Abstract
Objective: Splinting full-thickness cutaneous wounds in mice has allowed for a humanized model of wound healing. Delineating the epithelial edge and assessing time to closure of these healing wounds via macroscopic visualization have remained a challenge.
Approach: Double transgenic mice were created by crossbreeding K14-Cre and ROSAmT/mG reporter mice. Full-thickness excisional wounds were created in K14-Cre/ROSAmT/mG mice (n = 5) and imaged using both normal and fluorescent light on the day of surgery, and every other postoperative day (POD) until wound healing was complete. Ten blinded observers analyzed a series of images from a single representative healing wound, taken using normal or fluorescent light, to decide the POD when healing was complete. K14-Cre/ROSAmT/mG mice (n = 4) were subsequently sacrificed at the four potential days of rated wound closure to accurately determine the histological point of wound closure using microscopic fluorescence imaging.
Results: Average time to wound closure was rated significantly longer in the wound series images taken using normal light, compared with fluorescent light (mean POD 13.6 vs. 11.6, *p = 0.008). Fluorescence imaging of histological samples indicated that reepithelialization was complete at 12 days postwounding.
Innovation: We describe a novel technique, using double transgenic mice K14-Cre/ROSAmT/mG and fluorescence imaging, to more accurately determine the healing time of wounds in mice upon macroscopic evaluation.
Conclusion: The accuracy by which wound healing can be macroscopically determined in vivo in mouse models of wound healing is significantly enhanced using K14-Cre/ROSAmT/mG double transgenic mice and fluorescence imaging.
Keywords: : wound healing, mouse model, transgenic mice

Michael T. Longaker, MD, MBA
Introduction
Wound healing and repair involve three distinct but overlapping phases of inflammation, tissue formation, and tissue remodeling.1,2 The phases are dynamic and complex and cannot be replicated in vitro.3 Animal models have made a fundamental contribution to understanding the biological and cellular processes involved in wound healing, and in assessing therapeutic outcomes. An optimal animal wound healing model represents human wound healing as closely as possible. A variety of large animal models have been used for research into wound healing in the past, including rabbits, nonhuman primates, and pigs. Laboratory mice, however, are an economical and practical alternative. Mice can be purchased at low cost and are easy to house and maintain. Moreover, there are numerous mouse-specific reagents and transgenic variants making it possible to genetically manipulate mice for intricate mechanistic investigation. For these reasons, mice are one of the most commonly used laboratory animals for research.3–5
The use of mice for wound healing has been problematic in the past, due to the presence of a subcutaneous layer of striated muscle, the panniculus carnosus, found in mice but not humans. The only analogous muscle in humans is the platysma muscle in the neck. The panniculus carnosus is able to move independent of deeper muscles and allows wounds in mouse skin to heal primarily by contraction. Cutaneous wounds in humans, in contrast, heal through the production of granulation tissue and subsequent reepithelialization.6 This major mechanistic difference previously prohibited the extensive use of murine models for studying wound healing and investigating the efficacy of novel therapeutic regimens.
Clinical Problem Addressed
In 2004, Galiano et al. developed an elegant solution to this translational difference in wound healing phenotypes between mice and humans. By securing a silicone splint around full-thickness excisional wounds of mice, wound contraction is minimized, and mouse wound healing is “humanized.”4 Of the numerous wound healing models described, the silicone splinted full-thickness excisional wound model provides the best overall approximations of human wound healing, through reepithelialization, granulation tissue formation, and angiogenesis.2 Since its conception, this model has rapidly been adapted to study a variety of wound types and potential therapeutic treatments.7–10 The macroscopic inspection of healing wounds described in this model, however, is limited, specifically in the precise determination of the advancing edge of keratinocyte migration, and hence, the assessment of time for complete wound closure. Herein, we describe a novel methodology using double transgenic mice and fluorescence imaging to improve the accuracy of wound evaluation in the silicone splinted full-thickness excisional wound healing model.
Materials and Methods
Animals
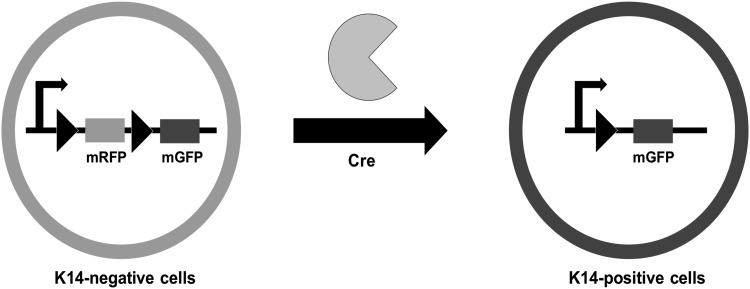
Double transgenic mice were created by crossbreeding ROSAmT/mG mice with K14-Cre mice. Both reporter mice variants were purchased from Jackson Laboratories (Bar Harbor, ME). Mice with the ROSAmT/mG gene act as a Cre activity reporter line with two-color fluorescence capability localized to all cell membranes. As seen in Fig. 1, the enhanced green fluorescent protein gene (mG) is preceded by the red fluorescent protein gene, tdTomato (mT) at the ROSA26 locus. mT is flanked on either side by loxP sites, which are targets of Cre recombinase activity. In the absence of Cre recombinase activity, red fluorescence expression via mT is widespread in all cells and tissues. mT sites are cleaved in cells that express Cre, causing loss of red fluorescence and emission of green fluorescence (mG) at the cell membrane.11 Mice that possess the Keratin 14 Cre (K14-Cre) transgene have Cre recombinase expression driven by a K14 promoter. This localizes Cre recombinase activity to the cells that express K14, which are primarily the ectodermal derivatives, including the keratinocytes of the skin, dental epithelium, and oral ectoderm.12 Through the recombinant excision of the floxed mT gene following crossbreeding of these two different strains, keratinocytes of the epidermis are effectively labeled with membrane-localized green fluorescent protein (GFP), and can be easily visualized with fluorescent light.
Figure 1.
Schematic of Cre-mediated excision of RFP in K14-Cre/ROSAmT/mG double transgenic mice. K14-positive cells possess Cre recombinase activity and thus express membrane-localized GFP, while K14-negative cells continue to express membrane-bound RFP. GFP, green fluorescent protein.
Homozygous K14-Cre mice and ROSAmT/mG mice were mated in the Stanford animal facility to create K14-Cre/ROSAmT/mG mice. Twelve-week-old female K14-Cre/ROSAmT/mG mice were used for this experiment (n = 9 total; n = 5 for serial imaging to evaluate wound healing and n = 4 for serial harvesting to evaluate histology). From weaning, mice were housed five animals per cage and were permitted to fully acclimatize to their environment with food and water ad libitum. Following creation of full-thickness excisional wounds, the mice were housed individually. All animals were treated humanely and protocols used were approved a priori by Stanford University's Administrative Panel on Laboratory Animal Care (Protocol No. 21308) according to National Institutes of Health and institutional guidelines.
Full-thickness excisional wounds
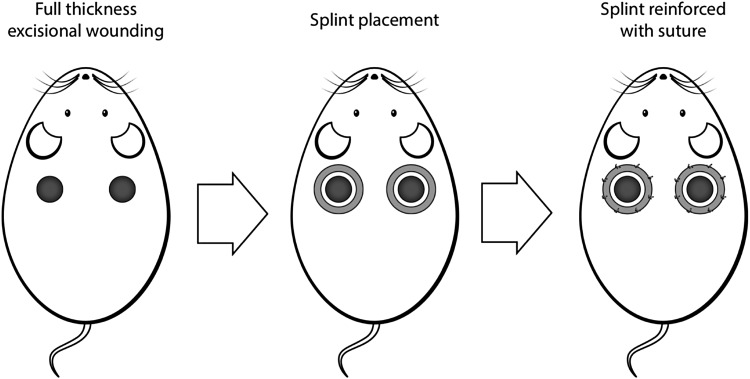
Full-thickness excisional wounds were created following the model described by Galiano et al.4 In brief, the mice were prepped by shaving the dorsum and applying a depilatory agent (Nair®; Church & Dwight Co, Princeton, NJ) to remove any remaining hair. Following anesthesia with isoflurane (isoflurane 2%, 2 L/min oxygen), two identical 6 mm circular wounds were created on the dorsum of each mouse, to either side of midline and just caudal to the scapula (Fig. 2). A 6 mm sterile punch biopsy tool was used as a stencil for the wound, and the actual full-thickness wound was excised using microscissors. Sterile donut shaped silicone splints were fixed to the surrounding wound edge using an immediate bonding cyanoacrylate adhesive (Krazy Glue, Elmer's, Inc., Columbus, OH) and eight simple interrupted 6-0 nylon sutures (Ethicon, Somerville, NJ). Wounds were covered with clear semiocclusive dressing (3M Tegaderm™, St. Paul, MN), which was changed on alternative days. Upon completion of surgery, the mice were placed under warming lamps and observed until they fully recovered from anesthesia. Images of both dorsal wounds were obtained on the day of surgery, postoperative day (POD) 0, and on alternate PODs subsequently, in both normal light, using a digital camera, and under fluorescent light, using a Nikon epifluorescence microscope (cyan GFP Ex436/20 Dm 455 Bar 480/40). Images were taken until POD 16, when wound healing was complete.
Figure 2.
Full-thickness excisional wound model with silicone splinting of wound.
The day of complete wound healing, as rated by blinded observers using light and fluorescent images (described in Wound analysis), ranged from POD 10 to 16. The experimental steps were repeated in four 12-week-old female K14-Cre/ROSAmT/mG mice. One mouse was harvested on POD 10, 12, 14, and 16 for histology to determine the accurate point of wound healing.
Histology
Mice were sacrificed with CO2 asphyxiation and cervical dislocation. The wound sites of harvested mice were excised to the hypodermis with microscissors and the tissues were immediately fixed in 4% paraformaldehyde at 4°C overnight. Samples were then rinsed in phosphate-buffered saline (PBS; Sigma-Aldrich, St. Louis, MO), and kept in 30% sucrose for 1 week at 4°C, before embedding in optimal cutting temperature (OCT) (OCT; Tissue-Tek, Sakura Finetek, Torrance, CA). OCT blocks were stored in −80°C overnight and were subsequently cut at 8–12 μM using a cryostat. Microscopy slides were stored at −20°C in the dark. Before microscopy, slides were brought to room temperature, washed in PBS, and mounted onto a slide in fluorescent mounting medium (Vector Laboratories, Inc., Burlingame, CA). Slides were imaged using a fluorescence microscope. mT and mG did not require immunostaining for visualization. A representative sample of 10 slides was taken of the center of each wound.
Wound analysis
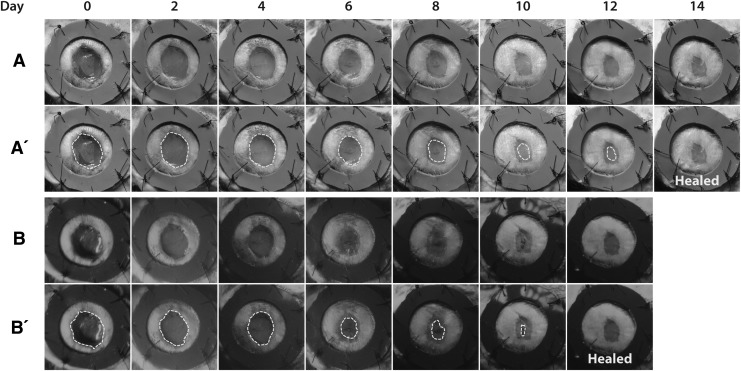
A representative paired wound healing image series from a single healing wound, in normal and fluorescent light, was chosen for analysis (Fig. 3). This paired series was presented to 10 blinded observers who had extensive experience in wound healing analysis in human and mouse models. Participants were asked to draw around the margins of epithelialization and to decide the POD of complete wound healing. Each participant rated both the normal light and fluorescent image series.
Figure 3.
The representative paired series of wounds of 12-week female K14-Cre/ROSAmT/mG mice using macroscopic (A) light imaging and (B) fluorescence imaging, which were presented to 10 blinded observers who determined the day of complete wound closure. Day of complete wound closure was determined to be 14 days in the light images, compared with 12 days in the fluorescence images. (A′) and (B′) are representative images from a blinded observer tracing the presumed open wound (yellow dotted line).
Statistical analyses
The Shapiro–Wilk test was used to assess normality. As the data were nonparametric, the Wilcoxon signed-rank test (two-tailed) was used to compare the difference in rated day of complete wound closure between wounds imaged using normal light and fluorescent imaging. A *p value less than 0.05 was considered statistically significant.
Results
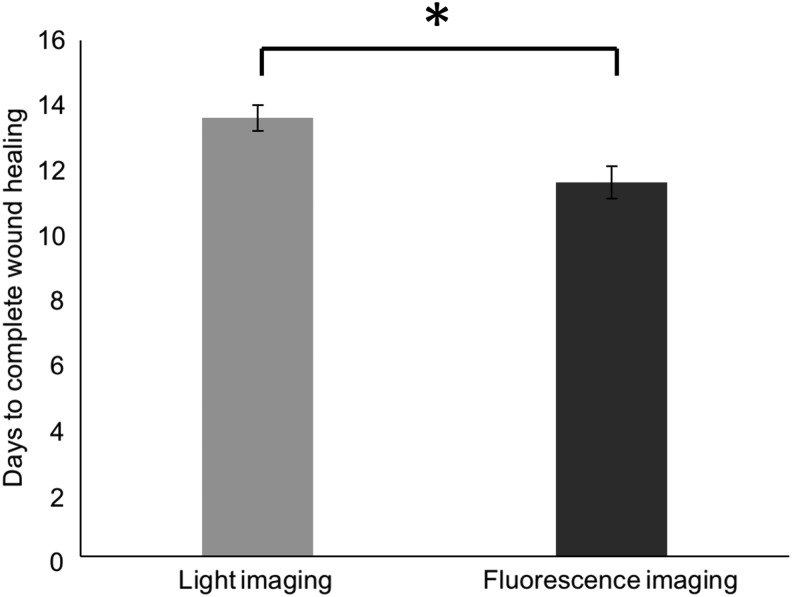
A series of images taken using normal light and fluorescence depicting mouse wounds in a splinted full-thickness excisional wound model (Fig. 3) were shown to blinded observers. The wounds were determined to be completely healed in a mean of 13.6 days (median 14 days) with a standard deviation of 1.58 using normal light images. Using fluorescence imaging, time to complete healing was a mean of 11.6 days (median 12 days) with a standard deviation of 1.26 (*p = 0.008, Table 1 and Fig. 4).
Table 1.
Day of determined wound closure
| Days to Complete Wound Healing on Macroscopic Evaluation Using | ||
|---|---|---|
| Participant No. | Light Imaging | Fluorescence Imaging |
| 1 | 14 | 12 |
| 2 | 12 | 10 |
| 3 | 14 | 12 |
| 4 | 16 | 14 |
| 5 | 12 | 12 |
| 6 | 14 | 10 |
| 7 | 12 | 10 |
| 8 | 16 | 12 |
| 9 | 12 | 12 |
| 10 | 14 | 12 |
| Mean | 13.6 | 11.6 |
| Standard deviation | 1.58 | 1.26 |
| Median | 14 | 12 |
| Range | 12–16 | 10–14 |
| p | 0.008* | |
Statistical analysis was conducted using the Wilcoxon signed-rank test (two-tailed) as data were not normally distributed.
Figure 4.
Blinded observers rated wound healing to occur at an earlier timepoint in the wound series taken under fluorescence (mean 11.6 ± 1.26, median 12, range 10–14) than wounds series taken under normal light (mean 13.6 ± 1.58, median 14, range 12–16). These timepoints were significantly different, as determined by Wilcoxon signed-rank test (*p = 0.008). Error bars represent the standard error of the mean (SEM).
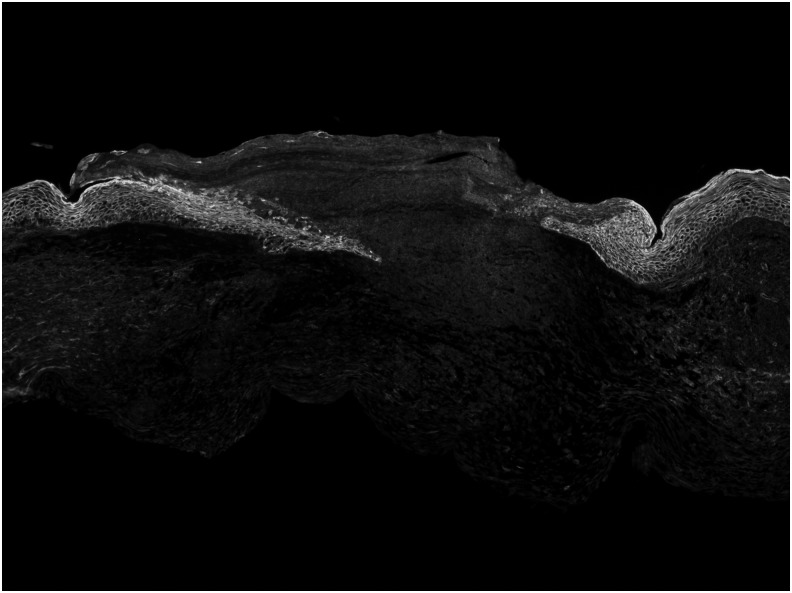
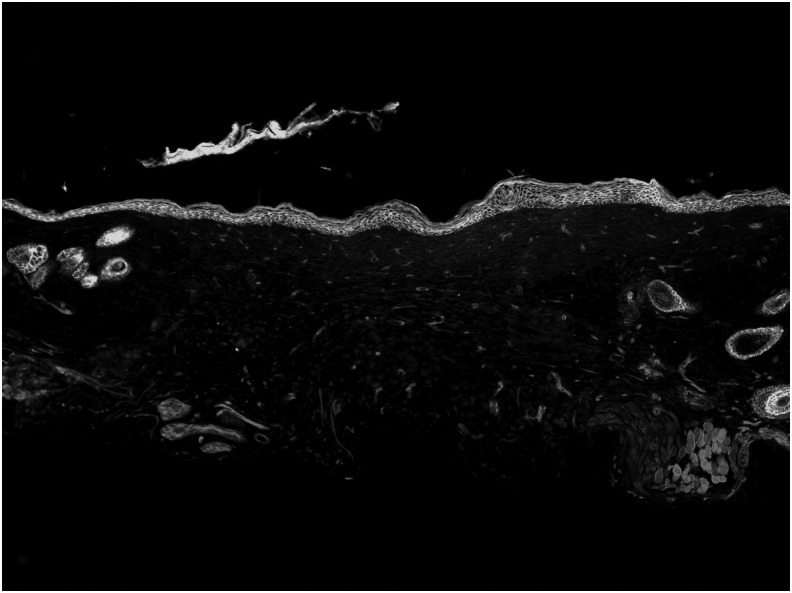
To determine the accuracy of time to complete closure, histology was performed on mice sacrificed at several timepoints. Fluorescence imaging of histological samples at POD 10 demonstrate incomplete reepithelialization as seen by a lack of continuity of the GFP-labeled keratinocytes (Fig. 5). On POD 12, microscopic fluorescence imaging demonstrates continuity of the GFP-labeled epithelium and a fully healed wound (Fig. 6). The findings from histology are consistent with results obtained from determination of wound closure using macroscopic fluorescence imaging.
Figure 5.
Fluorescence imaging of histological samples from the healing wound at postoperative day 10 reveals a lack of continuity of the GFP-expressing epithelial cells, and thus incomplete reepithelization. GFP = K14-positive cells, RFP = K14-negative cells.
Figure 6.
Fluorescence imaging of histological samples from the healing wound at postoperative day 12 reveals continuity of GFP-expressing epithelium, and thus a fully healed wound. GFP = K14-positive cells, RFP = K14-negative cells.
Discussion
The primary goal of animal wound healing models is to mimic human wound repair as closely as possible. The silicone splinted wound healing model, developed by Galiano et al.,4 has “humanized” the wound healing process in mice and increased the translational applicability of mouse models. The wound splinting technique minimizes the degree of wound contraction, a feature of healing murine, but not human, skin. Interpretation of the time to complete wound closure in these models, however, uses gross macroscopic inspection, which is difficult to accurately gauge. In normal light imaging, the pink color of healing wounds can appear visually indistinct from the pink of the scared healed mouse skin. This study describes a technique using double transgenic K14-Cre/ROSAmT/mG mice to selectively label epithelial keratinocytes, which express K14 in green, while labeling all other cells that do not express K14 in red. This can create more visually distinct wounds at different healing stages. The results indicate that the wound healing time estimated from macroscopic inspection of fluorescent images of K14-Cre/ROSAmT/mG mouse wounds more closely resembled the time to complete reepithelialization as assessed by fluorescence imaging of histological specimens.
Histological examination of wounds provides the most accurate estimate of wound healing time. It is not possible, however, to perform microscopy in vivo, and use of histology in investigating wound healing, a dynamic and constantly changing phenomena, is limited.13 Using K14-Cre/ROSAmT/mG mice with visualization of wounded sites in fluorescent light can increase the precision, by which wound healing can be assessed in mice in vivo on a macroscopic level. It is interesting to note that the wound healing time as determined by macroscopic inspection in normal light was estimated to be ∼2 days longer than the wound closure time determined using fluorescent light, which suggests that evaluation of wounds in normal light may lead to overestimations of wound closure times in other situations.
The limitations of this work include the fact that wounds were not imaged on every POD, that a fluorescent and light image series were taken from one representative wound, and a small sample of 10 blinded observers were asked to assess wound healing in these series. Nevertheless, fluorescence imaging adds significant strength and confirms the veracity of findings, illustrating the potential of this technique in experimental work. It is also noted that the technique described in this study uses transgenic K14-Cre/ROSAmT/mG mice, which limit its applicability to situations where transgenic mice are not required for other reasons. Lineage tracing of epidermal and dermal stem cells in the healing epidermis, for example, relies on the fluorescent properties of labeled cells.14,15 This technique will likely be best suited to situations where it is critical to accurately determine time to wound healing, for example, when testing the influence of certain pharmaceutical or environmental manipulations on wound healing.
Innovation
We present a technique using double transgenic mice and fluorescence imaging to provide an accurate and quantitative approach to measuring wound healing in mice macroscopically. This wound model should be used whenever possible, particularly when the accuracy of wound healing time is critical, such as in studies concerning new therapeutic interventions or the elucidation of further biochemical and mechanical properties of wound healing.
Key Findings
• The mouse splinted full-thickness excisional wound healing model provides a humanized model of wound repair.
• Determination of wound healing in this model is challenging.
• Using double transgenic mice and fluorescence imaging provides an accurate approach to measuring wound healing.
Abbreviations and Acronyms
- GFP
green fluorescent protein
- mT
red fluorescent protein gene, tdTomato
- OCT
optimal cutting temperature
- POD
postoperative day
Acknowledgments and Funding Sources
This work was supported, in part, by NIH grant K08 DE024269 (to D.C.W.), the Child Health Research Institute at Stanford University (to D.C.W.), NIH grant R01 GM087609 (to H.P.L.), a Gift from Ingrid Lai and Bill Shu in honor of Anthony Shu (to H.P.L.), the Hagey Laboratory for Pediatric Regenerative Medicine and Children's Surgical Research Program (to D.C.W., H.P.L., and M.T.L.), the Gunn/Olivier Fund (to M.T.L.). Additional funding was provided by the American Society of Maxillofacial Surgeons (ASMS)/Maxillofacial Surgeons Foundation (MSF) Research Grant Award (to M.S.H., H.P.L., and M.T.L.), the California Institute for Regenerative Medicine (CIRM) Clinical Fellow training grant TG2-01159 (to M.S.H.), the Stanford University School of Medicine Transplant and Tissue Engineering Fellowship Award (to M.S.H.), and the Sarnoff Cardiovascular Research Foundation (to W.X.H.).
Author Disclosure and Ghostwriting
No competing financial interests exist. The content of this article was expressly written by the authors listed. No ghostwriters were used to write this article.
About the Authors
Michael S. Hu, MD, MPH, MS, is a postdoctoral fellow at Stanford interested in stem cell biology and regenerative medicine. He is pursuing a career in craniofacial plastic surgery. Justin Cheng, BA, is a medical student interested in plastic surgery. Mimi R. Borrelli, MBBS, MSc, BSc, and Dre M. Irizarry, MD, are postdoctoral fellows interested in plastic surgery. Tripp Leavitt, MD, Elizabeth R. Zielins, MD, Dominik Duscher, MD, and Zeshaan N. Maan, MD, are plastic surgery residents. Graham G. Walmsley, MD, PhD, is a stem cell scientist working in venture capital. Wan Xing Hong, MD, MS, and Brad Stephan, MD, are general surgery residents. Alexander T.M. Cheung, BS, is a premedical student. Fereydoun Don Parsa, MD, FACS, is Chief of Plastic Surgery at the University of Hawai'i. Derrick C. Wan, MD, is an associate professor at Stanford. Geoffrey C. Gurtner, MD, and Hermann Peter Lorenz, MD, are professors at Stanford. Michael T. Longaker, MD, MBA is Professor of Surgery and Bioengineering at Stanford. He is the Director of Research for the Program in Regenerative Medicine, Children's Surgical Research, and Division of Plastic and Reconstructive Surgery. His extensive research experience includes wound healing, tissue engineering, and developmental/stem cell biology.
References
- 1.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med 1999;341:738–746 [DOI] [PubMed] [Google Scholar]
- 2.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature 2008;453:314–321 [DOI] [PubMed] [Google Scholar]
- 3.Wong VW, Sorkin M, Glotzbach JP, Longaker MT, Gurtner GC. Surgical approaches to create murine models of human wound healing. Biomed Res Int 2010;2011:969618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galiano RD, Michaels V, Dobryansky M, Levine JP, Gurtner GC. Quantitative and reproducible murine model of excisional wound healing. Wound Rep Regen 2004;12:485–492 [DOI] [PubMed] [Google Scholar]
- 5.Ansell DM, Campbell L, Thomason HA, Brass A, Hardman MJ. A statistical analysis of murine incisional and excisional acute wound models. Wound Rep Regen 2014;22:281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Ge J, Tredget EE, Wu Y. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat Protoc 2013;8:302–309 [DOI] [PubMed] [Google Scholar]
- 7.Cho H, Balaji S, Hone NL, et al. Diabetic wound healing in a MMP9-/-mouse model. Wound Rep Regen 2016;24:829–840 [DOI] [PubMed] [Google Scholar]
- 8.Park SA, Raghunathan VK, Shah NM, et al. PDGF-BB does not accelerate healing in diabetic mice with splinted skin wounds. PLoS One 2014;9:e104447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordts SC, Muthuramu I, Amin R, Jacobs F, De Geest B. The impact of lipoproteins on wound healing: topical HDL therapy corrects delayed wound healing in apolipoprotein E deficient mice. Pharmaceuticals 2014;7:419–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaels J, Churgin SS, Blechman KM, et al. db/db mice exhibit severe wound-healing impairments compared with other murine diabetic strains in a silicone-splinted excisional wound model. Wound Rep Regen 2007;15:665–670 [DOI] [PubMed] [Google Scholar]
- 11.The Jackson Laboratory. Mouse Strain Datasheet-007576. www.jax.org/strain/007576 (last accessed April29, 2017)
- 12.The Jackson Laboratory. Mouse Strain Datasheet-004782. www.jax.org/strain/004782 (last accessed April29, 2017)
- 13.Abe T, Fujimori T. Reporter mouse lines for fluorescence imaging. Dev Growth Differ 2013;55:390–405 [DOI] [PubMed] [Google Scholar]
- 14.Amoh Y, Li L, Katsuoka K, Penman S, Hoffman RM. Multipotent nestin-positive, keratin-negative hair-follicle bulge stem cells can form neurons. Proc Natl Acad Sci U S A 2005;102:5530–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amoh Y, Mii S, Aki R, et al. Multipotent nestin-expressing stem cells capable of forming neurons are located in the upper, middle and lower part of the vibrissa hair follicle. Cell Cycle 2012;11:3513–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]