Summary
The current study aimed to understand the developmental mechanisms regulating bud tip progenitor cells in the human fetal lung, which are present during branching morphogenesis, and to use this information to induce a bud tip progenitor-like population from human pluripotent stem cells (hPSCs) in vitro. We identified cues that maintained isolated human fetal lung epithelial bud tip progenitor cells in vitro and induced three-dimensional hPSC-derived organoids with bud tip-like domains. Bud tip-like domains could be isolated, expanded, and maintained as a nearly homogeneous population. Molecular and cellular comparisons revealed that hPSC-derived bud tip-like cells are highly similar to native lung bud tip progenitors. hPSC-derived epithelial bud tip-like structures survived in vitro for over 16 weeks, could be easily frozen and thawed, maintained multilineage potential, and successfully engrafted into the airways of immunocompromised mouse lungs, where they persisted for up to 6 weeks and gave rise to several lung epithelial lineages.
Keywords: lung, branching morphogenesis, progenitor, bud tip, human pluripotent stem cell, directed differentiation, organoid, lung organoid, SOX9, SOX2
Graphical Abstract
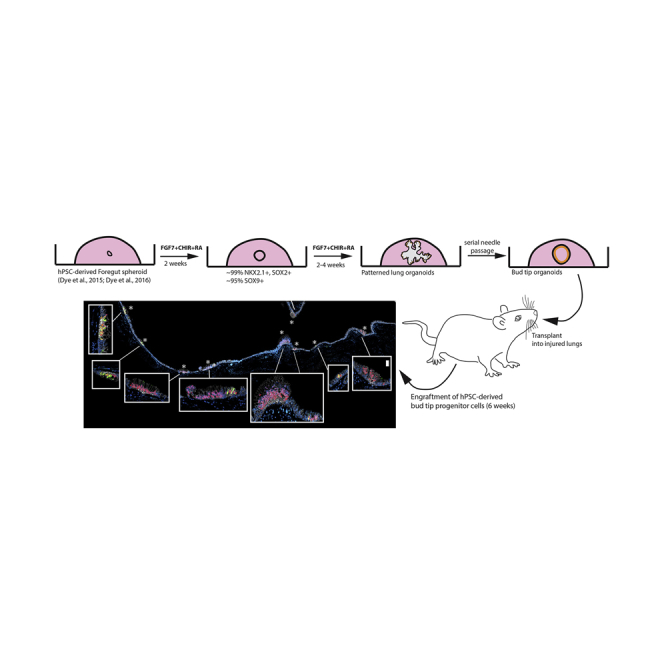
Highlights
-
•
Fgf7, CHIR-99021, and RA support growth of human lung bud tip progenitors in vitro
-
•
These factors induce lung bud tip progenitor-like organoids from hPSCs in vitro
-
•
hPSC-derived organoids are highly similar to human fetal bud tip progenitors
-
•
hPSC-derived bud tip organoids engraft into the injured murine airway
In this article, Jason Spence and colleagues identify a core signaling network that maintains isolated human epithelial lung bud tip progenitors in vitro and that induces a bud tip-like population from hPSCs. They show that iPSC-derived bud tip progenitors can engraft into an injured mouse airway, contribute to re-epithelialization, and differentiate into multiple lung epithelial lineages.
Introduction
During development, the lung undergoes branching morphogenesis, where a series of stereotyped epithelial bifurcations give rise to the branched, tree-like architecture of the adult lung (Metzger et al., 2008). A population of rapidly proliferating progenitor cells resides at the tips of the epithelium throughout the branching process (“bud tip progenitors”) (Rawlins et al., 2009). This population, which expresses Id2 and Sox9 in mice, has the capability to differentiate into both mature airway and alveolar cell types. At early stages of branching morphogenesis, this population of progenitors gives rise to proximal airway cells, while at later time points these progenitors give rise to alveolar cells (Rawlins et al., 2009).
Studies utilizing genetic mouse models have shown that lung branching morphogenesis and proximal-distal patterning are regulated by a series of complex mesenchymal-epithelial interactions that involve multiple signaling events, transcription factors, and dynamic regulation of the physical environment (Hines and Sun, 2014, Morrisey and Hogan, 2010, Varner and Nelson, 2014). These studies have identified major roles for several signaling pathways in branching, including Wnt, fibroblast growth factor, bone morphogenic protein, Sonic hedgehog, retinoic acid (RA), and Hippo signaling, among others. However, due to the complex and intertwined nature of these signaling networks, perturbations in one pathway often affect signaling activity of others (Hines and Sun, 2014, Morrisey and Hogan, 2010).
These developmental principles, learned from studying model organism development, have been used as a guide to successfully direct differentiation of human pluripotent stem cells into differentiated lung lineages and three-dimensional lung organoids (Miller and Spence, 2017) (Dye et al., 2016b). However, using this developmental information in a predictive manner to induce and maintain an epithelial bud tip progenitor cell population from hPSCs has remained elusive. For example, our own studies have shown that hPSCs can be differentiated into human lung organoids (HLOs) that possess airway-like epithelial structures and alveolar cell types; however, it was not clear if HLOs passed through a bud tip progenitor-like stage, mimicking all stages of normal development in vivo (Dye et al., 2015). More recent evidence from others has demonstrated that putative bud tip progenitor cells may be induced from hPSCs; however, these cells were rare and were not assessed in detail (Chen et al., 2017). Thus, generation of a robust population of bud tip progenitor cells from hPSCs would shed mechanistic light on how these cells are regulated, would provide a platform for further investigation into mechanisms of lung lineage cell fate specification, and would add a layer of control to existing directed differentiation protocols allowing them to pass through this developmentally important progenitor transition.
In the current study, we used isolated mouse epithelial bud tip cultures to identify conditions that maintained epithelial bud tip progenitors in vitro. These conditions were also tested using isolated human fetal epithelial bud tip progenitors cultured in vitro. We determined that FGF7 promoted an initial expansion of human epithelial bud tip progenitors, and that the addition of CHIR-99021 (a GSK3β inhibitor that acts to stabilize β-catenin) and all-trans RA (3-Factor conditions, herein referred to as “3F”) were required for growth/expansion of human fetal bud tips as epithelial progenitor organoids that maintained their identity in vitro.
When applied to hPSC-derived foregut spheroid cultures, we observed that 3F conditions promoted growth into larger organoid structures with a patterned epithelium that had airway-like and bud tip-like domains. Bud tip-like domains could be preferentially expanded into “bud tip organoids” using serial passaging. hPSC-derived bud tip organoids had a protein expression and transcriptional profile similar to human fetal progenitor organoids. Finally, we demonstrated that hPSC-derived epithelial bud tip organoids can engraft into an injured mouse airway and undergo multilineage differentiation. Taken together, these studies provide an improved mechanistic understanding of human lung bud tip progenitor cell regulation and establish a platform for studying the maintenance and differentiation of lung bud tip progenitor cells.
Results
Characterizing Human Fetal Lung Bud Tip Progenitors
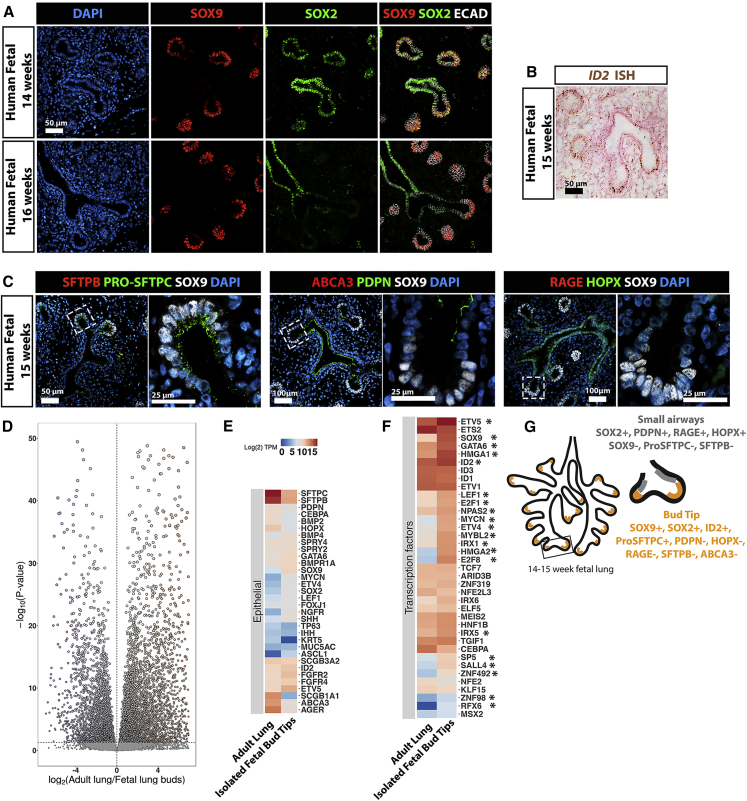
In mice, epithelial bud tip progenitor cells express several transcription factors, including Sox9, Nmyc, and Id2 (Chang et al., 2013, Moens et al., 1992, Okubo et al., 2005, Perl et al., 2005, Rawlins et al., 2009, Rockich et al., 2013). However, recent studies have suggested that significant differences exist between murine and human fetal bud tip progenitor cells (Danopoulos et al., 2017, Nikolić et al., 2017). To confirm and extend these recent findings, we carried out an immunohistochemical analysis using well-established protein markers that are present during mouse lung development (Figures 1A–1C and S1) on human lungs between 10 and 20 weeks of gestation. We also conducted RNA sequencing (RNA-seq) on freshly isolated epithelial lung bud tips, which were dissected to remove mesenchymal cells, to identify genes that were enriched in epithelial progenitors (Figures 1D and 1E). We note that our approach using manual and enzymatic dissection techniques were unlikely to yield pure epithelial cells, and likely possessed a small population of associated mesenchyme. Consistent with the developing mouse lung (Perl et al., 2005, Rockich et al., 2013), we observed that SOX9 is expressed in bud tip domains of the branching epithelium (Figures 1A and S1A). In contrast to the developing murine lung, we observed SOX2 expression in these bud tip progenitor domains until 16 weeks of gestation, at which time SOX2 expression was lost from this population (Figures 1A and S1A). We also observed expression of ID2 by in situ hybridization (Figures 1B and S1F), with expression becoming increasingly intense in the bud tips as branching progressed, up through 20 weeks gestation (Figure S1F). Bud tips expressed nearly undetectable levels of pro-SFTPC at 10 and 12 weeks, with low levels of expression detected by 14 weeks (Figure S1D). Pro-SFTPC expression became more robust by 15 and 16 weeks and continued to increase in the bud tips until 20 weeks (Figures 1C and S1D). SOX9+ bud tip cells were negative for several other lung epithelial markers including SFTPB, PDPN, RAGE, and HOPX (Figures 1C and S1C–S1E). We also examined expression of several proximal airway markers, including P63, acetylated tubulin (AC-TUB), FOXJ1, SCGB1A1, and MUC5AC, and noted that expression was absent from the epithelial bud tip progenitors (negative staining data not shown). We did observe that PDPN and HOPX were expressed in the transition zone/small airways directly adjacent to the SOX9+ bud tip domain at all time points examined (10–20 weeks of gestation), but that this region did not begin to express the AECI maker RAGE until 16 weeks of gestation (Figures 1C, S1B, S1C, and S1E). RNA-seq of isolated, uncultured human fetal epithelial lung bud tips (n = 2; 59 and 89 days gestation) supported protein-staining analysis of human fetal buds. Differential expression analysis to identify genes enriched in the human fetal bud tips (isolated human fetal epithelial bud tips versus whole adult lung) identified 7,166 genes that were differentially expressed (adjusted p value < 0.01; Figure 1D). A curated heatmap highlights genes corresponding to Figures 1A–1C and previously established markers of lung epithelial cells (Figure 1E). Human fetal bud tips have recently been shown to have enrichment for 37 transcription factors (Nikolić et al., 2017). In total, 20 of these 37 transcription factors were also enriched in our analysis (Figure 1F), and gene set enrichment analysis confirmed that this enrichment was highly significant (normalized enrichment score = −1.8, adjusted p value = 9.1 × 10−5). Combined, these data provided a profile of the protein and gene expression in human fetal lung buds prior to 15 weeks gestation (summarized in Figure 1G).
Figure 1.
Characterization of Bud Tip Progenitors from 14 to 16 Weeks of Human Fetal Lung Development
(A) Expression of SOX2 and SOX9 in human fetal lungs at 14 and 16 weeks of gestation. Scale bar represents 50 μm.
(B) Expression of ID2 in human fetal lungs at 15 weeks gestation as identified by in situ hybridization. Scale bar represents 50 μm.
(C) Expression of SOX9 along with pro-SFTPC, SFTPB, PDPN, HOPX, ABCA3, or RAGE in human fetal lung at 15 weeks. Scale bars represent 50 μm (low-magnification images) and 25 μm (high-magnification images).
(D) Volcano plot of differentially expressed genes identified by comparing isolated, uncultured human fetal bud tips (n = 2 biological replicates; 8 and 12 weeks gestation) with whole adult human lung (publically available dataset). A total of 7,166 genes were differentially expressed (adjusted p value < 0.01).
(E) Heatmap showing expression of genes known to be expressed lung epithelial cells in isolated human fetal bud tips and whole adult human lung.
(F) Heatmap showing expression of 37 human bud-tip enriched transcription factors (Nikolić et al., 2017) in isolated human fetal bud tips and whole adult human lung. Twenty of the 37 transcription factors (∗) were statistically significantly enriched in isolated fetal bud tips.
(G) Summary of markers expressed by bud tip cells in regions adjacent to the bud tips at 14–15 weeks gestation as identified by protein staining and in situ hybridization.
Murine Bud Tip Growth In Vitro
To establish an experimental framework that would allow us to efficiently work with rare/precious human tissue samples, we first conducted a low-throughput screen using mouse epithelial bud tips to identify factors that promoted tissue expansion and maintenance of SOX9 expression. Epithelial bud tips were isolated from lungs of embryonic day (E) 13.5 Sox9-eGFP mice and cultured in a Matrigel droplet (Figure S2A). Isolated Sox9-eGFP lung bud tips were shown to express GFP and to have the same level of Sox9 mRNA as their wild-type counterparts by qRT-PCR analysis (Figures S2B and S2C). Treating isolated E13.5 mouse bud tips with no growth factors (basal medium control) or individual factors identified from the literature as important for lung development showed that FGF7 robustly promoted growth, expansion, and survival of isolated buds for up to 2 weeks (Figure S2D). Interestingly, the same concentration of FGF7 and FGF10 had different effects on lung bud outgrowth, an observation that could not be overcome even when buds were exposed to a 50-fold excess of FGF10 (Figure S2E).
FGF7, CHIR-99021, and RA Are Sufficient to Maintain the Expression of SOX9 In Vitro
A lineage trace utilizing isolated epithelial bud tips from Sox9-CreER;Rosa26Tomato mice showed that FGF7 promoted outgrowth of Sox9+ distal progenitors cells (Figure S2F). However, Sox9 mRNA and protein expression were significantly reduced over time (Figures S2G and S2M).
We therefore sought to identify additional growth factors that could maintain expression of SOX9 in vitro. To do this, we grew bud tips in medium with all five factors included in our initial screen (“5F” media: FGF7, FGF10, BMP4, CHIR-99021, and RA), removed one growth factor at a time, and examined the effect on expression of Sox9 and Sox2 (Figures S2H–S2J). Bud tips were grown in 5F minus one factor for 2 weeks in culture. Removing any individual factor, with the exception of FGF7, did not affect the ability of isolated buds to expand (Figure S2H). qRT-PCR analysis showed that removing BMP4 led to a statistically significant increase in Sox9 mRNA expression levels when compared with other culture conditions (Figure S2I). Removing any other single factor did not lead to statically significant changes in Sox9 expression (Figure S2I). Sox2 gene expression was generally low in all culture conditions (Figure S2J).
Our data demonstrated that FGF7 is critical for in vitro expansion of isolated murine bud tips and removing BMP4 enhanced Sox9 expression. We therefore screened combinations of the remaining factors to determine a minimal set of factors that could maintain high Sox9 expression (Figures S2K–S2N). Cultured buds treated with 4-factor (“4F”) (FGF7, FGF10, CHIR-99021, and RA) or 3F conditions (FGF7, CHIR-99021, and RA) supported robust SOX9 protein and mRNA expression (Figures S2L–S2N), with no significant differences between Sox9 expression between these two groups. Therefore, a minimum set of three factors (FGF7, CHIR-99021, and RA) are sufficient to allow growth of mouse epithelial bud tip progenitor cells and to maintain Sox9 expression in vitro.
In Vitro Growth and Maintenance of Human Fetal Distal Epithelial Lung Progenitors
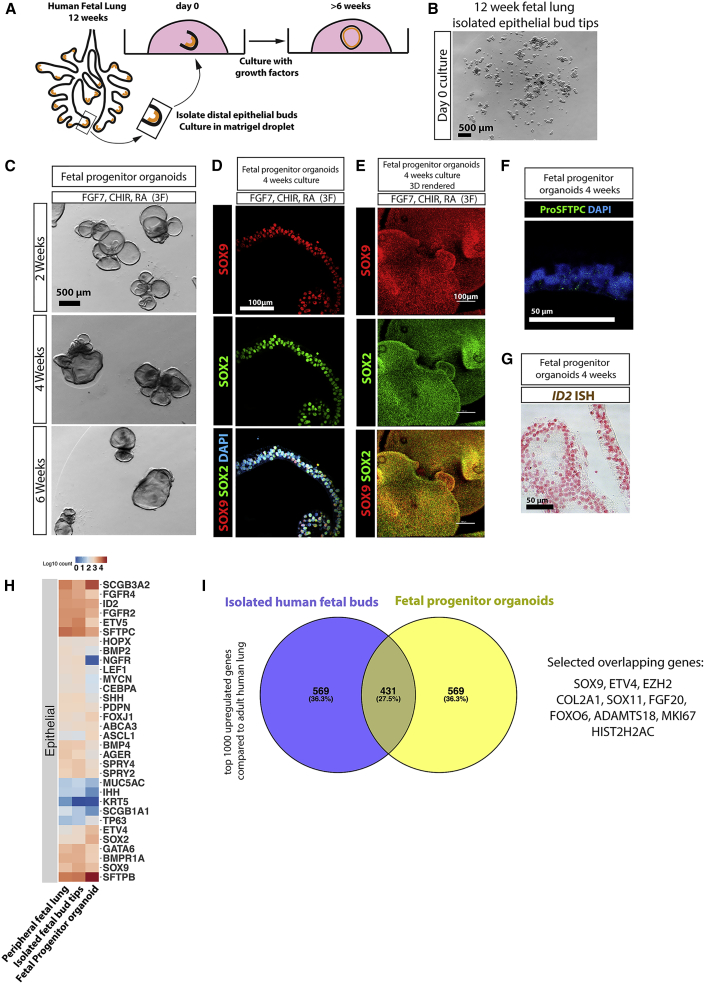
We asked if conditions supporting mouse bud tip progenitors also supported growth and expansion of human bud tip progenitors in vitro. Distal epithelial lung buds were enzymatically and mechanically isolated from the lungs of three different biological specimens at 12 weeks of gestation (84–87 days; n = 3) and cultured in a Matrigel droplet (Figures 2A and 2B). When human bud tips were cultured in vitro, we observed that FGF7 promoted an initial expansion of tissue in vitro after 2 and 4 weeks, but structures began to collapse by 6 weeks in culture (Figure S3A). All other groups tested permitted expansion and survival of buds as “fetal progenitor organoids” for 6 weeks or longer (Figures 2C and S3A). A description of the nomenclature for different tissues/samples/organoids used in this manuscript can be found in Table 1 of the Methods. Human fetal progenitor organoids exposed to 3F or 4F medium supported robust expression of the distal progenitor markers SOX9, SOX2, ID2, and NMYC (Figures S3B and S3C). In contrast, culture in only two factors (FGF7+CHIR-99021 or FGF7+RA) did not support robust bud tip progenitor marker expression (Figures S3B and S3C). qRT-PCR also showed that fetal progenitor organoids cultured in 3F or 4F medium expressed low levels of the proximal airway markers P63, FOXJ1, and SCGB1A1 when compared with FGF7-only conditions (Figures S3D and S3E).
Figure 2.
FGF7, CHIR-99021, and RA Are Sufficient to Maintain Isolated Human Fetal Bud Tip Progenitor Cells In Vitro
(A and B) Distal epithelial lung bud tips were isolated and cultured in Matrigel droplets. Scale bar represents 500 μm in (B).
(C). Whole-mount bright-field image of human fetal organoids grown in “3F” medium (FGF7, CHIR-99021, and RA) at 2, 4, and 6 weeks. Scale bar represents 500 μm.
(D) Protein staining of SOX2 and SOX9 in sections of fetal progenitor organoids grown in 3F medium. Scale bar represents 100 μm.
(E) Whole-mount staining, confocal z stack imaging, and 3D rendering of SOX2 and SOX9 in fetal progenitor organoids grown in 3F medium. Scale bar represents 100 μm.
(F) Pro-SFTPC after 4 weeks in culture in fetal progenitor organoids. Scale bars represent 50 μm.
(G) ID2 expression in fetal progenitor organoids after 4 weeks in culture as determined by in situ hybridization. Scale bar represents 50 μm.
(H) Heatmap showing expression of genes known to be expressed lung epithelial cells in the whole adult human lung (publically available dataset), in isolated human fetal bud tips (n = 2; 8 and 12 weeks gestation) and in fetal progenitor organoids (n = 2; both 12 weeks gestation, cultured for 2 weeks).
(I) Differential expression analysis (isolated fetal epithelial bud tips versus whole adult lung; fetal progenitor organoids versus whole adult lung) was used to identify the top 1,000 most highly upregulated genes from each comparison (log2 fold change < 0; adjusted p value < 0.05). Gene overlap was identified using a Venn diagram. Of all genes, 27.5% were common to both groups. A hypergeometric means test showed that the number of overlapping genes was highly significant (overlapping p value = 1.4 × 10−278).
Table 1.
Description of the Nomenclature Used in This Manuscript
| Sample Nomenclature | Description |
|---|---|
| Peripheral fetal lung | the distal/peripheral portion of the fetal lung (i.e., distal 0.5 cm) was excised from the rest of the lung using a scalpel. This includes all components of the lung (e.g., epithelial, mesenchymal, vascular). |
| Isolated fetal bud tip | the bud peripheral portion of the fetal lung was excised with a scalpel and subjected to enzymatic digestion and microdissection. The epithelium was dissected and separated from the mesenchyme, but a small amount of associated mesenchyme likely remained. |
| Fetal progenitor organoid | 3D organoid structures that arose from culturing isolated fetal epithelial bud tips. |
| Foregut spheroid | 3D foregut endoderm structure as described in Dye et al. (2015). Gives rise to patterned lung organoid (PLO) when grown in 3F medium. |
| Patterned lung organoid (PLO) | lung organoids that were generated by differentiating hPSCs, as described throughout the manuscript. |
| Bud tip organoid | organoids derived from PLOs, enriched for SOX2/SOX9 co-expressing cells, and grown/passaged in 3F medium. |
Collectively, our experiments demonstrated that FGF7, CHIR-99021, and RA represent a minimal set of additives required to maintain SOX9+/SOX2+ fetal progenitor organoids in vitro. Supporting mRNA expression data, we observed robust SOX9 and SOX2 protein expression as demonstrated by immunofluorescence in sections or by whole-mount staining after 4 weeks in culture (Figures 2D and 2E). Consistent with expression data in lung sections prior to 16 weeks gestation, we observed very weak pro-SFTPC protein expression in human bud tip progenitor organoids. Low levels of ID2 mRNA were also detected using in situ hybridization (Figures 2F and 2G). Protein staining for several markers was not detected in fetal progenitor organoids treated for 4 weeks in 3F medium, including P63, FOXJ1, SCGB1A1, MUC5AC, HOPX, RAGE, and SFTPB (n = 8; negative data not shown), consistent with human fetal epithelial bud tips prior to 16 weeks gestation (Figures S1A–S1E).
We also performed RNA-seq on tissue from the distal portion of fetal lungs (epithelium plus mesenchyme) (n = 3; 8, 12, and 18 weeks), on freshly isolated human epithelial bud tips (n = 2; 8, and 12 weeks) and on human fetal bud tip progenitor organoids grown in 3F medium for 4 weeks in culture (n = 2; 12 weeks, run with technical triplicates). Analysis revealed a high degree of similarity across samples when comparing epithelial gene expression (Figure 2H). In addition, we identified genes highly enriched in isolated fetal epithelial bud tips by conducting differential expression analysis on RNA-seq data. For this analysis, we compared whole human adult lung versus uncultured isolated lung buds (12 weeks gestation) and versus cultured fetal progenitor organoids (12 weeks gestation, cultured for 2 weeks). In these analyses, whole human lung was used as a “baseline,” with the assumption that average expression of any given gene would be low due to the heterogeneous mixture of cells pooled together in the sample, providing a good comparator to identify genes that were enriched in fetal tissue and organoid samples. The top 1,000 upregulated genes in bud tips and in fetal progenitor organoids were identified (log2 fold change < 0; adjusted p value < 0.05). When comparing upregulated genes, we observed that 431 (27.5%) of the genes were commonly upregulated in both fetal bud tips and cultured fetal organoids, which was highly statistically significant (p = 1.4 × 10−278 determined by a hypergeometric test) (Figure 2I).
FGF7, CHIR-99021, and RA Induce a Bud Tip Progenitor-like Population of Cells from hPSC-Derived Foregut Spheroids
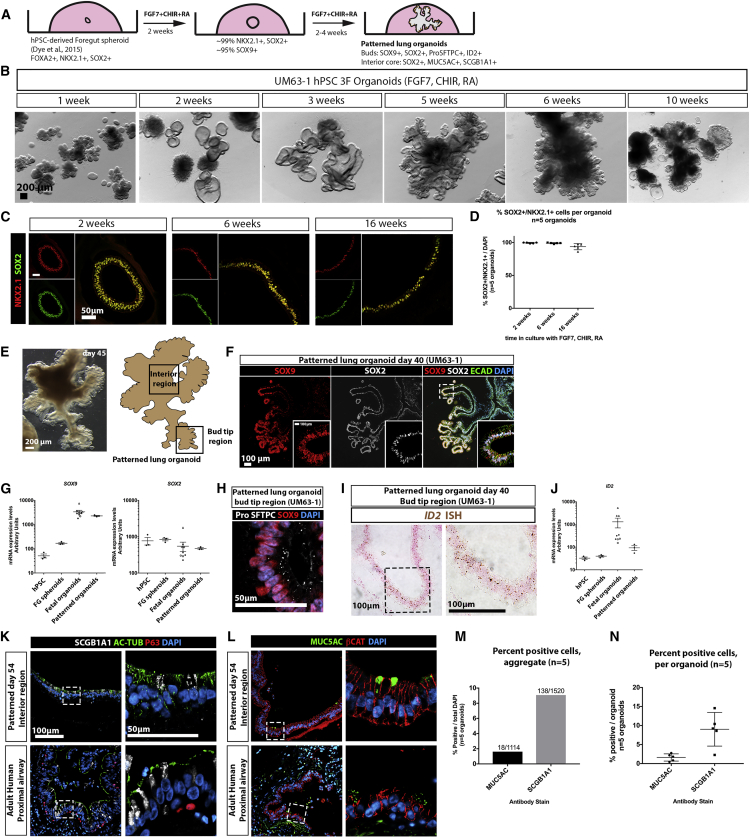
Given the robustness by which 3F medium (FGF7, CHIR-99021, and RA) supported mouse and human epithelial lung bud tip growth and maintenance in vitro, we sought to determine whether these culture conditions could promote an epithelial lung tip progenitor-like population from hPSCs. NKX2.1+ ventral foregut spheroids were generated as described previously (Dye et al., 2015, Dye et al., 2016a), and were cultured in a droplet of Matrigel and overlaid with 3F medium. Spheroids were considered to be “day 0” on the day they were placed in Matrigel (Figures 3A and 3B). Foregut spheroids cultured in 3F medium generated patterned organoids (hereafter referred to as “hPSC-derived patterned lung organoids”; PLOs) that grew robustly (Figures S4A and S4B), demonstrating predictable growth patterns (Figures S4C and S4D) and reproducible gene expression across several different embryonic and induced pluripotent stem cell lines (n = 4) (Figures 3B, S4G, and S4H). PLOs survived for over 16 weeks in culture, and could be frozen, thawed, and re-cultured (Figures S4E, S4E', and S4F).
Figure 3.
FGF7, CHIR-99021, and RA Generate Patterned Epithelial Lung Organoids from hPSCs
(A) Schematic of approach to derive patterned lung organoids (PLOs) from human pluripotent stem cells.
(B) Bright-field images of hPSC-derived foregut spheroids cultured in 3F medium (FGF7, CHIR-99021, and RA) and grown in vitro. Images taken at 2, 3, 5, 6, and 10 weeks. Images from a single experiment, representative of 6 experiments across 4 hPSC cell lines. Scale bars represent 200 μm.
(C) Immunostaining for NKX2.1 and SOX2 in PLOs at 2, 6, and 16 weeks. Scale bars represent 50 μm.
(D) Quantitative analysis of cells co-expressing NKX2.1 and SOX2 in PLOs at 2, 6, and 16 weeks, as shown in (B). Each data point (n = 5) represents a single organoid from one experiment, and the mean ± SEM is shown for each group. Data are representative of 6 experiments with 4 different cell hPSC lines.
(E) Bright-field image of a patterned lung organoid after 6 weeks (45 days) in culture, showing distal budded regions and interior regions, and a schematic representing a patterned lung organoid, highlighting bud tip region and interior regions. Scale bar represents 200 μm.
(F) PLOs co-stained for SOX9 and SOX2 protein expression by immunofluorescence. Inset: high magnification of boxed region. Scale bar represents 100 μm.
(G) qRT-PCR analysis of SOX9 and SOX2 in undifferentiated hPSCs (H9 hESC line, n = 3 independent samples from one experiment), in foregut spheroids (FG; n = 3 independent samples obtained from one experiment; each sample contains ≥25 pooled spheroids), fetal progenitor organoids (n = 3 biological replicates [12, 12, and 13 weeks] and 3 technical replicates each) and patterned lung organoids (n = 3 independent samples obtained from 1 experiment; each sample contained ≥3 pooled organoids). Data are representative of 6 experiments with 4 different cell hPSC lines.
(H) Pro-SFTPC and SOX9 co-expression in bud tip region of a PLO. Scale bars represent 50 μm.
(I) ID2 expression in PLOs after 40 days in vitro as determined by in situ hybridization. Scale bar represents 100 μm.
(J) qRT-PCR analysis of ID2 in undifferentiated hPSCs in undifferentiated hPSCs (H9 hESC line, n = 3 independent samples from 1 experiment), in foregut spheroids (FG; n = 3 independent samples obtained from 1 experiment; each sample contains ≥25 pooled spheroids), fetal progenitor organoids (n = 3 biological replicates [12, 12, and 13 weeks] with 3 technical replicates each) and patterned lung organoids (n = 3 independent samples obtained from 1 experiment; each sample contained ≥3 pooled organoids). Data are representative of 6 experiments with 4 different cell hPSC lines.
(K) Interior regions of patterned lung organoids (top) and adult human airway (bottom) co-stained for SCGB1A1, acetylated tubulin (AC-TUB) and P63. Scale bars represent 100 μm (left panels, low magnification) or 50 μm (right panels, high magnification).
(L) Interior regions of patterned lung organoids (top) and adult human airway (bottom) co-stained for MUC5AC and the epithelial marker β-catenin (βCAT). Scale bar in (K) also applies to images in (L); scale bars represent 100 μm (left panels, low magnification) or 50 μm (right panels, high magnification).
(M and N) Percent of cells expressing MUC5AC or SCGB1A1, plotted as aggregate data (number of cells positive in all organoids/total cells counted in all organoids) (M), and for each individual patterned lung organoid counted (number of cells positive in individual organoid/all cells counted in individual organoid) (N). N = 5 organoids for each graph, from a single experiment of organoids derived from UM63-1 hPSC line. Data are representative of 6 experiments across 4 hPSC lines.
At 2 and 6 weeks, PLOs co-expressed NKX2.1 and SOX2 in >99% of all cells (99.41% ± 0.82% and 99.06% ± 0.83%, respectively), while 16-week-old PLOs possessed 93.7% ± 4.6% NKX2.1+/SOX2+ cells (Figures 3C and 3D). However, PLOs at this later time point appeared less healthy (Figure 3B). Interestingly, no mesenchymal cell types were detected in PLO cultures at 2, 6, or 16 weeks by immunofluorescence (VIM, α-SMA, PDGFRα; negative immunostaining data not shown), and qRT-PCR analysis confirms very low expression of mesenchymal markers in PLOs generated from n = 3 hPSC lines (Figure S4H).
PLOs also exhibited regionalized proximal-like and bud tip-like domains (Figures 3E and 3F). In 100% of analyzed PLOs (n = 8), peripheral budded regions contained cells that co-stained for SOX9 and SOX2, whereas interior regions of the PLOs contained cells that were positive only for SOX2 (Figures 3E and 3F). PLOs expressed similar levels of SOX2 and SOX9 compared with fetal progenitor lung organoids (Figure 3G). Budded regions of PLOs also contained SOX9+ cells that weakly co-expressed pro-SFTPC, and ID2 based on in situ hybridization and qRT-PCR (Figures 3H–3J).
Interior regions of PLOs contained a small number of cells that showed positive immunostaining for the club cell marker SCGB1A1 (9%) and the goblet cell marker MUC5AC (1%), with similar morphology to adult human proximal airway secretory cells (Figures 3K–3N). No multiciliated cells were present, as AC-TUB was not localized to cilia (Figure 3K) and FOXJ1 staining was absent (data not shown). In addition, the basal cell marker P63 was absent from PLOs (Figure 3K), as was staining for markers of lung epithelial cell types including HOPX, RAGE, PDPN, ABCA3, SFTPB, and chromagranin A (CHGA) (negative data not shown).
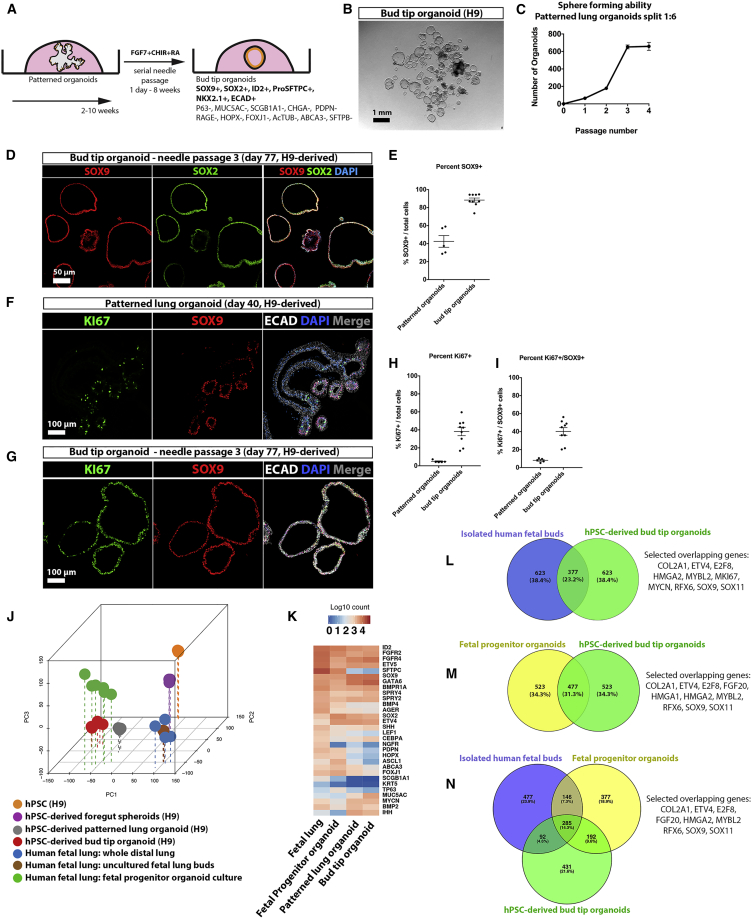
Expansion of Epithelial Tip Progenitor-like Cells from PLOs
PLOs gave rise to epithelial cysts after being passaged by mechanical shearing through a 27-gauge needle, followed by embedding in fresh Matrigel and growth in 3F medium (Figures 4A and 4B). PLOs were successfully needle passaged as early as 2 weeks, and as late as 10 weeks, with similar results. Needle-passaged cysts can be generated from hPSCs in as little as 24 days (9 days to generate foregut spheroids, 14 days to expand PLOs, and 1 day to establish cysts from needle-passaged organoids) (Figure 4A). Needle-passaged epithelium re-established small cysts within 24 hours and could be serially passaged every 7–10 days (Figure 4C). The cysts that formed after needle passaging were NKX2.1+ (Figure S5A) and cells co-expressed SOX2 and SOX9 (Figures 4D and 4E). Based on these protein-staining patterns, we refer to these cysts as “bud tip organoids.” Compared with PLOs, bud tip organoids possessed a much higher proportion of SOX9+ cells (42.5% ± 6.5%, n = 5; versus 88.3% ± 2.3%, n = 9; Figure 4E) and proliferating cells, as assessed by KI67 immunostaining (38.24% ± 4.7%, n = 9 for bud tip organoids versus 4.9% ± 0.6%, n = 5 for PLOs; Figures 4F–4I). In PLOs, we noted that proliferation was largely restricted to SOX9+ budded regions, but only a small proportion of SOX9+ cells were proliferating (8.1% ± 0.9%, n = 5), whereas bud tip organoids had a much higher proportion of proliferative SOX9+ cells (40.2% ± 4.3%, n = 9) (Figure 4I). Together, these data suggest that needle passaging enriches the highly proliferative bud tip regions of PLOs.
Figure 4.
Proliferative SOX9+/SOX2+ Progenitors Can Be Expanded from Patterned Lung Organoids
(A) Schematic of approach passage PLOs and expand bud tip organoids.
(B) Needle-sheared epithelial fragments were replated in a fresh Matrigel droplet that subsequently formed cystic structures, called “bud tip organoids.” Scale bar represents 1 mm.
(C) Quantitative assessment of organoid passaging and expansion. One single patterned lung organoid was needle passaged into 6 wells (passage 1), generating 75 new bud tip organoids in total (average 12.5 per well). A single well of the resulting bud tip organoids was then passaged into 6 new wells after 2 weeks in culture (1:6 split ratio), generating 200 new organoids in total (average 33 per well). This 1:6 passaging was carried out two additional times, every 1–2 weeks for up to 4 passages before growth plateaued. Three technical replicates were performed for expansion experiments using patterned lung organoids generated from the UM63-1 hPSC line; the graph plots the mean ± SEM. Data are representative of 6 experiments across 4 hPSC lines.
(D) Immunostaining for SOX9 and SOX2 in bud tip organoids. Scale bar represents 50 μm.
(E) Quantitation of the percent of SOX9+ cells in PLOs and bud tip organoids (number of SOX9+ cells/total cells). Each data point represents an independent organoid (n = 5 organoids) derived from a single experiment using H9 hPSC line and graphs indicate the mean ± SEM for each experimental group. Data are representative of 6 experiments across 4 hPSC lines.
(F and G) Immunostaining for KI67 and SOX9 in patterned lung organoids (F) and bud tip organoids (G). Scale bar represents 100 μm.
(H) Quantitation of the percent of all cells that were KI67+ in patterned and bud tip organoids (number of KI67+ cells/total cells).
(I) Quantitation of the percent of proliferating SOX9+ cells in patterned and bud tip organoids (number of KI67+/SOX9+ cells/total cells).
(H and I) Each data point represents an independent organoid (n = 5 patterned lung organoids and n = 9 bud tip organoids) derived from a single experiment using H9 hPSC line and graphs indicate the mean ± SEM for each experimental group. Data are representative of 6 experiments across 4 hPSC lines.
(J) Principal component analysis of RNA-seq data to compare the global transcriptome of hPSCs (n = 3 independent replicates of H9 hPSCs), foregut spheroids (n = 3 independent replicates of H9 hPSCs-derived spheroids. Each replicate contained ≥25 pooled spheroids), hPSC-derived patterned lung organoids (n = 3 independent replicates of H9 hPSCs-derived patterned lung organoids; each replicate contained n ≥ 3 pooled organoids), hPSC-derived bud tip organoids (n = 3 independent replicates of H9 hPSCs-derived bud tip organoids; each replicate contained n ≥ 3 organoids), whole peripheral (distal) fetal lung (n = 3; 8, 12, and 18 weeks), freshly isolated (uncultured) fetal lung buds (n = 2; 8 and 12 weeks gestation) and fetal progenitor organoids (n = 2; both 12 weeks gestation and cultured for 2 weeks). All biological replicates were run in technical triplicate.
(K) Heatmap showing expression of genes known to be expressed in lung epithelial cells in freshly isolated (uncultured) fetal lung buds and fetal progenitor organoids cultured for 2 weeks.
(L–N) Differential expression analysis of (1) isolated fetal bud tips versus whole adult lung, (2) fetal progenitor organoids versus whole adult lung, and (3) bud tip organoids versus whole adult lung was used to identify the top 1,000 most highly upregulated genes from each comparison (log2 fold change < 0; adjusted p value < 0.05). A Venn diagram illustrates common upregulated genes in (L) isolated bud tips and bud tip organoids, (M) fetal progenitor organoids and bud tip organoids, (N) isolated bud tips, fetal progenitor organoids, and bud tip organoids. (L) hypergeometric means test found that the shared gene overlap was highly significant (overlapping p value = 9.3e-901). (M) Hypergeometric means test found that the shared gene overlap was highly significant (overlapping p value = 1.2e-1,021). (N) A total of 285 overlapping genes were shared between the three groups. These genes represented 14.3% of all genes included in the comparison. A small subset of genes previously associated with human or mouse bud-tip progenitor cells are highlighted as “Selected overlapping genes” (L–N).
Bud tip organoids were further characterized using in situ hybridization and immunofluorescence (Figures S5B–S5D). Bud tip organoids exhibited protein-staining patterns consistent with 12–14 week fetal lungs, including the absence of HOPX, RAGE, PDPN, and ABCA3, while we did observe low ID2 expression (Figures S5B–S5D). Furthermore, no positive protein staining was detected for the proximal airway markers P63, FOXJ1, AC-TUB, MUC5AC, SCGB1A1, or CHGA (negative data not shown).
We then conducted unbiased analysis of bud tip organoids using RNA-seq to compare (1) hPSC-derived bud tip organoids; (2) whole peripheral (distal) human fetal lung tissue; (3) freshly isolated fetal bud tips and human fetal lung progenitor organoids; (4) undifferentiated hPSCs; and (5) hPSC-derived lung spheroids. Principal-component analysis and Spearman's correlation clustering revealed the highest degree of similarity between hPSC-derived bud tip organoids, PLOs, and human fetal organoids (Figures 4J and S5E). Interestingly, freshly isolated bud tips and whole distal human fetal lungs clustered together, while all cultured tissues (bud tip organoids, PLOs, and fetal progenitor organoids) clustered together, likely reflecting gene expression similarities driven by the tissue culture environment. This analysis highlights a high degree of molecular similarity between human fetal and hPSC-derived organoids grown in vitro (Figure 4J). A curated heatmap shows a high degree of similarity in gene expression of lung epithelial markers between the whole fetal lung, fetal progenitor organoids, PLOs, and bud tip organoids (Figure 4K).
Differential expression of RNA-seq data was also used to interrogate the relationship between tissues. We obtained upregulated genes from the following comparisons: (1) uncultured, freshly isolated lung buds (8 and 12 weeks gestation) versus whole adult lung tissue; (2) cultured fetal progenitor organoids (12 weeks gestation, cultured for 2 weeks) versus whole adult lung tissue; and (3) hPSC-derived bud tip organoids versus whole adult lung tissue. The top 1,000 upregulated genes in each of the three groups were identified (log2 fold change < 0; adjusted p value < 0.05), and overlapping genes were identified (Figures 4L and 4M). A hypergeometric means test found that overlap of enriched genes from isolated human bud tips and hPSC-derived bud tip organoids were highly significant, with 377 (23.2%) overlapping genes (p = 9.3e-901; Figure 4L). Overlap between cultured fetal progenitor organoids and hPSC-derived bud tip organoids was also highly significant, with 477 (31.3%) overlapping genes (p = 1.2e-1,021) (Figure 4M). Of note, when all three groups were compared, a core group of 285 common upregulated genes were identified, representing 14.3% of genes. Of these 285, several have been associated with human or mouse fetal bud tips (Nikolić et al., 2017, Rockich et al., 2013) (Figure 4N; COL2A1, ETV4, E2F8, FGF20, HMGA2, MYBL2, RFX6, SALL4, SOX9, and SOX11).
hPSC-Derived Bud Tip Organoids Maintain Multilineage Potential In Vitro and In Vivo
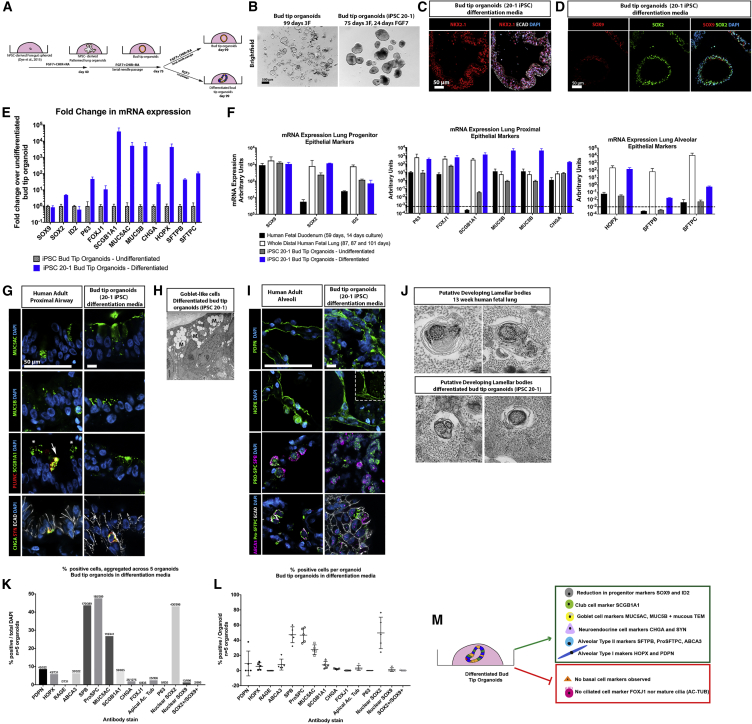
To demonstrate that hPSC-derived organoids have bona fide progenitor potential, we took approaches to differentiate them in vitro (Figure 5) and by transplanting them into injured mouse lungs in vivo (Figures 6 and S6). For these experiments, we used induced pluripotent stem cells (iPSC)-derived foregut spheroids to generate bud tip organoids (iPSC line 20-1 (iPSC20-1) [Spence et al., 2011]).
Figure 5.
Bud Tip Organoids Retain Multilineage Potential In Vitro
(A) Schematic of experimental setup. iPSC20-1 bud tip organoids were initially cultured in 3F medium and subsequently grown in 3F medium or medium containing FGF7 alone (“differentiation media”) for 24 days.
(B) Bright-field images of bud tip organoids growing in 3F medium (left) or FGF7-differentiation medium (right). Scale bar represents 500 μm.
(C) NKX2.1 immunofluorescence of bud tip organoids grown in FGF7-differentiation medium for 24 days. Scale bar represents 50 μm.
(D) SOX9 and SOX2 immunofluorescence of bud tip organoids grown in FGF7-differentiation medium for 24 days. Scale bar represents 50 μm.
(E) qRT-PCR analysis of bud tip organoids grown in 3F or FGF7-differentiation medium showing expression of several genes expressed in the lung epithelium. Data are shown as fold change relative to 3F-grown undifferentiated bud tip organoids. Each data point represents an independent sample (n = 3) obtained from one experiment derived from iPSC 20-1 hPSC line; each sample contained ≥3 pooled organoids. Data are representative of 5 experiments across 2 hPSC lines (iPSC20-1 and H9).
(F) qRT-PCR analysis of bud tip organoids grown in 3F or FGF7-differentiation medium, along with whole distal fetal lung and cultured whole fetal intestine as reference tissues, showing expression of several genes expressed in the lung epithelium. Data are shown as a.u. Values lower than 10−3 were considered undetected. Each data point in bud tip organoid groups represents an independent sample (n = 3) obtained from one experiment derived from iPSC 20-1 hPSC line; each sample contained ≥3 pooled organoids. Data are representative of 5 experiments across 2 hPSC lines (iPSC20-1 and H9). Whole human distal lung samples (n = 3) were taken from 87, 87, and 101 day gestation, and 3 independent samples of human fetal duodenum tissue were used as a comparison.
(G) Immunostaining for airway markers in the adult human lung, and in bud tip organoids grown in FGF7-differentiation medium. Markers are shown for goblet cells (MUC5AC, MUC5B), club cells (SCGB1A1, PLUNC), and neuroendocrine cells (synaptophysin [SYN], CHGA). Scale bars represents 50 μm.
(H) Transmission electron microscopy through a bud tip organoid grown in FGF7-differentiation medium showing mucus-filled vacuoles characteristic of goblet cells. Scale bar represents 100 nm.
(I) Immunostaining for alveolar markers in the adult human lung, and in bud tip organoids grown in FGF7-differentiation medium. Markers are shown for AECI cells (PDPN, HOPX) and AECII cells (pro-SFTPC, SFTPB, ABCA3). Scale bars represent 50 μm.
(J) Transmission electron microscopy of a human fetal lung at 13 weeks of gestation, and of a bud tip organoid grown in FGF7-differentiation medium showing immature lamellar bodies surrounded by monoparticulate glycogen, characteristic of immature AECII cells. Scale bars represent 100 nm.
(K and L) Quantitation of cell type markers in bud tip organoids grown in differentiation medium plotted as aggregate data (numbers at top of bars represent positive cells/total cells counted across five individual organoids) (H), and as individual data per organoid (number of positive cells per organoid) (I). Graphs indicate the mean ± SEM. Data are from a single differentiation experiment derived from iPSC 20-1 hPSC line and each data point represents an individual organoid (n = 5 organoids). Data are representative of five experiments across two hPSC lines (iPSC20-1 and H9).
(M) Summary of putative differentiated lung epithelial cell types generated in vitro.
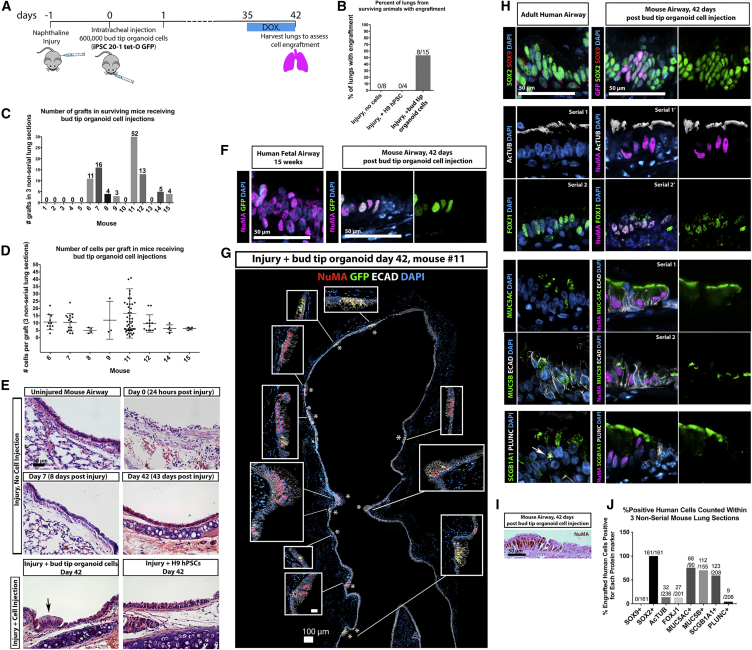
Figure 6.
Engraftment of hPSC-Derived Bud Tip Progenitor Organoid Cells into the Injured Mouse Airway
(A) Schematic of experimental design. Immunocompromised NSG male mice were injected with 300 mg/kg of naphthalene. Twenty-four hours post injury, mice were randomly assigned to receive an intratracheal injection of 600,000 single cells isolated from bud tip organoids generated from the iPSC 20-1 tet-O GFP line, undifferentiated H9 hPSCs, or no injection of cells. Doxycycline (1 mg/mL) was added to the drinking water for the final week to induce expression of the tet-O GFP construct. Lungs were analyzed 6 weeks after cell injection.
(B) Percent of lungs from surviving animals in each group exhibiting engraftment of human cells after 6 weeks, as determined by NuMA and GFP protein staining.
(C and D) Engraftment was assessed based on human-specific expression on NuMA and GFP in three independent histological sections from each surviving mouse. (C) The number of engraftment cell patches observed in 15 surviving animals in the group receiving bud tip organoid cells. (D) Quantitation of the number of human cells in each engrafted cell patch, in each mouse. Every data point represents the number of cells in a single patch of cells.
(E) H&E staining documenting the lung epithelial airway injury in control mice (injury, no cell injection), and in mice that received bud tip organoids, or undifferentiated iPSC injections. Engrafted patches of human cells were obvious (arrow), and were confirmed in serial sections using human-specific antibodies. Scale bar represents 50 μm for all images.
(F) Immunostaining for NuMA and GFP in human fetal lungs and in bud tip organoid transplanted lungs. Scale bar represents 50 μm.
(G) Bud tip organoid transplanted lung showing several images stitched together to generate a large panel demonstrating multiple sites of engraftment (denoted by asterisks), marked by NuMA and/or GFP, in the upper airway. High-magnification insets are shown. Scale bar represents 100 μm.
(H) Immunostaining of adult human lung tissue and bud tip organoid transplanted lung tissue showing immunostaining for several lung epithelial markers, including SOX9 and SOX2, multiciliated cell markers AC-TUB and FOXJ1, goblet cell markers MUC5AC and MUC5B, club cell markers SCGB1A1 and PLUNC. Arrow highlights a cell co-expressing SCGB1A1/PLUNC. Scale bar represents 50 μm.
(I) Bright-field microscopic image showing immunohistochemistry for NuMA in a patch of engrafted cells, counterstained using eosin to visualize tufts of multiple cilia. Scale bar represents 50 μm.
(J) Quantification of cell type markers in bud tip organoid transplanted lungs after 6 weeks. Data are plotted as aggregate data (numbers at top of bars represent positive cells/total cells counted across three engrafted lungs). Aggregated data from three non-serial sections for mouse 7, 11, and 12 is plotted.
In Vitro Differentiation
Since we were interested in investigating the potential of bud tip progenitor organoids to give rise to any lung epithelial lineage, we reasoned that withdrawing CHIR-99021 and RA may allow the cells to stochastically differentiate. Therefore, bud tip organoids were split into two treatment groups: 3F medium or FGF7-only for 23 days (Figure 5A). At the end of the experiment, control bud tip organoids maintained a clear, thin epithelium with a visible lumen, whereas FGF7-differentiated organoids appeared as dense cysts with mucus-like material inside of some organoids (Figure 5B). FGF7-differentiated organoids remained NKX2.1+/ECAD+ and SOX2+, signifying that they retained a lung epithelial identity, but showed decreased SOX9 protein expression (Figures 5C and 5D). Gene expression analysis using qRT-PCR to compare 3F (undifferentiated) versus FGF7-differentiated organoids showed significant increases of genes expressed in differentiated airway and alveolar epithelium (Figure 5E). When further compared with the whole distal portion of human fetal lung (positive control) and cultured human fetal duodenum (negative control), differentiated bud tip organoids showed a large increase relative to 3F in many genes associated with differentiated epithelial cells, with many genes trending toward levels seen in the whole fetal lung (Figure 5F).
Protein staining for airway and alveolar markers was carried out on human lung tissue samples as a reference and in FGF7-differentiated organoids (Figures 5G and 5I). We observed that cells within FGF7-differentiated organoids expressed proximal airway markers, including the goblet cell markers MUC5AC and MUC5B, the club cell markers SCGB1A1, and the neuroendocrine cell markers SNY and CHGA (Figure 5F). Of note, we observed that SCGB1A1 and PLUNC were co-expressed in only a small subset of club cells in the adult lung, whereas many club cells were marked only with SCGB1A1 (Figure 5G). We did not observe P63, FOXJ1, or cilia with AC-TUB staining in FGF7-differentiated organoids (negative data not shown). Transmission electron microscopy (TEM) also revealed cells with mucus-containing vesicles characteristic of goblet cells within FGF7-differentiated organoids (Figure 5H). Immunostaining for alveolar markers revealed cells positive for AECI markers PDPN and HOPX, as well as cells double-positive for AECII markers pro-SFTPC and SFTPB and pro-SFTPC and ABCA3 (Figure 5I). Of note, TEM also revealed putative lamellar bodies that were similar to those seen in 13 week human fetal lungs (Figure 5J). In both tissues, putative lamellar bodies were characteristically surrounded by what appears to be monoparticulate glycogen (Stahlman et al., 2000). Both iPSC-derived and human fetal tissue lamellar bodies appear to be immature, and did not possess the typical concentric lamellae of mature structures (Vanhecke et al., 2010) (Figure 5J). Based on both immunofluorescence and TEM data, it is most likely that the alveolar cell types observed in FGF7-differentiated organoids are reminiscent of an immature cell, and may require more specific conditions to further mature the primitive AECI and AECII cells.
Cells expressing cell-type-specific lung epithelial markers were quantitated within FGF7-differentiated organoids, showing that organoids possessed cells expressing the alveolar markers: PDPN, 8.6% of total cells; HOPX, 5.9% of total cells; SFTPB, ∼45% of total cells; pro-SFTPC, ∼45% of total cells; and ABCA3, 6.3% of total cells (Figures 5K and 5L). Many cells expressed airway markers: 43.2% of cells stained positive for nuclear SOX2, but not SOX9 (Figures 5K and 5L), 26.8% of cells stained positive for MUC5AC, while 7% of cells were positive for the club cell marker SCGB1A1, while FOXJ1 and P63 were absent (Figures 5K and 5L). In addition, 2.0% of cells expressed very clear co-staining for CHGA and synaptophysin, markers for neuroendocrine cells (Figures 5G, 5K, and 5L).
In Vivo Engraftment
We next sought to explore the differentiation potential of hPSC-derived bud tip progenitor organoids by transplanting them into the airways of injured mouse lungs (Figures 6 and S6). Recent studies have shown that damaging the lung epithelium promotes engraftment of adult human lung epithelial cells (Ghosh et al., 2017).
Interestingly, while we observed engrafted cells in the mouse airway 7 days post injury (Figures S6A–S6E), 79% of engrafted human cells were still co-labeled by nuclear SOX2 and nuclear SOX9 and remained highly proliferative (Figures S6F–S6J). Immunofluorescent staining showed that, of the small proportion of cells positive for only SOX2, 42.8% expressed the club cell marker SCGB1A1, and 25% expressed the goblet cell marker MUC5AC (Figures S6K–S6N), a large increase when compared with bud tip organoid controls (Figures S6O and S6P). Multiciliated cells were not observed (negative FOXJ1 staining in Figure S6K), nor were basal cells or alveolar cell markers (basal cells: P63, KRT5; AECI: HOPX, PDPN, RAGE; AECII: pro-SFTPC, SFTPB, ABCA3; negative staining not shown). Collectively, our data suggested that hPSC-derived lung bud tip progenitor-like cells can engraft into the injured airway; however, the majority of engrafted cells (79%; Figures S6F and S6G) retained expression of SOX9 after 7 days.
To further determine whether engrafted cells could differentiate into lung cell lineages, we carried out a long-term engraftment experiment where lungs were harvested 6 weeks after injection of cells (Figure 6). Mice received naphthalene injury 24 hr prior to intratracheal injection of 600,000 dissociated bud tip organoid cells derived from iPSCs (iPSC20.1 tet-O GFP) (Figure 6A). Doxycycline was given on the final week of the experiment to induce expression from the tet-O-GFP transgene in the iPSC-derived cells. Experimental cohorts included: injury plus no cell injection (n = 8 surviving animals); injury plus undifferentiated hPSC injection (n = 4 surviving animals); and injury plus bud tip organoid cell injection (n = 15 surviving animals) (Figure 6B). Of the 15 surviving mice that received bud tip organoid injections, 8 showed patches of NUMA+/GFP+ cells that engrafted into the airway (Figures 6C, 6D, 6F, and 6G). The number of engrafted cell patches (Figure 6C) and the number of cells per patch (Figure 6D) varied across individual mice. Lungs from animals in all experimental groups successfully recovered from the injury (Figure 6E). Engrafted human cells were not observed in the “no cell injection” nor the “undifferentiated hPSC injection” cohorts (Figure 6B), as determined by NUMA+/GFP+ staining (Figures 6F and 6G). While two mice that showed successful engraftment of bud tip organoid cells within the airway exhibited a total of four small patches of engrafted cells in the bronchioles, the overwhelming majority of engrafted cells were found in the trachea and primary/secondary bronchi of mice. The rare engrafted cells within the bronchioles stained positive for proximal cell markers SOX2, MUC5AC, or FOXJ1/AC-TUB (data not shown).
We conducted immunostaining for several lung epithelial markers to determine if engrafted tissue had differentiated, and we compared staining patterns with adult human lung tissue (Figure 6H). Immunostaining of the injury group that did not receive cells was also examined (Figures S6Q and S6R). We noted that engrafted human cells expressed low levels of NKX2.1, consistent with the human airway, where the majority of cells expressed very low levels of NKX2.1 (Figures S6S and S6T). Engrafted human cells (as determined by GFP or NuMA expression) also expressed SOX2, but not SOX9, suggesting that these cells had differentiated into an airway fate (Figure 6H). Consistent with this observation, we observed engrafted human cells that possessed multiple cilia and co-expressed NuMA and multiciliated cell markers, Ac-Tub, and FOXJ1 (Figure 6H), as determined in histological sections (Figure 6I). We also observed human cells expressing goblet cell markers MUC5AC and MUC5B. Finally, we observed human cells expressing the club cell marker SCGB1A1, but not PLUNC. Interestingly, we noted that PLUNC only marked a small subset of club cells in the human airway, suggesting that there is an underappreciated heterogeneity within this population in the human lung (Figure 6H). Quantitation of immunostaining confirmed that 100% of engrafted human cells were SOX2+, with roughly 75% of cells adopting a mucus-producing phenotype, ∼13% adopting a ciliated cell profile, and ∼0.5% exhibiting a neuroendocrine cell profile (Figure 6H). We did not find evidence that engrafted human cells expressed the basal cell marker P63 nor alveolar cell specific markers (negative data not shown).
Discussion
The ability to study human lung development, homeostasis, and disease is limited by our ability to carry out functional experiments in human tissues. This has led to the development of many different in vitro model systems using primary human tissue, and using cells and tissues derived from hPSCs (Dye et al., 2016b, Miller and Spence, 2017). Current approaches to differentiate hPSCs have used many techniques, including the stochastic differentiation of lung-specified cultures into many different lung cell lineages (Chen et al., 2017, Huang et al., 2013, Wong et al., 2012), fluorescence-activated cell sorting-based methods to purify lung progenitors from mixed cultures followed by subsequent differentiation (Gotoh et al., 2014, Jacob et al., 2017, Konishi et al., 2015, Longmire et al., 2012, McCauley et al., 2017), and by expanding three-dimensional ventral foregut spheroids into lung organoids (Dye et al., 2016a, Dye et al., 2015). Many or all, of these approaches rely on the differentiation of a primitive NKX2.1+ lung progenitor cell population, followed by subsequent differentiation into different cell lineages. However, during normal development, early NKX2.1-specified lung progenitors transition through a SOX9+ bud tip progenitor cell state on their way to terminal differentiation into both alveolar and airway cell fates (Branchfield et al., 2015, Rawlins et al., 2009). Despite the developmental importance of this bud tip progenitor, differentiation of a similar progenitor from hPSCs has remained elusive. For example, several studies have identified robust methods to sort and purify lung epithelial progenitor cells from a mixed population (Gotoh et al., 2014, Konishi et al., 2015, McCauley et al., 2017); however, whether or not these populations represent transition through an epithelial bud tip-like state is unknown. Thus, our findings demonstrating that hPSCs can be induced into a bud tip-like progenitor state represents an important step toward faithfully mimicking embryonic lung development in vitro. Our studies also suggest that the ability to induce, de novo, robust populations of cells from hPSCs that do not rely on specialized sorting or purification protocols represents biologically robust experimental findings. These findings can be used in a manner that predicts how a naive cell will behave in the tissue culture dish with a high degree of accuracy, and across multiple cell lines (D'Amour et al., 2005, Green et al., 2011, Spence et al., 2011).
Our studies also identified significant species-specific differences between the human and fetal mouse lung. Differences included both gene/protein expression differences, as well as functional differences when comparing how cells responded to diverse signaling environments in vitro. These mouse-human differences highlight the importance of validating observations made in hPSC-derived tissues by making direct comparisons with human tissues, as predictions or conclusions about human cell behavior based on results generated from the developing mouse lung may be misleading.
Our experimental findings, in combination with previously published work, have also raised new questions that may point to interesting avenues for future mechanistic studies to determine how specific cell types of the lung are generated. Previously, we have shown that lung organoids grown in high concentrations of FGF10 predominantly give rise to airway-like tissues, with a small number of alveolar cell types and a lack of alveolar structure, and these organoids also possess abundant mesenchymal cell populations (Dye et al., 2016a, Dye et al., 2015). Here, our results suggest that high concentrations of FGF10 alone do not play a major role in supporting robust growth of epithelial bud tip progenitor cells. We also note that lung organoids grown in high FGF10 possess P63+ basal-like cells (Dye et al., 2015), whereas bud tip organoids grown in 3F medium lack this population. These findings suggest that we still do not fully appreciate how various signaling pathways interact to control cell fate decisions or expansion of mesenchymal populations, and lay the groundwork for many future studies.
Taken together, our current work has identified a signaling network required for the induction, expansion, and maintenance of hPSC-derived lung epithelial bud tip progenitors. Simple needle passaging allowed us to expand a nearly homogeneous population of proliferative bud tip-like progenitor cells for over 16 weeks in culture, which remained multipotent in vitro and which were able to engraft into injured mouse lungs, terminally differentiate, and respond to systemic factors. The current study thus offers a robust and reproducible method to generate and maintain epithelial bud tip progenitors, which will facilitate future studies aimed at elucidating fundamental developmental mechanisms regulating human lung progenitor cells, and which may have applicability to regenerative medicine in the future.
Experimental Procedures
Mouse Models
All animal research was approved by the University of Michigan Committee on Use and Care of Animals. Lungs from Sox9-eGFP (MGI ID: 3844824), Sox9CreER; RosaTomato/Tomato (MGI ID: 5009223 and 3809523) (Kopp et al., 2011), or wild-type lungs from CD1 embryos (Charles River) were dissected at E13.5. Immunocompromised NSG mice (8- to 10-week-old) (Jackson Laboratory strain no. 0005557) were used for engraftment studies. Pilot studies identified that females were more sensitive to naphthalene and died at a higher rate, therefore male mice were used for engraftment experiments.
Human Lung Tissue
All research utilizing human tissue was approved by the University of Michigan institutional review board. Additional information can be found in the Supplemental Experimental Procedures.
Cell Lines and Culture Conditions
Mouse and Human Primary Cultures
Epithelial bud tips were isolated as essentially as described previously (del Moral and Warburton, 2010), and see Supplemental Experimental Procedures. Isolated mouse bud tips were cultured in 4–6 μL droplets of Matrigel, covered with medium, and kept at 37°C with 5% CO2. Isolated human fetal lung bud tips were cultured in 25–50 μL droplets of Matrigel, covered with medium, and kept at 37°C with 5% CO2. Culture medium was changed every 2–4 days.
Generation and Culture of hPSC-Derived Lung Organoids
The University of Michigan Human Pluripotent Stem Cell Research Oversight Committee approved all experiments using human embryonic and induced pluripotent stem cell (hESC, iPSC) lines. PLOs were generated from four independent pluripotent cell lines in this study: hESC line UM63-1 (NIH registry no. 0277) was obtained from the University of Michigan and hESC lines H9 and H1 (NIH registry no. 0062 and no. 0043, respectively) were obtained from the WiCell Research Institute. iPSC20.1 was described previously (Spence et al., 2011). ESC lines were routinely karyotyped to ensure normal karyotype and ensure the sex of each line (H9, XX; UM63-1, XX; H1, XY). All cell lines are routinely monitored for mycoplasma infection monthly using the MycoAlert Mycoplasma Detection Kit (Lonza). Stem cells were maintained as described previously (Spence et al., 2011), and ventral foregut spheroids were generated as described previously (Dye et al., 2016a, Dye et al., 2015). Following differentiation, free-floating foregut spheroids were collected from differentiated stem cell cultures and plated in a Matrigel droplet on a 24-well tissue culture grade plate. A summary of different tissue/sample types, nomenclature, and descriptions can be found in Table 1.
Culture Media, Growth Factors, and Small Molecules
Information for serum-free basal media, growth factors, and small molecules, and the in-house generation and isolation of human recombinant FGF10, including company information and catalog numbers, can be found in the Supplemental Experimental Procedures.
RNA-Seq and Bioinformatics Analysis
RNA-seq and alignment was carried out as published previously (Hill et al., 2017). Data analysis was performed using the R statistical programming language (http://www.R-project.org/) as described previously (Dye et al., 2015, Finkbeiner et al., 2015, Tsai et al., 2016). The complete sequence alignment, expression analysis, and all corresponding scripts can be found at https://github.com/hilldr/Miller_Lung_Organoids_2017.
Naphthaline Injury
Naphthaline (Sigma no. 147141) was dissolved in corn oil at a concentration of 40 mg/mL. Adult male NSG mice were chosen for these experiments because we observed improved recovery and survival following injury compared with female mice. Mice that were 8–10 weeks of age were given intraperitoneal injections at a dose of 300 mg/kg weight.
Intratracheal Injection of Fetal Progenitor Organoids and hPSC-Derived Bud Tip Organoid Cells into Immunocompromised Mouse Lungs
Generating Single Cells from Organoid Tissues
Two to three Matrigel droplets containing organoid tissues were removed from the culture plate and combined in a 1.5 mL Eppendorf tube with 1 mL of room temperature Accutase (Sigma no. A6964). The tube was laid on its side to prevent organoids from settling to the bottom. Tissue was pipetted up and down 15–20 times with a 1 mL tip every 5 min for a total of 20 min. Single cell status was determined by microscopic observation using a hemocytometer. Cells were diluted to a concentration of 500,000–600,000 cells per 0.03 mL in sterile PBS.
Intratracheal Injection of Cells
Injection of cells into the mouse trachea was performed as described previously (Badri et al., 2011, Cao et al., 2017). In brief, animals were anesthetized and intubated. Animals were given 500,000–600,000 single cells in 30–35 μL of sterile PBS through the intubation cannula.
RNA Extraction and qRT-PCR Analysis
These procedures were carried out as described previously (Chin et al., 2016, Finkbeiner et al., 2015), and see Supplemental Experimental Procedures. A list of primer sequences used can be found in Table S1.
Tissue Preparation, Immunohistochemistry, Electron Microscopy, and Imaging
These procedures were carried out as described previously (Dye et al., 2015, Rockich et al., 2013, Spence et al., 2009), and see Supplemental Experimental Procedures.
Quantification and Statistical Analysis
All plots and statistical analysis were done using Prism 6 software (GraphPad). For statistical analysis of qRT-PCR results, at least three biological replicates for each experimental group were analyzed and plotted with the SEM. If only two groups were being compared, a two-sided Student's t test was performed. In assessing the effect of length of culture with FGF7 on gene expression in mouse buds (Figure S2G), a one-way, unpaired ANOVA was performed for each individual gene over time. The mean of each time point was compared with the mean of the expression level for that gene at day 0 of culture. If more than two groups were being compared within a single experiment, an unpaired one-way ANOVA was performed followed by Tukey's multiple comparison test to compare the mean of each group with the mean of every other group within the experiment. For all statistical tests, a significance value of 0.05 was used. For every analysis, the strength of p values is reported in the figures according the following: p > 0.05, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. Details of statistical tests can be found in the figure legends.
Author Contributions
AJM and JRS designed experiments. ESW, VL and BRD provided critical expertise and experimental input. AJM, MSN, YA, AMC, SH, and FZ performed experiments and collected data. AJM, DRH and JRS analyzed and interpreted data. AJM and JRS wrote and prepared the manuscript.
Acknowledgments
J.R.S. is supported by the NIH-NHLBI (R01 HL119215). A.J.M. is supported by the NIH Cellular and Molecular Biology training grant at Michigan (T32 GM007315) and by the Tissue Engineering and Regeneration Training grant (DE00007057-40). The University of Washington Laboratory of Developmental Biology was supported by NIH award number 5R24HD000836 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. We thank Michael Ferguson for his preliminary experiments isolating and culturing human fetal epithelial bud tips. We apologize to those whose work we were unable to cite due to space limitations.
Published: December 14, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.11.012.
Accession Numbers
All raw RNA-seq data files have been deposited in the EMBL-EBI ArrayExpress database (ArrayExpress: E-MTAB-6023). Adult human RNA-seq samples representing bulk sequencing of whole-lung homogenates were obtained from the EMBL-EBI ArrayExpress repository (ArrayExpress: E-MTAB-1733) (Fagerberg et al., 2014).
Supplemental Information
References
- Badri L., Walker N.M., Ohtsuka T., Wang Z., Delmar M., Flint A., Peters-Golden M., Toews G.B., Pinsky D.J., Krebsbach P.H., Lama V.N. Epithelial interactions and local engraftment of lung-resident mesenchymal stem cells. Am. J. Respir. Cell Mol. Biol. 2011;45:809–816. doi: 10.1165/rcmb.2010-0446OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchfield K., Li R., Lungova V., Verheyden J.M., McCulley D., Sun X. A three-dimensional study of alveologenesis in mouse lung. Dev. Biol. 2015;409:429–441. doi: 10.1016/j.ydbio.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P., Aoki Y., Badri L., Walker N.M., Manning C.M., Lagstein A., Fearon E.R., Lama V.N. Autocrine lysophosphatidic acid signaling activates β-catenin and promotes lung allograft fibrosis. J. Clin. Invest. 2017;127:1517–1530. doi: 10.1172/JCI88896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.R., Martinez Alanis D., Miller R.K., Ji H., Akiyama H., McCrea P.D., Chen J. Lung epithelial branching program antagonizes alveolar differentiation. Proc. Natl. Acad. Sci. USA. 2013;110:18042–18051. doi: 10.1073/pnas.1311760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-W., Huang S.X., de Carvalho A.L.R.T., Ho S.-H., Islam M.N., Volpi S., Notarangelo L.D., Ciancanelli M., Casanova J.-L., Bhattacharya J. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017;19:542–549. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.M., Tsai Y.-H., Finkbeiner S.R., Nagy M.S., Walker E.M., Ethen N.J., Williams B.O., Battle M.A., Spence J.R. A dynamic WNT/β-CATENIN signaling environment leads to WNT-independent and WNT-dependent proliferation of embryonic intestinal progenitor cells. Stem Cell Reports. 2016;0:826–839. doi: 10.1016/j.stemcr.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Danopoulos S., Alonso I., Thornton M., Grubbs B., Bellusci S., Warburton D., Al Alam D. Human lung branching morphogenesis is orchestrated by the spatio-temporal distribution of ACTA2, SOX2 and SOX9. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017 doi: 10.1152/ajplung.00379.2017. ajplung.00379.2017–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Moral P.-M., Warburton D. Explant culture of mouse embryonic whole lung, isolated epithelium, or mesenchyme under chemically defined conditions as a system to evaluate the molecular mechanism of branching morphogenesis and cellular differentiation. Methods Mol. Biol. 2010;633:71–79. doi: 10.1007/978-1-59745-019-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A., Tsai Y.-H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4 doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B.R., Dedhia P.H., Miller A.J., Nagy M.S., White E.S., Shea L.D., Spence J.R., Rossant J. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. Elife. 2016;5:e19732. doi: 10.7554/eLife.19732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B.R., Miller A.J., Spence J.R. How to grow a lung: applying principles of developmental biology to generate lung lineages from human pluripotent stem cells. Curr. Pathobiol. Rep. 2016;4:47–57. doi: 10.1007/s40139-016-0102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Hill D.R., Altheim C.H., Dedhia P.H., Taylor M.J., Tsai Y.-H., Chin A.M., Mahe M.M., Watson C.L., Freeman J.J. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Reports. 2015;4:1140–1155. doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Ahmad S., White C.W., Reynolds S.D. Transplantation of airway epithelial stem/progenitor cells: a future for cell-based therapy. Am. J. Respir. Cell Mol. Biol. 2017;56:1–10. doi: 10.1165/rcmb.2016-0181MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh S., Ito I., Nagasaki T., Yamamoto Y., Konishi S., Korogi Y., Matsumoto H., Muro S., Hirai T., Funato M. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem Cell Reports. 2014;3:394–403. doi: 10.1016/j.stemcr.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.D., Chen A., Nostro M.-C., d'Souza S.L., Schaniel C., Lemischka I.R., Gouon-Evans V., Keller G., Snoeck H.-W. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011;29:267–272. doi: 10.1038/nbt.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D.R., Huang S., Nagy M.S., Yadagiri V.K., Fields C., Mukherjee D., Bons B., Dedhia P.H., Chin A.M., Tsai Y.-H. Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. Elife. 2017;6:e29132. doi: 10.7554/eLife.29132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines E.A., Sun X. Tissue crosstalk in lung development. J. Cell. Biochem. 2014;115:1469–1477. doi: 10.1002/jcb.24811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.X.L., Islam M.N., O'Neill J., Hu Z., Yang Y.-G., Chen Y.-W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J., Snoeck H.-W. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2013;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., Morley M., Hawkins F., McCauley K.B., Jean J.C., Heins H., Na C.-L., Weaver T.E., Vedaie M., Hurley K. Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Cell Stem Cell. 2017;21:472–488.e10. doi: 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S., Gotoh S., Tateishi K., Yamamoto Y., Korogi Y., Nagasaki T., Matsumoto H., Muro S., Hirai T., Ito I. Directed induction of functional multi-ciliated cells in proximal airway epithelial spheroids from human pluripotent stem cells. Stem Cell Reports. 2015;6:18–25. doi: 10.1016/j.stemcr.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp J.L., Dubois C.L., Schaffer A.E., Hao E., Shih H.P., Seymour P.A., Ma J., Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire T.A., Ikonomou L., Hawkins F., Christodoulou C., Cao Y., Jean J.C., Kwok L.W., Mou H., Rajagopal J., Shen S.S. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell. 2012;10:398–411. doi: 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of wnt signaling. Cell Stem Cell. 2017;20:844–857.e6. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger R.J., Klein O.D., Martin G.R., Krasnow M.A. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.J., Spence J.R. In vitro models to study human lung development, disease and homeostasis. Physiology (Bethesda) 2017;32:246–260. doi: 10.1152/physiol.00041.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens C.B., Auerbach A.B., Conlon R.A., Joyner A.L., Rossant J. A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev. 1992;6:691–704. doi: 10.1101/gad.6.5.691. [DOI] [PubMed] [Google Scholar]
- Morrisey E.E., Hogan B.L.M. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolić M.Z., Caritg O., Jeng Q., Johnson J.-A., Sun D., Howell K.J., Brady J.L., Laresgoiti U., Allen G., Butler R. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. Elife. 2017;6:e26575. doi: 10.7554/eLife.26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T., Knoepfler P.S., Eisenman R.N., Hogan B.L.M. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development. 2005;132:1363–1374. doi: 10.1242/dev.01678. [DOI] [PubMed] [Google Scholar]
- Perl A.-K.T., Kist R., Shan Z., Scherer G., Whitsett J.A. Normal lung development and function after Sox9 inactivation in the respiratory epithelium. Genesis. 2005;41:23–32. doi: 10.1002/gene.20093. [DOI] [PubMed] [Google Scholar]
- Rawlins E.L., Clark C.P., Xue Y., Hogan B.L.M. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich B.E., Hrycaj S.M., Shih H.-P., Nagy M.S., Ferguson M.A.H., Kopp J.L., Sander M., Wellik D.M., Spence J.R. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.R., Lange A.W., Lin S.-C.J., Kaestner K.H., Lowy A.M., Kim I., Whitsett J.A., Wells J.M. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev. Cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlman M.T., Gray M.P., Falconieri M.W., Whitsett J.A., Weaver T.E. Lamellar body formation in normal and surfactant protein B-deficient fetal mice. Lab. Invest. 2000;80:395–403. doi: 10.1038/labinvest.3780044. [DOI] [PubMed] [Google Scholar]
- Tsai Y.-H., Hill D.R., Kumar N., Huang S., Chin A.M., Dye B.R., Nagy M.S., Verzi M.P., Spence J.R. LGR4 and LGR5 function redundantly during human endoderm differentiation. Cell. Mol. Gastroenterol. Hepatol. 2016;2:648–662.e8. doi: 10.1016/j.jcmgh.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhecke D., Herrmann G., Graber W., Hillmann-Marti T., Mühlfeld C., Studer D., Ochs M. Lamellar body ultrastructure revisited: high-pressure freezing and cryo-electron microscopy of vitreous sections. Histochem. Cell Biol. 2010;134:319–326. doi: 10.1007/s00418-010-0736-4. [DOI] [PubMed] [Google Scholar]
- Varner V.D., Nelson C.M. Cellular and physical mechanisms of branching morphogenesis. Development. 2014;141:2750–2759. doi: 10.1242/dev.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.P., Bear C.E., Chin S., Pasceri P., Thompson T.O., Huan L.-J., Ratjen F., Ellis J., Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.