Abstract
Hsp100 polypeptide translocases are conserved AAA+ machines that maintain proteostasis by unfolding aberrant and toxic proteins for refolding or proteolytic degradation. The Hsp104 disaggregase from S. cerevisiae solubilizes stress-induced amorphous aggregates and amyloid. The structural basis for substrate recognition and translocation is unknown. Using a model substrate (casein), we report cryo-EM structures at near-atomic resolution of Hsp104 in different translocation states. Substrate interactions are mediated by conserved, pore-loop tyrosines that contact an 80 Å-long unfolded polypeptide along the axial channel. Two protomers undergo a ratchet-like conformational change that advances pore-loop-substrate interactions by two-amino acids. These changes are coupled to activation of specific ATPase sites and, when transmitted around the hexamer, reveal a processive rotary translocation mechanism and a remarkable structural plasticity of Hsp104-catalyzed disaggregation.
Hsp100 disaggregases are highly conserved stress responders that unfold and solubilize protein aggregates(1, 2). They are AAA+ (ATPases Associated with diverse cellular Activities) proteins that form hexameric rings, which couple ATP hydrolysis to polypeptide translocation through a central channel(1, 2). S. cerevisiae Hsp104 is powered by two distinct AAA+ domains per protomer and collaborates with the Hsp70 system to disaggregate and refold amorphous aggregates and amyloids such as Sup35 prions, thereby promoting stress tolerance and prion propagation(2).
Cooperative ATP hydrolysis by nucleotide-binding domains (NBD1 and NBD2) of Hsp104 requires conserved Walker A and B motifs, ‘sensor’ residues, and an Arg finger from the adjacent protomer(2, 3). The coiled-coil middle domain (MD) of Hsp104 mediates Hsp70 interactions and allosteric functions during hydrolysis and disaggregation(4–6). Conserved substrate-binding “pore loops” in the NBDs line the axial channel and contain essential Tyr residues that mechanically couple hydrolysis to translocation(7, 8). A recent structure of Hsp104 bound to the nonhydrolysable ATP analog, AMPPNP, identifies an “open”, spiral conformation with a ~30 Å-wide channel and an unusual heteromeric NBD1-NBD2 interaction that forms a flexible seam(9). High-resolution structures of the active hexamer have remained elusive and it is unknown how hydrolysis and conformational changes power disaggregation.
Here, we establish how Hsp104 binds and mechanically translocates substrates. Using the slowly hydrolysable ATP analog, ATPγS(10) and the substrate casein we determined Hsp104 structures to ~4.0 Å by cryo-EM. We identify pore-loop-substrate contacts and a rotary translocation mechanism involving a ratchet-like change that advances interactions along the substrate polypeptide by two amino acids. NBD1-ATP hydrolysis and substrate binding triggers a massive open to closed conformational change in the hexamer, thereby coupling substrate engagement and release to processive disaggregation.
Substrate-Bound Architecture of Hsp104
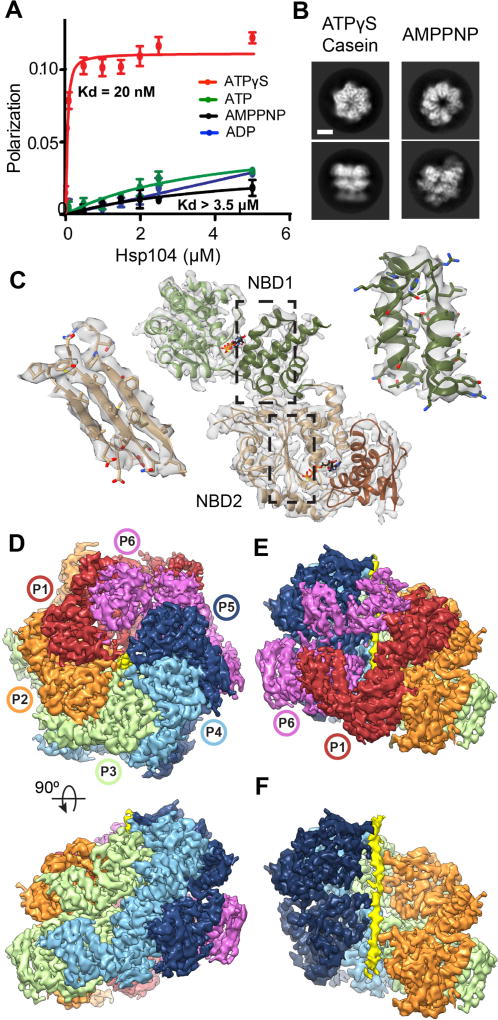
To capture the substrate-bound state of Hsp104, binding was assessed via fluorescence polarization (FP) using FITC-labeled casein, a model substrate that is actively translocated(4, 8). High-affinity interactions (Kd ~ 20nM) between Hsp104 and casein were identified in the presence of ATPγS but not ATP, AMPPNP, or ADP (Fig. 1A and S1). Two-dimensional (2D) reference-free classification of purified Hsp104:casein-ATPγS complexes revealed the hexamer undergoes a large rearrangement compared to the AMPPNP-bound open state(9) (Fig. 1B and S2A). An initial 3D structure refined to ~3.9 Å resolution (Fig. S2B and Table S1). However, a two-protomer site was identified to be flexible (Fig. S2C). Extensive 3D classification resolved these protomers and revealed two hexamer conformations, denoted: “closed” and “extended” (Fig. S2D). Refinement of the closed state yielded a 4.0 Å-resolution map with well-resolved protomers and axial channel (Fig. S2B and S3A–C). The AAA+ subdomains show side-chain features, enabling an atomic model to be built and refined using a homology model from the bacterial orthologue ClpB(11) (Fig. 1C and S3D–E) The AAA+ domains (protomers denoted P1–P6) form a near-symmetric closed double-ring (Figure 1D). Protomers P2–P5 are identical (RMSD = 2 Å) while the two mobile protomers, P1 and P6, each adopt different conformations (Fig. 1E and S3F). The outside hexamer diameter is ~115 Å (compared to 125 Å for the open state), and density for the AAA+ domains surround a ~10 Å-wide axial channel. Remarkably, the channel is partially occluded by a continuous, 80 Å strand of density that, based on the molecular model, is an unfolded portion of the substrate, casein (Fig. 1F).
Fig. 1. Substrate-bound Hsp104:casein closed complex.
(A) FITC-casein binding analysis, measured by fluorescence polarization in the presence of: ATPγS (red), ATP (green), AMPPNP (black), and ADP (blue) (values = mean ±SD, n=3). (B) Representative top and side-view 2D class averages comparing Hsp104-ATPγS:casein closed state and Hsp104-AMPPNP open state(9) (scale bar equals 50 Å). (C) Atomic model and segmented map of the AAA+ small (NBD1, green) and large subdomains (NBD2, brown). (D) Final reconstruction of Hsp104:casein segmented by protomers (P1–P6) and substrate (yellow). (E) Side view of the mobile protomer face (P1 and P6). (F) Channel view showing substrate polypeptide density (yellow).
Tyr Pore-Loop Contacts Along Substrate
In the closed state, pore-loop strands from both AAA+ domains become ordered compared to Hsp104-AMPPNP(9) and other structures(11, 12), and contact substrate in a right-handed spiral arrangement (Fig. 2A and S4). These regions are among the most highly resolved (< 4.0 Å, Fig. S3C), indicating bona fide interactions critical for translocation. The casein sequence could not be determined from the density, therefore a strand of 26 Ala residues was modeled. Hsp104 translocates unfolded polypeptides in the presence of ATPγS(8), therefore, the pore loop-substrate interactions likely adopt a fixed register during translocation. Alternatively, a specific region of casein may be uniformly trapped in the channel. Substrate contacts are made by five protomers (P1–P5), while protomer P6 breaks the helical arrangement and makes no direct contact (Fig. 2A and S4). Substrate density is not observed outside the channel, thus non-translocated portions of casein are likely disordered.
Fig. 2. Structural basis for substrate binding in the axial channel.
(A) Channel view of the map showing the extended poly-Ala strand modeled as the substrate (mesh) and pore loops with residues indicated. The P6 pore-loop regions not contacting substrate are indicated (circles) (B) Model and cryo-EM density showing P4 pore loop-substrate interactions mediated by Y257 (green) in the NBD1 and Y662 (green) and V663 in the NBD2 as well as lower pore loops that include residues K649 and Y650. (C) Spiral arrangement of the NBD1 and NBD2 canonical pore loops for P1–P5 contacting substrate and the disconnected position of the P6-NBD1 pore loop.
Conserved pore-loop tyrosines 257 and 662 in protomers P1–P5 directly contact substrate, potentially via the aromatic rings, which are positioned ~4–5 Å away from the backbone (Fig. 2B). Together with conserved V663 in NBD2, these residues contribute the majority of substrate interactions in the channel (Fig. S4). The lower loops (291–297 for NBD1 and 645–651 for NBD2) are also ordered and adjacent to the substrate, and K649 and Y650 for P2 and P4 appear to make contact (Fig. 2A–B).
The pore loops are separated by ~6–7 Å along the channel, making contact with approximately every second amino acid of the substrate (Fig. 2C). K256 and K258 in NBD1, which flank Y257, project towards neighboring loops, possibly stabilizing the spiral arrangement. Protomers P1 and P5 comprise the lowest and highest contact sites with the substrate and are separated by ~26 Å along the channel axis. Protomer P6 is between these sites, but disconnected from the substrate; its NBD1 pore loop is 13 Å away while its NBD2 pore loop was less resolved and unable to be modeled (Fig. 2C). Overall, this structure reveals that substrate interactions are mediated almost entirely by the conserved Tyr residues, establishing their direct role in coordinating substrate during translocation (1, 2).
Ratchet-Like States of Protomers
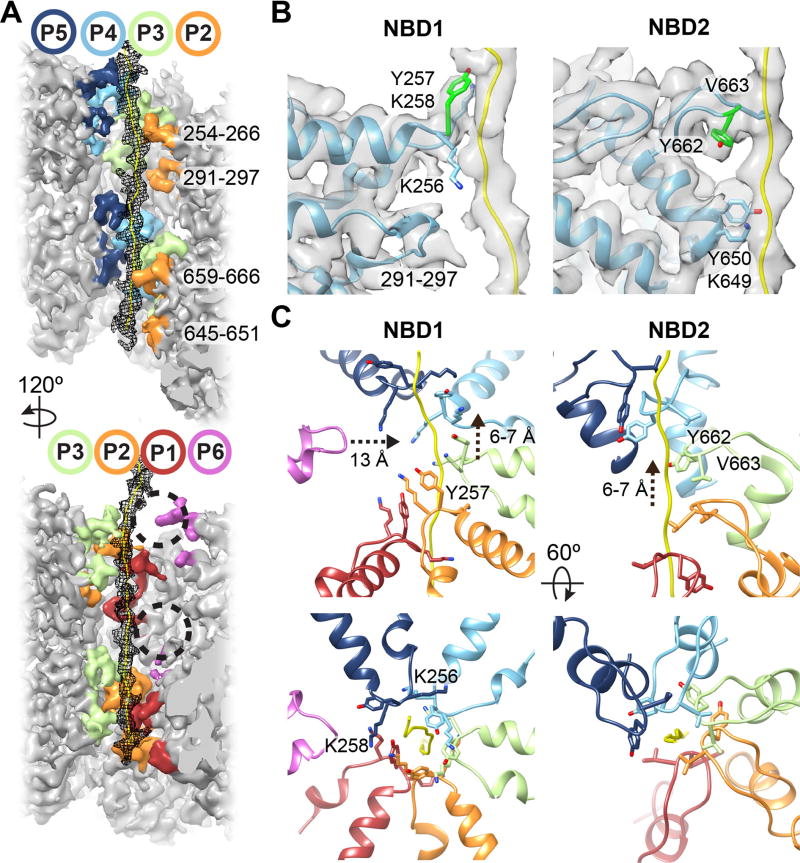
In addition to the closed state, our 3D classification analysis identified an ‘extended’ conformation of Hsp104:casein (Fig. S2D). Further classification and refinement resulted in a 4.1 Å resolution map from which an atomic model was determined (Fig. 3A, S2B and S5A–B). The structure reveals a substrate-bound hexamer with a different arrangement of the mobile protomers P1 and P6, which show distinct flexibility in the NBD1 and NBD2, respectively (Fig. 3A and S5C–D). Protomers P2–P5 are identical to the closed state (RMSD = ~0.8 Å) and density for the polypeptide substrate is slightly extended at the top of the channel, but overall similar and localizes to the same region (Fig. S5E).
Fig. 3. Hsp104:casein extended-state conformation advances substrate contacts.
(A) Cryo-EM reconstruction of Hsp104:casein identifying substrate (yellow) and an extended conformation of protomers P1 and P6. (B) Filtered map of P1 (red), and P6 (magenta) overlaid with the corresponding closed-state protomers (grey) following alignment to P4 in the hexamer. NBD conformational changes (arrows) resulting in extended-state interactions (black circles) with substrate (yellow) and the P2-NBD2 (orange) are shown. (C) Model and map of the P6 NBD1 and NBD2 pore loops showing change in the pore loop position (arrow) compared to the closed state (grey) for NBD1 and substrate contact by Y257 and Y662 (green). (D) Model and map of the NBD1 and NBD2 P1–P6 spiral of pore loop-substrate interactions.
The closed-extended state conformational differences for P1 and P6 are substantial (RMSD = 13.7 Å and 11.5 Å, respectively) and involve rotations of P1-NBD1 and P6-NBD2 (Fig. 3B, Fig. S6A). P6 rotates towards the channel axis and the pore loops become well-ordered and directly contact substrate. The P6 pore-loop tyrosines, Y257 and Y662, directly contact substrate similarly as the other protomers (Fig. 3C). Conversely, P1-NBD2 rotates counterclockwise, releasing its interaction with P6 to contact P2-NBD2 but maintains contact with the substrate (Fig. 3B).
Remarkably, P6-Y257 becomes positioned at the topmost contact site along the polypeptide, advancing interactions by 2 amino acids (~7 Å) compared P5-Y257 (Fig. 3D and S6B). These changes bring P6 pore loops in register to form a two-turn right-handed spiral of contacts. Each pore loop rotates ~60° and rises ~6–7 Å, enabling evenly spaced Tyr-substrate interactions across a 74 Å-length of the channel. Together, the extended and closed states reveal a ratchet-like conformational change of the hexamer that yields a two-amino acid translocation step (Movie S1). While other conformations may exist that were not resolved, the extended and closed states predominate the dataset (Fig S2D), therefore, these changes are likely critical for orchestrating substrate binding and release steps during translocation.
Coordinated Nucleotide Pockets
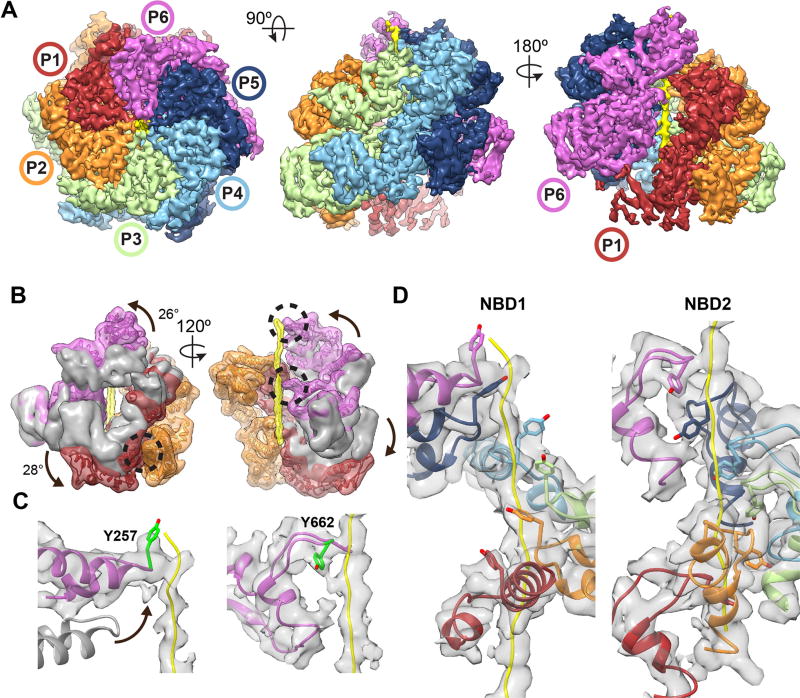
NBD1 and NBD2 nucleotide pockets were examined to determine how nucleotide state is coupled to substrate interactions. P3–P5 nucleotide pockets are identical with well-resolved density and a bound ATPγS (Fig. S7A). In NBD1, R334 from the clockwise neighboring promoter contacts the γ-phosphate, establishing this residue as the Arg finger(13). R333 is adjacent the α/β-phosphates, acting potentially as a sensor residue considering that the NBD1 does not contain a cis sensor 2 motif(3). In the NBD2 pocket, the Arg finger, R765, interacts with the γ-phosphate while the sensor 2, R826 in the cis protomer, is positioned adjacent the α/β-phosphates (Fig. S4A). Thus, for P3–P5, which make well-defined contacts with the substrate in both states, NBD1 and NBD2 are primed for ATP hydrolysis.
Conversely, the mobile promoter (P6, P1, and P2) NBDs (Fig. S7B), are in different active and inactive configurations, based on the position of Arg fingers and nucleotide density. For the P6 protomer, both NBDs appear inactive in the closed state (Fig. 4A) with P5-R334 ~12 Å away and P5-R765 ~6 Å from the respective NBD1 and NBD2 γ-phosphate in P6. In contrast, both P5-R334 and P5-R765 in the extended state are identified to contact the respective γ-phosphates directly, indicating these sites are in an active configuration. Notably, the closed-extended conformational change results in both nucleotide-pocket activation and substrate contact by P6 (Fig. 3B).
Fig. 4. Extended-state activation of the nucleotide pockets is coupled to translocation.
(A) Map and model of the P6, P1 and P2 nucleotide pockets. Arg-fingers, NBD1-R334 and NBD2-R765, are shown (green) with γ-phosphate contact indicated (*) for the active sites and distances shown for the inactive sites. Sensor 2 residues NBD1-R333 and the NBD2-R826 are shown (cyan). (B) Rotary translocation model showing closed-to-extended states resulting in active (green), inactive (red) and unbound/inactive (grey dash) states of the NBDs. Pore loop spiral (grey gradient) is shown contacting substrate (yellow). Arg-finger contact and NBD activation is depicted by the interlocking contact.
P1 contacts substrate at the lowest position in the hexamer and appears inactive in both states (Fig. 4A). In the closed state, P6-R334 is ~11 Å away and P6-R765 is ~21 Å from the respective γ-phosphate in P1. These Arg residues are further separated from the nucleotide pockets in the extended state, with R333, R334, and R765 more than 30 Å away (Fig. 4A). For P2, NBD1 is in an active configuration in both states. However, P2-NBD2 switches from inactive in the closed state to active in the extended state due to the P1-NBD2 conformational change that brings R765 adjacent the γ-phosphate (Fig. 4A). Finally, based on difference maps, density for nucleotide is present in all sites, but appears reduced at certain sites: NBD1 in P6 and P1 for the closed state, and the NBD2 in P1 for both states, indicating partial occupancy or a post-hydrolysis state (Fig. S7C).
The NBD states along with substrate interactions are depicted in a schematic to explain how active site rearrangements and the closed-extended conformational changes drive substrate translocation (Fig. 4B). In the closed state, substrate is bound by five protomers with four NBD1 sites and three NBD2 sites in an active (ATP) configuration. By contrast, in the extended state, substrate is bound by six protomers with five NBD1 sites and five NBD2 sites in an active state. Importantly, the ‘off’ protomer (P6) that is unbound to substrate in the closed state becomes active in the extended state and binds substrate at the next position. The protomer counterclockwise from this position is in the lowest ‘down’ position (P1) and remains inactive in both states, but undergoes a rotation in the extended state that activates NBD2 of the neighboring protomer.
These results suggest a rotary-type translocation mechanism whereby four protomers remain bound to substrate in a similar configuration with the NBDs primed for hydrolysis, while two protomers at the transition site between the lowest and highest position undergo conformational changes that alter substrate interactions. Given the right-handed spiral of pore loops, position of the ‘up’ and ‘down’ protomers, and NBD1-NBD2 direction of translocation, peptide movement could occur through a counterclockwise cycling of these closed and extended states. Based on this model the inactive protomer in the ‘down’ position could release substrate and re-engage in the ‘up’ position, thereby advancing translocation by a two-amino acid step. Transmitting these changes counterclockwise would enable the hexamer to advance processively along the polypeptide during translocation (Movie S2). Some variability in the step size, potentially to accommodate bulky residues, could be achieved by conformational changes in the extended-state protomer in the ‘up’ position that shift its pore-loop contact. These results parallel the right-handed substrate interactions and rotary-driven hydrolysis models for DNA/RNA helicases, including AAA+ (E1, DnaA and MCM)(3, 14, 15) and RecA (Rho)(16) families. Thus, this ratcheting mechanism may be conserved among many ATP-driven translocases.
Allosteric Control by the MD
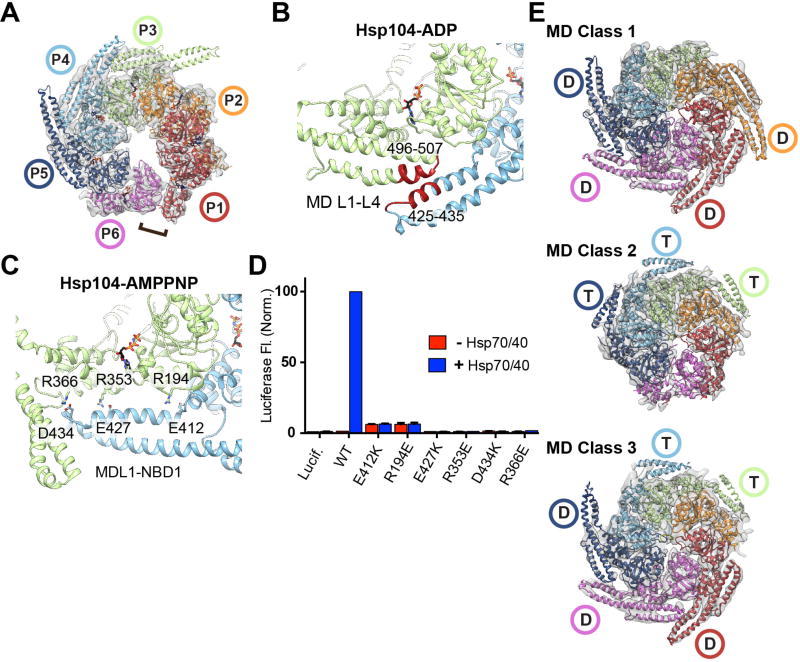
The Hsp104-AMPPNP(9) structure revealed an open spiral conformation that is substantially different from the closed, substrate-bound states characterized here. Furthermore, a MD-NBD1 interaction was identified that suggested an allosteric control mechanism. To further explore the MD and the role of nucleotide in the Hsp104 conformational cycle we determined the cryo-EM structure of Hsp104 incubated with ADP to 5.6 Å resolution (Fig. 5A, S8A–C). The reconstruction reveals an identical AAA+ arrangement compared to Hsp104-AMPPNP (RMSD = 2.5 Å) involving a left-handed spiral architecture defined by a ~10 Å rise per protomer and a heteromeric AAA+ interaction between P6-NBD1 and P1-NBD2.
Fig. 5. Nucleotide-specific MD conformations identified around the substrate-bound hexamer.
(A) Cryo-EM map and model of Hsp104-ADP in the open spiral conformation,, with the NBD1-NBD2 heteromeric interaction shown (bracket). (B) MD-MD (L1–L3) interaction interface identified in the Hsp104-ADP reconstruction (red). (C) MD L1-NBD1 interaction identified in the Hsp104-AMPPNP structure(9), showing putative salt-bridge contacts analyzed by mutagenesis. (D) Luciferase reactivation measured by fluorescence following incubation with Hsp104 wildtype or indicated mutants in the absence (red) or presence (blue) of Hsp70/40 system. Values are normalized to wt+Hsp70/40 and represent the mean ±SD (n=4). (E) Final reconstructions and models of Hsp104:casein following MD classification analysis identifying the ADP-state (D) or ATP-state (T) conformation.
The MD is resolved for three protomers (P3–P5) and identified to be in a crisscross equatorial arrangement stabilized by contacts between the first (L1) and third (L3) helices (Fig. 5B). This arrangement is similar to previous structures(11, 12, 17) but strikingly different to the Hsp104-AMPPNP structure, in which the same MD helix L1 region makes contact across NBD1 of the clockwise protomer (Fig. 5C). Comparison of these ATP- and ADP-state MD conformations reveals a substantial, ~30° rotation around position 409 at the NBD1-MD junction (Fig. S8D and Movie S3). While hexamers exclusively-bound to ATP or ADP are likely rare in vivo, these data reveal that the MD adopts two nucleotide-specific conformations that reflect pre- and post-hydrolysis states.
Sites that comprise the MD-MD interactions in Hsp104-ADP are critical for function(4, 5, 12, 13). However, the AMPPNP-specific MD L1-NBD1 interaction has not been characterized. Therefore, single charge-reversal mutations were introduced to disrupt three putative L1-NBD1 salt bridges(9): E412-R194, E427-R353, and D434-R366 (Fig. 5C). These mutants exhibit robust ATPase activity (Fig. S8E). However, they are unable to reactivate denatured firefly luciferase aggregates in vitro (Fig. 5D) or confer thermotolerance in vivo (Fig. S8E), supporting a functional role for the L1-NBD1 interaction.
To determine MD conformations in the casein-bound complex, additional 3D sub-classification analysis and refinement was performed on the Hsp104 closed-state data without applying a mask (Fig. S9A). Three classes with distinct MD arrangements were identified: MD Class 1, Class 2 and Class 3, which refined to 6.7–6.9 Å (Fig. 5E, Fig. S9B). For these maps, the AAA+ core and substrate density are identical to the closed-state structure. In the MD Class1, density corresponding to the MD coiled-coil is identified for 4 protomers (P1, P2, P5 and P6), revealing an ADP-state, crisscross arrangement around the P6-P1 mobile-protomer face (Fig. 5E). Notably, the NTDs for all protomers are also resolved in this class, revealing that they interact together in an alternating, triangular arrangement with the polypeptide strand oriented asymmetrically in the channel, towards the P3 and P5 NTDs (Fig. S10).
For MD Class 2, density corresponding to MD helices L1 and L2 is identified for protomers P3–P5 revealing L1 is positioned across the clockwise protomer, indicating an ATP-state conformation. (Fig. 5E). For Class 3 both MD conformations are identified: P1, P5, and P6 adopt the ADP-state, while P3 and P4 are in the ATP-state (Fig. 5E). This classification captures specific MD conformations that are in remarkable agreement with our analysis of the nucleotide pockets (Fig. 4). Thus, in an actively translocating hexamer, MD conformational changes likely propagate around the hexamer in accordance with nucleotide state. The MD could function to lock the ATP state for protomers that are in contact with substrate (P2–P5) and transition to a release, ADP-state towards the mobile face, thereby allosterically tuning the closed and extended conformational changes that advance substrate.
NBD1-Driven Rearrangement Engages Substrate
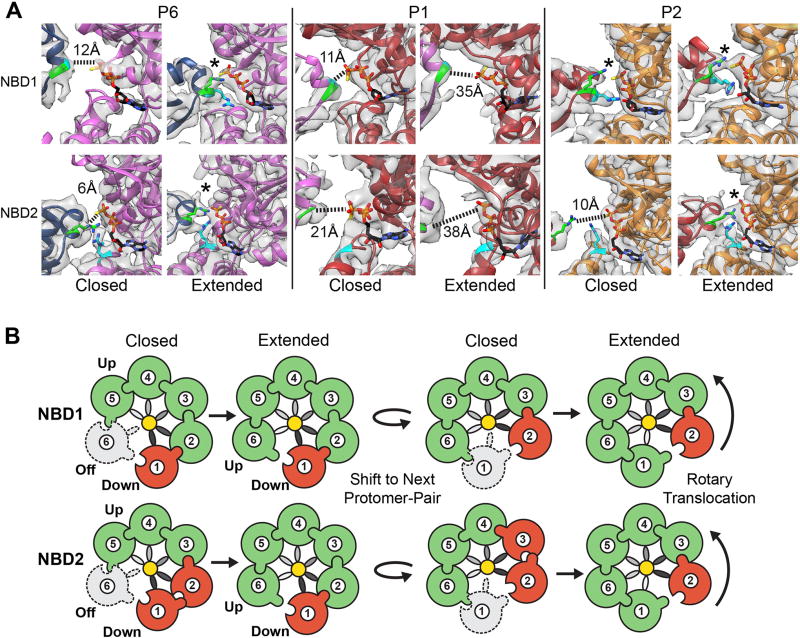
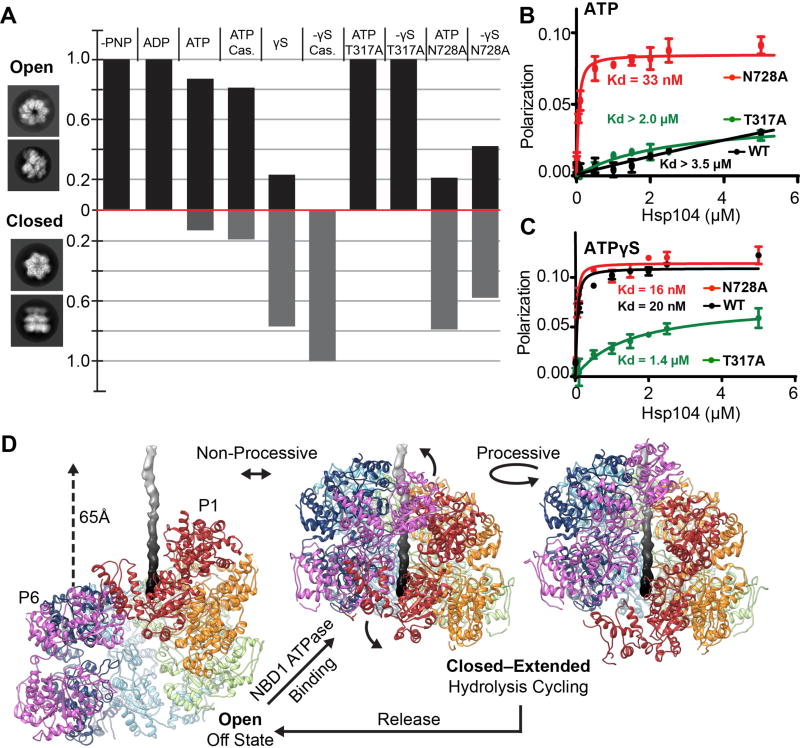
To determine how the open and closed states may function together, cryo-EM datasets of wildtype and mutant Hsp104 incubated with different nucleotides were analyzed by 2D and 3D classification methods (Fig. 6A, and Fig. S11A). As expected, AMPPNP and ADP datasets classify with 100% of the data matching the open conformation. With ATP incubations, alone and with substrate, Hsp104 primarily adopts the open conformation (>80%), however a notable fraction (10–20%) are in the closed state. A 3D reconstruction of the Hsp104-ATP was determined to 6.7 Å and is identical to the AMPPNP and ADP-bound structures (cross-correlation = 0.98) (Fig. S11B). Thus, during active hydrolysis (Fig S8E) the open state is favored, however both conformations exist in equilibrium. With ATPγS, nearly 80% of hexamers are in the closed state (Fig 6A), which increases to 100% with casein, demonstrating that ATPγS and substrate together trigger complete conversion to the closed state.
Fig. 6. Nucleotide-state and NBD function in the open-closed conformations and model for the disaggregation cycle.
(A) The fraction of open and closed conformations, determined by 3D classification analysis, is shown for wildtype Hsp104 and sensor 1 ATPase mutants (T317A in NBD1 and N728A in NBD2) following incubation with indicated nucleotides and substrate. (B,C) FITC-casein (60 nM) binding to Hsp104 wildtype (black) and mutants: T317A (green) and N728A (red) in the presence of (2 mM) (B) ATP or (C) ATPγS. Values represent mean ±SD (n=3). (D) Proposed models for non-processive and processive modes of translocation involving open to closed conformational change upon substrate engagement/release and ratchet-like open-to-extended protomer changes that occur around the hexamer during cycles of ATP hydrolysis.
The sensor 1 ATPase mutants(18), T317A in NBD1 and N728A in NBD2, were investigated to determine the role of NBD1 and NBD2 function (Fig. 6A). In contrast to wildtype, T317A incubated with ATP or ATPγS classifies with 100% matching the open state, indicating that a hydrolysis-active NBD1 promotes the closed conformation. Conversely, ~80% and 60% of N728A hexamers match the closed state in the presence of ATP and ATPγS, respectively. In casein-binding experiments, N728A binds with high affinity (Kd ~33nM), while wildtype and T317A show weak binding (Kd > 2µM) (Fig. 6B) in the presence of ATP. With ATPγS, both wildtype and N728A bind with a high affinity (Kd ~16–20nM), whereas T317A has a reduced affinity, in comparison (Kd ~1.4µM) (Fig. 6C). Thus, Hsp104 exists in open-closed conformational equilibrium that is differentially controlled by NBDs. Hydrolysis by NBD1 promotes the closed state, while hydrolysis by NBD2 favors the open state. Substrate binding and the open-closed conformational change are coupled and likely driven by NBD1 function, whereas NBD2 may be important for substrate release steps.
Massive conformational changes are required to transition between the open and closed states (Fig. 6D and Movie S4). In the open state, protomer P1 is in the topmost position and the hexamer adopts a left-handed spiral with P6 ~50 Å below, along the axial channel. Upon conversion to the closed state, P6 shifts by ~65 Å, and rotates towards the channel by ~60° (Fig. S12), resulting in a right-handed spiral and a channel that has narrowed by ~10 Å. Considering that substrate interactions and hydrolysis by NBD1 are critical for the closed state (Figure 6A–C), we propose that this large conformational change drives substrate binding and release steps of the cycle (Fig 6D). When the conformational change is modeled with substrate, nearly 30 residues can be translocated into the channel. Additionally, similar open ‘lock-washer’ conformations are populated by other translocases(19–22), which may represent a conserved ‘off-state’.
Disaggregation involves nonprocessive and processive mechanisms(23, 24). Cycling between the open and closed states may enable nonprocessive bind/release “pulling” events. Alternatively, the two different substrate-bound states suggest a processive mechanism whereby two protomers undergo ratchet-like conformational changes that enable substrate binding and release steps to occur while the hexamer remains engaged. This rotary-like mechanism could drive disaggregation when coupled to stepwise cycles of ATP hydrolysis around the ring (Movie S2). Such a cooperative mechanism could enable dissolution of more stable aggregates or amyloids(23). While this mechanism contrasts with ‘stochastic’ models proposed for ClpX(1), Hsp104 may exhibit different conformational cycles tuned to different substrates(23). Remarkably, both extended and closed states reveal a precise 6–7 Å separation of the pore loop-substrate contacts. A two-amino acid step involving conformational changes at the spiral interface would continually maintain this register during translocation. This pore-loop spacing is observed in related AAA+ rings(25–27), and likely represents a conserved feature of translocases. While additional states are likely involved, our structures reveal the remarkable structural plasticity of Hsp104, which enables adaptable mechanisms of protein disaggregation(2, 23).
Supplementary Material
Acknowledgments
The authors thank J. Smith and L. Rice for discussion of the manuscript and F. DiMaio for help with Rosetta. M.J. is supported by a Target ALS Springboard Fellowship. E.S. is supported by an AHA predoctoral fellowship and NIH grant T32GM008275. E.C. is supported by NIH grant T32GM008076. M.T. is supported by NIH grants K12GM081259 and K22NS09131401. K.M. is supported by an NSF Graduate Research Fellowship (DGE-1321851). J.S. is supported by NIH grant R01GM099836, a Muscular Dystrophy Association Research Award (MDA277268), the Life Extension Foundation, the Packard Center for ALS Research at Johns Hopkins University, and Target ALS. D.S. is supported by NIH grants: R01GM109896, R01GM077430, and R01GM110001A. Cryo-EM maps and models are deposited in the Protein Data Bank: Hsp104:casein closed state (EMDB: 8697, PDB: 5VJH), extended state (EMDB: 8746, PDB: 5VYA), and middle domain conformation (EMDB: 8745, PDB: 5VY9) and Hsp104-ADP (EMDB: 8744, PDB: 5VY8).
Footnotes
References
- 1.Olivares AO, Baker TA, Sauer RT. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol. 2016;14:33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sweeny EA, Shorter J. Mechanistic and Structural Insights into the Prion-Disaggregase Activity of Hsp104. J Mol Biol. 2016;428:1870–1885. doi: 10.1016/j.jmb.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 4.Jackrel ME, et al. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oguchi Y, et al. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat Struct Mol Biol. 2012;19:1338–1346. doi: 10.1038/nsmb.2441. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, et al. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc Natl Acad Sci U S A. 2013;110:8513–8518. doi: 10.1073/pnas.1217988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 8.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 9.Yokom AL, et al. Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat Struct Mol Biol. 2016;23:830–837. doi: 10.1038/nsmb.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyle SM, et al. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 12.Carroni M, et al. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife. 2014;3:e02481. doi: 10.7554/eLife.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wendler P, et al. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 15.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuck A, et al. Structural basis for the disaggregase activity and regulation of Hsp104. Elife. 2016;5 doi: 10.7554/eLife.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyubimov AY, Costa A, Bleichert F, Botchan MR, Berger JM. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc Natl Acad Sci U S A. 2012;109:11999–12004. doi: 10.1073/pnas.1209406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 21.Sledz P, et al. Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A. 2013;110:7264–7269. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M, et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeSantis ME, et al. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li T, et al. Escherichia coli ClpB is a non-processive polypeptide translocase. Biochem J. 2015;470:39–52. doi: 10.1042/BJ20141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wehmer M, Sakata E. Recent advances in the structural biology of the 26S proteasome. Int J Biochem Cell Biol. 2016;79:437–442. doi: 10.1016/j.biocel.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20:781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torrente MP, Castellano LM, Shorter J. Suramin inhibits Hsp104 ATPase and disaggregase activity. PLoS One. 2014;9:e110115. doi: 10.1371/journal.pone.0110115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redwan EM, Xue B, Almehdar HA, Uversky VN. Disorder in milk proteins: caseins, intrinsically disordered colloids. Curr Protein Pept Sci. 2015;16:228–242. doi: 10.2174/1389203716666150224145900. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Zheng S, Agard DA, Cheng Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J Struct Biol. 2015;192:174–178. doi: 10.1016/j.jsb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiMaio F, et al. Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biter AB, Lee S, Sung N, Tsai FT. Structural basis for intersubunit signaling in a protein disaggregating machine. Proc Natl Acad Sci U S A. 2012;109:12515–12520. doi: 10.1073/pnas.1207040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sweeny EA, et al. The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol Cell. 2015;57:836–849. doi: 10.1016/j.molcel.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.