D approaches ℓP from below. For w ≪ ℓP, the parameter α decreases below unity before g = ℓP is reached, and for small α the precise nature of the local conformations is irrelevant. All that matters is that the ideal part is a diffusion process with exponentially decaying correlations of vx(t). Similarly, the local probability of collision is still (ε/2)ℒ2(x)dx, because each segment pair collides independently. The latter assumption eventually breaks down at since the transversal segment coordinates become correlated. But up to this point Eq. (4) is valid, as is Eq. (5).
Second, observables that are dimensionless in the telegraph model can only depend on α, Eq. (7), in the limit of large L. This combination α is plotted in Fig. 3(d). It turns out that αD2/(ℓPw) depends only on D/ℓP [36]. Now consider the average extension X and the variance about that average σ2. In the telegraph model these quantities have units of (position) and (position)2, and for large values of L they must be proportional to L. We therefore conclude that the data must collapse as
| (8) |
Here fX and fσ are universal scaling functions that depend only on α. We can numerically compute the form of these functions simply by simulating the telegraph model [36].
We have compared our theory to direct numerical simulations (DNS) of three-dimensional, confined, self-avoiding wormlike chains [39] using the PERM algorithm [44,45]. Figures 4(a) and 4(b) show that our theory quantitatively captures the DNS results for all values of w/ℓP tested [36], up to w/ℓP = 0.4. This agreement is remarkable, as the theory assumes weak self-avoidance, w ≪ ℓP.
FIG. 4.
One-parameter scaling of the mean extension X and the extension variance σ2. Comparison of one-parameter theory (solid black lines) to results of three-dimensional direct numerical simulations (DNS) (a), (b) and experiments (c), (d). DNS: crosses. The DNS method [39,46] is described in the Supplemental Material [36]. Experiments (same as in Fig. 1): ▽ [25], square [26], filled square [27], circle [28], filled circle [29], triangle [30], filled triangle [31], inverted filled triangle [32], diamond [10], filled diamond [33], pentagon [34], and filled pentagon [35]. The details of these experiments and the selection of experimental data sets are described in the Supplemental Material [36]. In addition, the predicted scalings for the mean extension, X ~ α1/3, and for the extension variance, σ2 ~ α−3, are indicated. The color bar shows the range of ℓP/w for DNS (top) and experiments (bottom). The dashed lines in (b) show theoretical predictions from Eq. (10) for ℓP/w = 12 (dashed blue line) and 36 (dashed red line). See Fig. S-2 in Supplemental Material [36] for the telegraph-model results as a function of channel width D.
Figures 4(c) and 4(d) show the comparison between the results of the experiments summarized in Fig. 1 and our theory. The theory not only collapses the experimental data, but provides good quantitative agreement, in particular with the most recent experiments [10,33,34]. There is some scatter of the experimental data around the theoretical curve, but this is expected because the nanofluidic experiments are quite difficult to control.
In the limit α ≪ 1 our theory allows us to map the problem to an uncorrelated weakly self-avoiding diffusion process [43,47]. This follows from the fact that the correlation function in the telegraph model, Eq. (3), decays to zero before the next collision occurs, for α ≪ 1. As a result, the ideal random walk is simply diffusive, with diffusion constant . This has two consequences.
First, for α ≪ 1 observables depend on v0 and r only through the combination 𝒟. Since the extension is linear in L for large L, we deduce that X/L can only depend on ε and 𝒟 in this limit. Since 𝒟 has units of (position)2/(time) while ε has units of (position)/(time)2 in the telegraph model, we see that the only possible combination is X/L ∝ (ε𝒟)1/3. This gives X/(La)=fX(α)∝α1/3, explaining the power law in Figs. 4(a) and 4(c). For the variance we conclude that σ2/L ∝ 𝒟, independent of ε [Figs. 4(b) and 4(d)]. Alternatively, we can deduce these scalings by a mean-field argument, indicating that fluctuations of ℒ are negligible when α ≪ 1. Assuming that ℒ ~ T/X, we find for the extension distribution , whereupon d log P/dX = 0 yields X/(v0T) ~ α1/3. For the variance we obtain .
Second, we can use the exact mathematical results derived in Ref. [47] to deduce the prefactors:
| (9a) |
| (9b) |
as α → 0. The constraints for cX and cσ are rigorously proven mathematical bounds [47].
Now consider the limit of large α. The extension X tends to La in this limit [11], since the frequency of hairpins tends to zero. The variance decays as σ2 ~ α−3, as Fig. 4 shows. To deduce this power law, we estimate the variance of the strongly extended polymer as (number of hairpins)× (hairpin extension)2. To determine the number of hairpins, note that the expected number of collisions for a hairpin of contour length h is ~αh/g. The resulting hairpin is therefore likely to survive the collision check only if h is of the order g/α or smaller. This requires a second switch of direction within the length g/α. This occurs with probability (g/α)/g = α−1, so that the number of hairpins is (L/g)α−1. To obtain the hairpin extension we multiply its contour length h ~ g/α by its alignment a, so that the typical hairpin extension becomes ~ga/α. Therefore, σ2 ∝ (L/g)α−1(ga/α)2 = Lga2α−3 for large α.
For very large values of α, the theory fails [Fig. 4(b)] because hairpins are so rare that alignment fluctuations (not included in the telegraph model) dominate the variance [11]. This correction is taken into account simply by adding the variance in the extreme Odijk limit [48]:
| (10) |
Here dσ is a universal constant. By fitting the solid line in Fig. 4(b) for α > 10, we find dσ ≈ 0.09. We observe excellent agreement between this refined theory and the simulation data for ℓP/w = 12. For the stiffer polymers (ℓP/w = 36), still longer contour lengths are required to reach the large-L limit and to reduce the statistical error.
Finally, we show that our theory contains scaling laws derived earlier as particular asymptotic limits. In very narrow channels, a ≈ 1 and 〈sin θ〉 ≈ (D/ℓP)1/3. Using these approximations in Eqs. (5) and (6) gives
| (11) |
where C ≈ 1.95 is a constant [36]. The parameter ξ appears in Odijk’s scaling theory [11] and the extension scales as X ~ ξ1/3 [11] in this special limit. In wide channels, for , we have that and g ≈ ℓP. Using diffusion approximations for the distribution of the polymer in the channel [36] gives
| (12) |
This is the result of Ref. [15], implying the same scaling X ~ D−2/3 that Odijk’s scaling arguments [11] predict in this asymptotic limit. At first glance it is perhaps surprising that the two distinct limits Eqs. (11) and (12) are described by the same random-walk process. After all, the three-dimensional polymer conformations are entirely different in the two regimes, described by invoking deflection segments, hairpins, and blobs. Our universal theory, by contrast, rests on the fact that the macroscopic statistics of a weakly interacting random walk do not depend on the microscopic details of the process [49].
We can also conclude that the DNA experiments shown in Fig. 1 cannot obey the scalings X ~ D−2/3 or X ~ ξ1/3 because the experiments do not satisfy the strong inequalities D ≫ ℓP or D ≪ ℓP (see Fig. S-5 in the Supplemental Material [36]), and therefore do not reach the asymptotic limits required for these power laws to emerge. Our theory shows, and Fig. 4(a) confirms, that X ~ α1/3 for small values of α. But the parameter α depends upon D and ℓP in an intricate way via Eq. (7), in general not in a power-law fashion.
In summary, we have shown that DNA confinement in nanochannels can be modeled by a telegraph process, collapsing all of the data in terms of a scaling variable α. Our theory brings to the fore universal properties of confined polymers in a good solvent in a way that is obscured by the prevailing scaling theories [9,11,12,14,15,41]. The distinction between deflection segments, hairpins, and blobs, which leads to the need to define separate regimes, is not necessary. Rather, the statistics of the confined polymer chain for adopt a universal behavior at sufficiently long length scales, independent of the details of the microscopic model.
Supplementary Material
Acknowledgments
We thank Daniel Ödman for helping us to uncover an error in the simulations of the telegraph model. This work was supported by VR Grant No. 2013-3992 and by the National Institutes of Health (R01-HG006851). D. G. acknowledges the support of a Doctoral Dissertation Fellowship from the University of Minnesota. Computational resources were provided by the Minnesota Supercomputing Institute, and by C3SE and SNIC.
References
- 1.Shaqfeh ESG. J Non-Newtonian Fluid Mech. 2005;130:1. [Google Scholar]
- 2.Latinwo F, Schroeder CM. Soft Matter. 2011;7:7907. doi: 10.1039/c1sm05298e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reisner W, Pedersen JN, Austin RH. Rep Prog Phys. 2012;75:106601. doi: 10.1088/0034-4885/75/10/106601. [DOI] [PubMed] [Google Scholar]
- 4.Dorfman KD, King SB, Olson DW, Thomas JDP, Tree DR. Chem Rev. 2013;113:2584. doi: 10.1021/cr3002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai L, Renner CB, Doyle PS. Adv Colloid Interface Sci. 2016;232:80. doi: 10.1016/j.cis.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, Kwok PY. Nat Biotechnol. 2012;30:771. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo K, Dhingra DM, Odijk T, de Pablo JJ, Graham MD, Runnheim R, Forrest D, Schwartz DC. Proc Natl Acad Sci USA. 2007;104:2673. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaeli Y, Ebenstein Y. Nat Biotechnol. 2012;30:762. doi: 10.1038/nbt.2324. [DOI] [PubMed] [Google Scholar]
- 9.Daoud M, de Gennes P. J Phys France. 1977;38:85. [Google Scholar]
- 10.Reinhart WF, Reifenberger JG, Gupta D, Muralidhar A, Sheats J, Cao H, Dorfman KD. J Chem Phys. 2015;142:064902. doi: 10.1063/1.4907552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odijk T. Phys Rev E. 2008;77:060901(R). doi: 10.1103/PhysRevE.77.060901. [DOI] [PubMed] [Google Scholar]
- 12.Muralidhar A, Tree DR, Dorfman KD. Macromolecules. 2014;47:8446. [Google Scholar]
- 13.Wang Y, Tree DR, Dorfman KD. Macromolecules. 2011;44:6594. doi: 10.1021/ma201277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai L, van der Maarel JRC, Doyle PS. Macromolecules. 2014;47:2445. [Google Scholar]
- 15.Werner E, Mehlig B. Phys Rev E. 2014;90:062602. doi: 10.1103/PhysRevE.90.062602. [DOI] [PubMed] [Google Scholar]
- 16.Werner E, Mehlig B. Phys Rev E. 2015;91:050601. doi: 10.1103/PhysRevE.91.050601. [DOI] [PubMed] [Google Scholar]
- 17.Onsager L. Ann NY Acad Sci. 1949;51:627. [Google Scholar]
- 18.Odijk T. J Polym Sci, Polym Phys Ed. 1977;15:477. [Google Scholar]
- 19.Skolnick J, Fixman M. Macromolecules. 1977;10:944. [Google Scholar]
- 20.Dobrynin AV. Macromolecules. 2005;38:9304. [Google Scholar]
- 21.Trizac E, Shen T. Europhys Lett. 2016;116:18007. [Google Scholar]
- 22.Stigter D. Biopolymers. 1977;16:1435. doi: 10.1002/bip.1977.360160705. [DOI] [PubMed] [Google Scholar]
- 23.Bustamante C, Marko JF, Siggia ED, Smith S. Science. 1994;265:1599. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh CC, Balducci A, Doyle PS. Nano Lett. 2008;8:1683. doi: 10.1021/nl080605+. [DOI] [PubMed] [Google Scholar]
- 25.Reisner W, Morton KJ, Riehn R, Wang YM, Yu Z, Rosen M, Sturm JC, Chou SY, Frey E, Austin RH. Phys Rev Lett. 2005;94:196101. doi: 10.1103/PhysRevLett.94.196101. [DOI] [PubMed] [Google Scholar]
- 26.Reisner W, Beech JP, Larsen NB, Flyvbjerg H, Kristensen A, Tegenfeldt JO. Phys Rev Lett. 2007;99:058302. doi: 10.1103/PhysRevLett.99.058302. [DOI] [PubMed] [Google Scholar]
- 27.Thamdrup LH, Klukowska A, Kristensen A. Nanotechnology. 2008;19:125301. doi: 10.1088/0957-4484/19/12/125301. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C, Zhang F, Van Kan JA, Van Der Maarel JRC. J Chem Phys. 2008;128:225109. doi: 10.1063/1.2937441. [DOI] [PubMed] [Google Scholar]
- 29.Utko P, Persson F, Kristensen A, Larsen NB. Lab Chip. 2011;11:303. doi: 10.1039/c0lc00260g. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y, Kim KS, Kounovsky KL, Chang R, Jung GY, DePablo JJ, Jo K, Schwartz DC. Lab Chip. 2011;11:1721. doi: 10.1039/c0lc00680g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner E, Persson F, Westerlund F, Tegenfeldt JO, Mehlig B. Phys Rev E. 2012;86:041802. doi: 10.1103/PhysRevE.86.041802. [DOI] [PubMed] [Google Scholar]
- 32.Gupta D, Sheats J, Muralidhar A, Miller JJ, Huang DE, Mahshid S, Dorfman KD, Reisner W. J Chem Phys. 2014;140:214901. doi: 10.1063/1.4879515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta D, Miller JJ, Muralidhar A, Mahshid S, Reisner W, Dorfman KD. ACS Macro Lett. 2015;4:759. doi: 10.1021/acsmacrolett.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iarko V, Werner E, Nyberg LK, Müller V, Fritzsche J, Ambjörnsson T, Beech JP, Tegenfeldt JO, Mehlig K, Westerlund F, Mehlig B. Phys Rev E. 2015;92:062701. doi: 10.1103/PhysRevE.92.062701. [DOI] [PubMed] [Google Scholar]
- 35.Alizadehheidari M, Werner E, Noble C, Reiter-Schad M, Nyberg LK, Fritzsche J, Mehlig B, Tegenfeldt JO, Ambjornsson T, Persson F, Westerlund F. Macromolecules. 2015;48:871. [Google Scholar]
- 36.See Supplemental Material at http://link.aps.org/supplemental/10.1103/PhysRevLett.119.268102 which includes Ref. [37], for details on the telegraph model, idealpolymer simulations, direct numerical simulations, and discussion of the experimental data.
- 37.Werner E, Westerlund F, Tegenfeldt JO, Mehlig B. Macromolecules. 2013;46:6644. [Google Scholar]
- 38.Cifra P. J Chem Phys. 2009;131:224903. doi: 10.1063/1.3271830. [DOI] [PubMed] [Google Scholar]
- 39.Tree DR, Wang Y, Dorfman KD. Phys Rev Lett. 2013;110:208103. doi: 10.1103/PhysRevLett.110.208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persson F, Utko P, Reisner W, Larsen NB, Kristensen A. Nano Lett. 2009;9:1382. doi: 10.1021/nl803030e. [DOI] [PubMed] [Google Scholar]
- 41.Odijk T. Macromolecules. 1983;16:1340. [Google Scholar]
- 42.Odijk T. J Chem Phys. 2006;125:204904. doi: 10.1063/1.2400227. [DOI] [PubMed] [Google Scholar]
- 43.van der Hofstad R, den Hollander F, König W. Probab Theory Relat Fields. 2003;125:483. [Google Scholar]
- 44.Grassberger P. Phys Rev E. 1997;56:3682. [Google Scholar]
- 45.Prellberg T, Krawczyk J. Phys Rev Lett. 2004;92:120602. doi: 10.1103/PhysRevLett.92.120602. [DOI] [PubMed] [Google Scholar]
- 46.Tree DR, Muralidhar A, Doyle PS, Dorfman KD. Macromolecules. 2013;46:8369. doi: 10.1021/ma401507f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Hofstad R. J Stat Phys. 1998;90:1295. [Google Scholar]
- 48.Burkhardt TW, Yang Y, Gompper G. Phys Rev E. 2010;82:041801. doi: 10.1103/PhysRevE.82.041801. [DOI] [PubMed] [Google Scholar]
- 49.Grosberg AY, Khokhlov AR. Statistical Physics of Macromolecules. AIP Press; New York: 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.