Abstract
Purpose
Mammalian central nervous system axons fail to regenerate after injury. Contributing factors include limited intrinsic growth capacity and an inhibitory glial environment. Inflammation-induced optic nerve regeneration (IIR) is thought to boost retinal ganglion cell (RGC) intrinsic growth capacity through progrowth gene expression, but effects on the inhibitory glial environment of the optic nerve are unexplored. To investigate progrowth molecular changes associated with reactive gliosis during IIR, we developed an imaging mass spectrometry (IMS)-based approach that identifies discriminant molecular signals in and around optic nerve crush (ONC) sites.
Methods
ONC was performed in rats, and IIR was established by intravitreal injection of a yeast cell wall preparation. Optic nerves were collected at various postcrush intervals, and longitudinal sections were analyzed with matrix-assisted laser desorption/ionization (MALDI) IMS and data mining. Immunohistochemistry and confocal microscopy were used to compare discriminant molecular features with cellular features of reactive gliosis.
Results
IIR increased the area of the crush site that was occupied by a dense cellular infiltrate and mass spectral features consistent with lysosome-specific lipids. IIR also increased immunohistochemical labeling for microglia and macrophages. IIR enhanced clearance of lipid sulfatide myelin-associated inhibitors of axon growth and accumulation of simple GM3 gangliosides in a spatial distribution consistent with degradation of plasma membrane from degenerated axons.
Conclusions
IIR promotes a robust phagocytic response that improves clearance of myelin and axon debris. This growth-permissive molecular remodeling of the crush injury site extends our current understanding of IIR to include mechanisms extrinsic to the RGC.
Keywords: optic nerve regeneration, inflammation, lipidomics, optic nerve crush, microglia
All visual information transmitted from the retina to the brain travels through axons of the retinal ganglion cells (RGCs), and vision loss can occur from injury to these projections. Most central nervous system axons fail to regenerate after injury, and restoration of vision through repair or regeneration of neural connections in the visual system is an important goal in neurobiology. Low intrinsic growth capacity and an inhibitory glial environment contribute to failure of RGC axon regeneration.1,2
Inflammation-induced optic nerve regeneration (IIR) was initially described after the incidental observation that intraocular injections that violate the crystalline lens are associated with increased RGC axon regeneration.3,4 This effect was caused by an acute inflammatory response provoked by the presence of lens material in the vitreous and can be achieved without lens injury by intravitreal injection of a yeast cell wall preparation (Zymosan A),5 especially when combined with a cell-permeant analogue of cyclic adenosine monophosphate (CPT-cAMP).6 The invading immune cells activate progrowth RGC gene expression programs,7 and IIR is thought to work primarily through such RGC-intrinsic mechanisms.8,9 Recent evidence suggests that IIR also supports remyelination,10 but additional effects on the optic nerve environment extrinsic to the RGC remain largely unexplored.
IIR models and other strategies such as gene deletion11 and enhancement of neuronal electrical activity12 improve intrinsic growth capacity and have led to identification of several candidate proregenerative factors. These include protein phosphatases and kinases, transcription factors, extracellular matrix proteins, cytokines, and growth factors.1 Discovery-based approaches that focus on the optic nerve injury site could provide new insights about RGC-extrinsic influences on axon regeneration. This might enhance our ability to translate RGC-intrinsic targets into human treatment because combinatorial approaches to optic nerve regeneration with various RGC-intrinsic and -extrinsic targets have shown potential in animal models.10–16 Despite evidence that a wide array of lipid molecular species could play important roles7,17–28 in optic nerve regeneration, to our knowledge this has never been studied with a formal lipidomics approach.
Matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry (IMS) is a molecular imaging method that generates chemical maps of thin tissue sections from spatially resolved mass spectra.29–37 We developed a MALDI IMS–based approach that identifies discriminant spectral features of optic nerve crush (ONC) sites to screen for progrowth molecular changes associated with reactive gliosis during IIR and selected experimental conditions that produce lipid-enriched mass spectra. To our knowledge this is the first study that has applied IMS to an optic nerve injury and regeneration paradigm.
Methods
ONC and Intravitreal Injections
All animal procedures were performed in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and policies of the UCLA Animal Research Committee. A rat model of IIR was established by intravitreal injection of a yeast cell wall preparation (Zymosan A)5 plus a CPT-cAMP analogue6 immediately after ONC. Ten-week-old male Fischer rats were deeply anesthetized with inhaled isoflurane, and the eyes were treated with topical anesthetic (proparacaine HCl 0.5% ophthalmic) and cycloplegic (tropicamide 0.5% ophthalmic) agents to reduce pain and assist with visualization of intravitreal injections. The left optic nerve was exposed by blunt dissection through a temporal, fornix-based conjunctival incision and crushed for 10 seconds with self-closing forceps (Dumoxel #N5; Dumont, Montignez, Switzerland). Absence of injury to the retinal vascular supply was confirmed by funduscopic examination. Intravitreal injections (5 μL) of PBS vehicle or a suspension of finely ground, sterilized28 Zymosan A (Z4250, 12.5 mg/mL; Sigma-Aldrich Corp., St. Louis, MO, USA) plus CPT-cAMP (C3912, 100 μM; Sigma-Aldrich Corp.) were performed with a pulled glass pipette attached to a Hamilton syringe on a manual micromanipulator. Injections were made 2 mm posterior to the limbus, and care was taken to prevent lens injury, choroidal hemorrhage, or retinal detachment. Absence of these intraocular adverse events was confirmed by fundoscopic examination. Conjunctival incisions were closed with 8-0 vicryl sutures, and petrolatum ophthalmic ointment was applied to the ocular surface. Experimental groups consisted of naïve controls, ONC plus intravitreal Zymosan/CPT-cAMP (ONC+ZC), and ONC plus intravitreal vehicle (ONC+Veh) (one nerve per animal).
Immunohistochemistry
Animals were euthanized by intraperitoneal sodium pentobarbital injection followed by transcardial perfusion with ice-cold PBS plus 4% paraformaldehyde (4% PFA). Eyes and attached optic nerves were removed en bloc with orbital contents, the corneas were pierced with a 30-gauge needle, and the tissue was placed in 4% PFA for 2 hours at room temperature (RT). A razor was then used to make an oblique incision through the clear cornea and extended through the sclera to the equator, and the tissue was post-fixed overnight at 4°C. The orbital tissues were dissected away from the globe and attached optic nerve, and the globes were bisected through the same plane as the corneal incision and without transecting the optic nerve head (ONH). Vannas scissors were used to cut around the ONH, and the nerve (with attached small square of peripapillary tissue) and bisected eyecups were cryoprotected overnight at 4°C in 30% sucrose.
Cryoprotected tissue was embedded in optimal cutting temperature compound and flash frozen in liquid nitrogen. Longitudinal sections of nerve or cross sections of retina (14 μm) were cut with a cryostat, mounted on plus-charged glass microscope slides, and permeabilized for 30 minutes at RT in TBS plus 0.25% Tween 20 (0.25% TBST). The sections were then blocked with 10% normal donkey serum in TBS for 1 hour at RT and incubated overnight at 4°C with gentle shaking in 0.1% TBST plus 2% BSA (2% BSA-0.1% TBST) and selected primary antibodies. The sections were rinsed with 0.1% TBST and incubated for 1 hour at RT with Hoechst 33258 in 2% BSA-0.1% TBST plus appropriate combinations of cyanine- and Alexa Fluor–conjugated donkey secondary antibodies. Finally, the sections were rinsed with 0.1% TBST and coverslipped with an aqueous mounting medium. Primary antibodies were rabbit polyclonal anti-GAP43 (ab16053, 1:250; Abcam, Cambridge, MA, USA), rabbit polyclonal anti-Iba1 (019-19741, 1:200; Wako Chemicals USA, Richmond, VA, USA), and chicken polyclonal anti-GFAP (ab4674, 1:1000; Abcam).
Confocal Microscopy and Image Analysis
Images of immunostained tissues were obtained at 10× or 40× primary magnification with a confocal microscope (Fluoview FV1000; Olympus, Center Valley, PA, USA). Identical microscope, laser, and software settings were used for any groups that would subsequently be compared. Data were exported from Fluoview software as eight-bit gray value images and loaded in ImageJ software (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA). Growth-associated protein 43 (GAP43) is neuronal phosphoprotein that is involved in structural plasticity and is only expressed in RGCs during axon growth.38–42 Growth of GAP43-positive axons beyond the site of crush injury in longitudinal sections of optic nerve is a measure of bona fide axon regeneration.3,5,42,43 For longitudinal sections of optic nerve, the crush sites were identified by both the end of the wave front of GAP43 staining and the increased nuclear density visualized with Hoechst 33258.28 Beginning at the crush site, linear regions of interest (ROIs) were drawn perpendicular to the long axis of the nerve at 100-μm intervals. The eight-bit data from the GAP43 channel was converted to binary using a uniform threshold for all images. The tally of nonzero pixels was divided by the total number of pixels in each corresponding linear ROI so that GAP43 immunopositivity could be expressed as a percentage. Four to five nerves per treatment condition were sampled, and the results from two to four representative sections per nerve were averaged. ROIs were also drawn around the crush core region in images of Hoechst-stained serial sections of optic nerve. The area occupied by the hypercellular crush core was calculated for each section. Four nerves per treatment condition were sampled, and the results from four representative sections per nerve were averaged. In all image analysis procedures, averages for individual nerves (as opposed to values per section) were used as data points for subsequent statistical analysis.
Sample Preparation and IMS
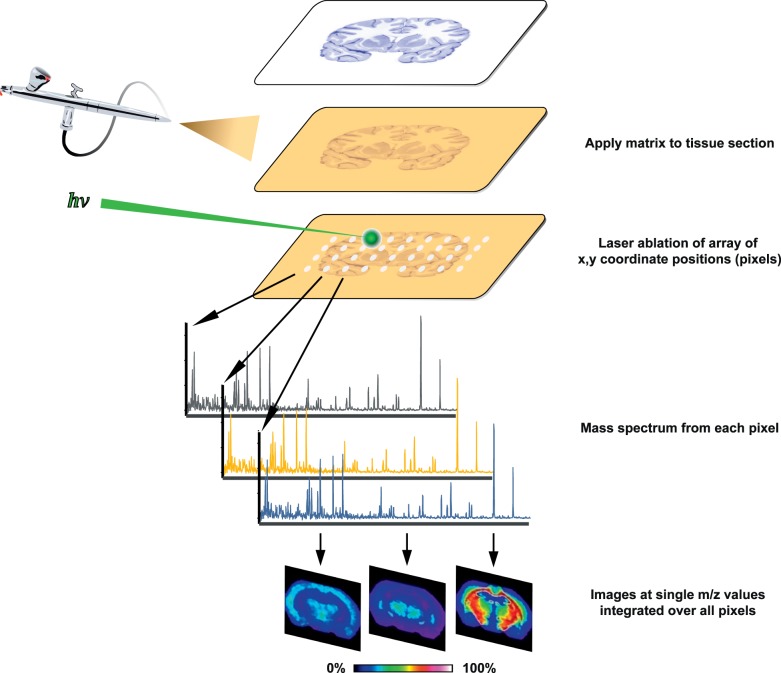
To screen molecular changes of the optic nerve lipidome during IIR, we employed MALDI IMS. An outline of the workflow of MALDI IMS is given in Figure 1 and described below.29–37 Briefly, thin sections of tissue are mounted on a conductive MALDI target plate, coated with an appropriate chemical matrix that determines which analytes will be ionized, and placed inside a MALDI ion source. In MALDI, laser shots are fired at discrete positions on the target to generate plumes of ions that can be accelerated into a mass analyzer. In IMS, laser applications are rastered across the tissue surface so that each resulting mass spectrum is coregistered with a planar coordinate (pixel). Intensity maps created from ions of interest can be used to relate the signal to an underlying histologic feature.
Figure 1.
MALDI IMS process overview. Thin tissue sections are mounted on MALDI target plates and coated with chemical matrix that assists laser desorption and ionization of tissue matrix. As a laser is rastered through a predefined array of coordinate locations (pixels), each location is associated with a unique mass spectrum. Ion signal intensities are encoded as different hues in a color lookup table, and images are created by plotting signal intensities of ions of interest by pixel location. Data-mining approaches are used to discover discriminant spectral features.
Animals were euthanized by CO2 inhalation, and the eyes and attached optic nerves were quickly dissected from the orbits. The globes were pierced with a 27-gauge, metal-hub hypodermic needle just inferior to the optic nerve, and the nerve tissue was straightened along the needle. The tissue was frozen by immersing the metal base of the needle in liquid nitrogen. The tissue on the end of the needle was then immersed in embedding solution (2.7% carboxymethyl cellulose) and placed on a precooled weigh boat floating on liquid nitrogen. This process was repeated until sufficient frozen embedding material surrounded the sample. Samples were mounted onto a cryostat chuck and aligned based on the orientation of the hypodermic needle.44 Twelve-micrometer sections were cut and flattened with a polytetrafluoroethylene-coated slide on top of 45 × 45-mm indium tin oxide-coated glass MALDI targets before thaw mounting. The samples were coated with a chemical matrix of 1,5-diaminonaphthalene via sublimation with an in-house device30,45 (110°C for 15 minutes). Data were acquired in negative ion mode from three or four nerves per time point and treatment condition with a 15-μm pixel size using a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (Bruker SolariX; Bruker Daltonics, Billerica, MA, USA). Mass range was 260 to 1300 with 500 laser shots per pixel. The instrument is equipped with a modified smart-beam laser operated at 2 kHz with a Gaussian laser beam profile at 355 nm. All data sets had mass resolving powers greater than 200,000 at m/z (mass-to-charge ratio) 400. Data preprocessing steps were performed with FTMS Processing (Bruker Daltonics, Billerica, MA, USA) and included spectral alignment through internal calibration of well-known lipids common to all spectra and selection of peaks with signal-to-noise ratio greater than 3. After mass spectral data acquisition, the 1,5-diaminonaphthalene MALDI matrix was removed with a methanol wash, and the tissue was stained with hematoxylin and eosin (H&E). The tissue was then scanned at 20× primary magnification using a microscope (Nikon Eclipse 90i: Nikon Instruments, Inc., Melville, NY, USA).
Data Mining, Tentative Lipid Identification, and Statistical Analysis
Preprocessed mass spectra were loaded into software (SCiLS lab 2016b; SCiLS, Bremen, Germany) and overlaid with high-resolution optical images of corresponding tissue sections that were stained with H&E after MALDI IMS acquisitions. ROIs were manually drawn around the perimeter of each nerve, and the mass spectral data contained within these regions were segmented with the k-means clustering algorithm in SCiLS lab software. k-Means cluster analysis is commonly used to reveal latent molecular features contained in large MALDI IMS data sets,46 and we used this approach to define ROIs that correspond to the hypercellular core of the crush injury site. In IMS, pixels within the same cluster have similar mass spectra at a multivariate level. The cluster map image allows for rapid comparison of the molecular histology revealed by IMS to the classical histology revealed by H&E. The subset of resulting segments that corresponded to the dense cellular infiltrate at the center of each crush site, as observed on postacquisition H&E, was used to define ROIs for subsequent receiver-operator characteristic (ROC) analysis.46 The areas of these crush center ROIs were also recorded and averaged. For ROC analysis, crush center ROIs from the ONC+Veh and ONC+ZC groups were pooled as a single class for comparison with naïve controls. Discriminant m/z values for ROC analysis were defined as those with an area under the ROC curve greater than 0.75. Images of selected discriminant m/z values were overlaid on postacquisition H&E optical images, and ROIs were manually drawn to calculate the tissue area occupied by (or deficient of) those mass spectral signals.
Exact masses for all entries in the Lipid MAPS Structure Database (LMSD) were obtained with LMSDSearch.php scripts47,48 and cross-checked against discriminant m/z values. Observed m/z values were assumed to represent deprotonated anions, and tentative lipid identifications were made on the basis of mass accuracy if the observed m/z matched an exact mass to less than 3.0 parts per million. For all lipid identifications discussed below, these criteria returned only one match, or a small set of isomers from the same LMSD subclass. Elucidation of m/z values colocalized with selected m/z images was performed with Pearson's correlation analysis,49 and statistical significance was defined as P = 0.05. Only statistically significant correlations were considered in the colocalization analysis. ROI area and axon regeneration measurements were analyzed with statistical software (Minitab; Minitab, Inc., State College, PA, USA) with Student's t-test and displayed as box plots or mean ± SEM. P values less than 0.05 were considered statistically significant.
Results
Crush Site Hypercellularity Is Increased During IIR
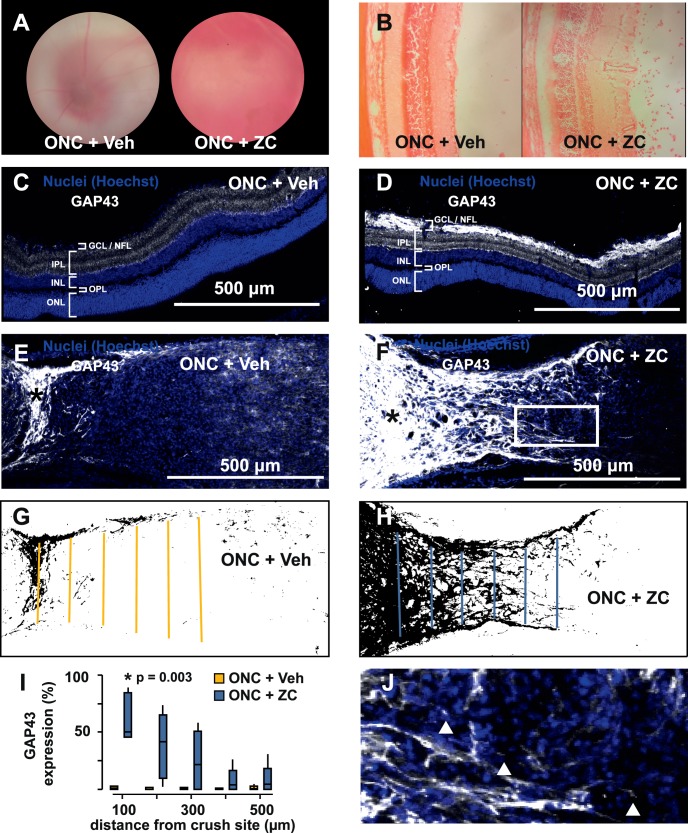
Intravitreal Zymosan A and CPT-cAMP injection provoked a prominent inflammatory infiltrate in the posterior segment (Figs. 2A, 2B) and increased nerve fiber layer staining for GAP43 when combined with ONC+ZC (Figs. 2C, 2D). Compared to the vehicle-injected group (ONC+Veh), optic nerves from the ONC+ZC group had significantly increased GAP43 immunopositivity beyond the crush site and contained many growth cone–like structures by day 14 (Figs. 2E–J).
Figure 2.
Establishment of an inflammation-induced optic nerve regeneration model. (A) Hazy fundus view 3 days after ONC immediately followed by intravitreal injection of Zymosan A plus CPT-cAMP (ONC+ZC) is caused by an inflammatory infiltrate that is evident on H&E (B). (C, D) GAP43 is elevated in the ganglion cell layer and nerve fiber layer of the ONC+ZC group (day 14). (E, F) The optic nerve distal to the crush site (*) has increased GAP43 expression in the ONC+ZC group (day 14). (G, H) Binarized versions of the images in E and F and linear ROIs drawn at 100-μm intervals were used to quantify GAP43 expression. (I) GAP43 expression as a function of distance from the crush site. (J) Expanded view of the inset box from panel F shows examples of growth cone–like structures that tip GAP43-positive processes in the ONC+ZC group (triangles). ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL/NFL, ganglion cell layer/nerve fiber layer.
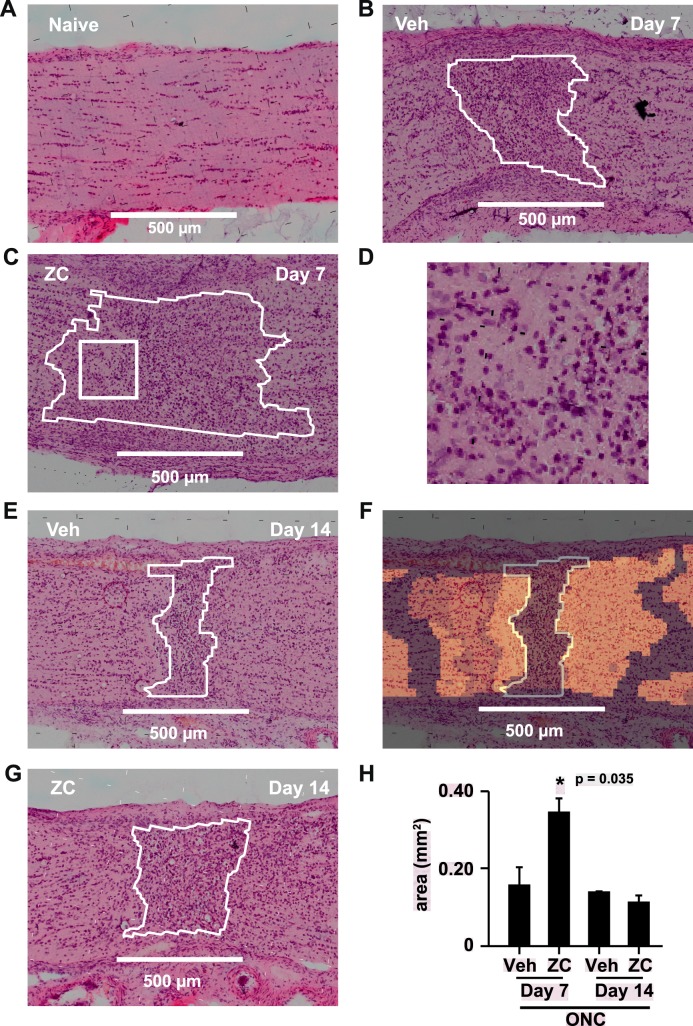
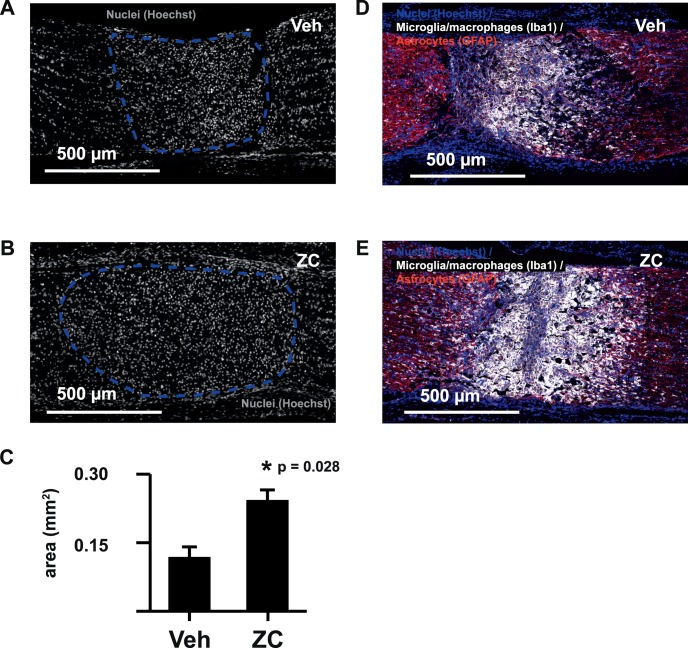
k-Means clustering of mass spectral data acquired by MALDI IMS resulted in symmetric, cross-sectional segments centered on the hypercellular crush injury core (Fig. 3F). Seven days after crush, the area occupied by the hypercellular core of the crush site was significantly increased by IIR (Figs. 3B, 3C, 3H). Close inspection of postacquisition optical images of H&E-stained tissue sections demonstrated that the dense cellular infiltrate contained many cells that resembled histiocytes with abundant, eosinophilic cytoplasm and round, eccentrically located nuclei (Fig. 3D). We confirmed IIR-induced increases in the size of the crush core region by quantifying the area occupied by densely packed Hoechst-stained nuclei (Figs. 4A–C), and the cells in this region were strongly labeled by the microglia/macrophage marker Iba1 (Figs. 4D, 4E).
Figure 3.
k-Means cluster analysis of the crush core region. (A) Postacquisition H&E image of a longitudinal section of naïve control nerve. Inset: quantification of crush center areas obtained by k-means cluster analysis. (B, C, E, G) Representative examples of postacquisition H&E-stained sections and corresponding crush center ROIs (white outlines) identified by k-means clustering. (D) Expanded view of the square drawn within the crush center ROI outline in panel C. Many crush site cells resemble histiocytes with round, eccentrically located nuclei and abundant eosinophilic cytoplasm. (F) k-means cluster analysis of the mass spectral data associated with the section of nerve from E produces symmetric segments of nerve centered about the crush site. Regions of tissue with the same color in the cluster map image overlay are partitioned in the same segment and have similar spectral characteristics. (H) Quantification of the size of the hypercellular crush core region by k-means segmentation.
Figure 4.
IIR enhances microglia/macrophage accumulation at the crush site. The size of the hypercellular crush core region (dashed lines in A and B) demonstrated by Hoechst nuclear stain is significantly increased by IIR at day 7 (C). The cellular infiltrate occupying the crush core region strongly labels with microglia/macrophage marker Iba1 (D, E).
IIR Enhances Lysosome-Specific Lipid Signatures at the Crush Site
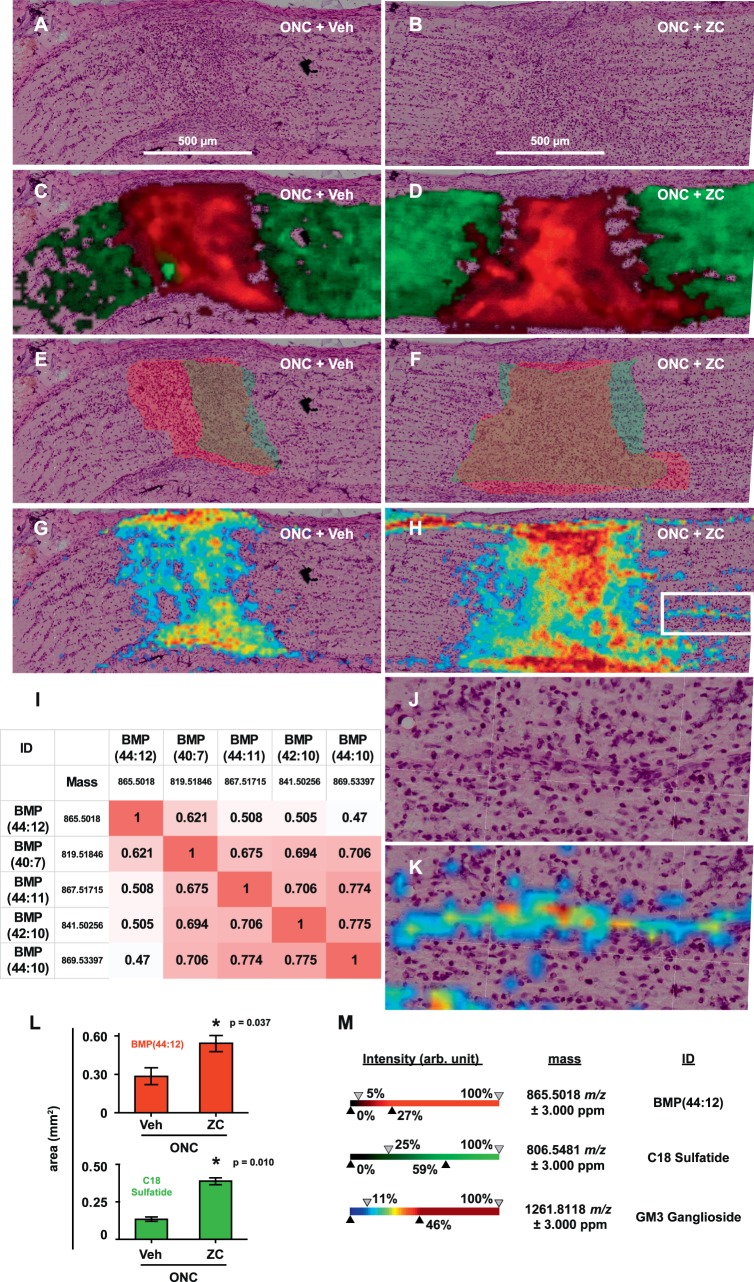
ROC analysis revealed which mass spectral peaks effectively discriminate the hypercellular core of day 7 crush sites from naïve nerves. Approximately 450 discriminant ions were identified and ranked in descending order of area under the ROC curve (auROC; Supplementary Material). Three of the top 20 discriminant ions (all with auROC > 0.998) matched lysosome-specific50,51 bis(monoacylglycero)phosphates (BMPs). The most abundant of these (m/z 865.5018) matched a BMP containing 44 carbons and 12 desaturated positions in its fatty acyl moieties (BMP[44:12]; Figs. 5C, 5D). With Pearson's correlation analysis, we determined which other m/z values were colocalized with m/z 865.5018 and found that the top 10 m/z values (ranked by Pearson's correlation coefficient) matched other polyunsaturated BMPs or their 13C isotopes (Fig. 5I). Consistent with our observations of crush center areas from k-means clustering and Iba1 immunohistochemistry, the tissue area occupied by m/z 865.5018 was significantly increased by IIR at day 7 (Figs. 5C–F, 5L).
Figure 5.
IIR remodels the optic nerve lipidome. (A, B) Representative postacquisition H&E images of longitudinal sections from the ONC+Veh and ONC+ZC groups (day 7) without overlays. (C, D) Intensity map (m/z image) overlays for selected ions m/z 865.5018 (red) and 806.5481 (green). These were tentatively identified as BMP(44:12) and C18:0 sulfatide, respectively. (E, F) ROIs corresponding to the tissue area occupied by (or deficient of) m/z 865.5018 (red) and 806.5481 (green), respectively. (G, H) overlays of the m/z image for 1261.8118 (GM3 ganglioside). (I) Correlation matrix for m/z 865.5018. Highly correlated ions that matched 13C isotopes of the indicated BMP species were omitted for clarity. (J, K) Expanded views of the boxed area in H, with and without m/z image overlay for 1261.8118. (L) Summary of ROI area data for BMP(44:12) and C18 sulfatide. (M) Ion intensity scales for m/z images in C, D, G, H, and K.
IIR Reduces Lipid Sulfatides and Increases Simple GM3 Gangliosides
We next searched for discriminant m/z values that match lipid constituents known to be enriched in myelin and axoplasmic membranes. We found matches for several lipid sulfatides (sulfoglycosphingolipids; Supplementary Spreadsheet 1) that comprise a major proportion of myelin membranes and inhibit axon growth in vitro and in vivo.28,52–56 An ion with m/z 806.5481 matched the common C18:0 sulfatide ([3′-sulfo]Galβ-Cer[d18:1/18:0]) and was cleared from a significantly larger area of the crush site during IIR (Figs. 5C–F, 5L). Simple GM3 gangliosides are formed via lysosomal degradation of more complex gangliosides that are highly enriched in neuronal plasma membranes.57–59 We found that a variety of GM3 gangliosides accumulated in the crush site (Figs. 5G, 5H; Supplementary Material). In the ONC+ZC group, but not the ONC+Veh group, GM3 gangliosides also accumulated in a linear spatial distribution localized to organized columns of unknown cell types adjacent to the crush center (Figs. 5J, 5K).
Discussion
IIR is primarily attributed to progrowth gene expression programs in RGCs that interact with an inflammatory infiltrate in the vitreous.3,5–7,9,16,60–62 Recent evidence suggests that some features of the optic nerve glial environment can also be favorably impacted by IIR, as in the case of remyelination.10 The fact that phagocytic cells can support axon regeneration by modifying the nonpermissive optic nerve glial environment has been appreciated for many years,63–67 but the potential of posterior segment inflammation to enhance the phagocytic injury response in the optic nerve per se has not been addressed.9,68 We found that IIR enhances the optic nerve phagocytic response to crush injury and produces lipid molecular changes that reflect clearance of myelin and axon debris. This extends current understanding of IIR to include mechanisms extrinsic to RGCs. RGC-extrinsic and -intrinsic mechanisms of axon regeneration are increasingly recognized as intimately linked,69 and our results support the hypothesis that IIR is no exception. However, Ahmed et al.68 previously showed that an inflammatory reaction produced by direct injection of Zymosan into the optic nerve is neuroprotective but not axogenic. One potential explanation for this apparent discrepancy is that net effect of inflammation on axon regeneration could depend on the location of the inciting inflammatory stimulus within the visual pathway.
An enlarged region of crush site hypercellularity with associated Iba1 immunopositivity and increased lysosome-specific lipid signatures contained in characteristic mass spectra support the concept that IIR incites a more robust phagocytic response in the injured nerve. IIR-enhanced clearance of lipid sulfatide from the day 7 crush site could be reflective of broader efficiencies of myelin debris removal, and specific reduction of lipid sulfatides improves axon regeneration during IIR.28 Simple GM3 gangliosides normally represent a small fraction of brain gangliosides in adults. These molecular species are precursors to complex gangliosides that comprise the vast majority of this class.57–59 Complex gangliosides are highly enriched in axoplasmic membranes where they promote axon-myelin stability by binding myelin-associated glycoprotein. They are formed by sequential addition of monosaccharides to an oligosaccharide chain bound to a sphingolipid backbone.57–59 Complex gangliosides expressed on the axon surface are degraded by sequential removal of monosaccharide residues by lysosomal hydrolases,57,59,70 so accumulation of simple GM3 gangliosides after IIR is consistent with enhanced phagocytosis and degradation of degenerated axon debris. As with myelin debris clearance, this would promote a growth-permissive environment.71,72 Linear accumulations of simple GM3 gangliosides that track away from the crush site and localize to well-organized columns of cells are distinct characteristics of the crush border zone in IIR. The cellular character of these tissue features is still unknown; one possibility is that these could be phagocytic cells that have tracked between degenerating axon bundles.
Important goals of future work include understanding how IIR promotes an enhanced phagocytic response to injury within the optic nerve per se and whether this enhanced response makes a significant contribution to axon regeneration relative to RGC-intrinsic mechanisms. One limitation of this study is that we did not definitively determine whether the Iba1+ cells are primarily activated microglia73 or both microglia and macrophages, and this increases the number of possibilities. Compromise of the blood–nerve barrier is normally limited to the immediate vicinity of the crush injury site,74–76 and it is possible that IIR increases the proportion of the tissue with a leaky blood–nerve barrier. This would enhance tissue access for circulating monocytes and opsonins. Microglia have substantially increased rates of mitosis after crush injury alone,73 and it is possible that IIR further stimulates microglial mitosis and activation. For example, we made several tentative identifications of bioactive lipid-signaling molecules that regulate cellular proliferation and survival (Supplementary Material).25,77,78
We employed an IMS approach to discover unexpected changes in the molecular content and spatial distribution of the ONC lipidome during IIR. While previous studies have defined the spatial organization of lipids in naïve optic nerve, ours addresses injury and regeneration-induced molecular remodeling of the tissue. We used this approach to show that inflammation in the posterior segment increases the inflammatory response in the nerve per se, beyond that observed in crush injury alone. Associated discriminant mass spectral features were suggestive of growth-permissive remodeling of the crush injury site. However, the molecular mechanisms that govern this process and the degree to which it ultimately influences the regenerative outcome remain unknown. To our knowledge, this is the first formal lipidomics approach taken to study any form of optic nerve regeneration and represents a potentially powerful new mode of molecular discovery. MALDI IMS is well-suited for tissue lipidomic profiling, but the technology is versatile and our general approach to molecular discovery in optic nerve regeneration or other forms of optic nerve injury need not be limited to lipids.
Supplementary Material
Acknowledgments
The authors thank M. Alec Marin for helpful discussion and Shannan R. Eddington for technical assistance.
Supported by Research to Prevent Blindness, the UCLA EyeSTAR Program, National Institutes of Health/NIGMS Grant 5P41GM103391-07-8142 (RMC), and the Frank Stein and Paul S. May Grant for Innovative Glaucoma Research (Glaucoma Research Foundation) (DTS).
Disclosure: D.T. Stark, None; D.M.G. Anderson, None; J.M.K. Kwong, None; N.H. Patterson, None; K.L. Schey, None; R.M. Caprioli, None; J. Caprioli, None
References
- 1. He Z, Jin Y. . Intrinsic control of axon regeneration. Neuron. 2016; 90: 437– 451. [DOI] [PubMed] [Google Scholar]
- 2. Yiu G, He ZG. . Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006; 7: 617– 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leon S, Yin YQ, Nguyen J, Irwin N, Benowitz LI. . Lens injury stimulates axon regeneration in the mature rat optic nerve. J Neurosci. 2000; 20: 4615– 4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fischer D, Pavlidis M, Thanos S. . Cataractogenic lens injury prevents traumatic ganglion cell death and promotes axonal regeneration both in vivo and in culture. Invest Ophthalmol Vis Sci. 2000; 41: 3943– 3954. [PubMed] [Google Scholar]
- 5. Yin YQ, Cui Q, Li YM,et al. Macrophage-derived factors stimulate optic nerve regeneration. J Neurosc. 2003; 23: 2284– 2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kurimoto T, Yin YQ, Omura K,et al. Long-distance axon regeneration in the mature optic nerve: contributions of oncomodulin, cAMP, and pten gene deletion. J Neurosci. 2010; 30: 15654– 15663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fischer D, Petkova V, Thanos S, Benowitz LI. . Switching mature retinal ganglion cells to a robust growth state in vivo: gene expression and synergy with RhoA inactivation. J Neurosci. 2004; 24: 8726– 8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benowitz LI, He ZG, Goldberg JL. . Reaching the brain: advances in optic nerve regeneration. Exp Neurol. 2017; 287: 365– 373. [DOI] [PubMed] [Google Scholar]
- 9. Andereggen L, Trakhtenberg EF, Yin YQ, Benowitz LI. . Inflammation and Optic Nerve Regeneration. Hoboken: John Wiley & Sons, Inc.; 2015: 189– 204. [Google Scholar]
- 10. Marin MA, de Lima S, Gilbert H-Y, Giger RJ, Benowitz L, Rasband MN. . Reassembly of excitable domains after CNS axon regeneration. J Neurosci. 2016; 36: 9148– 9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun F, Park KK, Belin S,et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011; 480: 372– 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lim JHA, Stafford BK, Nguyen PL,et al. Neural activity promotes long-distance, target-specific regeneration of adult retinal axons. Nat Neurosci. 2016; 19: 1073– 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho KS, Chen DF. . Promoting optic nerve regeneration in adult mice with pharmaceutical approach. Neurochem Res. 2008; 33: 2126– 2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cho KS, Yang L, Lu B,et al. Re-establishing the regenerative potential of central nervous system axons in postnatal mice. J Cell Sci. 2005; 118: 863– 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Lima S, Habboub G, Benowitz LI. . Combinatorial therapy stimulates long-distance regeneration, target reinnervation, and partial recovery of vision after optic nerve injury in mice. : Goldberg JL, Trakhtenberg EF, . Axon Growth and Regeneration, Pt 2. San Diego: Elsevier Academic Press, Inc.; 2012: 153– 172. [DOI] [PubMed] [Google Scholar]
- 16. de Lima S, Koriyama Y, Kurimoto T,et al. Full-length axon regeneration in the adult mouse optic nerve and partial recovery of simple visual behaviors. Proc Natl Acad Sci U S A. 2012; 109: 9149– 9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strochlic L, Dwivedy A, van Horck FP, Falk J, Holt CE. . A role for S1P signalling in axon guidance in the Xenopus visual system. Development. 2008; 135: 333– 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Argaw A, Duff G, Zabouri N,et al. Concerted action of CB1 cannabinoid receptor and deleted in colorectal cancer in axon guidance. J Neurosci. 2011; 31: 1489– 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cherif H, Argaw A, Cecyre B,et al. Role of GPR55 during axon growth and target innervation. eNeuro. 2015; 2: ENEURO.0011-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campbell DS, Holt CE. . Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001; 32: 1013– 1026. [DOI] [PubMed] [Google Scholar]
- 21. Campbell DS, Holt CE. . Apoptotic pathway and MAPKs differentially regulate chemotropic responses of retinal growth cones. Neuron. 2003; 37: 939– 952. [DOI] [PubMed] [Google Scholar]
- 22. Guy AT, Nagatsuka Y, Ooashi N,et al. Glycerophospholipid regulation of modality-specific sensory axon guidance in the spinal cord. Science. 2015; 349: 974– 977. [DOI] [PubMed] [Google Scholar]
- 23. Stark DT, Bazan NG. . Synaptic and extrasynaptic NMDA receptors differentially modulate neuronal cyclooxygenase-2 function, lipid peroxidation, and neuroprotection. J Neurosci. 2011; 31: 13710– 13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joly S, Pernet V. . Sphingosine 1-phosphate receptor 1 is required for retinal ganglion cell survival after optic nerve trauma. J Neurochem. 2016; 138: 571– 586. [DOI] [PubMed] [Google Scholar]
- 25. Yung YC, Stoddard NC, Mirendil H, Chun J. . Lysophosphatidic acid signaling in the nervous system. Neuron. 2015; 85: 669– 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koriyama Y, Takagi Y, Chiba K,et al. Requirement of retinoic acid receptor beta for genipin derivative-induced optic nerve regeneration in adult rat retina. PLoS One. 2013; 8: e71252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kato S, Matsukawa T, Koriyama Y, Sugitani K, Ogai K. . A molecular mechanism of optic nerve regeneration in fish: The retinoid signaling pathway. Prog Retin Eye Res. 2013; 37: 13– 30. [DOI] [PubMed] [Google Scholar]
- 28. Winzeler AM, Mandemakers WJ, Sun MZ, Stafford M, Phillips CT, Barres BA. . The lipid sulfatide is a novel myelin-associated inhibitor of CNS axon outgrowth. J Neurosci. 2011; 31: 6481– 6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anderson DM, Mills D, Spraggins J, Lambert WS, Calkins DJ, Schey KL. . High-resolution matrix-assisted laser desorption ionization-imaging mass spectrometry of lipids in rodent optic nerve tissue. Mol Vis. 2013; 19: 581– 592. [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson DM, Ablonczy Z, Koutalos Y,et al. High resolution MALDI imaging mass spectrometry of retinal tissue lipids. J Am Soc Mass Spectrom. 2014; 25: 1394– 1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. . MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007; 4: 828– 833. [DOI] [PubMed] [Google Scholar]
- 32. Wenke JL, Rose KL, Spraggins JM, Schey KL. . MALDI imaging mass spectrometry spatially maps age-related deamidation and truncation of human lens aquaporin-0. Invest Ophthalmol Vis Sci. 2015; 56: 7398– 7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grey AC, Schey KL. . Age-related changes in the spatial distribution of human lens alpha-crystallin products by MALDI imaging mass spectrometry. Invest Ophthalmol Vis Sci. 2009; 50: 4319– 4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han J, Schey KL. . MALDI tissue imaging of ocular lens alpha-crystallin. Invest Ophthalmol Vis Sci. 2006; 47: 2990– 2996. [DOI] [PubMed] [Google Scholar]
- 35. Zemski Berry KA, Gordon WC, Murphy RC, Bazan NG. Spatial organization of lipids in the human retina and optic nerve by MALDI imaging mass spectrometry. J Lipid Res. 2014; 55: 504– 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caprioli RM, Farmer TB, Gile J. . Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997; 69: 4751– 4760. [DOI] [PubMed] [Google Scholar]
- 37. Grey AC, Crouch RK, Koutalos Y, Schey KL, Ablonczy Z. . Spatial localization of A2E in the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2011; 52: 3926– 3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaden H, Stuermer CAO, Bahr M. . GAP-43 immunoreactivity and axon regeneration in retinal ganglion-cells of the rat. J Neurobiol. 1994; 25: 1570– 1578. [DOI] [PubMed] [Google Scholar]
- 39. Moya KL, Benowitz LI, Jhaveri S, Schneider GE. . Changes in rapidly transported proteins in developing hamster retinofugal axons. J Neurosci. 1988; 8: 4445– 4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meiri KF, Pfenninger KH, Willard MB. . Growth-associated protein, gap-43, a polypeptide that is induced when neurons extend axons, is a component of growth cones and corresponds to pp46, a major polypeptide of a subcellular fraction enriched in growth cones. Proc Natl Acad Sci U S A. 1986; 83: 3537– 3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Doster SK, Lozano AM, Aguayo AJ, Willard MB. . Expression of the growth-associated protein gap-43 in adult-rat retinal ganglion-cells following axon injury. Neuron. 1991; 6: 635– 647. [DOI] [PubMed] [Google Scholar]
- 42. Berry M, Carlile J, Hunter A. . Peripheral nerve explants grafted into the vitreous body of the eye promote the regeneration of retinal ganglion cell axons severed in the optic nerve. J Neurocytol. 1996; 25: 147– 170. [DOI] [PubMed] [Google Scholar]
- 43. Ng TF, So KF, Chung SK. . Influence of peripheral nerve grafts on the expression of GAP-43 in regenerating retinal ganglion cells in adult hamsters. J Neurocytol. 1995; 24: 487– 496. [DOI] [PubMed] [Google Scholar]
- 44. Anderson DMG, Lambert W, Calkins DJ,et al. Imaging MS of rodent ocular tissues and the optic nerve. Methods Mol Biol. 2017; 1618: 15– 27. [DOI] [PubMed] [Google Scholar]
- 45. Hankin JA, Barkley RM, Murphy RC. . Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007; 18: 1646– 1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jones EA, Deininger S-O, Hogendoorn PCW, Deelder AM, McDonnell LA. . Imaging mass spectrometry statistical analysis. J Proteomics. 2012; 75: 4962– 4989. [DOI] [PubMed] [Google Scholar]
- 47. Sud M, Fahy E, Cotter D,et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007; 35: D527– 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fahy E, Subramaniam S, Murphy RC,et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009; 50: S9– S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McDonnell LA, van Remoortere A, van Zeijl RJ, Deelder AM. . Mass spectrometry image correlation: quantifying colocalization. J Proteome Res. 2008; 7: 3619– 3627. [DOI] [PubMed] [Google Scholar]
- 50. Hullin-Matsuda F, Luquain-Costaz C, Bouvier J, Delton-Vandenbroucke I. . Bis(monoacylglycero)phosphate, a peculiar phospholipid to control the fate of cholesterol: implications in pathology. Prostaglandins Leukot Essent Fatty Acids. 2009; 81: 313– 324. [DOI] [PubMed] [Google Scholar]
- 51. Gallala HD, Sandhoff K. . Biological function of the cellular lipid BMP-BMP as a key activator for cholesterol sorting and membrane digestion. Neurochem Res. 2011; 36: 1594– 1600. [DOI] [PubMed] [Google Scholar]
- 52. Obrien JS, Sampson EL. . Lipid composition of normal human brain–gray matter white matter and myelin. J Lipid Res. 1965; 6: 537– 544. [PubMed] [Google Scholar]
- 53. Xiao SY, Finkielstein CV, Capelluto DGS. . The enigmatic role of sulfatides: new insights into cellular functions and mechanisms of protein recognition. : Capelluto DGS, . Lipid-Mediated Protein Signaling. New York, NY: Springer; 2013: 27– 40. [DOI] [PubMed] [Google Scholar]
- 54. Takahashi T, Suzuki T. . Role of sulfatide in normal and pathological cells and tissues. J Lipid Res. 2012; 53: 1437– 1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jeon SB, Yoon HJ, Park SH, Kim IH, Park EJ. . Sulfatide, a major lipid component of myelin sheath, activates inflammatory responses as an endogenous stimulator in brain-resident immune cells. J Immunol. 2008; 181: 8077– 8087. [DOI] [PubMed] [Google Scholar]
- 56. Eckhardt M. . The role and metabolism of sulfatide in the nervous system. Mol Neurobiol. 2008; 37: 93– 103. [DOI] [PubMed] [Google Scholar]
- 57. Schnaar RL. . Brain gangliosides in axon-myelin stability and axon regeneration. FEBS Lett. 2010; 584: 1741– 1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vyas AA, Patel HV, Fromholt SE,et al. Gangliosides are functional nerve cell ligands for myelin-associated glycoprotein (MAG), an inhibitor of nerve regeneration. Proc Natl Acad Sci U S A. 2002; 99: 8412– 8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sandhoff K, Harzer K. . Gangliosides and gangliosidoses: principles of molecular and metabolic pathogenesis. J Neurosci. 2013; 33: 10195– 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kurimoto T, Yin YQ, Habboub G,et al. Neutrophils express oncomodulin and promote optic nerve regeneration. J Neurosci. 2013; 33: 14816– 14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yin Y, Cui Q, H-y, Gilbert et al. Oncomodulin links inflammation to optic nerve regeneration. Proc Natl Acad Sci U S A. 2009; 106: 19587– 19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yin YQ, Henzl MT, Lorber B,et al. Oncomodulin is a macrophage-derived signal for axon regeneration in retinal ganglion cells. Nat Neurosci. 2006; 9: 843– 852. [DOI] [PubMed] [Google Scholar]
- 63. Lotan M, Schwartz M. . Cross talk between the immune system and the nervous system in response to injury: implications for regeneration. FASEB J. 1994; 8: 1026– 1033. [DOI] [PubMed] [Google Scholar]
- 64. Lazarov-Spiegler O, Solomon AS, Zeev-Brann AB, Hirschberg DL, Lavie V, Schwartz M. . Transplantation of activated macrophages overcomes central nervous system regrowth failure. FASEB J. 1996; 10: 1296– 1302. [DOI] [PubMed] [Google Scholar]
- 65. David S, Bouchard C, Tsatas O, Giftochristos N. . Macrophages can modify the nonpermissive nature of the adult mammalian central nervous system. Neuron. 1990; 5: 463– 469. [DOI] [PubMed] [Google Scholar]
- 66. Frank M, Wolburg H. . Cellular reactions at the lesion site after crushing of the rat optic nerve. Glia. 1996; 16: 227– 240. [DOI] [PubMed] [Google Scholar]
- 67. Blaugrund E, Duvdevani R, Lavie V, Solomon A, Schwartz M. . Disappearance of astrocytes and invasion of macrophages following crush injury of adult rodent optic nerves: implications for regeneration. Exp Neurol. 1992; 118: 105– 115. [DOI] [PubMed] [Google Scholar]
- 68. Ahmed Z, Aslam M, Lorber B, Suggate EL, Berry M, Logan A. . Optic nerve and vitreal inflammation are both RGC neuroprotective but only the latter is RGC axogenic. Neurobiol Dis. 2010; 37: 441– 454. [DOI] [PubMed] [Google Scholar]
- 69. Lutz AB, Barres BA. . Contrasting the glial response to axon injury in the central and peripheral nervous systems. Dev Cell. 2014; 28: 7– 17. [DOI] [PubMed] [Google Scholar]
- 70. Werth N, Schuette CG, Wilkening G, Lemm T, Sandhoff K. . Degradation of membrane-bound ganglioside GM2 by β-hexosaminidase A: stimulation by gm2 activator protein and lysosomal lipids. J Biol Chem. 2001; 276: 12685– 12690. [DOI] [PubMed] [Google Scholar]
- 71. Tanaka T, Ueno M, Yamashita T. . Engulfment of axon debris by microglia requires p38 MAPK activity. J Biol Chem. 2009; 284: 21626– 21636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hosmane S, Tegenge MA, Rajbhandari L,et al. Toll/interleukin-1 receptor domain-containing adapter inducing interferon-beta mediates microglial phagocytosis of degenerating axons. J Neurosci. 2012; 32: 7745– 7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Qu J, Jakobs TC. . The time course of gene expression during reactive gliosis in the optic nerve. PLoS One. 2013; 8: e67094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vargas ME, Barres BA. . Why is Wallerian degeneration in the CNS so slow? Ann Rev Neurosci. 2007; 30: 153– 179. [DOI] [PubMed] [Google Scholar]
- 75. Brosius Lutz A, Barres BA. Contrasting the glial response to axon injury in the central and peripheral nervous systems. Dev Cell. 2014; 28: 7– 17. [DOI] [PubMed] [Google Scholar]
- 76. Vargas ME, Watanabe J, Singh SJ, Robinson WH, Barres BA. . Endogenous antibodies promote rapid myelin clearance and effective axon regeneration after nerve injury. Proc Natl Acad Sci U S A. 2010; 107: 11993– 11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kihara Y, Mizuno H, Chun J. . Lysophospholipid receptors in drug discovery. Exp Cell Res. 2015; 333: 171– 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kihara Y, Maceyka M, Spiegel S, Chun J. . Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br J Pharmacol. 2014; 171: 3575– 3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.