Summary
Enhancers instruct spatio-temporally specific gene expression in a manner tightly linked to higher-order chromatin architecture. Critical chromatin architectural regulators condensin I and condensin II play non-redundant roles controlling mitotic chromosomes. But aspects of interphase condensin loading and function, or in transcription regulation remain unclear. Here we report that both condensin complexes exhibit an unexpected, dramatic estrogen-induced recruitment to estrogen receptor α (ER-α)-bound eRNA+ active enhancers in interphase breast cancer cells, exhibiting non-canonical interaction with ER-α via its DNA-binding domain (DBD). Condensins positively regulate ligand-dependent enhancer activation, at least in part, by recruiting an E3 ubiquitin ligase, HECTD1, to modulate the binding of enhancer-associated coactivators/corepressors, including p300 and RIP140, permitting full eRNA transcription, formation of enhancer:promoter looping and the resultant coding gene activation. Collectively, our results reveal an important, unanticipated transcriptional role of interphase condensins in modulating estrogen-regulated enhancer activation and coding gene transcriptional program.
Graphical abstract

INTRODUCTION
Enhancers empower the genome with a precise control of temporally and spatially necessary gene expression patterns (Plank and Dean, 2014). The recent discovery of pervasive transcription of enhancer RNAs (eRNAs) revealed enhancers themselves as transcription units (Kim et al., 2010). eRNA levels correlate highly with enhancer activities (Andersson et al., 2014; Hah et al., 2013; Kim et al., 2010; Melgar et al., 2010; Wang et al., 2011; Wu et al., 2014), and both enhancer transcription and transcripts were found to contribute to enhancer function (Hsieh et al., 2014; Kaikkonen et al., 2013; Lai et al., 2013; Lam et al., 2013; Li et al., 2013a; Melo et al., 2013; Mousavi et al., 2013; Pnueli et al., 2015; Schaukowitch et al., 2014), adding an important layer of understanding into the fundamental mechanisms underlying enhancer action (Lam et al., 2014). However, the molecular mechanisms control the appropriate transcriptional output of enhancers and subsequent activation of coding genes remain elusive.
The long-range nature of enhancer functions tightly connects their regulation to chromatin architectures (Plank and Dean, 2014). Cohesin has recently been shown to positively regulate transcription by modulating enhancer function and enhancer-promoter looping (Kagey et al., 2010; Li et al., 2013a; Schmidt et al., 2010), raising the possibility that other architectural complexes important in mitosis/meiosis, particularly condensins, might as well assume critical roles on enhancers and/or in transcription regulation (A.J. et al., 2010; Hirano, 2012). Condensins are highly conserved multi-subunit complexes containing structural maintenance of chromosome (SMC) proteins. Together with two other such SMC-containing complexes - cohesin and SMC5/SMC6 complexes, they contribute to the formation, maintenance and dynamics of eukaryotic chromosome architecture (A.J. et al., 2010; Hirano, 2012; Jeppsson et al., 2014). In vertebrates, two related condensin I and II pentameric complexes (Figure 1A), exhibiting similar topological structures (A.J. et al., 2010; Hirano, 2012), play non-overlapping but critical roles for chromosome packing in mitosis (Green et al., 2012; Hirano, 2012; Ono et al., 2003). Compared to roles in mitosis, less is known about condensin functions in interphase. Condensin I was originally considered mainly cytoplasmic during interphase, whereas condensin II has been recognized to exhibit a nuclear localization, thought to concentrate on chromatin until prophase (Hirano, 2012; Ono et al., 2003). In particular, it remains largely unclear where condensin I and condensin II are localized in the interphase chromatin, how do they get recruited and exert their functions, if any, in transcription regulation.
Figure 1. Estrogen-induced loading of condensins to ER-α-bound active enhancers.
(A) A cartoon diagram showing the subunit constituents of the condensin I and condensin II complexes. (B) Chromatin fractionation followed by Western blots showing the localization of condensin subunits in MCF-7 cells upon E2 or ICI treatment. (C) Venn diagram showing the genome-wide ChIP-Seq peak numbers of NCAPG and NCAPH2, and their overlap with that of ER-α in E2-treated MCF-7 cells. (D) Heatmaps showing ChIP-Seq data of condensin I (NCAPG, NCAPH, NCAPG (Y.K.)) and condensin II (NCAPH2) together with p300, RNA Pol II, active histone marks H3K4me2 and H3K27Ac on active enhancers (n=1,248) in MCF-7 cells (−/+E2, with scales indicated. The map was sorted vertically by the binding intensity of ER-α. (E) A snapshot of the UCSC genome browser (hg18) showing the ChIP-Seq tracks of condensins subunits, ER-α, input control, and GRO-Seq (+ and − denote the transcription of two strands) in TFF1 locus (signals under ±E2 treatment are represented by two colours). (F,G) Profile plots showing normalized ChIP-Seq or GRO-Seq tag intensities (±E2) of ER-α, NCAPG, NCAPH2, p300, RNA Pol II and eRNAs on the active enhancer group (n=1,248) compared to "primed enhancers" (n=5,763), see Figure S2A for other features of these two groups. TFF1e: TFF1 enhancer (an intronic enhancer localized in the TMPRSS3 gene). (H) Hierarchical cluster analysis showing the correlation between the E2-induced recruitment of the interrogated transcription factors and histones modifications on the 1,248 active enhancers. Pairwise Pearson correlation coefficients (PCC, scaled on top of the heatmap) between samples are shown. The heatmap with red-green gradient denotes the fold (log2) of induction in response to E2 (scale indicated). All heatmaps and profiles are centered on ER-α binding sites in +E2 situation. Also see Figures S1 and S2, Table S1.
In this study, we found that, surprisingly, multiple condensin I and condensin II subunits are rapidly, specifically and strongly recruited to ER-α-bound functionally active enhancers in response to estrogen stimulation in human breast cancer cells. The loading of interphase condensins to these active enhancers was likely achieved by interaction with ER-α via the DNA-binding domain (DBD) of the latter. Mechanistically, condensins were required for full ligand-activated eRNA transcription, at least in part based on its recruitment of an E3 ubiquitin ligase HECTD1, which modulates proper recruitment of transcriptional coactivators and corepressors via ubiquitinating and dismissing a specific corepressor RIP140. This hierarchical control then licenses RNA polymerase II (Pol II) loading to enhancers, eRNA transcription and enhancer:promoter chromosomal interactions, together lead to up-regulation of target coding genes. Our current data has thus identified an unexpected, enhancer-based important function of condensin complexes in regulated transcriptional control, which is likely to be required for at least some other classes of DNA binding transcription factors in diverse cell types.
RESULTS
Loading of interphase condensin I and II to ER-α binding sites in breast cancer cells
About 10–30% and ~50% of condensin I and condensin II proteins, respectively, were found as chromatin-associated in MCF-7 breast cancer cells, consistent with findings in other cell types (Heale et al., 2006; Ono et al., 2003) (Figure 1B). Following 3–4 days of culture in serum-deficient ”stripping” medium, MCF-7 cells were largely (~80–95%) blocked in the G0/G1 phase(Villalobos et al., 1995) (Figure S1A), in contrast to the status without stripping (~42% G0/G1, Figure S3A). Even double thymidine block did not further enrich cells in G1/S (Figure S1A), providing an ideal cell line model to study potential interphase functions of condensins. ChIP-Seq with an antibody against the NCAPG subunit of condensin I identified 2,916 peaks genome-wide in cells cultured in the absence of estrogen treatment (i.e. 100nM of 17-β-estradiol or E2), strikingly increasing to 7,292 peaks 1hr after E2 treatment (Figure 1C and Figure S1C). NCAPG in majority bound to intergenic (55%) and intronic regions (36%), with only ~4.6% located on RefSeq gene promoters (Figure S1B). Remarkably, ~77% (5,623/7,292) of all NCAPG binding peaks overlapped with those of ER-α (Figure 1C); and ESR1/ERE was the most enriched motif for all the NCAPG binding sites by HOMER motif analysis (Heinz et al., 2010) (Figure S1D). Analogous experiments for condensin II (i.e. NCAPH2), revealed similar enrichment to intergenic/intronic regions (Figure 1C), remarkable gain of peaks after E2 treatment (3,636 to 10,192) (Figure S1E) and high overlap with ER-α binding (Figure 1C and Figure S1F). Specificity of the antibodies was confirmed as the knockdown of the mRNAs encoding these two proteins by small interfering RNAs (siRNAs) resulted in dramatic reduction of their binding (Figure S1G). ChIP-Seq by a specific antibody against NCAPH, another subunit of condensin I (Figure 1A), despite being less robust, also yielded predominantly intergenic and intronic locations, consistent with NCAPG results (Figure S1B), overlapping ER-α binding sites (Figure 1D,E and Figure S1H). E2 treatment caused a switch of the motif enriched on NCAPH-bound intergenic sites to ERE (Figure S1I). These ChIP-Seq results were yet further confirmed by using a second antibody from independent source against NCAPG (NCAPG (Y.K.), a generous gift from K. Yokomori) (Figure 1D,E). A representative UCSC browser screenshot is shown in Figure 1E, with the results confirmed by ChIP-qPCRs on several regions (Figure S1G,J,K).
Interphase condensins preferentially enrich to ER-α-bound, eRNA+ active enhancers
Previous work from our lab (Li et al., 2013a) and others (Hah et al., 2013) established that a subgroup of ER-α/H3K27Ac co-bound enhancers (n=1,248, Table S1), exhibiting E2-upregulated eRNAs, high intensity of ER-α binding and close proximity to estrogen target genes, constitute the major E2-activated functional enhancers in MCF-7 cells, referred to as E2-induced "active enhancers" or “eRNA+ enhancers” (Figure S2A). Besides these active enhancers (n=1,248), ER-α/H3K27Ac co-bound sites contained another group, which we referred to as non-active/“ primed" enhancers, displaying no significant eRNA induction, a lower ER-α binding intensity and lack of Pol II or p300 increase in response to E2 (n=5,763, Figure S2A); these "primed" enhancers also exhibited higher levels of H3K27me3 (data not shown).
ChIP-Seq analyses showed that both condensin I (NCAPG) and condensin II (NCAPH2) were strongly induced by E2 to bind the active enhancers (Figure 1D,E,F and Figure S2B), but not the primed cohort (Figure S2A and Figure 1G), and their binding on active enhancers was highly correlated with that of ER-α (Figure S2C). By analysing published ChIA-PET datasets, we found that the ER-α binding sites involved in chromosomal looping (Fullwood et al., 2009) exhibited stronger recruitment of NCAPG and NCAPH2 than those without looping (Figure S2D), further suggesting condensins’ enrichment on functional enhancers. When we compared the binding of condensins at active enhancers to that of other transcription factors/cofactors and histone marks (listed in Supplemental Experimental Procedures), we found that NCAPG and NCAPH2 exhibited the most dramatically induced recruitment in response to E2, similar to that of SRC2 and SRC3, the classical ER-α coactivators (CoA), and ER-α itself (Figure S2E). A hierarchical cluster analysis (HCA) of the E2-induced binding of these factors indicated that condensins exhibited the highest correlation with ER-α (Figure 1H). Together, these data reveal that both condensin I and II complexes are preferentially recruited to ER-α-regulated, functionally active enhancers upon E2 treatment; and exhibited the most dramatic induction in response to ligand.
As a comparison, we also examined NCAPG recruitment in mitotic MCF-7 cells. The binding of NCAPG to ER-α-bound sites was partially diminished in asynchronized MCF-7 cells that contain ~30% mitotic cells (Figure S3A vs. Figure S1A), as exemplified by TFF1 locus (Figure S3B, purple peaks, vs. Figure 1E), which further decreased to very minimal levels in mitosis-enriched cells (Figure S3B, pink peaks). This was observed on most of the active enhancers (Figure S3C vs. Figure 1D, indicating that the observed binding of condensins to active enhancers represent interphase events.
Condensins are loaded by ER-α to active enhancers in response to E2
To test for potential interactions between condensin complexes and ER-α, we first performed gel filtration using MCF-7 nuclear extract and found highly-overlapped co-fraction profiles of them in majority between ~1–1.5MDa, noting that some subunits (e.g. SMC2 and NCAPH2) and ER-α itself also present in additional fractions (Figure 2A). Co-immunoprecipitation (IP) experiments revealed that specific condensin subunits co-precipitate with ER-α (Figure 2B,C and Figure S3D). Reciprocally, both the endogenous and overexpressed ER-α can pull-down multiple condensin subunits (Figure 2D and Figure S3E), and these interactions were unlikely bridged by DNA (Figure 2C). ChIP-Western result suggests that NCAPG/ER-α interaction takes place on chromatin (Figure 2E). Importantly, condensin/ER-α interaction was not disrupted when lysine 539 of ER-α was mutated to alanine (i.e. L539A, Figure 2D), a mutation that precludes ligand-dependent binding of LxxLL-containing nuclear receptor coactivators (CoAs) and corepressors (CoRs)(Ruff et al., 2000), as exemplified by its failure to bind SRC3 or RIP140 (Figure 2D). By contrast, the DNA binding domain (DBD) of ER-α exhibited the strongest association with condensins (Figure S3F). These data suggested that condensins interact with ER-α in a non-canonical manner, very distinctive from those “LxxLL”-containing coregulators. ICI 182780 (ICI), a down-regulator of ER-α, completely abolished E2-induced condensin recruitment to active enhancers, suggesting the recruitment depends on ER-α, likely via their direct interaction (Figure 2F). Condensin protein levels were not obviously altered by E2 or ICI (Figure 1B). No direct interaction between condensin I and II complexes was detected in our co-IP experiments (Figure S3G). Consistent with this, a mass spectrometry experiment following NCAPG IP showed that NCAPG pulls down condensin I subunits and SMCs, but not condensin II constituents (Figure S3H and Table S2). Interestingly, several E3 ubiquitin ligases, as well as ubiquitin itself, were found to co-IP with NCAPG (Figure S3H and Table S2), suggesting that condensins may associate with unappreciated partners in breast cancer cells.
Figure 2. ER-α interacts with condensins.
(A) Gel filtration of E2-treated MCF-7 nuclear extracts followed by Western blots showing the elution profiles of target proteins as indicated, with Histone H4 as a control. Red asterisks denote irrelevant bands. (B) Co-IP in the whole cell lysate followed by Western blots showing that endogenous condensin subunits co-precipitated with ER-α (without Benzonase). (C) The presence of Benzonase or Ethidium Bromide (EB) did not cause detectable change of condensin/ ER-α interaction. (D) Co-IP followed by Western blots showing that overexpressed FLAG-tagged ER-α pulls down condensin subunits (Benzonase added). The L539A mutant of ER-α interacts with condensin subunits, but not with canonical coregulators SRC3 and RIP140. (E) ChIP-Western data showing that two antibodies against NCAPG pull-down ER-α. (F) ChIP-Seq profile plots (centered on ER-α binding peaks in +E2 situation) showing the binding to active enhancers of both condensin I (NCAPG) and condensin II (NCAPH2) in presence of E2 or ICI treatments. (G) Venn diagram showing the genome-wide overlap of ChIP-Seq peaks of condensin I (i.e. NCAPG) and condensin II (i.e. NCAPH2) in E2-liganded MCF-7 cells. (H) Two-step ChIP-qPCR results are shown using antibodies against condensin I and II (NCAPG and NCAPH2) and ER-α in liganded cells; experiment was repeated 3 times; "p" and "e" after gene names denote promoter and enhancer, respectively; Data are represented as mean ± s.d.. *p<0.05, **p<0.01, ***p<0.001, (Two-tailed students' T-test). IP/Co-IP experiments were performed in MCF-7 cells unless otherwise indicated. Also see Figure S3.
Condensin I and II co-localized on 5,253 sites in the genome (Figure 2G), and most of these co-bound regions are occupied by ER-α (~94%). This high overlap differs from their largely non-overlapping chromatin localization in mitosis (Green et al., 2012; Hirano, 2012; Ono et al., 2003). Two-step ChIP experiment followed by qPCR revealed that ER-α co-occupied chromatin regions were simultaneously bound by condensins (Figure 2H). Interestingly, NCAPH2 and NCAPG also reciprocally re-ChIP each other (Figure 2H). These data suggested that, although condensin I and II do not directly interact (Ono et al., 2003) (Figure S3G), they simultaneously co-occupy ER-α-bound active enhancers. But these data could not prove if both condensin I, II and ER-α all bind the same regions simultaneously. Knockdown of NCAPH subunit reduced the binding of NCAPG to ER-α binding sites (Figure S3J), suggesting that the binding of condensins happens as a complex; it also decreased the protein levels of the whole condensin I complex (Figure S3I). Condensin I knockdown (i.e. siNCAPG) did not apparently affect either the binding or complex stability of condensin II (Figure S3I,K). These data together suggested that it is unlikely to have a mix-and-match condensin complex recruited to enhancers in MCF-7 cells, but we could not exclude the possibility of any sub-stoichiometric condensin complex formed in the MCF-7 cells, and/or only present in certain genomic regions.
Condensins are required for E2-activated gene and eRNA transcriptional program
We used at least two different siRNAs to effectively knockdown multiple condensin subunits (Figure S3I and S4A–C), which resulted in significantly dampened E2 activation of multiple coding genes interrogated, as shown by RT-qPCR results of TFF1, FOXC1, SMAD7, SIAH2 and PGR (Figure 3A and Figure S4D) expression. This did not involve any changes of ER-α mRNA or protein levels (Figure S4E,F). Results from global run-on sequencing (GRO-Seq) confirmed this inhibition (Figure 3B,C). Upon depletion of either NCAPG or NCAPD3, the E2-induced fold-change (E2-FC) of all E2-upregulated coding genes is significantly reduced (Figure 3C). E2 target genes exhibited a dramatic transcriptional attenuation upon NCAPG knockdown, as shown by their whole-gene profiles (red vs. purple line, Figure S4G). Correspondingly, RNA Pol II loading was also decreased over their gene bodies (Figure S4H). Either NCAPG or NCAPD3-activated gene groups showed a high overlap with E2-upregulated targets (Figure 3D), suggesting that condensins are widely involved in E2-dependent gene activation program. In support of this, ESR1 appeared as a top Gene Ontology (GO) term for NCAPG-activated gene cohorts (Figure S4I). In addition, the two condensin complexes regulated a partially overlapping category of coding genes (Figure 3D), consistent with their partially overlapped chromatin localization (Figure 2G). Representative browser images of GRO-Seq and Pol II ChIP-Seq are shown for TFF1 locus (Figure 3B). Together, these data indicate that condensins play an important role in activating the expression of estrogen target genes, acting at the level of transcription.
Figure 3. Condensin I and condensin II control ER-α-regulated gene activation in a partially overlapping manner.
(A) RT-qPCR data showing the expression of TFF1 and FOXC1 mRNA levels in the wild-type or in cells with condensin I or condensin II knockdown ("m"" after gene names denotes mRNA). (n=5). KD, knockdown. Data are represented as mean ± s.d.. *p<0.05, **p<0.01, ***p<0.001, (Two-tailed students' T-test). (B) Genome browser image showing the results of GRO-Seq and Pol II ChIP-Seq at TFF1 locus in the presence of condensin knockdown (siNCAPG or siNCAPD3) vs siCTL transfected cells. (TFF1e, highlighted area). (C) Boxplots showing the E2-induced fold changes (E2-FC) of all the E2-upregulated coding genes in the siCTL group and siNCAPG or siNCAPD3 groups. P values were calculated by two tailed students' T-test. (D) Venn diagram showing the overlap of gene groups regulated by condensin I, condensin II, and E2, as calculated from GRO-Seq data. Also see Figures S4 and S5.
Flow cytometry experiment revealed no significant change of cell cycle after condensin depletion in MCF-7 cells (Figure S5A), excluding the possibility that the observed transcription inhibition was caused by indirect effects. This is consistent with previous results that single condensin knockdown did not obviously impact mitotic index (Ono et al., 2003).
Condensins regulate full eRNA activation and enhancer:promoter looping
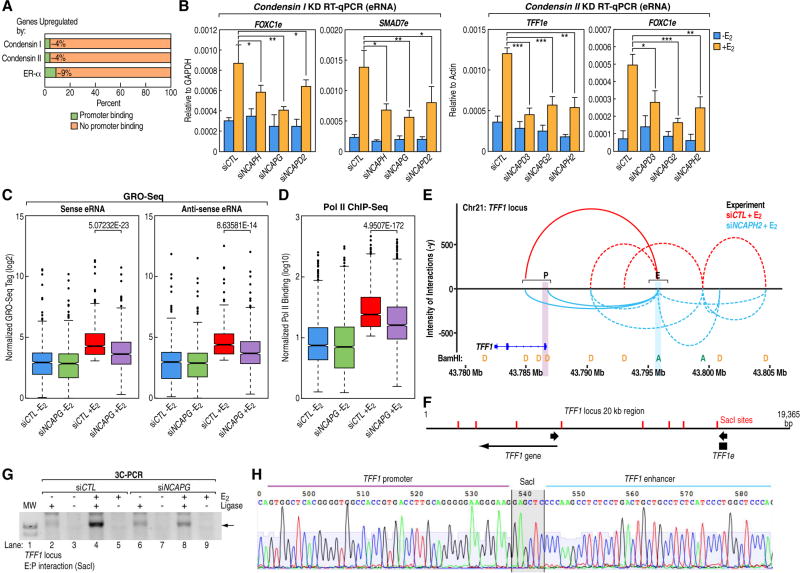
Promoter binding was a rare event for either condensin I, II or ER-α, for the coding genes they regulate (Figure 4A). This made us to focus on their possible roles acting on enhancers, given their strong enrichment there. We first tested if condensins have any roles in controlling eRNA transcription, a key mark of active enhancers (Andersson et al., 2014; Hah et al., 2013; Kim et al., 2010; Lam et al., 2014; Wang et al., 2011; Wu et al., 2014). RT-qPCR results revealed clear inhibition of eRNAs when specific condensin subunits were knocked down (Figure 4B), accompanied by reduced Pol II loading (Figure S5B). Genome-wide analyses of GRO-Seq (Figure 4C and Figure S5C) and RNA Pol II ChIP-Seq (Figure 4D) confirmed the quantitative but significant inhibition of enhancer transcription upon depletion of condensins. Screen shoots of these data for TFF1 and FOXC1 enhancers are shown (Figure 3B and Figure S5D). Interestingly, eRNA levels are higher from enhancers neighbouring to genes regulated by condensin I or II, or both, as compared to those next to genes only regulated by E2 (Figure S5E), consistent with the possibility that condensins function through modulating highly transcribing enhancers.
Figure 4. Condensins are needed for full eRNA activation and enhancer:promoter looping.
(A) Bar graphs showing the percentage (green colored) of the RefSeq genes up-regulated by condensin I, II or ER-α that possesses direct promoter-binding of the respective factor. (B) RT-qPCR data indicating the levels of representative E2-induced eRNAs when either condensin I or condensin II subunit was knocked down ("e"" after gene names denotes eRNA). (n=5). KD, knockdown. Data are represented as mean ± s.d.. *p<0.05, **p<0.01, ***p<0.001, (Two-tailed students' T-test). (C) Boxplots of normalized GRO-Seq tags showing levels of E2-induced eRNAs on the active enhancers in siCTL or siNCAPG transfected cells, from both sense and antisense directions. (Two-tailed students' T-test). (D) Boxplot of normalized ChIP-Seq tags showing RNA Pol II recruitment to active enhancers in same group of cells as in panel C. (Twotailed students' T-test). (E) Results from 3D-DSL assay showing detected chromatin interactions in the displayed region of TFF1 locus in the presence of either siCTL or siNCAPH2 (blue) (plotted by GGbio, see methods). The normalized intensities of interaction counts are plotted on the y-axis, and the x-axis depicts coordinates from UCSC browser (Hg19). Interaction data are overlaid with positions of the Donor (D, yellow) and Acceptor (A, green) BamHI sites. The pertinent E:P interactions are shown in solid lines and other interactions are shown in dotted lines. The interactions are quantitatively coded by height. Highlighted areas denote E: enhancer, P: promoter. More details can be found in the Supplemental Experimental Procedures. (F,G) A 3C-PCR assay showing the intensities of a specific E:P looping in the TFF1 locus upon either siCTL or siNCAPG treatment (±E2). The positions of SacI sites and the 3C primers are indicated in panel F. The arrow in panel G points to specific PCR product. Control 3C samples without T4 ligase are shown in G. More details can be found in the Supplemental Experimental Procedures. MW, molecular weight. (H) Sanger sequencing of the 3C-PCR product from panel G (arrow) showing that the ligated fragment comprises regions from TFF1 promoter and enhancer. Also see Figure S5 and Table S4.
In accord with the role/contribution of eRNA to the formation of enhancer:promoter looping (Hsieh et al., 2014; Lai et al., 2013; Li et al., 2013a; Pnueli et al., 2015), we found that the knockdown of a condensin subunit (siNCAPH2) inhibited enhancer:promoter (E:P) contact frequency in the TFF1 locus by 3D-DSL assays (Figure 4E, Supplemental Experimental Procedures). The effect is specific for the interrogated E:P looping (solid lines, Figure 4E), as there is no clear change of other interactions surrounding the acceptors sites (dotted lines, Figure 4E). The reduced E:P looping was confirmed by 3C-PCR assays using another restriction enzyme SacI (Figure 4F). A PCR product migrating at the predicted size (i.e. ~1.1kb) was detected only in ligated samples (Figure 4G), and exhibited clear reduction upon depletion of a condensin subunit (Figure 4G, lane 4 vs. 8). Sanger sequencing confirmed the identity of this PCR product (Figure 4H). Similar reduction of specific E:P looping was found in the FOXC1 locus by both 3D-DSL and 3C-PCR (Figure S5F–I), with concomitant decrease of the eRNA (Figure 4B and Figure S5D). As a control, a condensin-independent gene GATA3 did not exhibit obvious change of its E:P looping (Figure S5J). These data suggest that condensins play an important role during estrogen-induced enhancer activation, including allowing Pol II recruitment, eRNA transcription and E:P looping.
Condensins maintain appropriate CoA:CoR binding on active enhancers
Previous studies have revealed sequential/dynamic cofactor recruitment to classical ER-α-bound sites (Metivier et al., 2003; Shang et al., 2000), preceding Pol II loading. To examine at which step condensins act, we performed ChIPs for pioneer factor FOXA1, several conventional CoAs and CoRs, and ER-α itself, upon knockdown of condensin subunits. This resulted in no significant change of either FOXA1 or ER-α binding at active enhancers (Figure 5B and Figure S6A–C). However, E2-induced increment of p300 binding was markedly inhibited, as indicated by both ChIP-qPCR and ChIP-Seq (Figure 5A,B). Reduction was also observed for recruitment of other CoAs including SRC1, SRC3 and TIP60 (Figure S6D–F). As many of these CoAs possess histone acetyltransferase activity, it is consistent that the histone H3 lysine 27 acetylation (H3K27Ac) displayed a significant decrease on the active enhancers, corroborating the conclusion that enhancer activation was compromised (Figure 5C). The consequence also included quantitative decrease of MED1 binding, consistent with its role in both transcription activation and E:P looping (Hsieh et al., 2014; Kagey et al., 2010; Lai et al., 2013)(Figure S6G). Interestingly, loading of condensins seems to be a downstream event during enhancer activation subsequent to the trans-recruitment/assembly of the “MegaTrans” complex (Liu et al., 2014), as demonstrated by the reduction of condensin binding upon dual knockdown of RARα/RARϒ (Figure S6I), while RARα binding was unaltered when condensin was knocked down (Figure S6H). A representative genome browser image of aforementioned ChIP-Seq results is shown for the TFF1 locus (Figure 5D). To appreciate the extend of condensin effects, transient knockdown of p300 and NCAPG followed by RT-qPCR were performed, which revealed comparable inhibition of E2-target coding genes and eRNAs (Figure S6J), although partial effect of p300 knockdown could be attributed to the reduced level of ER-α itself (Figure S6J). We also tested condensins knockdown on binding of two CoRs, RIP140 and CtBP1 (Watson et al., 2012; White et al., 2005). Interestingly, while RIP140 displayed an E2-induced binding on several ER-α/condensin co-bound sites (Figure 5E), its recruitment became further augmented when condensin was depleted (Figure 5E). Knockdown of RIP140 increased transcription of several interrogated eRNAs by RT-qPCR (Figure S6K), confirming its role as a CoR. Intriguingly, a concomitant increase of CtBP1 binding was detected (Figure S6L). These observed changes of CoA and CoR binding should not be attributed to alterations of their protein amounts (Figure S6M). These data together suggest that condensins license enhancer activation by maintaining a fine balance of E2-dependent recruitment of CoAs and CoRs.
Figure 5. Condensins license appropriate coactivator (CoA) and corepressor (CoR) recruitment during enhancer activation.
(A,B) ChIP-qPCR (n=3) and ChIP-Seq profile data showing the effects of condensin knockdown on binding of p300 or ER-α. (C) ChIP-Seq profile plots showing the levels of H3K27Ac histone modification on active enhancers in control or knockdown conditions as indicated. (D) Genome browser image showing the binding of p300 and deposition of H3K27Ac, as well as ER-α in TFF1 locus in the presence or absence of NCAPG or NCAPD3. (E) ChIP-qPCR results of RIP140 showing its levels of recruitment to indicated ER-α binding sites upon siCTL or siNCAPG treatment. (n=3). Profiles in panels B and C are centered on ER-α binding sites in +E2 situation. Experiments were repeated as indicated; Data are represented as mean ± s.d.; *p<0.05, **p<0.01, ***p<0.001, (Two-tailed students' Ttest). Also see Figure S6.
Condensin-dependent recruitment of HECTD1 licenses enhancer activation
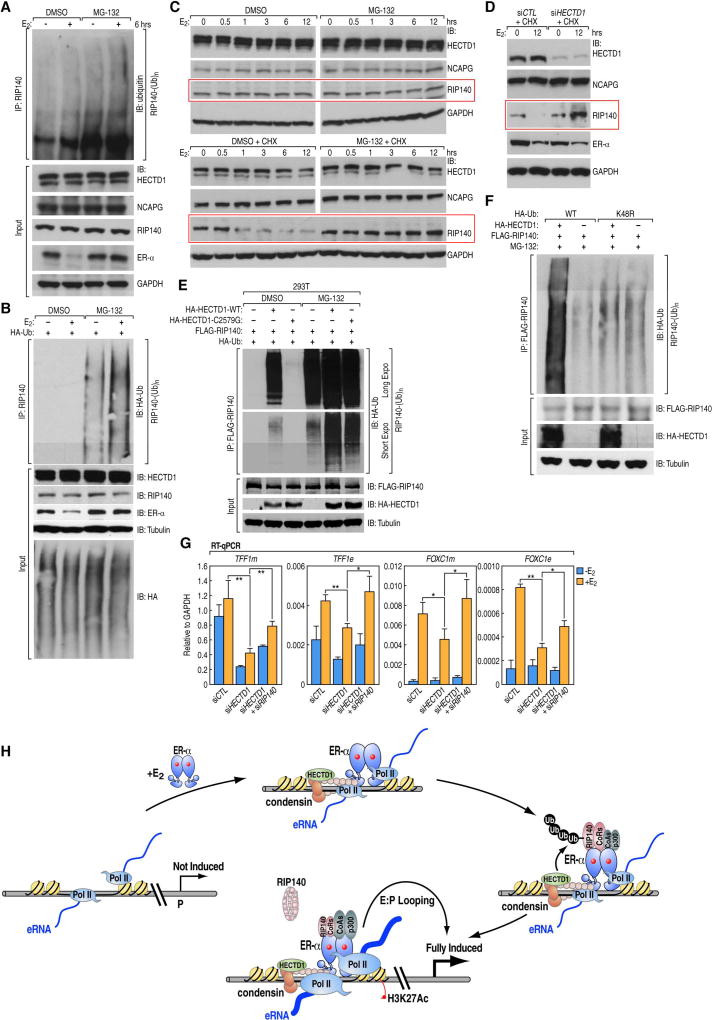
We next explored the finding that some E3 ubiquitin ligases co-immunoprecipitated with NCAPG in mass spectrometry (Figure S3H). We were particularly intrigued by a potential importance of HECTD1 (Sarkar and Zohn, 2012; Zhou et al., 2012), a member of the HECT family, which possesses several cofactors for nuclear receptors (Nawaz et al., 1999; Sun et al., 2014). Co-IP experiments confirmed the interaction between condensin subunits and HECTD1 in MCF-7 cells (Figure 6A and Figure S7A). Like condensins, HECTD1 interacted with the DBD of ER-α, and the interaction was independent of the ER-α L539 residue (Figure 6B and Figure S7B). HECTD1 elution profile in gel filtration coincided well with those of condensins (Figure 2A). Mapping of interacting domains showed that the C terminus and a central fragment of HECTD1 interact with condensin subunit NCAPH (Figure S7C,D). ChIP-qPCR using two different commercial antibodies against HECTD1, despite different affinities, both revealed an E2-induced binding to several condensin/ER-α binding sites (Figure S7E and data not shown). ChIP-Seq using one of these HECTD1 antibodies identified 3,274 peaks genome-wide in liganded MCF-7 cells, about ~45% and ~41% of which overlapping the sites of ER-α and NCAPG, respectively (Figure S7F). Heatmap analysis revealed the presence of HECTD1 and its E2-induction on the eRNA+ active enhancers (Figure 6C), as exemplified by the TFF1 locus (Figure 6D). Furthermore, HECTD1 and NCAPG binding exhibited high correlation (Figure 6E). This is an interesting observation consistent with the finding that active ubiquitination and protein proteolysis events are enriched on active enhancers (Catic et al., 2013). When we knocked down NCAPG, the binding of HECTD1 on interrogated ER-α/condensin co-bound sites was significantly reduced (Figure 6F), but the HECTD1 protein level was not affected (Figure S6M), suggesting that HECTD1 was recruited in a condensin-dependent manner. Similar to condensin knockdown, depletion of HECTD1 caused an inhibition of p300 recruitment and increase of RIP140 binding to ER-α-regulated sites (Figure 6G,H). This was accompanied by reduced transcription of eRNAs and coding genes in response to E2 (Figure 6I). To elucidate if the E3 ligase activity is important for HECTD1 function, a “rescue” experiment were performed, which showed that an HA-tagged mouse wild-type HECTD1 (mHECTD1-WT) expression plasmid could rescue, at least in part, the eRNA inhibition resulted from HECTD1 knockdown in MCF-7 cells (Figure 6J and Figure S7G, H). In contrast, a HECTD1 mutant (C2579G) that is defective of E3 ligase activity (Sarkar and Zohn, 2012) failed to produce rescue (Figure 6J and Figure S7G,H). These results together indicate that the condensin-dependent recruitment of HECTD1 is needed for full activation of eRNA transcription.
Figure 6. Condensin-dependent recruitment of HECTD1 is required for E2-induced eRNA activation.
(A) Endogenous co-IP followed by Western blots showing condensin and HECTD1 interaction using indicated antibodies. (Long Expo and Short Expo represent exposure time). Long and Short Expo indicate the lengths of exposure time. (B) Similar to experiment in Figure 2D, this panel shows the Western blots of HECTD1 following co-IPs in 293T cells transfected with FLAG-tagged ER-α or its L539A mutant. Benzonase was added. (C) A heatmap showing HECTD1 ChIP-Seq result centered at ER-α-binding active enhancers with a scale as indicated. (D) A representative genome browser image from TFF1 locus showing HECTD1 binding to ER-α binding sites (TFF1e, TFF1 enhancer). (E) ChIP-seq intensity plots ranked by NCAPG peaks showing the binding of NCAPG (red) and HECTD1 (green) at active enhancers, with the Pearson correlation coefficient shown. (F) ChIP-qPCR results showing the binding of HECTD1 with and without siNCAPG treatment (+E2); (n=3). (G,H) ChIP-qPCR results showing the recruitment of p300 and RIP140 to interrogated regions in control cells or siHECTD1 transfectants. (n=3). (I) RT-qPCR data showing the expression levels of interrogated mRNAs and eRNAs in cells with and without siHECTD1 knockdown (n=4). (J) RT-qPCR data showing the levels of interrogated eRNAs upon HECTD1 knockdown and/or rescues by overexpression of wild type mouse HECTD1 (mHECTD1-WT) or its catalytically defective mutant (mHECTD1-C2579G) (Sarkar and Zohn, 2012); (n=3). Experiments were repeated as indicated; Data are represented as mean ± s.d.; *p<0.05, **p<0.01, (Two-tailed students' T-test). Also see Figure S7.
HECTD1 polyubiquitinates and dismisses RIP140 from active enhancers
Previously, RIP140 was found polyubiquitinated in macrophages for proper inflammatory gene transcription (Ho et al., 2012). Considering that the absence of HECTD1 augmented RIP140 binding on active enhancers, we sought to test if RIP140 might be dismissed by, or a direct substrate of HECTD1. Polyubiquitination (Ubn) of RIP140 could be detected in MCF-7 cells after E2 treatment, and was enhanced by MG-132, a proteasome inhibitor (Figure 7A,B). E2 treatment did not alter the total protein levels of RIP140 in MCF-7 cells, until the addition of translation inhibitor cycloheximide (CHX) during E2 stimulation (Figure 7C). A likely explanation for this is that E2 induced polyubiquitination of RIP140 and hence its degradation, but this was counteracted by new RIP140 synthesis. When HECTD1 was knocked down in presence of CHX, the E2-triggered RIP140 reduction was disrupted (Figure 7D), suggesting HECTD1 is the enzyme responsible for RIP140 turnover. In vivo ubiquitination assays were performed in 293T cells by ectopically co-expressing Flag-tagged RIP140 and HA-tagged wild-type HECTD1 or C2579G mutants, together with either wild-type or K48R mutant ubiquitin. Wild-type but not C2579G mutant of HECTD1 promoted RIP140 polyubiquitination (Figure 7E) in the presence of wild-type ubiquitin but not K48R mutant (Figure 7F). Functionally, the reduced activation of E2 target genes and eRNAs due to siHECTD1 could be at least partially rescued by RIP140 knockdown (Figure 7G). These data together suggest that RIP140 could be one of the functional substrates of HECTD1 during E2-induced enhancer activation. A model diagram is shown in Figure 7H to depict the role of condensins and HECTD1 in regulating the E2-regulated coactivator/corepressor recruitment and enhancer activation.
Figure 7. Evidences suggesting RIP140 as a polyubiquitination substrate of HECTD1.
(A,B) Immunoblotting with indicated antibodies showing IP results using a native RIP140 antibody in wild type MCF-7 cells (A) or cells transfected with a HA-Ub plasmid (B), revealing the poly-ubiquitination ((Ub)n) of RIP140. (C) Western blots showing total protein levels of HECTD1, NCAGP and RIP140 in MCF-7 cells upon different lengths of E2 treatment with or without cycloheximide (CHX, 10ug/ml) or MG132. Red boxes highlight the changes of RIP140. (D) Western blots showing that knockdown of HECTD1 in MCF-7 cells blocked the reduction of RIP140 protein levels after E2 treatment, in the presence of CHX. Levels of other interrogated proteins are shown, too. (E,F) Western blots with indicated antibodies showing results from in vivo ubiquitination assays by ectopically co-expressing FLAG-tagged RIP140 and HA-tagged HECTD1 or C2579G mutants in the presence of wild-type or K48R mutant ubiquitin. (Long Expo and Short Expo represent exposure time). (G) RT-qPCR data showing the expression levels of interrogated mRNAs and eRNAs upon HECTD1 knockdown with or without co-depletion of RIP140. (n=3). Data are represented as mean ± s.d.; *p<0.05, **p<0.01, (Two-tailed students' T-test). Biochemical experiments were performed in MCF-7 cells unless otherwise indicated. (H) A proposed model of the role(s) of condensins on estrogen-regulated active enhancers.
DISCUSSION
Enhancer binding of condensin I and condensin II
Where are interphase condensin I and condensin II on the chromatin? Our current study reveals a previously unsuspected interphase chromatin loading of condensin I and condensin II to the ER-α-bound active enhancers, in a rapid, simultaneous manner in response to estrogen stimulus in human cancer cells. Their binding represents probably the most robust signature of the eRNA+ active enhancers than other interrogated factors or marks, distinctive from other chromatin structural molecules (e.g. cohesin). This dramatic enhancer enrichment is quite surprising especially for mammalian condensin I as it was considered to display low nuclear/chromatin abundance in interphase (Hirano, 2012). Intriguingly, chromatin-associated proteins levels of condensins do not change by E2 treatment, implying that the enhancer-bound portion was re-distributed from other regions, reminiscent of the relocation of cohesin on yeast chromatin after initial loading (Lengronne et al., 2004).
The quite high co-localization between the two condensin complexes in interphase is distinct from their "non-overlapping" localization in mitosis (Hirano, 2012; Ono et al., 2003), thus extending observations in other organisms (D'Ambrosio et al., 2008; Kim et al., 2013; Kranz et al., 2013) or a recent study in murine stem cells reporting the presence of condensin II on (super-) enhancers (Dowen et al., 2013). Interestingly, condensin II is not enriched at enhancers in Drosophila (Van Bortle et al., 2014), raising a possibility that their enhancer-based roles represent evolved functions. Importantly, our data provide insight into the poorly understood process of condensin loading to chromatin, that they are recruited to regulatory elements by interacting with a transcription factor (i.e. ER-α). Of course, it is also possible that this initial recruitment by ER-α allows subsequent direct association of condensin with enhancer DNA by other strategies, such as topological entrapment (Piazza et al., 2014).
Condensins activate eRNAs and coding genes
Functionally, GRO-Seq data reveal that condensins activate gene expression, and that they act at the level of transcription. These results are rather unexpected as condensins were long considered to "condense" chromatin, which supposedly might attenuate transcription, as exemplified by the roles of condensin-like dosage compensation complex (DCC) in X chromosome gene repression in C.elegans (A.J. et al., 2010). Interestingly, even in DCC-defective mutant worms, while expression of X genes increases, many autosomal genes seem to be reduced (Jans et al., 2009). Mechanistically, condensins modulate the activation of ER-α-bound enhancers by regulating the balanced recruitment of coactivators vs. corepressors. In turn, these events license RNA Pol II binding, eRNA transcription and enhancer full activation. The effects of condensin knockdown on eRNA transcription were comparable to those observed with sip300, but appeared quantitative, likely suggesting certain redundancy or yet unknown mechanisms underlying eRNA transcription. On the basis of a role of eRNA in enhancer:promoter looping formation and gene activation (Hsieh et al., 2014; Lai et al., 2013; Li et al., 2013a), we suggest that, the modulation of eRNAs by condensins is an important component of the full activation of coding target genes in response to regulatory signals. Consistent with this, condensin depletion reduces the intensity/stability of interrogated enhancer:promoter loopings. But our data could not clearly define if the looping defect is completely or partially a consequence of reduced eRNA levels. Indeed, there could well be a possibility that condensins directly control higher-order chromatin architecture, potentially by regulating topological domain borders (Hirano, 2012; Jeppsson et al., 2014; Van Bortle et al., 2014). This role is actually reported for Drosophila NCAPH2 by a recent study (Li et al., 2015).
Condensins and ubiquitination machinery
Mitotic condensins are thought to play structural roles in regulating chromatin, possibly through ATPase or DNA super-coiling activities (Hirano, 2012; Hudson et al., 2008; St-Pierre et al., 2009), or topological entrapment of chromosomes (Cuylen et al., 2013; Hirano, 2012). But in interphase, their regulatory mechanism on gene expression/transcription is rather elusive. Our data suggest that, at least in part, condensins exert interphase actions via recruiting specific ubiquitination machinery to control enhancer activation. These data provide insight into the long-observed dynamic/cyclic recruitment of CoAs (e.g. p300 and SRC3) and CoRs (e.g. RIP140) to nuclear receptors (Foulds et al., 2013; Metivier et al., 2003; Shang et al., 2000). In accord with this finding, a recent genome-wide study of transcription factor ubiquitination revealed that active protein turnover by ubiquitination is required for gene regulation and often takes place on active enhancer regions (Catic et al., 2013).
Condensins/HECTD1 and cancer
Finally, the functions of condensins reported here in breast cancer cells have important disease implications. Indeed, mutations or altered expression of condensin subunits are associated with several cancer types (Emmanuel et al., 2011; Leiserson et al., 2014; Murakami-Tonami et al., 2014; Ryu et al., 2007); HECTD1 expression is elevated in ER-α-positive breast cancer patients in Oncomine databases (Figure S7I), and is important for ER-α-negative breast cancer cell invasion and metastasis (Li et al., 2013b). Given the important roles of condensins acting on tightly regulated specific genomic regions (e.g. enhancers), we are tempted to propose that dysregulated temporal or spatial loading of interphase condensins may lead to aberrant gene expression, possibly underlies human cancers.
The current study therefore serves to support an enhancer-based important function exerted by interphase condensins on a specific ligand-activated transcriptional program, and extend our knowledge of eRNA transcription and enhancer function.
EXPERIMENTAL PROCEDURES
Cell culture, RNA extraction and RT-qPCR
These experiments were carried out as previously described (Li et al., 2013a). For RT-qPCRs, primer sequences are listed in Table S3. More details can be found in Supplemental Experimental Procedures.
Antibodies and siRNAs
Information of antibodies and siRNAs are included in Supplemental Experimental Procedures. Generally, two rounds of siRNA transfection were performed (Figure S4A), with efficiency of knockdown tested for every experiment performed.
3C and 3D-DSL
The procedures of 3C and 3D-DSL followed previous methods (Li et al., 2013a). 3C-PCRs were performed using SacI restriction enzyme, using primers pre-tested for their efficiency and linearity (primers listed in Table S3). For results presented, 30 cycles of PCR were performed, and lower cycles gave similar results. 3D-DSL was performed using BamHI (for 3C step), and followed by DSL using oligonucleotides listed in Table S4. More details can be found in Supplemental Experimental Procedures.
ChIP-qPCR, two-step ChIP, ChIP-Seq and GRO-Seq
Two-step ChIP was conducted following a published method (Ross-Innes et al., 2010). Other ChIP, ChIP-Seq and GRO-Seq experiments were performed as previously reported (Li et al., 2013a). More details can be found in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
The authors are grateful to Janet Hightower for assistance with figure preparation, to Dr. Majid Ghassemian from the UCSD Biomolecular/Proteomics Facility for assistance in mass spectrometry. Drs. Kyoko Yokomori and Irene Zohn kindly provided an NCAPG (Y.K.) antibody, and the HECTD1 plasmids, respectively. We thank Dr. Arshad Desai (UCSD) for critical reading of the manuscript, and Dr. Yan Zhang (La Jolla Institute for Allergy and Immunology), Dr. Weijie Lan (UCSD), Marian Chuang and Audrey Hong for experimental contributions. W. Li and W.Liu were supported by a Breast Cancer Research Program (BCRP) Postdoctoral Fellowship award from Department of Defence (DoD, BC110381) and a Susan G. Komen postdoctoral fellowship (PDF12229881), respectively. M.G.R. is an investigator with the Howard Hughes Medical Institute. This work was supported by grants from NIH to M.G.R. (DK018477, DK039949, DK074868, HL065445, NS034934, CA173903, and GM104459).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
ChIP-Seqs and GRO-Seqs datasets are deposited in GEO databases (GSE62229).
Supplemental information includes seven Supplemental Figures and corresponding legends, four Supplemental Tables, Supplemental Experimental Procedures, and Supplemental References.
AUTHOR CONTRIBUTIONS
M.G.R. and W.Li. conceived the project. W.Li, S.O. performed experiments associated with condensin I and HECTD1 with contribution from X.Z. and X.H.. Y.H. conducted condensin II related experiments with contributions from W.Liu. S.O. and W.Li contributed GRO-Seq experiments. Q.M. conducted bioinformatic analyses with contributions from D.M. (condensin II) and B.T. (3D-DSL). Technical assistance was provided by Z.L., X.S. (3D-DSL), A.Y.C. (Flow cytometry) and X.H.. K.O. and J.Z. performed deep-sequencing. W.Li and M. G. R. wrote the manuscript with contributions from Y.H. and S.O. All the authors declare no conflict of interests.
All the authors declare no conflict of interests.
References
- AJ W, AF S, BJ M. Condensin and cohesin complexity: the expanding repertoire of functions. Nature reviews. Genetics. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catic A, Suh CY, Hill CT, Daheron L, Henkel T, Orford KW, Dombkowski DM, Liu T, Liu XS, Scadden DT. Genome-wide map of nuclear protein degradation shows NCoR1 turnover as a key to mitochondrial gene regulation. Cell. 2013;155:1380–1395. doi: 10.1016/j.cell.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Metz J, Hruby A, Haering CH. Entrapment of chromosomes by condensin rings prevents their breakage during cytokinesis. Dev Cell. 2013;27:469–478. doi: 10.1016/j.devcel.2013.10.018. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Bilodeau S, Orlando DA, Hubner MR, Abraham BJ, Spector DL, Young RA. Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem cell reports. 2013;1:371–378. doi: 10.1016/j.stemcr.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel C, Gava N, Kennedy C, Balleine R, Sharma R, Wain G, Brand A, Hogg R, Etemadmoghadam D, George J, et al. Comparison of expression profiles in ovarian epithelium in vivo and ovarian cancer identifies novel candidate genes involved in disease pathogenesis. PloS one. 2011;6 doi: 10.1371/journal.pone.0017617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds CE, Feng Q, Ding C, Bailey S, Hunsaker TL, Malovannaya A, Hamilton RA, Gates LA, Zhang Z, Li C, et al. Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol Cell. 2013;51:185–199. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood M, Liu M, Pan Y, Liu J, Xu H, Mohamed Y, Orlov Y, Velkov S, Ho A, Mei P, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L, Kalitsis P, Chang T, Cipetic M, Kim J, Marshall O, Turnbull L, Whitchurch C, Vagnarelli P, Samejima K, et al. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. Journal of cell science. 2012;125:1591–1604. doi: 10.1242/jcs.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko C, Kraus W. Enhancer transcripts mark active estrogen receptor binding sites. Genome research. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale JT, Ball AR, Jr, Schmiesing JA, Kim JS, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin Y, Laslo P, Cheng J, Murre C, Singh H, Glass C. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PC, Tsui YC, Feng X, Greaves DR, Wei LN. NF-kappaB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat Immunol. 2012;13:379–386. doi: 10.1038/ni.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C-L, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney C, Lee G-SM, Chen S, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D, Ohta S, Freisinger T, Macisaac F, Sennels L, Alves F, Lai F, Kerr A, Rappsilber J, Earnshaw W. Molecular and genetic analysis of condensin function in vertebrate cells. Molecular biology of the cell. 2008;19:3070–3079. doi: 10.1091/mbc.E08-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Gladden J, Ralston E, Pickle C, Michel A, Pferdehirt R, Eisen M, Meyer B. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes & development. 2009;23:602–618. doi: 10.1101/gad.1751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson K, Kanno T, Shirahige K, Sjogren C. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nature reviews. Molecular cell biology. 2014;15:601–614. doi: 10.1038/nrm3857. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen M, Spann N, Heinz S, Romanoski C, Allison K, Stender J, Chun H, Tough D, Prinjha R, Benner C, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Zhang T, Wong NC, Davidson N, Maksimovic J, Oshlack A, Earnshaw WC, Kalitsis P, Hudson DF. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun. 2013;4:2537. doi: 10.1038/ncomms3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray J, Costa A, Bear D, Wu J, Harmin D, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz AL, Jiao CY, Winterkorn LH, Albritton SE, Kramer M, Ercan S. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol. 2013;14:R112. doi: 10.1186/gb-2013-14-10-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom U, Cesaroni M, Beringer M, Taatjes D, Blobel G, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M, Cho H, Lesch H, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen M, Kim A, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends in biochemical sciences. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Vandin F, Wu H, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2014 doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lyu X, Hou C, Takenaka N, Nguyen HQ, Ong CT, Cubenas-Potts C, Hu M, Lei EP, Bosco G, et al. Widespread Rearrangement of 3D Chromatin Organization Underlies Polycomb-Mediated Stress-Induced Silencing. Mol Cell. 2015;58:216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen A, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013a;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou Q, Sunkara M, Kutys ML, Wu Z, Rychahou P, Morris AJ, Zhu H, Evers BM, Huang C. Ubiquitylation of phosphatidylinositol 4-phosphate 5-kinase type I gamma by HECTD1 regulates focal adhesion dynamics and cell migration. J Cell Sci. 2013b;126:2617–2628. doi: 10.1242/jcs.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Merkurjev D, Yang F, Li W, Oh S, Friedman MJ, Song X, Zhang F, Ma Q, Ohgi KA, et al. Enhancer Activation Requires trans-Recruitment of a Mega Transcription Factor Complex. Cell. 2014;159:358–373. doi: 10.1016/j.cell.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melgar MF, Collins FS, Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome biology. 2010;12 doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo C, Drost J, Wijchers P, van de Werken H, de Wit E, Oude Vrielink J, Elkon R, Melo S, Léveillé N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager G, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Molecular cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami-Tonami Y, Kishida S, Takeuchi I, Katou Y, Maris J, Ichikawa H, Kondo Y, Sekido Y, Shirahige K, Murakami H, et al. Inactivation of SMC2 shows a synergistic lethal response in MYCN-amplified neuroblastoma cells. Cell cycle (Georgetown, Tex.) 2014;13 doi: 10.4161/cc.27983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z, Lonard D, Smith C, Lev-Lehman E, Tsai S, Tsai M, O'Malley B. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Molecular and cellular biology. 1999;19:1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers M, Neuwald A, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Piazza I, Rutkowska A, Ori A, Walczak M, Metz J, Pelechano V, Beck M, Haering CH. Association of condensin with chromosomes depends on DNA binding by its HEAT-repeat subunits. Nat Struct Mol Biol. 2014;21:560–568. doi: 10.1038/nsmb.2831. [DOI] [PubMed] [Google Scholar]
- Plank JL, Dean A. Enhancer Function: Mechanistic and Genome-Wide Insights Come Together. Molecular cell. 2014;55:5–14. doi: 10.1016/j.molcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Rudnizky S, Yosefzon Y, Melamed P. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin alpha-subunit gene. Proc Natl Acad Sci U S A. 2015;112:4369–4374. doi: 10.1073/pnas.1414841112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes C, Stark R, Holmes K, Schmidt D, Spyrou C, Russell R, Massie C, Vowler S, Eldridge M, Carroll J. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes & development. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff M, Gangloff M, Wurtz JM, Moras D. Estrogen receptor transcription and transactivation: Structure-function relationship in DNA- and ligand-binding domains of estrogen receptors. Breast cancer research : BCR. 2000;2:353–359. doi: 10.1186/bcr80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu B, Kim D, Deluca A, Alani R. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PloS one. 2007;2 doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Zohn I. Hectd1 regulates intracellular localization and secretion of Hsp90 to control cellular behavior of the cranial mesenchyme. The Journal of cell biology. 2012;196:789–800. doi: 10.1083/jcb.201105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA Facilitates NELF Release from Immediate Early Genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie P, Ross-Innes C, Hurtado A, Brown G, Carroll J, Flicek P, Odom D. A CTCF-independent role for cohesin in tissue-specific transcription. Genome research. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauvé V, Ratsima H, D'Amours D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Molecular cell. 2009;34:416–426. doi: 10.1016/j.molcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Sun T, Wang X, He HH, Sweeney CJ, Liu SX, Brown M, Balk S, Lee GS, Kantoff PW. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene. 2014;33:2790–2800. doi: 10.1038/onc.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Nichols MH, Li L, Ong C-TT, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome biology. 2014;15 doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos M, Olea N, Brotons JA, Olea-Serrano MF, Almodovar JMRd, Pedraza V. The E-screen assay: a comparison of different MCF7 cell stocks. Environmental Health Perspectives. 1995;103 doi: 10.1289/ehp.95103844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen M, Ohgi K, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Schwabe JW. Nuclear hormone receptor co-repressors: structure and function. Molecular and cellular endocrinology. 2012;348:440–449. doi: 10.1016/j.mce.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Yore MM, Deng D, Spinella MJ. Limiting effects of RIP140 in estrogen signaling: potential mediation of anti-estrogenic effects of retinoic acid. The Journal of biological chemistry. 2005;280:7829–7835. doi: 10.1074/jbc.M412707200. [DOI] [PubMed] [Google Scholar]
- Wu H, Nord AS, Akiyama JA, Shoukry M, Afzal V, Rubin EM, Pennacchio LA, Visel A. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS genetics. 2014;10 doi: 10.1371/journal.pgen.1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bi D, Lin Y, Chen P, Wang X, Liang S. Shotgun proteomics and network analysis of ubiquitin-related proteins from human breast carcinoma epithelial cells. Molecular and cellular biochemistry. 2012;359:375–384. doi: 10.1007/s11010-011-1031-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.