Abstract
The PURA gene encodes Pur-alpha, a 322 amino acid protein with repeated nucleic acid binding domains that are highly conserved from bacteria through humans. PUR genes with a single copy of this domain have been detected so far in spirochetes and bacteroides. Lower eukaryotes possess one copy of the PUR gene, whereas chordates possess 1–4 PUR family members. Human PUR genes encode Pur-alpha (Pura), Pur-beta (Purb) and two forms of Pur-gamma (Purg). Pur-alpha is a protein that binds specific DNA and RNA sequence elements. Human PURA, located at chromosome band 5q31, is under complex control of three promoters. The entire protein coding sequence of PURA is contiguous within a single exon. Several studies have found that overexpression or microinjection of Pura inhibits anchorage-independent growth of oncogenically transformed cells and blocks proliferation at either G1-S or G2-M checkpoints. Effects on the cell cycle may be mediated by interaction of Pura with cellular proteins including Cyclin/Cdk complexes and the Rb tumor suppressor protein. PURA knockout mice die shortly after birth with effects on brain and hematopoietic development. In humans environmentally induced heterozygous deletions of PURA have been implicated in forms of myelodysplastic syndrome and progression to acute myelogenous leukemia. Pura plays a role in AIDS through association with the HIV-1 protein, Tat. In the brain Tat and Pura association in glial cells activates transcription and replication of JC polyomavirus, the agent causing the demyelination disease, progressive multifocal leukoencephalopathy. Tat and Pura also act to stimulate replication of the HIV-1 RNA genome. In neurons Pura accompanies mRNA transcripts to sites of translation in dendrites. Microdeletions in the PURA locus have been implicated in several neurological disorders. De novo PURA mutations have been related to a spectrum of phenotypes indicating a potential PURA syndrome. The nucleic acid, G-rich Pura binding element is amplified as expanded polynucleotide repeats in several brain diseases including fragile X syndrome and a familial form of amyotrophic lateral sclerosis/fronto-temporal dementia. Throughout evolution the Pura protein plays a critical role in survival, based on conservation of its nucleic acid binding properties. These Pura properties have been adapted in higher organisms to the as yet unfathomable development of the human brain.
Keywords: Pura (Pur-alpha), Purb, Pur-beta, Purg, Pur-gamma-A and Purgamma-B, AIDS, myelodysplastic syndrome, acute myelogenous leukemia, AML, progressive multifocal leukoencephalopathy, PML, polyomavirus JC, JCV, fragile X syndrome, FXS, FMR1, amyotrophic lateral sclerosis, ALS, C9ORF72, dementia
1. Introduction
PURA is the gene encoding the sequence-specific single-stranded nucleic acid-binding protein, Pur-alpha (Bergemann et al., 1992; Ma et al., 1994). Pur-alpha (Pura) is a member of the Pur family of proteins, which consists of four members: Pur-alpha (GenBank M96684.1; GI:190749), Pur-beta (Purb) (GenBank AY039216.1; GI:14906267) (Bergemann et al., 1992) and two forms of Pur-gamma (Purg) (Variant A, GenBank AF195513.2; Variant B, GenBank AY077841) (Liu and Johnson, 2002). The GenBank entries noted here are the first of these gene coding sequences to be recorded. Convention for human genes stipulates that the gene, PURA, is in all capital letters and Italicized. The protein, Pur-alpha, is denoted as the same alphabetic symbol as the gene but not Italicised, with the first letter capitalized, and can be found listed as Pura (to be used here), Purα, Pur-alpha, Puralpha and Pur alpha. The Pura product of the PURA gene is present in every human cell type examined.
An amino acid domain constituting the major portion of each Pur family member is extraordinarily conserved in sequence from bacteria through humans. This signature Pur domain has adapted from serving a function critical in bacterial species survival to serving one or more functions related to mammalian species survival (Johnson, 2003). The coding region of PURA in humans has no introns, as is true of bacterial genes. This is also true of human PURA family members, PURB and PURG. There are introns in the non-coding region of PURA (Wortman et al., 2010) and PURG (Liu and Johnson, 2002), which may reflect their complex regulatory mechanisms in higher organisms. Sequence conservation in PURA pertains specifically to the signature Pura domain. In the human encoded protein this is a sequence of 58 residues (174 nt) repeated three times (Fig. 1). In bacteroides and spirochete Pura, this motif is present once. In the encoded Pura protein the amino terminus of this repeat contains a sequence of 26 residues (78 nt) that is particularly well conserved throughout species. The Pur domain consists of a Class 1 region of basic and aromatic residues followed by a Class 2 region of acidic and hydrophobic residues. This Pura domain formation and sequence are readily recognizable in humans and spirochetes. Other bacterial species have ORF sequences somewhat similar to the Pur domain but not so clearly recognizable. These signature Pur domains differ slightly from each other within each Pur protein and among the three different human PUR gene coding sequences.
Figure 1. Comparison of the signature Pur domain in similar proteins of diverse species from spirochetes to humans.
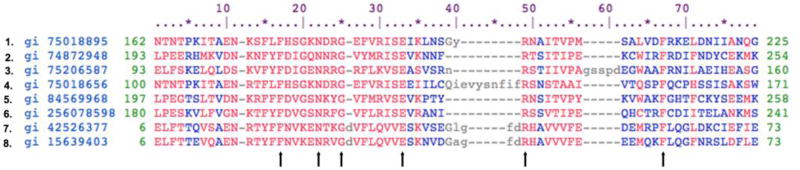
The sequences shown were aligned to genomic databases using the CDD/SPARCLE protocol with a 2-bit color identity (Marchler-Bauer et al., 2017) The sequences presented are from eight of the most diverse species containing the Pur domain. The species as numbered at left are: 1. Caenorhabditis elegans; 2. Drosophila melanogaster; 3. Arabadopsis thaliana; 5. Caenorhabditis elegans, repeat 2; 5. Homo sapiens Pura, repeat 3; 6. Schistosoma mansoni; 7. Treponema denticola; 8. Treponema pallidum. Note that all the Pur domains shown from these species align with human repeat 3 (Line 5). C. elegans, D. melanogaster and A. thaliana also have three repeats. The bacterial sequences shown have only one Pur domain. Several amino acids evolutionarily identically conserved in these domains are indicated by arrows at bottom.
The human gene hPURA is located at chromosome 5, band q31 (Ma et al., 1995; Lezon-Geyda et al., 2001). The four Pur family members are transcribed from three loci on separate chromosomes. Purg forms are exceptional in that both the A form (Purg-A) and B form (Purg-B) are transcribed from the same gene locus (Liu and Johnson, 2002). There is only one PURG promoter, one transcription start site and one PURG protein N-terminal coding sequence. Through an unusual transcription mechanism, however, two coding sequences are utilized to derive different C termini. This mechanism involves transcription extending through its PURG-A termination point and continuing to produce a 38 kb intron, which, for Purg-A, would be 3′ to the coding sequence. Upon splicing of this intron the Purg-B coding sequence is created. Although the two Purg proteins differ in their C termini, they possess three identical signature Pur domains (Liu and Johnson, 2002).
Pura is the protein family member most readily extracted from virtually every human tissue based on its preferential binding to a single-stranded DNA element (Bergemann and Johnson, 1992; Bergemann et al., 1992). Purb is less readily extracted from most human tissues. It has been most thoroughly studied in human pulmonary myofibroblasts and mouse fibroblasts as a repressor of smooth muscle actin gene transactivation (Kelm et al., 1999; Hariharan et al., 2014). Purb is expressed in the myocardium during heart failure in rabbits (Gupta et al., 2003). Currently little is known about the tissue distribution of Purg. Based on use of a human multiple tissue cDNA panel, Purg is at relatively low levels in most adult tissues. The data suggest a role for Purg-A up-regulation in tumorigenesis. Human Purg-B mRNA has thus far been detected in certain fetal and tumor cells and in human testis (Liu and Johnson, 2002). Purg is expressed at high levels in mouse embryonic brain (Khalili et al., 2003).
In higher eukaryotes, Pura is the most widely expressed of the Pur protein family members. Based on expression patterns derived from gene atlas data (see PURA gene Wikipedia), highest levels were found in the prefrontal cortex, CD4 and CD8 T cells, natural killer cells, prostate and olfactory bulb (https://commons.wikimedia.org/wiki/File%3APBB_GE_PURA_204021_s_at_fs.png) (Su et al., 2004). In mouse embryo brain Pura levels are very low, Purg being the predominant family member. Purg levels decrease as the embryo developes, and Pura levels increase to peak in the brain shortly after mouse birth. At that time homozygous PURA knockout mice die (Khalili et al., 2003). Although Pura is virtually ubiquitous, its levels within a given cell can vary greatly depending on cell cycle stage. Pura levels in mouse NIH 3T3 cells decline at the onset of S-phase and peak at mitosis (Barr and Johnson, 2001).
The signature Pur domain is detected in PUR genes of certain bacteria but apparently not in all bacterial species. It has thus far not been detected in archaebacteria. It has been detected in spirochetes and bacteroides (Fig. 1). The PUR genes of bacterial species encode only one signature Pur domain. It is not cogent to identify the bacterial PUR domain as most similar to that of any single PUR gene of higher eukaryotes. Certain lower eukaryotes, including C. elegans (Witze et al., 2009) also possess only one PUR gene. Others, including Danio rerio (zebrafish, GenBank AY450553.1) (Penberthy et al., 2004) and Drosophila melanogaster (GenBank AF021259.1) possess one PUR gene with three Pur domains, just as each human PUR gene does (Fig. 2). One cannot identify any one of the human Pur proteins, e.g., Pura, as having descended directly from a bacterial Pur. Nonetheless, because the signature Pur domain of certain bacteria is so strongly conserved throughout evolution, because these repeats comprise the majority of amino acids of all human Pur proteins, and because these bacteria, such as bacteroides, were evolutionarily present vastly earlier than were eukaryotes, it is logical to consider the Pur proteins, including Pura, as ancient proteins.
Figure 2. Sequence comparison of human Pur proteins, Pura, Purb and two forms of Purg.
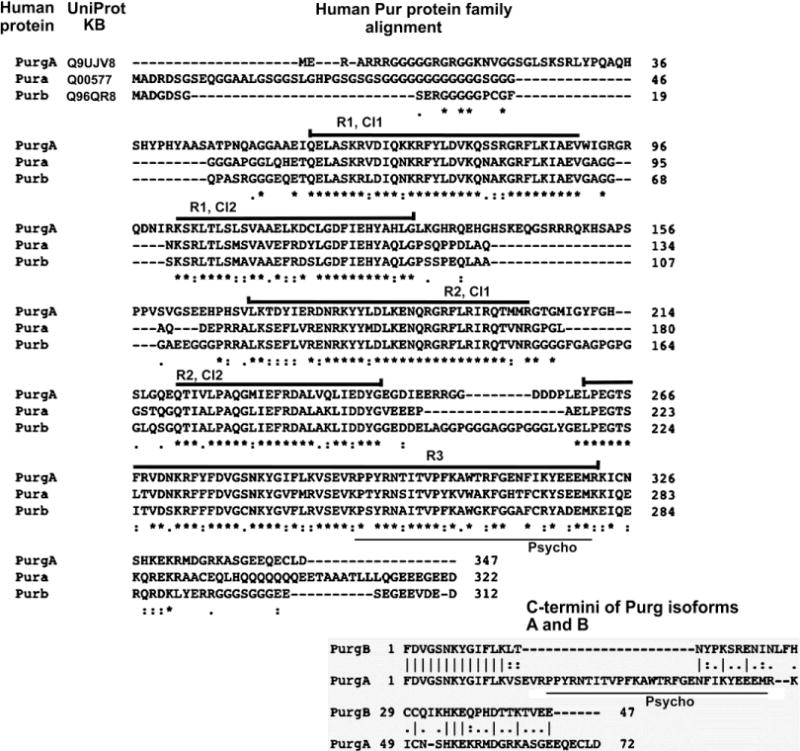
All four of the human protein Pur family members are aligned using the Clustal Omega algorithm. Pura, Purb and Purg-A are each derived from a distinct gene locus. Their entire sequences are aligned at top. Stars indicate amino acid identities; one dot indicates a substitution; two dots indicate a conservative substitution. Purg-B is transcribed from the Purg-A gene locus. Transcription of Purg-B, however, passes through the Purg-A stop point producing a 38 kb intron that is spliced out to make Purg-B mRNA. It is only the C-termini that differ between Purg-A and Purg-B. Sequences of these are presented at the bottom. Vertical lines indicate identities, and 2 dots indicate conservative substitutions. The Psycho motif is lightly underlined in both top and bottom alignments. In Pura it binds the Rb protein, among others (Ma et al., 1994; Johnson et al., 1995), and it is conserved in evolution. It is eliminated in Purg-B. Class 1 (Cl1) and Class2 (Cl2) segments of Pura repeats (R) 1, 2 and 3 are as indicated and heavily underlined.
2. Regulation of PURA gene transcription
The distinct but evolutionarily conserved roles of Pura suggest the possibility of PURA gene expression regulated differently depending on organism-dependent intracellular and external signals. Cytomegalovirus infection of mouse NIH 3T3 cells leads to a rapid decrease in levels of PURA mRNA and mouse Pura protein. Transcription of hPURA is regulated from three distinct and widely-separated transcription start sites (TSS) (Wortman et al., 2010). Each of these TSS is strongly homologous to a similar site in mouse chromosomal DNA. A mouse promoter had been identified for a start site that was subsequently determined to be TSS I (Muralidharan et al., 2001). Transcripts from TSS I and II are characterized by the presence of large and overlapping 5′-UTR introns terminated at the same splice receptor site (Wortman et al., 2010). Transcription at TSS I, most distal to the PURA coding sequence, and TSS III, located within 80 bp of the translational start codon, are upregulated by E2F1, CAAT and NF-Y binding elements (Darbinian et al., 2006; Wortman et al., 2010). Transcription at TSS II, located about 1 kb upstream of the coding sequence, is downregulated by consensus binding elements for interferon regulatory factors (IRFs), particularly IRF3 (Wortman et al., 2010). It is evident that hPURA is regulated by complex control mechanisms distinct from those of many other human genes as well as of those of bacteria.
3. Pura protein: evolutionary conservation of a DNA and RNA binding domain
3.1 Evolution of PURA and the Pur protein family
Pura was the first protein discovered in higher organisms that binds ssDNA in a sequence-specific manner. Human Pura comprises 322 amino acids. Pur-alpha binds a purine-rich strand by contacting the G residues in both ss and double-stranded (ds) DNA. It prefers to bind the sequence (G2–4N1–3)n, where N is not G. N may be any nucleotide, and n represents the number of repetitions of this small sequence. N may appear as many as three times in this element (Bergemann et al., 1992; Wortman et al., 2005). Pura binds both DNA and RNA (Gallia et al., 1999; Gallia et al., 2000). Mouse Pura differs from human Pura by only three residues, including two Gly residues in the N-terminal poly-Gly stretch (Wortman et al., 2005).
The evolutionary conservation of the Pur domain points to its importance for the survival of multiple species. This is intriguing because there is a vast difference in Pura functions in lower organisms versus humans. For example, Pur-alpha plays a role in gene transcription and RNA translation (Gallia et al., 2001; Johnson et al., 2006; White et al., 2009), processes that are coupled in bacteria, but which are in different cell compartments in mammals. In addition, Pura is essential for development of the brain in mammals (Khalili et al., 2003; Hokkanen et al., 2012; Johnson et al., 2013). In humans Pura functions as a transcriptional activator, as an RNA transport protein, in mRNA translation and as a regulator of DNA replication in the cell cycle (Johnson, 2003; White et al., 2009; Johnson et al., 2013). Pura functions regulating the smooth muscle actin gene in myofibroblasts involve interaction with family member Pur-beta (Kelm et al., 1997; Hariharan et al., 2014).
Coupling of Pura’s ability to bind nucleic acids to it ability to interact with regulatory and transport proteins may unite the requirements for Pura in all organisms. The constant in all Pura mechanisms is its ability to bind a single- strand of DNA or RNA inducing separation of double strands resulting in polynucleotide structural changes. The nucleic acid binding domains of Pura are most strongly conserved throughout species. Associated with this constant feature is the Pura ability to interact with other proteins. Pura can confer on other proteins, which themselves do not bind to polynucleotides, the ability to interact with specific nucleic acid sequences. Examples of this type of adapter protein function are Pura binding to the HIV-1 Tat and TAR RNA (Chepenik et al., 1998) as well as binding to CDK protein kinases and DNA (Barr and Johnson, 2001; Liu and Johnson, 2002). The protein binding domains of Pura are less conserved throughout evolution than are the domains for nucleic acid binding. These features of Pura highlight an unusual functional role of Pura: It serves a sequence-specific nucleic acid binding function critical for survival of many species while allowing different proteins to interact with these sequences in different species and cell types.
The best example of Pura functional interaction with both nucleic acid and protein may be its role in propagation of the AIDS virus, HIV-1. Pura binds to the HIV-1 protein, Tat (Krachmarov et al., 1996), which binds to the nascent viral transcripts at an RNA element called TAR to stimulate replication of the HIV-1 genome (Chepenik et al., 1998). Pura binds its specific single-stranded sequence element adjacent to the Tat binding site in TAR and stimulates transcription. Pura also binds Cyclin T1, a phosphorylation regulatory partner of Cdk9, necessary for Tat activity (White et al., 2009). This protein-RNA complex can stimulate phosphorylation of RNA polymerase II, thereby enhancing processivity of HIV-1 transcription (Garber et al., 1998a; Garber et al., 1998b; Bieniasz et al., 1999). Much of what we know regarding Pura activities in gene transcription and regarding Pura binding to mRNA has been gleaned from these studies on HIV-1 Tat. Little is known, however, about HIV-1 infection of hippocampal neurons, and much of what we know regarding Pura activities in mRNA transport has been derived from studies of such neurons. A potential functional pathway of Pura-RNA association, from transcriptional stimulation to mRNA translation in dendritic cytoplasm can now be delineated as studied in hippocampal neurons.
3.2 Nucleic acid binding and unwinding functions of Pura throughout evolution
Mutational analyses and structural studies of Pura have provided insight into the mechanisms of its nucleic acid binding and unwinding. The three signature Pur domains of human and mouse Pura (hPura and mPura) are 100% identical in amino acid sequence between the species, and the entire human and mouse proteins differ by only three residues. Each of the three repeated Pur domains (repeats I-III in Fig. 3) can be subdivided into a Class 1, basic and aromatic section and a Class 2, acidic and hydrophobic section. This subdivision is useful because each Class 1 section is separated from each Class 2 by a non-conserved amino acid sequence of variable length (Fig. 3). Single-stranded DNA binding analyses of point mutations in mouse Pura have identified residues in the Class 1 sections that are important for both binding and unwinding (Wortman et al., 2005). A highly conserved arginine at position 71 of mPura (72 of hPura), near the beginning of each Class 1 repeat section, is critical for binding. X-ray crystallography from a truncated version of Drosophila Pura indicates that the structure of each signature Pur domain comprises four beta strands that form one sheet followed by an alpha helix in the configuration N-ββββα-C (Graebsch et al., 2009). It had previously been demonstrated that mutation of Arg71 to Glu of mouse Purα (Arg72 of human Purα, see Fig. 2) inhibits ssDNA binding and dsDNA strand separation (Wortman et al., 2005). This Arg occurs with Lys as Arg-Lys corresponding to RK 152–153 of Pur repeat 2 and KR 229–230 of repeat 3 (Fig. 2). A basic amino acid in this position of the duo, preceding aromatic residues, is a defining feature of repeated Pur domains from bacteria through humans. Additional mutations have added evidence that the β-sheets and connecting residues of Pur domains are the interaction surfaces for ss-nucleic acid binding (Graebsch et al., 2009). Comparison of Figs. 1 and 2 shows that the helical region of human Pur repeat 3, containing the psycho motif and an amphipathic helix (Ma et al., 1994), is less conserved among repeats than is the beta-sheet region. Fig. 3 illustrates certain properties of beta-sheets and helical repeats in Pur proteins. These helical motifs are involved in protein binding, and the crystallography data suggest that repeat 3 is specifically involved in intermolecular protein-protein interactions (Graebsch et al., 2010; Johnson et al., 2013). These investigators used a truncated version of Drosophila Pura containing repeated Pura domains but lacking the N- and C-terminal regions. Such truncated Pura from either human or Drosophila retains ability to bind linear, partially dsDNA (Weber et al., 2016). It has been reported, however, that Pura lacking the C-terminal region cannot bind linear duplex DNA unless the DNA has a single-strand entry point. A bend or distortion in supercoiling would provide such entry. The physiological binding site of Pura upstream of the c-MYC gene has such a bend (Bergemann and Johnson, 1992). The C-terminal truncated Pura binds well to negatively supercoiled plasmid DNA (Wortman et al., 2005). It is notable that chromosomal DNA is primarily negatively supercoiled.
Figure 3. Diagram of reported Pura nucleic acid-binding and protein-binding regions in relation to signature Pur domains and to structural features from crystallographic studies. A. Box diagram map of nucleic acid and protein binding regions.
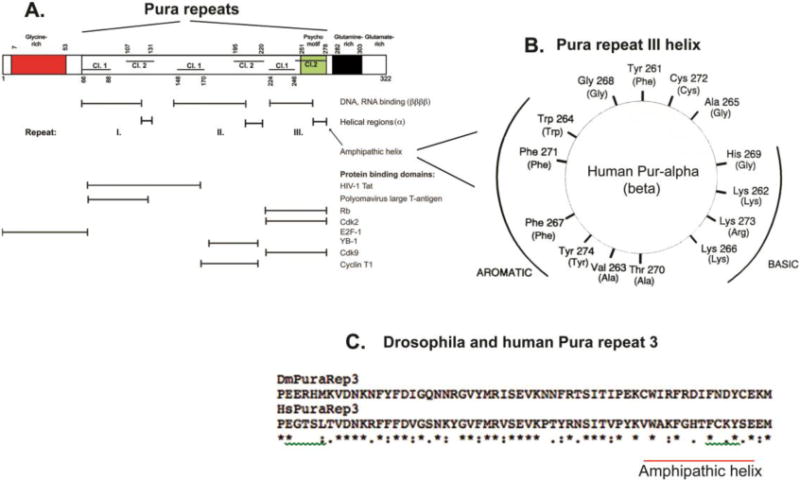
Red indicates the region containing long tracts of poly-Gly. Structure: not determined. Green indicates the psycho motif, which is present in Repeat III of eukaryotic organisms and in the only repeat of bacterial Pura (Figure 1). It is present in several other DNA replicative proteins (Ma et al., 1994). Structure: alpha helix (a). Black indicates a Gln-rich region in mammalian Pura proteins. Structure: not determined. Horizontal lines in the boxes denote the Class 1 (lower lines) and Class 2 (upper lines) portions of each Pur repeat. Note that sequences of variable length separate each repeat and the portions of Repeats I and II. X-ray crystallographic analyses of Drosophila Pura reveal that the N-terminal sequence of each repeat, comprising all of Class 1 and a part of Class 2, is structured as four beta-strands comprising a beta sheet, reported to be primarily involved in nucleic acid binding (Graebsch et al., 2009; Weber et al., 2016). The C-terminal 14–15 amino acids of each repeat are in an alpha helical structure (a) primarily involved in protein binding. Horizontal lines with vertical stops below the map denote indicated Pura regions. Positions of reported protein binding are from (White et al., 2009). B. Human Pura amphipathic helix. The wheel is a 2-dimensional depiction looking down the axis of the Repeat III 14 amino acid helix. Aromatic residues are present on one side of the helix and basic residues on the other. For comparison, the homologous region of Purb is included on the wheel. Purb has a very similar amphipathic helix, as shown by the residues in parentheses. C. Alignment comparison of human vs. Drosophila Repeat III. Alignment is with the Clustal Omega algorithm. The human Pura sequence is from Genbank M96684.1. Drosophila melanogaster is from Genbank AF021259.1. The entire gene coding sequence of human Pura is shown compared to that of Drosophila in Supplementary Fig. 1S. In Fig. 3C, stars indicate amino acid identities; one dot indicates a substitution; two dots indicate a conservative substitution. Beta sheet and helical structural regions are conserved. The helical regions in the psycho motifs differ. The human Rb binding site is altered in Drosophila. Drosophila lacks a double-sided amphipathic helix, but residues on the aromatic side of the helix are strongly conserved.(Figure 3B from Bergemann et al., 1992, Mol Cell Biol, 12, 5673–5682 used with permission.)
It may be a special property of Pura among known transcriptional regulatory proteins that it binds to both DNA and RNA to regulate transcription and that it can then remain bound to the resulting mRNA transcript as it becomes processed in the cytoplasm (Li et al., 2001; Johnson et al., 2006). Along with cytoplasmic mRNA, Pura in neurons binds to specific long, non-coding RNA (LncRNA) molecules, BC1 in mice and rats and a cognate, BC200, in humans (Kobayashi et al., 2000; Johnson et al., 2006). RNA can mediate binding of Pura to other proteins (Gallia et al., 1999). It may be that the function of these two LncRNAs is to mediate the binding of Pura to specific proteins active in mRNA transport and translation in neuronal processes.
The association of Pur proteins with different types of RNA molecules may be an adaptive mechanism of higher organisms to utilize the nucleic acid binding and unwinding properties of Pura, well-honed through evolution, to facilitate specific neuronal aspects of gene expression. In rat hippocampal neurons Pura is visualized in granular complexes (Kanai et al., 2004), where it has been shown to be present with specific mRNA molecules (Johnson et al., 2006), Fig. 4A, -B, -D. The Pura-containing granules are localized to sites of mRNA translation at junctions of dendritic branching (Fig. 4B). In rat hippocampal neurons the Pura-mRNA granules are seen exclusively in dendrites and not in axons, as determined using marker antibodies for dendritic or axonal proteins (Johnson et al., 2006), Fig. 4C. It is not known whether other neurons display this specific Pura localization to dendrites. Pura has been reported to bind to 18S ribosomal RNA and to inhibit translation of luciferase mRNA in vitro (Gallia et al., 2001). It is not known whether inhibition of translation is a general activity of Pura with bound mRNA or whether it is specific for this in vitro system. In any case, the combined results of several laboratories agree that Pura stimulates gene transcription in the nucleus, binds to the resulting mRNA transcript, accompanies this mRNA to the cytoplasm, remains bound during transport of this RNA over relatively long distances and functions at specific sites of mRNA translation. Pura is thus potentially unique in that it functions at every step of the anabolic pathway of expression of a given gene.
Figure 4. Pura visualized by laser confocal microscopy with cytoplasmic proteins in rat hippocampal neuronal dendrites.
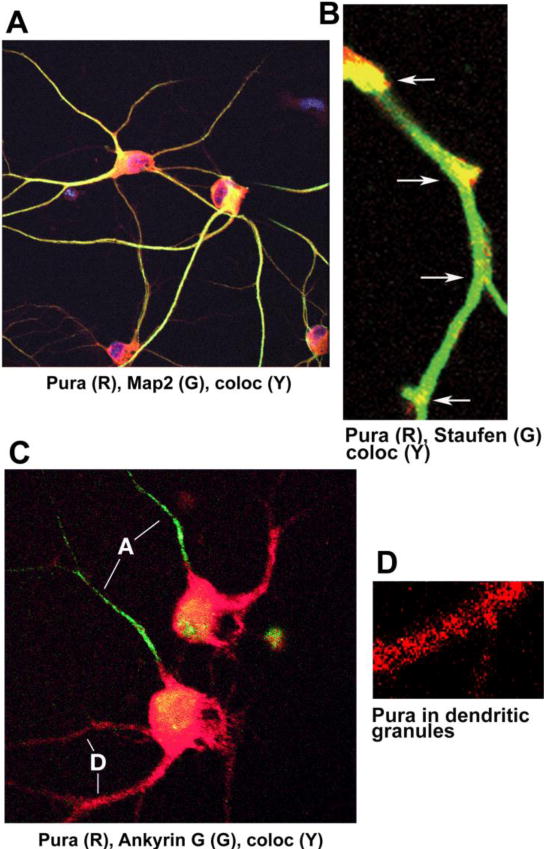
Pura localizations with proteins exemplifying aspects of dendritic mRNA transport are shown. A. Pura (red) colocalized with microtubule-associated protein 2 (Map2, green). Colocalization (yellow) is seen primarily in long neuronal processes. Map2 is a protein associated with microtubules, along which Pura- and RNA-containing granules are transported via a kinesin motor. B. Pura colocalized with Staufen in a hippocampal neuronal dendrite. Colocalized Pura (yellow) is seen at dendritic branch junctions. These junctions are reportedly sites of dendritic mRNA translation. C. Pura localized specifically in rat hippocampal neuronal dendrites, not in axons. Ankyrin G (green) is a protein marker specifically located in axons. A single green axon protrudes from each hippocampal neuron. Pura (red) is visualized in multiple protruding dendrites. Pura is not significantly present with ankyrin G in the axon. D. Granular nature of structures containing Pura (red) in a rat hippocampal neuronal dendrite. Pura is detected in such structures together with multiple proteins, including Purb, FMRP and Staufen together with long non-coding RNA BC1 (BC200 in humans).(Portions of Figures 5 & 6 from Johnson et al., 2006, J Neurosci Res, 83:6, 929–943 used with license from Journal of Neuroscience Research.)
4. Inhibitory effects of Pura on oncogenesis
Excess Pura inhibits proliferation of cancer cells (Stacey et al., 1999; Barr and Johnson, 2001; Darbinian et al., 2001). Overexpression of Pura inhibits proliferation and anchorage-independent colony formation of Ras-transformed NIH 3T3 cells (Barr and Johnson, 2001). Pura inhibition of proliferation of these cells is seen whether Pura is expressed following transfection (Barr and Johnson, 2001) or microinjected (Stacey et al., 1999). Pura may alter the cell cycle by binding to the retinoblastoma tumor suppressor gene product, Rb (Johnson et al., 1995). A hypophosphorylated form of Rb becomes hyperphosphorylated by Cyclin D/Cdk4 and Cyclin E/Cdk2, mediating release of transcription factor E2F1, which governs several processes leading to the G1-S transition and initiation of cellular DNA replication. Phosphorylation of Rb by Cyclin A/Cdk2 occurs throughout progression of S-phase. Pura specifically binds to hypophosphorylated Rb (Johnson et al., 1995; Itoh et al., 1998). The Pura effect on cancer cell proliferation is cell cycle dependent, inhibition occurring at G1-S or at G2-M checkpoints (Stacey et al., 1999). Regulation of the G2-M checkpoint is mediated by Cyclin B/Cdk1. Pura specifically binds to the Cdk1, Cdk2 and Cdk4 moieties of the respective Cyclin/Cdk complexes (Barr and Johnson, 2001) The cell cycle effects of Pura on oncogenic growth may thus be examples of Pura interaction with cyclin-dependent protein kinases either directly (Barr and Johnson, 2001; Liu et al., 2005) or indirectly through interaction with tumor suppressor Rb (Johnson et al., 1995).
5. Chromosomal alterations at the PURA locus in myelodysplastic syndrome and acute myelogenous leukemia
The localization of PURA to chromosome band 5q31 has suggested its involvement in at least one form of cancer. 5q31 is a locus frequently deleted in myelodysplastic syndrome (MDS), a potentially fatal blood disorder that may progress to acute myelogenous leukemia (AML) (Lezon-Geyda et al., 2001). Del (5q) MDS is pathologically distinct from most MDS cases and has different treatment options. Fluorescence in situ hybridization (FISH) analysis of chromosomes from patients with MDS was used to demonstrate the statistical link between this disease and deletion of PURA, at 5q31. Approximately 30% of all MDS cases possess abnormalities at 5(q). Of these, more than 90% show a deletion of PURA at 5q31. 23% show a 5q translocation, with one break point occurring in or near the PURA locus. Despite the proximity of such breaks to PURA, there is currently no evidence for mutations in the PURA coding sequence. All of the deletions noted are heterozygous, and thus haploid insufficiency likely leads to a loss of protective effect of Pura. This would imply codominant expression of both PURA alleles. PURA is not the only gene deleted in del(5q) MDS, although it is well within the most frequently deleted region (Fig. 5). Nor is any gene, including PURA, known to be the sole deletion at 5q31 in MDS. One possibility is that haploinsufficiency of several genes at 5q31 contributes to del(5q) MDS. In MDS and AML deletions of PURA are not genetically inherited but are apparently environmentally acquired. The acquired disposition of PURA to haploinsufficiency may be facilitated by other cellular agents. Translation of Pura is downregulated by certain cellular microRNAs in monocytes. These miRNAs can modulate the differentiation-dependent susceptibility of monocytic cells to HIV-1 infection (Shen et al., 2012). PURA is a potential target of miR-144, and increased expression of this miRNA may contribute to PURA downregulation in monocytes. Another chromosome disorder linked to MDS is monosomy of chromosome 7. PURB, the gene for Pur-beta, is located at 7p13. Pur-beta deletions have been visualized independently of monosomy 7 in MDS. Pur-beta deletions and translocations have each been visualized in MDS. Furthermore, simultaneous deletion of PURA at 5q31 and PURB at 7p13 is linked to enhanced progression of MDS to AML (Lezon-Geyda et al., 2001).
Figure 5. Map of PURA and nearby genes located on Chromosome 5, band 31.
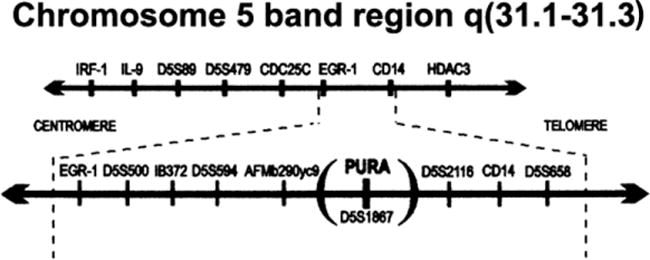
Location of the PURA gene in relation to other genes and markers that have been mapped to band region 5q31. The order of the markers shown is adopted from bacmaps at the LBNL Human Genome Center. Other genes mapped to chromosome 5 band region q31 encode proteins that include: interferon regulatory factor 1, IRF1; interleukin-9, IL-9; a CDC2 phosphatase, CDC25C; early growth response-1 zinc-finger protein, EGR-1; myelomonocytic differentiation antigen, CD14; and a histone deacetylase, HDAC3. (Figure from Lezon-Geyda et al., 2001, Leukemia, 15:6, 954–962 used with permission.)
6. Role of PURA in brain development, neuronal function and disease
The first report of PURA gene disorders in the brain came from its genetic inactivation in the mouse (Khalili et al., 2003). The PURA homozygous knockout mice experience movement disorders and die shortly after birth. Histological analysis of brain specimens from these homozygous mice reveal severe defects in brain layer development and tissue wasting. Although they survive, heterozygous PURA knockout mice exhibit seizure-like disorders (Khalili et al., 2003). Recently, Pur-alpha heterozygous deletion has been linked to memory deficits in the mouse (Barbe et al., 2016). In rat hippocampal neurons, the Pura RNA transport complex contains a number of proteins including Pur-beta and fragile X mental retardation proteins, and is driven by kinesin (Kobayashi et al., 2000; Kanai et al., 2004). This complex transports mRNAs to sites of translation at junctions of neuronal processes (Johnson et al., 2006). In association with microtubule binding protein, Map2, transport with Pura is exclusively detected in dendrites (Johnson et al., 2006).
6.1. Pura in Progressive Multifocal Leukoencephalopathy (PML)
In the brain, protein Pura plays a role in diseases involving glial cells as well as neurons. Pura is known to participate in the pathogenesis of PML, a degeneration of the oligodendroglial cells, which form the nerve sheath in the CNS (Chen et al., 1995; Krachmarov et al., 1996; White et al., 2009). PML is caused by the polyomavirus, JCV. Although the vast majority of adults are seropositive for JCV, the virus, at low infection levels in kidney epithelial tissue and bone marrow, rarely causes any symptoms. Under certain immunosuppressive conditions, however, JCV undergoes genetic changes in its non-coding replication/transcription control region and gains the ability to infect oligodendrocytes in the brain (Johnson et al., 2015). In this case Pura acts by altering both gene expression (Chen et al., 1995; Krachmarov et al., 1996; Gallia et al., 1998; White et al., 2009; Sariyer et al., 2016) and replication of JCV (Chen et al., 1995; Krachmarov et al., 1996; Wortman et al., 2000; Daniel et al., 2001; Reiss and Khalili, 2003; Daniel et al., 2004). The types of immune suppression activating JCV in glial cells include HIV-1 infection. There is a documented interaction between Pura, the HIV-1 protein, Tat, and a regulatory DNA sequence in JCV (Krachmarov et al., 1996; Daniel et al., 2001). Tat, produced by HIV-1- infected cells, can freely traverse cell membranes to act upon JCV in oligodendrocytes. Tat acts together with Pura to activate JCV DNA replication and at high concentrations to inhibit replication (Chang et al., 1996; Daniel et al., 2001; Daniel et al., 2004).
6.2. Pura in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD)
Alterations in Pura expression have been detected in central nervous system diseases not of known viral etiology. Pura is linked to ALS, otherwise known as Lou Gehrig’s disease. ALS is a neural disease of unknown etiology involving both the brain and spinal cord, resulting in progressive loss of muscle control. ALS is associated to varying degrees with FTD. There are several contributing causes of ALS/FTD. The familial form is most commonly caused by an expansion of a hexanucleotide repeat derived from the nucleotide sequence GGGGCC (Chio et al., 2012; Majounie et al., 2012; Xu et al., 2013).This expanded polynucleotide repeat occurs in the first intron of the open reading frame of a gene on chromosome 9, C9ORF72. This particular hexanucleotide repeat expansion is denoted as the C9 repeat or the C9 HRE and this form of ALS is referred to as C9 ALS. Whereas non-diseased people possess a few hexanucleotide repeats, people with ALS or FTD have been reported to possess from hundreds to thousands of such repeats (Ling et al., 2013). Pura binds similar G-rich repeats very tightly in either DNA or RNA (Bergemann and Johnson, 1992; Chepenik et al., 1998; Tretiakova et al., 1998; Gallia et al., 2000). Pura makes contact with the G-rich strand of its binding element, separating the two DNA strands. For example, Pura strand-separates human telomeric DNA, which is a repeat of the hexanucleotide TTAGGG, remaining bound only to the G-rich strand (Wortman et al., 2005). In ALS, Pura may bind the C9 HRE or its ss RNA transcripts (Xu et al., 2013; Rossi et al., 2015). As a consequence of this binding, Pura may influence the synthesis of so-called RAN translation products of this transcript repeat. RAN translation denotes Repeat-Associated, Non-AUG translation. This is aberrant because most protein translation begins at the methionine codon AUG. Such RAN translation of the transcribed C9 repeat would yield products comprising long dipeptide repeats (Ash et al., 2013; Cleary and Ranum, 2013; Flores et al., 2016). For example, RAN translation of the (GGGGCC)n repeat expansion could produce, depending on the start point within a codon, dipeptides G-A, G-P or G-R. Translation of the C-rich strand would produce additional dipeptides P-A and P-R. The dipeptides are reported to be neurotoxic, with potential differences in toxicity among the dipeptide repeats (Flores et al., 2016). Due to its documented effects on translation in neurons, it is conceivable that binding of Pura to the C9 HRE can influence the outcome of RAN translation. Reduction of Pura protein levels exacerbates certain cellular characteristics of C9 ALS. In a Drosophila model of ALS addition of Pura helps restore certain normal characteristics in cultured cells expressing the C9 HRE and rescues neurons from degeneration (Xu et al., 2013). Pura corrects neuronal changes that result from defects in the gene, FUS, that lead to ALS (Daigle et al., 2016). The mechanistic pathways of Pura function in ALS are not currently known. Peptides based on Pur sequences may help shed light on these pathways. Peptides mimicking specific domains within Pura may be able to influence important activities of the protein in ALS as well as in certain other expanded repeat diseases.
6.3. Mechanisms of Pura actions at expanded polynucleotide repeats
In discussing Pura mechanisms of action, it is useful to consider the different diseases to which these actions might apply. Several diseases have been found to involve expanded polynucleotide repeats as a causative factor. At least seven neurological disorders result from trinucleotide repeat expansion: two fragile X syndromes, FRAXA (Verkerk et al., 1991; Houdayer et al., 2002) and FRAXE (Knight et al., 1994; Murray et al., 2000), X-linked spinal and bulbar muscular atrophy (SBMA) (La Spada et al., 1994; LaFevre-Bernt and Ellerby, 2003; Sahashi et al., 2015), myotonic dystrophy (Mulders et al., 2009), Huntington’s disease (La Spada et al., 1994; Kay et al., 2015), spinocerebellar ataxia type 1 (SCA1) (Orr et al., 1993; Andres et al., 2004), and dentatorubral-pallidoluysian atrophy (DRPLA) (Koide et al., 1994; La Spada et al., 1994; Evers et al., 2011). Of these diseases, Huntington’s and DRPLA each possess a repeat of the triplet CAG, which is not a canonical Pur binding element although it may bind Pura to some extent. We shall discuss here one example of a neurological disease that does possess a canonical Pur binding element, fragile-X syndrome.
Fragile X syndrome linked to the FRAXA locus is the most common inherited genetic disease accounting for mental retardation, and it is believed to be primarily caused by the expansion of an unstable CGG repeat in the first exon of the FMR1 gene on the X chromosome (Fry and Loeb, 1994; Houdayer et al., 2002). The (GGC)n repeat is a classic Pur binding element, to which Pura binds with an extremely high affinity (Bergemann and Johnson, 1992; Wortman et al., 2005). Bound tightly to its element, Pura could conceivably influence interactions among polynucleotides. For example, the G and C repeat, in either Fragile X-associated tremor/ataxia syndrome (FXTAS) or C9 ALS, is subject to the formation of G-quartets, formations in which G-residues are mutually bonded in non-helical secondary structures. These highly stable structures could hinder gene replication, transcription or, when present in RNA, gene translation. On the other hand, G-quartets on one strand could free up a C-rich second strand for interactions. G-quartets are capable of blocking the passage of DNA- or RNA-polymerases (Rhodes and Lipps, 2015). Pura binds to the FMR1 rCGG repeats to modulate repeat-mediated neurodegeneration in a Drosophila model of FRXAS (Jin et al., 2007). A role for G-quartets in the pathology of C9 ALS has been described (Fratta et al., 2012; Ishiura and Tsuji, 2015; Maizels, 2015; Zhou et al., 2015). Pura binding to its element induces the formation of secondary structures (Wortman et al., 2005), but Pura influence specifically on G-quartets has not been examined. Pura tightly bound to its element could also influence the formation of RNA-DNA hybrid structures. RNA hybridized to double-stranded DNA forms a stable structure (R-loop) in which the complementary strand of DNA is displaced. Such R-loops could hinder gene expression in a similar fashion to G-quartets. Because Pura can unwind double-stranded polynucleotides (Wortman et al., 2005), it could facilitate the formation of G-quartets by freeing up the G-rich strand to fold back upon itself. Double-stranded DNA unwinding by Pura could also facilitate formation of R-loops by forming single stranded regions into which RNA complementary to either DNA strand can insert. Pura can bind to the C9 HRE G-rich repeat containing G-quartets, as either DNA or RNA, and may even unwind the quartets. Thus Pura, through its ability to sequence-specifically bind polynucleotides, can regulate products derived from either of the C9 HRE complementary strands. While there is evidence that the described interactions of Pura with the expanded polynucleotide repeats can occur, many other consequences of Pura binding remain possible.
6.4. Protein-protein interactions of Pura in expanded polynucleotide repeat diseases and potential effects on regulation of the PURA gene
In addition to its nucleic acid binding properties, Pura may play a role in FRX or ALS through protein-protein interactions. The protein FMR1 is colocalized with Pura in rat hippocampal neurons (Johnson et al., 2006). As distinct from Map2, colocalization of FMR1 with Pura is seen in all neuronal processes, including axons (Johnson et al., 2006). Both FMR1 and Pura have been reported as components of neuronal RNA transport granules (Kanai et al., 2004). Binding of Pura to other cellular proteins could directly affect expression of the PURA gene. Purα binds to a GC/GA-rich sequence within its own promoter and inhibits gene expression (Muralidharan et al., 2001). Similarly, binding of Pura to RNA of an expanded polynucleotide repeat could affect expression of the PURA gene. In both cases, the mechanism of action may be that the binding of Pura to a cellular constituent might reduce effective intracellular Pura levels. This reduction would trigger feedback stimulation of the PURA gene, but it is not known whether this could compensate for the sequestration of Pura. Sequestration of Pura by the C9 expanded repeat has previously been suggested as a pathological mechanism in C9 ALS and FTD (Xu et al., 2013). Expanded rGGGGCC repeats could sequester specific RNA-binding proteins, including Pura, from their normal functions, ultimately leading to cell death. The findings of Xu and colleagues suggest that Pura interaction with the expanded rGGGGCC repeats could cause neurodegeneration. Pura may be a factor in the pathogenesis of both ALS and FTD.
6.5. Mutations of the PURA gene in neurological diseases and brain development
Several groups have now reported that a variety of brain disorders are associated with microdeletions or de novo mutations localized to the PURA gene at human chromosome locus 5q31 (Brown et al., 2013; Hunt et al., 2014; Lalani et al., 2014; Tanaka et al., 2015). This association had been termed the 5q31.3 microdeletion syndrome (Shimojima et al., 2011; Hosoki et al., 2012), and it localized to a chromosome region of overlap centered on PURA. Subsequent reports described specific de novo mutations in PURA as responsible for certain severe neurological phenotypes of the 5q31.3 microdeletion syndrome (Hunt et al., 2014; Lalani et al., 2014; Tanaka et al., 2015). The mutations observed affect the Pura protein, and they include frameshifts, inframe deletions, missense and nonsense mutations. Although the phenotype of PURA microdeletion in humans is variable, common traits among them include neurodevelopmental delays, learning disabilities, hypotonia, early feeding difficulties, speech and walking defects and seizure-like movements (Hunt et al., 2014). Analyses of de novo mutations in family trios are now among the strongest evidence for a causative role of PURA in disease. Their relation to a proposed PURA syndrome is likely (Hunt et al., 2014). The issue of whether or how these mutations are causal (e.g., are they a sole cause or a significant contributor?) will benefit from more thorough characterization of molecular consequences of these mutations in rodents and humans. It is notable that certain features of human de novo PURA deletions, such as developmental delays and seizure-like movements, have also been ascribed to heterozygous PURA deletions in the mouse (Khalili et al., 2003). Such studies in humans and mice are best interpreted together to clarify phenotypes and their underlying molecular causes.
It is important to distinguish the described PURA syndrome from the previously described 5q31.3 microdeletion syndrome. One potential distinction, based on recent definitions, is that PURA syndrome is diagnosed by a PURA pathogenic sequence variant in a proband (representing 90% of such PURA-related neurodevelopmental disorders). In contrast the 5q31.3 microdeletion syndrome is caused by a non-recurrent genomic 5q31.3 deletion, which may encompass all or a part of PURA (10% of affected individuals) (Reijnders et al., 2017).
The definition of the term, PURA syndrome, in the previous paragraph must be considered in conjunction with effects of PURA gene knockouts in mice. Homozygous deletion of PURA is lethal in mice (Khalili et al., 2003), and brain development at time of death shows defects in dendritic connections in the hippocampus and cerebellum accompanied by mislamination of these brain regions (Johnson et al., 2006; Johnson et al., 2013). Analysis of cognitive deficiencies in PURA heterozygous mice reveals that neuronal losses in the hippocampus and cerebellum are accompanied by gait ataxia and significant memory deficits (Barbe et al., 2016). PURA microdeletions affecting the brain may be compared to the larger deletions observed in the blood diseases MDS and AML (Lezon-Geyda et al., 2001). These mutations arise specifically in the blood cell lineage and can involve deletion of PURA flanking sequences as well as of PURA. The chromosome alterations in MDS and AML are believed to be induced by exposure of an individual to an environmental agent targeting myeloid cell development. Benzene has specifically been identified as inducing mutations at 5q31 (Stillman et al., 2000). As is the case in 5q31-related MDS and AML, haploinsufficiency of Pura may play a role in the brain PURA microdeletions, although the role of various mutated Pura proteins remains to be determined.
7. The PURA gene conundrum
Fascination with the PURA gene stems from the curious duality of its function throughout evolution. On one hand, PUR genes are among the most conserved of all genes, being present in certain species of bacteria and in all, or nearly all, eukaryotic organisms through humans. In humans PURA is expressed in every cell type, albeit to different degrees and at different times. Such extensive expression over such extensive lineage implies that PURA serves a primitive function that is critical for species survival in a wide range of cellular life. On the other hand, PURA expression serves one of the least primitive of functions, one that is essential for that most distinctive and recently evolved milestone in higher metazoans: development of the human brain. How can the duality of PURA function in a most broadly primitive requirement for survival be reconciled with its required function in a most modern, species specific and organ specific facet of evolution?
The resolution of this conundrum resides in part in the sequence of the Pura protein and part in the sequence of the PURA gene itself. The encoded protein sequence of PURA reveals two functionally and evolutionarily distinct components of the protein. The first is a very highly conserved motif of 55–70 amino acids in various species. As discussed in section 1.2, this is the canonical Pur domain, the conservation of which is illustrated in Fig. 1. Human Pura contains three repeats of this domain. DNA- and RNA-binding studies have determined that Pur domains are functional nucleic-acid binding domains. X-ray crystallographic and database analyses show that Pur domains share a fold with the Whirly class of nucleic-acid binding proteins (Graebsch et al., 2009). The third Pur repeat is involved in dimerization of Pur proteins. Mutation of a highly conserved phenylalanine in Pura abolishes double-stranded DNA unwinding without a complete loss of Pura DNA binding (Weber et al., 2016). As shown in Fig. 1, it is the nucleic acid binding Pur repeated domains that comprise the most highly conserved regions of Pura. The second distinct component of Pura is the less-conserved remainder of the Pura protein surrounding the repeats. These approximately 110 amino acids, are largely involved in protein-protein binding (White et al., 2009). This analysis suggests that the conserved Pura nucleic acid binding and dimerization domains are essential for a function common to nearly all cell survival, whereas the less conserved protein binding regions of Pura are subject to evolutionary adaptation to new functions in diverse organisms.
The unusual sequence and structure of the hPURA gene provide evidence for the strict conservation coupled with the evolutionary adaptability that help resolve the PURA conundrum. Functions of Pura in organisms of different evolutionary status are related through an ability to interact with nucleic acids (Jin et al., 2007; Graebsch et al., 2009; Witze et al., 2009; Aumiller et al., 2012; Xu et al., 2013; Weber et al., 2016). These related but distinct Pura roles in organisms of different phyla suggest a need for expression regulated differently depending on extracellular, environmentally induced signals.
8. Conclusions and research considerations
PURA is the gene encoding Pur-alpha, Pura, a sequence-specific single-stranded DNA and RNA binding protein. Pura has distinct amino acid domains that bind polynucleotides and, simultaneously, several proteins regulating gene function. The nucleic acid binding domains of Pura serve a function that is critical for most cellular life, and these domains are highly conserved throughout evolution. The Pura nucleic acid binding domains have evolved to function in processes essential for development of the mammalian hematopoietic and central nervous systems. Mutations in PURA and alterations in Pura activity have now been implicated in multiple neurological diseases. Notably, deficiencies of Pura protein function have been implicated as a pathological factor of certain expanded polynucleotide repeat diseases, including amyotrophic lateral sclerosis, ALS. It will be enlightening to recognize any functional defect in Pura connecting these neurological diseases.
The present review highlights two directions in which investigations of PURA, the gene and gene product, are likely to proceed to yield important findings. They are: 1) Determination of the mechanism of action of Pura. What basic function of the signature Pur domain, repeated three times in hPura, is so important for life preservation that this domain is conserved from bacteria through humans? Studies of Pura action will necessitate studies of Pur family members Pur-beta and Pur-gamma, the genes of which (PURB and PURG) are regulated separately at different chromosomal loci; and 2) Illumination of the specialized regulatory structure of PURA and functions of Pura that have made this protein essential for achieving a pinnacle of evolution, the development of the mammalian central nervous system. Elucidation of the pathways of function of Pura may provide entry points to that vast uncharted frontier of molecular biological research: the working of the human brain.
Supplementary Material
Figure 1S. Comparison of Pura complete sequences from Human (H. sapiens, GB M96684.1) and Drosophila (D. melanogaster, GB AF021259.1). Alignment is with the Clustal Omega algorithm. Stars indicate amino acid identities; one dot indicates a substitution; two dots indicate a conservative substitution.
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series–a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by the National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The corresponding Gene Wiki entry for this review can be found here: https://en.wikipedia.org/wiki/PURA
REFERENCES CITED
- Andres AM, Soldevila M, Lao O, Volpini V, Saitou N, Jacobs HT, Hayasaka I, Calafell F, Bertranpetit J. Comparative genetics of functional trinucleotide tandem repeats in humans and apes. J Mol Evol. 2004;59:329–39. doi: 10.1007/s00239-004-2628-5. [DOI] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumiller V, Graebsch A, Kremmer E, Niessing D, Forstemann K. Drosophila Pur-alpha binds to trinucleotide-repeat containing cellular RNAs and translocates to the early oocyte. RNA Biol. 2012;9:633–43. doi: 10.4161/rna.19760. [DOI] [PubMed] [Google Scholar]
- Barbe MF, Krueger JJ, Loomis R, Otte J, Gordon J. Memory deficits, gait ataxia and neuronal loss in the hippocampus and cerebellum in mice that are heterozygous for Pur-alpha. Neuroscience. 2016;337:177–190. doi: 10.1016/j.neuroscience.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr SM, Johnson EM. Ras-induced colony formation and anchorage-independent growth inhibited by elevated expression of Puralpha in NIH3T3 cells. J Cell Biochem. 2001;81:621–38. doi: 10.1002/jcb.1099. [DOI] [PubMed] [Google Scholar]
- Bergemann AD, Johnson EM. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol. 1992;12:1257–65. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergemann AD, Ma ZW, Johnson EM. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol Cell Biol. 1992;12:5673–82. doi: 10.1128/mcb.12.12.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Analysis of the effect of natural sequence variation in Tat and in cyclin T on the formation and RNA binding properties of Tat-cyclin T complexes. J Virol. 1999;73:5777–86. doi: 10.1128/jvi.73.7.5777-5786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N, Burgess T, Forbes R, McGillivray G, Kornberg A, Mandelstam S, Stark Z. 5q31.3 Microdeletion syndrome: clinical and molecular characterization of two further cases. Am J Med Genet A. 2013;161A:2604–8. doi: 10.1002/ajmg.a.36108. [DOI] [PubMed] [Google Scholar]
- Chang CF, Gallia GL, Muralidharan V, Chen NN, Zoltick P, Johnson E, Khalili K. Evidence that replication of human neurotropic JC virus DNA in glial cells is regulated by the sequence-specific single-stranded DNA-binding protein Pur alpha. J Virol. 1996;70:4150–6. doi: 10.1128/jvi.70.6.4150-4156.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NN, Chang CF, Gallia GL, Kerr DA, Johnson EM, Krachmarov CP, Barr SM, Frisque RJ, Bollag B, Khalili K. Cooperative action of cellular proteins YB-1 and Pur alpha with the tumor antigen of the human JC polyomavirus determines their interaction with the viral lytic control element. Proc Natl Acad Sci USA. 1995;92:1087–91. doi: 10.1073/pnas.92.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepenik LG, Tretiakova AP, Krachmarov CP, Johnson EM, Khalili K. The single-stranded DNA binding protein, Pur-alpha, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene. 1998;210:37–44. doi: 10.1016/s0378-1119(98)00033-x. [DOI] [PubMed] [Google Scholar]
- Chio A, Restagno G, Brunetti M, Ossola I, Calvo A, Canosa A, Moglia C, Floris G, Tacconi P, Marrosu F, Marrosu MG, Murru MR, Majounie E, Renton AE, Abramzon Y, Pugliatti M, Sotgiu MA, Traynor BJ, Borghero G, Consortium, S ALS/FTD phenotype in two Sardinian families carrying both C9ORF72 and TARDBP mutations. J Neurol Neurosurg Psychiatry. 2012;83:730–3. doi: 10.1136/jnnp-2012-302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet. 2013;22:R45–51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle JG, Krishnamurthy K, Ramesh N, Casci I, Monaghan J, McAvoy K, Godfrey EW, Daniel DC, Johnson EM, Monahan Z, Shewmaker F, Pasinelli P, Pandey UB. Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity. Acta Neuropathol. 2016;131:605–20. doi: 10.1007/s00401-015-1530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel DC, Kinoshita Y, Khan MA, Del Valle L, Khalili K, Rappaport J, Johnson EM. Internalization of exogenous human immunodeficiency virus-1 protein, Tat, by KG-1 oligodendroglioma cells followed by stimulation of DNA replication initiated at the JC virus origin. DNA Cell Biol. 2004;23:858–67. doi: 10.1089/dna.2004.23.858. [DOI] [PubMed] [Google Scholar]
- Daniel DC, Wortman MJ, Schiller RJ, Liu H, Gan L, Mellen JS, Chang CF, Gallia GL, Rappaport J, Khalili K, Johnson EM. Coordinate effects of human immunodeficiency virus type 1 protein Tat and cellular protein Puralpha on DNA replication initiated at the JC virus origin. J Gen Virol. 2001;82:1543–53. doi: 10.1099/0022-1317-82-7-1543. [DOI] [PubMed] [Google Scholar]
- Darbinian N, Gallia GL, King J, Del Valle L, Johnson EM, Khalili K. Growth inhibition of glioblastoma cells by human Pur(alpha) J Cell Physiol. 2001;189:334–40. doi: 10.1002/jcp.10029. [DOI] [PubMed] [Google Scholar]
- Darbinian N, White MK, Khalili K. Regulation of the Pur-alpha promoter by E2F-1. J Cell Biochem. 2006;99:1052–63. doi: 10.1002/jcb.20872. [DOI] [PubMed] [Google Scholar]
- Evers MM, Pepers BA, van Deutekom JC, Mulders SA, den Dunnen JT, Aartsma-Rus A, van Ommen GJ, van Roon-Mom WM. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS One. 2011;6:e24308. doi: 10.1371/journal.pone.0024308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores BN, Dulchavsky ME, Krans A, Sawaya MR, Paulson HL, Todd PK, Barmada SJ, Ivanova MI. Distinct C9orf72-Associated Dipeptide Repeat Structures Correlate with Neuronal Toxicity. PLoS One. 2016;11:e0165084. doi: 10.1371/journal.pone.0165084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc Natl Acad Sci U S A. 1994;91:4950–4. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallia GL, Darbinian N, Jaffe N, Khalili K. Single-stranded nucleic acid-binding protein, Pur alpha, interacts with RNA homologous to 18S ribosomal RNA and inhibits translation in vitro. J Cell Biochem. 2001;83:355–63. doi: 10.1002/jcb.1247. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Darbinian N, Tretiakova A, Ansari SA, Rappaport J, Brady J, Wortman MJ, Johnson EM, Khalili K. Association of HIV-1 Tat with the cellular protein, Puralpha, is mediated by RNA. Proc Natl Acad Sci U S A. 1999;96:11572–7. doi: 10.1073/pnas.96.20.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallia GL, Johnson EM, Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallia GL, Safak M, Khalili K. Interaction of the single-stranded DNA-binding protein Puralpha with the human polyomavirus JC virus early protein T-antigen. J Biol Chem. 1998;273:32662–9. doi: 10.1074/jbc.273.49.32662. [DOI] [PubMed] [Google Scholar]
- Garber ME, Wei P, Jones KA. HIV-1 Tat interacts with cyclin T1 to direct the P-TEFb CTD kinase complex to TAR RNA. Cold Spring Harb Symp Quant Biol. 1998a;63:371–80. doi: 10.1101/sqb.1998.63.371. [DOI] [PubMed] [Google Scholar]
- Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998b;12:3512–27. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graebsch A, Roche S, Kostrewa D, Soding J, Niessing D. Of bits and bugs–on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure. PLoS One. 2010;5:e13402. doi: 10.1371/journal.pone.0013402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graebsch A, Roche S, Niessing D. X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface. Proc Natl Acad Sci U S A. 2009;106:18521–6. doi: 10.1073/pnas.0907990106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Sueblinvong V, Raman J, Jeevanandam V, Gupta MP. Single-stranded DNA-binding proteins PURalpha and PURbeta bind to a purine-rich negative regulatory element of the alpha-myosin heavy chain gene and control transcriptional and translational regulation of the gene expression. Implications in the repression of alpha-myosin heavy chain during heart failure. J Biol Chem. 2003;278:44935–48. doi: 10.1074/jbc.M307696200. [DOI] [PubMed] [Google Scholar]
- Hariharan S, Kelm RJ, Jr, Strauch AR. The Puralpha/Purbeta single-strand DNA-binding proteins attenuate smooth-muscle actin gene transactivation in myofibroblasts. J Cell Physiol. 2014;229:1256–71. doi: 10.1002/jcp.24564. [DOI] [PubMed] [Google Scholar]
- Hokkanen S, Feldmann HM, Ding H, Jung CK, Bojarski L, Renner-Muller I, Schuller U, Kretzschmar H, Wolf E, Herms J. Lack of Pur-alpha alters postnatal brain development and causes megalencephaly. Hum Mol Genet. 2012;21:473–84. doi: 10.1093/hmg/ddr476. [DOI] [PubMed] [Google Scholar]
- Hosoki K, Ohta T, Natsume J, Imai S, Okumura A, Matsui T, Harada N, Bacino CA, Scaglia F, Jones JY, Niikawa N, Saitoh S. Clinical phenotype and candidate genes for the 5q31.3 microdeletion syndrome. Am J Med Genet A. 2012;158A:1891–6. doi: 10.1002/ajmg.a.35439. [DOI] [PubMed] [Google Scholar]
- Houdayer C, Lourdaux J, Billette de Villemeur T, Royer-Legrain G, Bahuau M, Bonnefont JP, Feldmann D, Couderc R. Simple fluorescent PCR assay for discriminating FRAXA fully mutated females from normal homozygotes. Genet Test. 2002;6:135–9. doi: 10.1089/10906570260199410. [DOI] [PubMed] [Google Scholar]
- Hunt D, Leventer RJ, Simons C, Taft R, Swoboda KJ, Gawne-Cain M, study, D.D.D. Magee AC, Turnpenny PD, Baralle D. Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet. 2014;51:806–13. doi: 10.1136/jmedgenet-2014-102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura H, Tsuji S. Epidemiology and molecular mechanism of frontotemporal lobar degeneration/amyotrophic lateral sclerosis with repeat expansion mutation in C9orf72. J Neurogenet. 2015;29:85–94. doi: 10.3109/01677063.2015.1085980. [DOI] [PubMed] [Google Scholar]
- Itoh H, Wortman MJ, Kanovsky M, Uson RR, Gordon RE, Alfano N, Johnson EM. Alterations in Pur(alpha) levels and intracellular localization in the CV-1 cell cycle. Cell Growth Differ. 1998;9:651–65. [PubMed] [Google Scholar]
- Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–64. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM. The Pur protein family: clues to function from recent studies on cancer and AIDS. Anticancer Res. 2003;23:2093–100. [PubMed] [Google Scholar]
- Johnson EM, Chen PL, Krachmarov CP, Barr SM, Kanovsky M, Ma ZW, Lee WH. Association of human Pur alpha with the retinoblastoma protein, Rb, regulates binding to the single-stranded DNA Pur alpha recognition element. J Biol Chem. 1995;270:24352–60. doi: 10.1074/jbc.270.41.24352. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Daniel DC, Gordon J. The pur protein family: genetic and structural features in development and disease. J Cell Physiol. 2013;228:930–7. doi: 10.1002/jcp.24237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, Khalili K, Winckler B, Gordon J. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res. 2006;83:929–43. doi: 10.1002/jnr.20806. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Wortman MJ, Lundberg PS, Daniel DC. Orderly Steps in Progression of JC Virus to Virulence in the Brain. Brain Disord Ther. 2015;4 doi: 10.4172/2168-975X.S2-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–25. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kay C, Collins JA, Skotte NH, Southwell AL, Warby SC, Caron NS, Doty CN, Nguyen B, Griguoli A, Ross CJ, Squitieri F, Hayden MR. Huntingtin Haplotypes Provide Prioritized Target Panels for Allele-specific Silencing in Huntington Disease Patients of European Ancestry. Mol Ther. 2015;23:1759–71. doi: 10.1038/mt.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelm RJ, Jr, Cogan JG, Elder PK, Strauch AR, Getz MJ. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle alpha-actin promoter. J Biol Chem. 1999;274:14238–45. doi: 10.1074/jbc.274.20.14238. [DOI] [PubMed] [Google Scholar]
- Kelm RJ, Jr, Elder PK, Strauch AR, Getz MJ. Sequence of cDNAs encoding components of vascular actin single-stranded DNA-binding factor 2 establish identity to Puralpha and Purbeta. J Biol Chem. 1997;272:26727–33. doi: 10.1074/jbc.272.42.26727. [DOI] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, Amini S, Gordon J. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol. 2003;23:6857–75. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Voelckel MA, Hirst MC, Flannery AV, Moncla A, Davies KE. Triplet repeat expansion at the FRAXE locus and X-linked mild mental handicap. Am J Hum Genet. 1994;55:81–6. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Agui K, Kamo S, Li Y, Anzai K. Neural BC1 RNA associates with pur alpha, a single-stranded DNA and RNA binding protein, which is involved in the transcription of the BC1 RNA gene. Biochem Biophys Res Commun. 2000;277:341–7. doi: 10.1006/bbrc.2000.3683. [DOI] [PubMed] [Google Scholar]
- Koide R, Ikeuchi T, Onodera O, Tanaka H, Igarashi S, Endo K, Takahashi H, Kondo R, Ishikawa A, Hayashi T, et al. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nat Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- Krachmarov CP, Chepenik LG, Barr-Vagell S, Khalili K, Johnson EM. Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Pur alpha. Proc Natl Acad Sci U S A. 1996;93:14112–7. doi: 10.1073/pnas.93.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR, Paulson HL, Fischbeck KH. Trinucleotide repeat expansion in neurological disease. Ann Neurol. 1994;36:814–22. doi: 10.1002/ana.410360604. [DOI] [PubMed] [Google Scholar]
- LaFevre-Bernt MA, Ellerby LM. Kennedy’s disease. Phosphorylation of the polyglutamine-expanded form of androgen receptor regulates its cleavage by caspase-3 and enhances cell death. J Biol Chem. 2003;278:34918–24. doi: 10.1074/jbc.M302841200. [DOI] [PubMed] [Google Scholar]
- Lalani SR, Zhang J, Schaaf CP, Brown CW, Magoulas P, Tsai AC, El-Gharbawy A, Wierenga KJ, Bartholomew D, Fong CT, Barbaro-Dieber T, Kukolich MK, Burrage LC, Austin E, Keller K, Pastore M, Fernandez F, Lotze T, Wilfong A, Purcarin G, Zhu W, Craigen WJ, McGuire M, Jain M, Cooney E, Azamian M, Bainbridge MN, Muzny DM, Boerwinkle E, Person RE, Niu Z, Eng CM, Lupski JR, Gibbs RA, Beaudet AL, Yang Y, Wang MC, Xia F. Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet. 2014;95:579–83. doi: 10.1016/j.ajhg.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezon-Geyda K, Najfeld V, Johnson EM. Deletions of PURA, at 5q31, and PURB, at 7p13, in myelodysplastic syndrome and progression to acute myelogenous leukemia. Leukemia. 2001;15:954–62. doi: 10.1038/sj.leu.2402108. [DOI] [PubMed] [Google Scholar]
- Li Y, Koike K, Ohashi S, Funakoshi T, Tadano M, Kobayashi S, Anzai K, Shibata N, Kobayashi M. Pur alpha protein implicated in dendritic RNA transport interacts with ribosomes in neuronal cytoplasm. Biol Pharm Bull. 2001;24:231–5. doi: 10.1248/bpb.24.231. [DOI] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Barr SM, Chu C, Kohtz DS, Kinoshita Y, Johnson EM. Functional interaction of Puralpha with the Cdk2 moiety of cyclin A/Cdk2. Biochem Biophys Res Commun. 2005;328:851–7. doi: 10.1016/j.bbrc.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Liu H, Johnson EM. Distinct proteins encoded by alternative transcripts of the PURG gene, located contrapodal to WRN on chromosome 8, determined by differential termination/polyadenylation. Nucleic Acids Res. 2002;30:2417–26. doi: 10.1093/nar/30.11.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZW, Bergemann AD, Johnson EM. Conservation in human and mouse Pur alpha of a motif common to several proteins involved in initiation of DNA replication. Gene. 1994;149:311–4. doi: 10.1016/0378-1119(94)90167-8. [DOI] [PubMed] [Google Scholar]
- Ma ZW, Pejovic T, Najfeld V, Ward DC, Johnson EM. Localization of PURA, the gene encoding the sequence-specific single-stranded-DNA-binding protein Pur alpha, to chromosome band 5q31. Cytogenet Cell Genet. 1995;71:64–7. doi: 10.1159/000134065. [DOI] [PubMed] [Google Scholar]
- Maizels N. G4-associated human diseases. EMBO Rep. 2015;16:910–22. doi: 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Chromosome, A.L.S.F.T.D.C. French research network on, F.F.A. Consortium, I. Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–30. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, Wieringa B, Wansink DG. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106:13915–20. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan V, Sweet T, Nadraga Y, Amini S, Khalili K. Regulation of Puralpha gene transcription: evidence for autoregulation of Puralpha promoter. J Cell Physiol. 2001;186:406–13. doi: 10.1002/1097-4652(2000)9999:999<000::AID-JCP1039>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Murray A, Ennis S, Youings SA, Sharrock AJ, Lewis C, Pound MC, Macpherson JN, Dennis NR, Morton NE, Jacobs PA. Stability and haplotype analysis of the FRAXE region. Eur J Hum Genet. 2000;8:583–9. doi: 10.1038/sj.ejhg.5200504. [DOI] [PubMed] [Google Scholar]
- Orr HT, Chung MY, Banfi S, Kwiatkowski TJ, Jr, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LP, Zoghbi HY. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–6. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- Penberthy WT, Zhao C, Zhang Y, Jessen JR, Yang Z, Bricaud O, Collazo A, Meng A, Lin S. Pur alpha and Sp8 as opposing regulators of neural gata2 expression. Dev Biol. 2004;275:225–34. doi: 10.1016/j.ydbio.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Reijnders MRF, Leventer RJ, Lee BH, Baralle D, Selber P, Paciorkowski AR, Hunt D. PURA-Related Neurodevelopmental Disorders. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 2017. [PubMed] [Google Scholar]
- Reiss K, Khalili K. Viruses and cancer: lessons from the human polyomavirus, JCV. Oncogene. 2003;22:6517–23. doi: 10.1038/sj.onc.1206959. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–37. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S, Serrano A, Gerbino V, Giorgi A, Di Francesco L, Nencini M, Bozzo F, Schinina ME, Bagni C, Cestra G, Carri MT, Achsel T, Cozzolino M. Nuclear accumulation of mRNAs underlies G4C2-repeat-induced translational repression in a cellular model of C9orf72 ALS. J Cell Sci. 2015;128:1787–99. doi: 10.1242/jcs.165332. [DOI] [PubMed] [Google Scholar]
- Sahashi K, Katsuno M, Hung G, Adachi H, Kondo N, Nakatsuji H, Tohnai G, Iida M, Bennett CF, Sobue G. Silencing neuronal mutant androgen receptor in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2015;24:5985–94. doi: 10.1093/hmg/ddv300. [DOI] [PubMed] [Google Scholar]
- Sariyer IK, Sariyer R, Otte J, Gordon J. Pur-Alpha Induces JCV Gene Expression and Viral Replication by Suppressing SRSF1 in Glial Cells. PLoS One. 2016;11:e0156819. doi: 10.1371/journal.pone.0156819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CJ, Jia YH, Tian RR, Ding M, Zhang C, Wang JH. Translation of Pur-alpha is targeted by cellular miRNAs to modulate the differentiation-dependent susceptibility of monocytes to HIV-1 infection. FASEB J. 2012;26:4755–64. doi: 10.1096/fj.12-209023. [DOI] [PubMed] [Google Scholar]
- Shimojima K, Isidor B, Le Caignec C, Kondo A, Sakata S, Ohno K, Yamamoto T. A new microdeletion syndrome of 5q31.3 characterized by severe developmental delays, distinctive facial features, and delayed myelination. Am J Med Genet A. 2011;155A:732–6. doi: 10.1002/ajmg.a.33891. [DOI] [PubMed] [Google Scholar]
- Stacey DW, Hitomi M, Kanovsky M, Gan L, Johnson EM. Cell cycle arrest and morphological alterations following microinjection of NIH3T3 cells with Pur alpha. Oncogene. 1999;18:4254–61. doi: 10.1038/sj.onc.1202795. [DOI] [PubMed] [Google Scholar]
- Stillman WS, Varella-Garcia M, Irons RD. The benzene metabolite, hydroquinone, selectively induces 5q31- and -7 in human CD34+CD19- bone marrow cells. Exp Hematol. 2000;28:169–76. doi: 10.1016/s0301-472x(99)00144-7. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka AJ, Bai R, Cho MT, Anyane-Yeboa K, Ahimaz P, Wilson AL, Kendall F, Hay B, Moss T, Nardini M, Bauer M, Retterer K, Juusola J, Chung WK. De novo mutations in PURA are associated with hypotonia and developmental delay. Cold Spring Harb Mol Case Stud. 2015;1:a000356. doi: 10.1101/mcs.a000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretiakova A, Gallia GL, Shcherbik N, Jameson B, Johnson EM, Amini S, Khalili K. Association of Puralpha with RNAs homologous to 7 SL determines its binding ability to the myelin basic protein promoter DNA sequence. J Biol Chem. 1998;273:22241–7. doi: 10.1074/jbc.273.35.22241. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–14. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Weber J, Bao H, Hartlmuller C, Wang Z, Windhager A, Janowski R, Madl T, Jin P, Niessing D. Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha. Elife. 2016;5 doi: 10.7554/eLife.11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Johnson EM, Khalili K. Multiple roles for Puralpha in cellular and viral regulation. Cell Cycle. 2009;8:1–7. doi: 10.4161/cc.8.3.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witze ES, Field ED, Hunt DF, Rothman JH. C. elegans pur alpha, an activator of end-1, synergizes with the Wnt pathway to specify endoderm. Dev Biol. 2009;327:12–23. doi: 10.1016/j.ydbio.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman MJ, Hanson LK, Martinez-Sobrido L, Campbell AE, Nance JA, Garcia-Sastre A, Johnson EM. Regulation of PURA gene transcription by three promoters generating distinctly spliced 5-prime leaders: a novel means of fine control over tissue specificity and viral signals. BMC Mol Biol. 2010;11:81. doi: 10.1186/1471-2199-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman MJ, Johnson EM, Bergemann AD. Mechanism of DNA binding and localized strand separation by Pur alpha and comparison with Pur family member, Pur beta. Biochim Biophys Acta. 2005;1743:64–78. doi: 10.1016/j.bbamcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Wortman MJ, Krachmarov CP, Kim JH, Gordon RG, Chepenik LG, Brady JN, Gallia GL, Khalili K, Johnson EM. Interaction of HIV-1 Tat with Puralpha in nuclei of human glial cells: characterization of RNA-mediated protein-protein binding. J Cell Biochem. 2000;77:65–74. doi: 10.1002/(sici)1097-4644(20000401)77:1<65::aid-jcb7>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, Jin P. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110:7778–83. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Liu C, Geng Y, Zhu G. Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci Rep. 2015;5:16673. doi: 10.1038/srep16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1S. Comparison of Pura complete sequences from Human (H. sapiens, GB M96684.1) and Drosophila (D. melanogaster, GB AF021259.1). Alignment is with the Clustal Omega algorithm. Stars indicate amino acid identities; one dot indicates a substitution; two dots indicate a conservative substitution.


