Abstract
Gram-negative Photorhabdus bacteria have a dual lifestyle: they are mutualists of Heterorhabditis nematodes and are pathogens of insects. Together, this nematode–bacterium partnership has been used to successfully control a wide range of agricultural insect pests. Photorhabdus produce a diverse array of small molecules that play key biological roles in regulating their dual roles. In particular, several secondary metabolites (SM) produced by this bacterium are known to play a critical role in the maintenance of a monoxenic infection in the insect host and are also known to prevent contamination of the cadaver from soil microbes and/or predation by arthropods. A few of the SM this bacteria produce have been isolated and identified, and their biological activities have also been tested in laboratory assays. Over the past two decades, analyses of the genomes of several Photorhabdus spp. have revealed the presence of SM numerous gene clusters that comprise more than 6% of these bacteria genomes. Furthermore, genome mining and characterization of biosynthetic pathways, have uncovered the richness of these compounds, which are predicted to vary across different Photorhabdus spp. and strains. Although progress has been made in the identification and function of SM genes and gene clusters, the targeted testing for the bioactivity of molecules has been scarce or mostly focused on medical applications. In this review, we summarize the current knowledge of Photorhabdus SM, emphasizing on their activity against plant pathogens and parasites. We further discuss their potential in the management of agricultural pests and the steps that need to be taken for the implementation of Photorhabdus SM in pest management.
Keywords: agricultural pests, bioactivity, genomes, Photorhabdus, secondary metabolites
In the United States alone, plants are subject to attack by more than 50,000 different pathogens, primarily fungi, viruses, bacteria, and nematodes. Although a variety of chemical and other management tools are available, none is ideal with respect to environmental safety, efficacy, and/or costs (Pimentel et al., 1992, 2005; Pimentel and Greiner, 1997; Foster and Mourato, 2000; Roberts et al., 2003). Current chemical (including antibiotic) solutions increasingly lose their effectiveness because of emerging resistance (Hillocks, 2012). Moreover, the recent banning of several chemical nematicides and the loss of methyl bromide from the pest-control market compels the need for new and environmentally friendly methods to enhance current management systems (Kerry, 1998; Chitwood, 2003).
A promising approach to develop novel control methods is to study microorganisms and the SM they produce with biological activity against plant parasites and pathogens (Lange and Sanchez Lopez, 1996; Hattori, 2001; Webster et al., 2002; Stroebel, 2003; Berdy, 2005; Teasdale et al., 2009; Donadio et al., 2012). Indeed, many microorganisms are known to benefit plants by successfully controlling soil-borne pathogens, including plant-parasitic nematodes and plant-pathogenic microbes (Chen and Dickson, 1998; Kerry, 1998; Sikora and Hoffmann-Hergarten, 1993; Siddiqi and Mahmood, 1999). In addition, the bioactive metabolites from many of these beneficial microbes have shown to represent a valuable resource for the discovery of medical drugs and agricultural agents (Webster et al., 2002; Berdy, 2005). For example, SM of several Pseudomonas spp. are known to protect plants from diseases caused by various soil-borne pathogenic fungi (Haas et al., 1992). In particular, two SM, hydrogen cyanide and 2,4-diacetylphloroglucinol produced by Pseudomonas fluorescens strain CHA0, have been demonstrated to suppress tobacco black root rot (Voisard et al., 1989).
Many insect-pathogenic bacteria are also known to produce several natural compounds SM with broad biological activities. For instance, the screening of many microbial extracts from Bacillus spp., including Bacillus thuringiensis among others, has revealed an extraordinary large and structurally diverse number of natural compounds with antimicrobial, antiviral, immunosuppressive, and antitumor activities (Sansinenea and Ortiz, 2011). More recently, lipopeptides (surfactins, iturins, and fengycins) produced by Bacillus spp. have been shown to play a key role in protecting plants against a wide range of phytopathogens, including bacteria, fungi, and oomycetes (Ongena and Jacques, 2008). Furthermore, these molecules are also known to take part in beneficial interactions of Bacillus species with plants by stimulating host defense mechanisms (Mukherjee and Das, 2005).
Another insect-pathogenic bacterium, Serratia marcescens, has been proven to produce a variety of SM and other biomolecules with antimicrobial and antiprotozoal activities that are crucial for its success in the diverse environments they thrive (Williams, 1973; Kurtz et al., 1981). For example, prodigiosin, which is considered a multifaceted SM pigment, has been reported to have antifungal and immunosuppressive activities (Thomson et al., 2000).
Over the past two decades, a growing interest has been put in investigating SM produced by the bacterial symbionts of entomopathogenic nematodes (Steinernematidae and Heterorhabditidae), including Xenorhabdus and Photorhabdus spp. Many of the bioactive metabolites produced by these bacteria are perceived to have potential for their application in agriculture and medical arenas (Webster et al., 2002; Brachmann et al., 2007; Bode, 2009, 2011). Presently, many laboratories are dedicated to the study of small molecule SM natural products from these bacteria. However, despite these efforts, only a small fraction of the existing SM diversity has been examined or bioassayed for their value in controlling agricultural pests. In this review, we focus on Photorhabdus bacteria and summarize the research that has been done in relation to SM and their potential against agriculture pathogens and parasites.
The Dual Lifestyle of Photorhabdus
Photorhabdus are gram-negative bacteria that are pathogens to insects and have a mutualistic relationship with Heterorhabditis nematodes (Heterorhabditidae). The bacteria reside in the intestine of the only free-living stage of the nematodes, also known as the 3rd stage infective juvenile (IJ). IJ invade a susceptible insect host, seeking the hemolymph. Once in the hemocoel, the IJ release their symbionts. The released bacteria contribute to the killing of the insect host and grow to high density in the resulting cadaver. Photorhabdus are essential for nematode growth and development, presumably both by serving as a direct food source and by supplying nutrients through degradation of the insect carcass (Akhurst, 1982; Akhurst and Dunphy, 1993). When nematode numbers become high and nutrients become limiting in the insect cadaver, nematode progeny reassociate with bacteria and differentiate into the colonized, nonfeeding IJ form that emerges into the soil to forage for a new host. Because of their insecticidal capabilities, entomopathogenic nematode–symbiont bacteria pairs have been successfully implemented in biological control and integrated pest management programs worldwide (Kaya and Gaugler, 1993; Gaugler, 1999; Grewal et al., 2005).
This nematode–bacterium–insect system is also viewed as a tractable model system amenable to study the physiological, chemical, structural, and developmental aspects of beneficial symbiotic associations and their differences from pathogenic associations (Burnell and Stock, 2000; Goodrich-Blair and Clarke, 2007; Stock and Goodrich-Blair, 2008). Photorhabdus symbionts must evade the immune system of the insect; kill the insect host; and repel invading scavengers and other competitors, including other bacteria, fungi, nematodes, amoebae, insects, and even birds (Waterfield et al., 2009). Conversely, during the phase of mutualistic association with Heterorhabditis nematodes, Photorhabdus must refrain from producing toxic metabolites and has to evade the immune system of the nematode and avoid being used as a food source. These contrasting but very significant challenges associated with the different phases of Photorhabdus life cycle are met by the production of proteinaceous toxins, extracellular enzymes, and crystalline inclusion proteins, in addition to biosynthesizing a large variety of small-molecule SM with various biological activities. The switch between the pathogenic and the mutualistic phases in the life cycle of this bacterium (and by implication, the regulation of the production of their virulence factors and SM products) is presently being investigated (Somvanshi et al., 2012; Clarke, 2016).
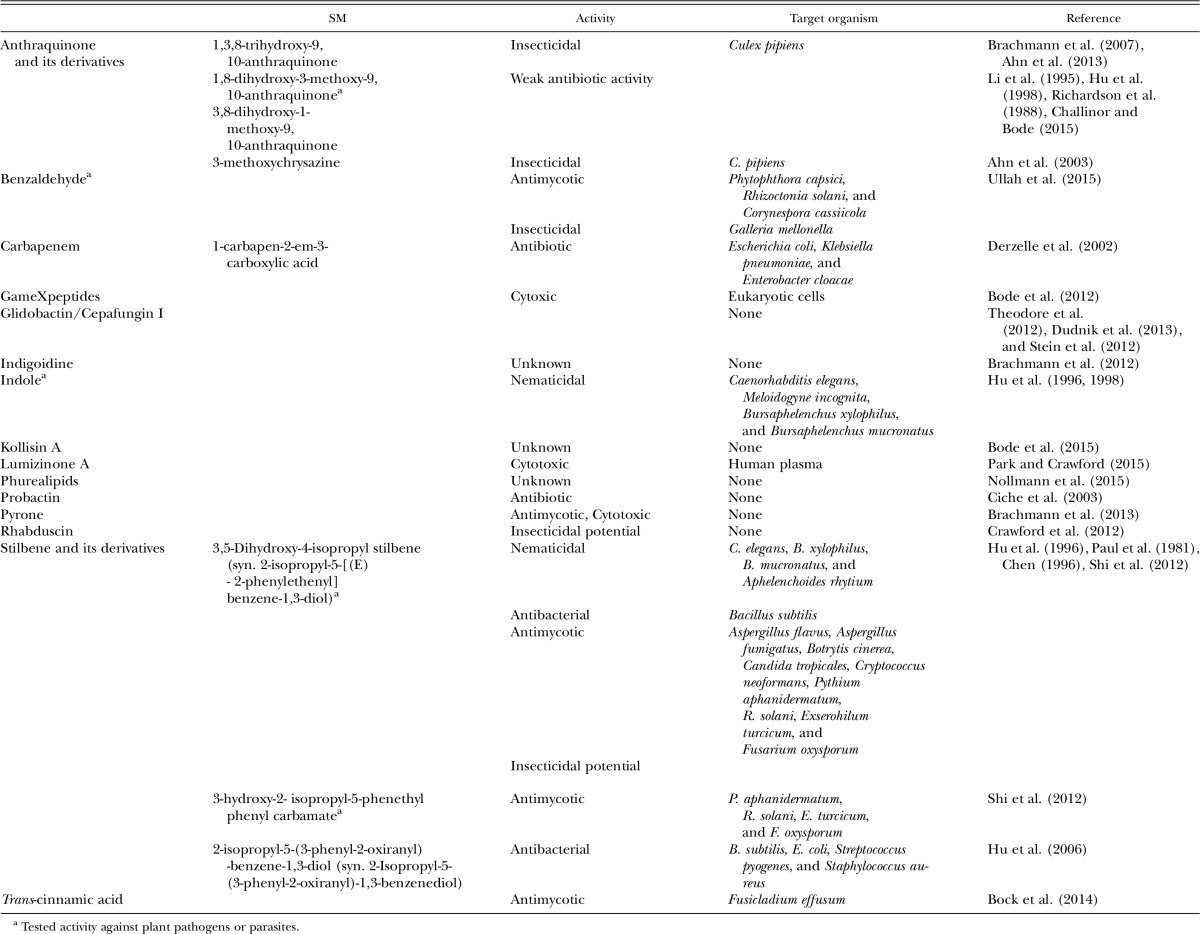
Importantly, numerous bioactive SM belonging to diverse chemical classes have been reported (Table 1). Their biological activity is very diverse and includes antibiotic (Akhurst, 1982; McInerney et al., 1991a, 1991b) and nematicidal activities (Hu and Webster, 1995; Hu et al., 1996, 1998, 1999; Han and Ehlers, 1999), among others. Thus, Photorhabdus bacteria are now viewed as a rich source of novel classes of pharmacologically active compounds showing exciting biological activities.
Table 1.
Presently identified Photorhabdus secondary metabolites (SM) and their biological activity.
Nematicidal Activity
Early studies by Hu and Webster (1995) showed that cell-free culture filtrates of several Photorhabdus spp. and/or strains were toxic to second-stage juveniles (J2) of the root-knot nematode, Meloidogyne incognita, as well as to fourth-stage juveniles (J4), as well as to adults of the pine wilt nematode, Bursaphelenchus xylophilus. In another study, Han and Ehlers (1999) tested the trans-specific nematicidal activity of Photorhabdus luminescens culture filtrates isolated from two Heterorhabditis species, Heterorhabditis bacteriophora H06 strain and Heterorhabditis indica LN2 strain against other entomopathogenic nematode species that were deprived of their native symbionts. The authors concluded that these filtrates had toxic effects on nonsymbiotic nematodes (trans-specific activity) and suggested they may have an impact on competitive interactions when one insect host is infected by different nematode species.
More recently, Orozco et al. (2016) showed that crude extracts of Photorhabdus l. sonorensis (CH35 strain) inhibited M. incognita J2 and that mortality of nematodes was concentration dependent. The authors also tested crude extracts on nontarget species, including Steinernema carpocapsae and Caenorhabditis elegans, demonstrating a very low nematicidal activity against them (Orozco et al., 2016).
Until now, only two SM molecules, a stilbene derivative (3,5-dihydroxy-4-isopropylstilbene) and indole, had been found to have nematicidal activity (Hu et al., 1996). Interestingly, Photorhabdus is the only organism outside the plant kingdom to produce this stilbene. Hu et al. (1996) found that a stilbene derivative (3,5-Dihydroxy-4-isopropylstilbene) at a concentration of 200 µg/ml was toxic to a selection of bacterial- and fungal-feeding nematodes, including C. elegans, B. xylophilus, Bursaphelenchus mucronatus, and Aphelenchoides rhytium. However, the authors showed that at the same concentration, this SM had no toxicity to second-stage juveniles of then root-knot nematode, M. incognita (Hu et al., 1996). This study clearly showed that SM may have different effects on different nematode species and highlights the need to further test the bioactivity of stilbene derivatives against other plant parasitic nematodes.
Another compound, isolated from several Photorhabdus strains with shown nematicidal activity is indole (Hu et al., 1996, 1998). This molecule had activity against three plant parasitic nematode species: M. incognita, B. xylophilus, and B. mucronatus at concentrations greater than 200 µg/ml. Specifically, indole caused high levels of paralysis of M. incognita and Bursaphelenchus spp. at a concentration of 100 to 300 µg/ml.
Both stilbene and indole also inhibited egg hatching of M. incognita (Hu and Webster, 1995; Hu et al., 1999). Another molecule with nematicidal activity was 2-stilbenol, which showed to be more toxic to B. xylophilus than stilbene, causing 100% mortality at 6.25 to 12.5 µg/ml (Hu et al., 1996).
Antibacterial Activity
During its life cycle, Photorhabdus produce several broad-spectrum antibiotics, which are secreted into the insect hemolymph when the bacteria enter the stationary-phase condition, preventing the putrefaction of the infected cadaver (Webster et al., 2002). Early studies conducted by Poinar et al. (1980) showed that P. luminescens inhibited Bacillus cereus subsp. rnycoides and Bacillus subtilis. In addition, Akhurst (1982) demonstrated that both in vivo and in vitro cultures of a selection of P. luminescens strains had activity against several bacteria species including the plant pathogen Erwinia carotovora.
Paul et al. (1981) were the first to isolate and structurally characterized two antibiotic molecules from P. luminescens pure cultures. Specifically, the authors reported two trans-stilbene derivatives, compounds V and VI. Compound V was isolated as 20% of the extract of H. bacteriophora and identified them as, 3,5-dihydroxy-4-isopropyl-trans-stilbene. Compound VI which was isolated as 6% of the extract was characterized as 3,5-dihydroxy-4-ethyl-trans-stilbene.
Later studies by J. Webster’s team found that hydroxylated stilbene congeners, produced by Photorhabdus spp., also display antibiotic activities. Specifically, Chen (1996) reported that 2-isopropyl-5-(3-phenyl-2-oxiranyl)-benzene-1,3-diol (syn. 2-Isopropyl-5-[3-phenyl-2-oxiranyl]-1,3-benzenediol) stilbene had antibiotic effect against the gram-positive B. subtilis. In addition, Hu et al. (2006) also isolated, identified, and chemically synthesized a novel antimicrobial compound, 1,2-isopropyl-5-(3-phenyl-oxiranyl)-benzene-1,3-diol (syn. 2-Isopropyl-5-(3-phenyl-2-oxiranyl)-1,3-benzenediol) from the larvae of Galleria mellonella infected with the Heterorhabditis megidis 90–P. luminescens C9 complex. This molecule was tested against several clinical isolates of gram-negative and gram-positive bacteria, showing potent antimicrobial activity at different minimum inhibitory concentrations ranging from 6.25 to 12.5 μg/ml, to 100.0 μg/m. However, these SM have not yet been evaluated against plant pathogenic bacteria.
Photorhabdus are also known to produce several other SM compounds with broad-spectrum antibiotic properties (McInerney et al., 1991a, 1991b; Maxwell et al., 1994; Webster et al., 2002; Eleftherianos et al., 2007; Waterfield et al., 2009; Inman and Holmes, 2012; Clarke, 2016). One of them is carbapenem molecule, which is a prominent class of β-lactam antibiotic, which was first identified and characterized by Derzelle et al. (2002). Many naturally occurring carbapenems have been reported thus far, mostly all originated from streptomycetes and with wide broad-spectrum activity against many gram-positive and gram-negative bacteria. Carbapenems have gained clinical prominence to treat infections with rapidly spreading multidrug-resistant human pathogens such as methicillin-resistant Staphylococcus aureus (Coulthurst et al., 2005; Huber et al., 2009). Yet, the activity of these SM against plant pathogenic microbes has not been elucidated.
Photorhabdus, also produce 1-carbapen-2-em-3-carboxylic acid, which has shown antibiotic activity against Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae (Derzelle et al., 2002). However, at present, no studies have been undertaken to test the activity of this molecule against phytopathogenic bacteria.
Anthraquinone molecules are considered a rarity in gram-negative bacteria, but have been identified in Photorhabdus. Recent studies have shown that Type II polyketide synthase enzymes are involved in the production of these SM (Brachmann et al., 2007). Two anthraquinone derivatives have been identified in P. luminescens, 1,3,8-trihydroxy-9,10-anthraquinone and two of its monomethyl ether derivatives, 1,8-dihydroxy-3-methoxy-9,10-anthraquinone and 3,8-dihydroxy-1-methoxy-9,10-anthraquinone (Richardson et al., 1988; Sztaricskai et al., 1992; Li et al., 1995; Hu et al., 1998). Although the antibiotic activity of these SM has been reported as ‘weak’ (Richardson et al., 1988; Challinor and Bode, 2015), further studies should be conducted to consider a wider repertoire of bacteria species and/or strains.
Another compound, a catecholate siderophore (photobactin) was reported to play a role in antibiosis in the insect cadaver by sequestering iron from invading microbes (Ciche et al., 2003). Furthermore, a few studies showed that extracts of several Photorhabdus spp. have bactericidal activity against Erwinia amylovora, the causative agent of fire blight disease (Hevesi et al., 2004; Hu et al., 1996). More recently, Uma et al. (2010) showed the potential of P. luminescens to control two important plant pathogens: Xanthomonas and Pseudomonas. Despite this knowledge, no efforts have been placed on the testing of purified Photorhabdus SM on phytopathogenic bacteria.
Antimycotic Effects
Inside the insect host, Photorhabdus spp. produce antimycotic compounds that help prevent invasion of the cadaver by soil fungal competitors (Chen et al., 2008; Webster et al., 2002). Many studies have been undertaken to assess the antimycotic properties of crude cell-free extracts. For example, Chen et al. (2008) were the first to test the activity of P. luminescens (from H. megidis) against 32 species of fungi from a range of habitats. Their study showed that cell filtrates of this bacterium completely inhibited the growth of various plant pathogenic fungi spp., including Botrytis cinerea, Ceratocystis ulmi, Ceratocystis dryocoetidis, Mucor piriformis, Pythium coloratum, Pythium ultimum, and Trichoderma pseudokingii, among others.
Shapiro-Ilan et al. (2009) also evaluated crude extracts of several Photorhabdus spp. and strains against a selection of fungal plant pathogens, including Glomerella cingulata, Phomopsis sp., Phytophthora cactorum, and Fusicladosporium effusum, which are fungal or oomycete pathogens of pecan, and Monilinia fructicola, a fungal pathogen of peach, considering in planta and in vitro assays. Their study showed that the crude extracts had moderate effect of the tested fungi.
Similarly, San-Blas et al. (2012) reported strong antifungal effects of metabolites from Venezuelan strains of Photorhabdus, against the causative agent of the cacao frosty pod rot disease, Moniliophthora roreri. In another study, Hazir et al. (2016) evaluated the potency of cell-free supernatants of three Photorhabdus strains, Photorhabdus temperata, P. luminescens (VS) and P. luminescens (K122) against several plant pathogenic fungi. Their results demonstrated that these cell-free culture filtrates can inhibit the vegetative growth of a variety of economically important phytopathogenic fungi, including Fusicladium carpophilum (peach scab), F. effusum (pecan scab), M. fructicola (brown rot), G. cingulata (anthracnose), and Armillaria tabescens (root rot). Recently, Orozco et al (2016) evaluated crude extracts of P. l. sonorensis against two fungal pathogens, Fusarium oxysporum (f.sp. asparagi) and Alteraria alternata, based on in vitro inhibition assays. Although the extracts inhibited growth of these fungi, they were considered to have moderate to low effect.
A few efforts have also been placed into the testing of specific SM molecules. For instance, Li et al. (1995) were the first ones to report the activity of specific metabolites, including 3,5-dihydroxy-4-isopropylstilbene, against several fungal spp. of medical and agricultural importance, including Aspergillus flavus, Aspergillus fumigatus, B. cinerea, Candida tropicales, and Cryptococcus neoformans. The authors showed that this SM had strong fungicidal activity and suggested that it may probably function as an antagonistic agent preventing the insect cadaver to be attacked by fungal saprobes.
Shi et al. (2012) isolated and identified seven metabolites from P. temperata SN259 strain. Three of these compounds were depicted as novel stilbene derivative molecules. The activity of these metabolites was evaluated against four plant pathogenic fungi, including Pythium aphanidermatum, Rhizoctonia solani, Exserohilum turcicum, and F. oxysporum in in vitro assays. Two stilbene derivatives, 3-hydroxy-2-isopropyl-5-phenethyl phenyl carbamate and 2-isopropyl-5-([E]-2-phenylethenyl) benzene-1,3-diol (syn. 3,5-dihydroxy-4-isopropyl stilbene), showed strong inhibition against P. aphanidermatum with an effective concentration (EC50) values of 2.8 and 2.7 μg/ml, respectively.
Bock et al. (2014) also purified and identified trans-cinnamic acid from P. luminescens and tested it against a pecan fungal pathogen, Fusicladium effusum showing an inhibitory activity at a concentration of 148 to 200 μg/ml.
Ullah et al. (2015) isolated and characterized benzaldehyde, an aromatic aldehyde, isolated from P. temperata M102. The authors tested the activity of this molecule against three fungal plant pathogens, Phytophthora capsici, R. solani, and Corynespora cassiicola, demonstrating its ability to inhibit their growth in in vitro assays.
Insecticidal Activity
A few studies have demonstrated the insecticidal activity of cell-free filtrates of various Photorhabdus spp. and/or strains. For example, Shrestha and Lee (2012) tested the oral toxicity of P. l. laumondii (TT01 strain) crude extracts against the sweet potato whitefly, Bemisia tabaci, and adults, showing they were completely lethal at 60 hr posttreatment.
Orozco et al. (2016) also showed that crude extracts of P. l. sonorensis have insecticidal activity against the corn earworm Helicoverpa zea. Their study showed that the crude extracts had low insecticidal activity, killing only 11% to 37 % of H. zea neonates.
It is well recognized that Photorhabdus produce a wide array of toxins that contribute to the killing of an insect host (Waterfield et al., 2009). But in addition, many SM molecules, including stilbene derivatives and anthraquinone derivatives, genistine (a furan derivative), and a phenol derivative, have been identified from in vitro cultures, showing insecticidal activity (Eleftherianos et al., 2007; Chalabaev et al., 2008).
Photorhabdus anthraquinone has been reported to have bird and ant deterrent properties (Baur et al., 1998). Ahn et al. (2013) also demonstrated that two anthraquinone molecules, 1,3-dimethoxy-8-hydroxy-9,10-anthraquinone and 3-methoxychrysazine, isolated from P. temperata had mosquitocidal activity against Culex pipiens larvae.
Eleftherianos et al. (2007) also reported that stilbenes are involved in intercepting the immune system of the insects by inhibiting phenoloxidase (PO), an insect enzyme involved in the production of melanotic nodules, using Manduca sexta larvae as the model system. Crawford et al. (2012) also reported that the production of another molecule, rhabduscin, an amidoglycosyl- and vinyl-isonitrile-functionalized tyrosine derivative in P. luminescens, has immunosuppressive activity against PO.
Ullah et al. (2015) showed that another compound, benzaldehyde, exhibited insecticidal activity against G. mellonella larvae in a dose-dependent manner. For example, the authors reported that at a 8 mM concentration, they observed 100% insect mortality at 108 hr after injection. Furthermore, in vivo assays showed that benzaldehyde also has an effect on the insect immune response by inhibiting PO activity and nodule formation.
Culture Conditions and SM Production
Several studies have shown that culture conditions play a critical role in determining the quantity and quality of SM molecules produced by Photorhabdus. For example, Hu et al. (1998) showed that two stilbene derivatives, 5-dihydroxy-4-isopropylstilbene and 3,5-dihydroxy-4-ethylstilbene, can be isolated from extracts collected from nematode–bacterium infected cadaver cultures. However, only 3,5-dihydroxy-4-ethylstilbene was only isolated from the extracts derived from Photorhabdus-only culture broth. Similarly, these authors showed that three anthraquinone pigments were isolated when extracts were obtained from nematode-bacterium infected G. mellonella cadavers.
Moreover, Hu et al. (1996, 1998) reported that stilbene was commonly produced in G. mellonella cadavers infected by different Photorhabdus spp. In particular, they showed that stilbene produced by P. luminescens C9 strain (bacterial symbiont of H. megidis 90 strain). Specifically, it was observed that after 24 hr of nematode infection, the stilbene derivative, 3,5-dihydroxy-4-isopropylstilbene, produced by P. luminescens C9, increased rapidly at 2 to 5 d postinfection and remained at a level of 3,000 to 3,600 μg/g wet larvae for about 21 d and decreased gradually thereafter. The authors concluded that the early production and continued presence of a relatively large amount of 3,5-dihydroxy-4-isopropylstilbene in the infected cadavers support the hypothesis that the antibiotics produced by Photorhabdus help minimize competition from other microorganisms and prevent the putrefaction of the nematode-infected insect cadaver (Hu et al., 1996). Following up on these findings, Hu et al. (1998) also studied the metabolic composition of the Photorhabdus–Heterorhabditis–Galleria interaction and found that stilbene, 3,5-dihydroxy-4-ethylstilbene, and several anthraquinone derivatives were major metabolic components of G. mellonella cadavers infected by H. megidis 90.
Recently, Orozco et al. (2016) also showed that fewer signals were detected in thin layer chromatography analyses when extracts were obtained from in vitro cultures. These results confirm the premise that synthesis of Photorhabdus metabolites is dependent not only on the conditions and the signals the bacteria encounter in the insect cadaver or under in vitro conditions, but it is also depended on the presence and/or the absence of their nematode hosts.
Photorhabdus Genome and SM Discovery
Natural products have traditionally been identified from a top–down perspective, but more recently genomics- and bioinformatics-guided bottom–up approaches have provided powerful alternative strategies. High throughput sequencing has become a fast and affordable approach for investigating bacteria as a source of novel toxins, metabolites, and enzymes for use in agriculture and pharmaceutical applications (Duchaud et al., 2003; fFrench-Constant et al., 2003, 2007; Kontnik et al., 2010; Edwards and Holt, 2013).
Genome mining is another valuable approach to identify the genes and/or gene clusters involved in the production of new antibiotics and the discovery of cryptic products. For example, genomic analysis of P. luminescens TT01 strain (genome size: 5.5 Mb) revealed that nearly 6% of its genome is involved in the production of SM (Duchaud et al., 2003; Clarke, 2008). The reported SM proportion in this bacterium is higher than the 3.8% observed in Streptomyces coelicolor, the model organism for studying SM production, suggesting that Photorhabdus has either the same or higher total coding capacity.
Mining of Photorhabdus genomes has also revealed the presence of several genes involved in the production of insecticidal proteins and hydrolytic enzymes, including proteases, lipases, and chitinases. In addition, many gene clusters involved in the biosynthesis of SM, including isopropylstilbenes, ethyl stilbenes, anthraquinones, siderophore photobactin, and carbapenems, have been identified setting the path for further research on their roles and functionality in the dual life cycle of this bacterium.
Recent studies have also identified more than 20 loci in the genomes of Photorhabdus, which are thought to be involved in the synthesis of antibiotic peptide molecules (Bode, 2009). However, until now, only three loci have been characterized, including those involved in the synthesis of stilbene, carbapenem, and anthraquinone (Brachmann et al., 2007; Joyce et al., 2008; Derzelle et al., 2002). For example, Brachmann et al. (2007) identified the biosynthesis gene cluster plu4186–plu4194 that is responsible for the production of anthraquinone in P. luminescens (TT01 strain). This pigment is produced by proteins, which are encoded in the 9-gene antA-I locus (Brachmann et al., 2007). Genes at both ends of this locus (plu4185 and plu4195) were also predicted to encode transcriptional regulators, although the role for these genes in the regulation of anthraquinone production remains obscure.
Easom and Clarke (2008) showed that there is another transcriptional regulator, HdfR, which was originally identified during a screen for Photorhabdus mutants that were unable to colonize their nematode host. The gene acts as a repressor of antA-I expression and anthraquinone production. However, the role of this SM during nematode colonization was not verified.
Joyce et al. (2008) described the biosynthesis pathways of stilbene, a multipotent molecule with roles in both the pathogenic and mutualistic lifestyles. The authors reported the proteins involved in stilbene production are encoded in genes that are located in at least four different genetic loci. It was also hypothesized that these different loci may be regulated independently from each other and that the flux toward stilbene synthesis may involve both.
Derzelle et al. (2002) identified and characterized a cluster of eight genes (named cpmA to cpmH), which were found to be responsible for the production of a carbapenem-like antibiotic P. luminescens strain TT01. This gene cluster is apparently different in several aspects from car operons in other bacteria. This study also showed that cpm mRNA peaks during the exponential growth phase of the bacterium and is regulated by a Rap/Hor homolog identified in the P. luminescens genome. Marker-exchange mutagenesis of this gene resulted in a decrease of antibiotic production. Furthermore, Derzelle et al. (2002) also showed that regulation of the cpm operon also influences the luxS-like signaling mechanism of quorum sensing.
Brachman et al. (2009) reported that many SM loci are cryptic in Photorhabdus and therefore the genes are apparently not expressed under normal laboratory conditions. For example, it has been shown that indigoidine synthetase is functional but silent in P. luminescens (Brachman et al., 2009). For example, through heterologous gene expression of indC in E. coli and considering a promoter exchange approach, the authors demonstrated that indigoidine can be produced in Photorhabdus.
Conclusions
It is now widely accepted that the intensive use of chemical pesticides for control of plant parasitic nematodes and other plant pests (including bacteria and fungi) has led to severe negative environmental impacts (1988; Pimentel et al., 1992; Foster and Mourato, 2000; Stickle, 2003). The EPA is in the process of reviewing the use of organophosphate and carbamate pesticides with the intention of phasing out obsolete and toxic chemicals, as has already happened with methyl bromide—a soil fumigant that had been widely used for controlling plant-parasitic nematodes and other soil-borne pathogens. Furthermore, resistance to both antibiotics such as streptomycin and oxytetracycline, presently licensed for use used in crop protection, has been spreading rapidly in plant pathogenic bacteria of economic significance (Vidaver, 2002). Thus, there is a great need for the discovery and use of pesticides that are not only highly effective but also have low toxicity and reduced impact in the environment (Pearce and Koundouri, 2003).
In this respect, the use of microorganisms and their natural products has emerged as a promising alternative for more rational and safe crop management. Furthermore, modern omics, genetics, and biochemical tools have made substantial contributions toward the unraveling of the biological activity of microbial SM (Demain and Fang, 2000). Furthermore, current automated bioinformatics platforms have enabled the prediction of gene pathways involved in the production of SM natural products and also to understand the function of these molecules in the biology of the microorganisms that produce them.
Over the past two decades, significant progress has been made in the mining of Photorhabdus genomes. Comparison across different species and strains has revealed the presence of several conserved biosynthesis gene clusters that are involved in the biosynthesis of diverse natural products (Tobias et al., 2016). Furthermore, these bioactive small molecules have been shown to play key roles in the pathogenic and mutualistic phases of this bacterium.
Despite this progress, there are yet many critical steps that must be pursued before their consideration of Photorhabdus SM as agricultural pesticides. Most of what is known regarding the bioactivity of this bacterium’s SM is based on structure comparisons and/or similarities with molecules of known antimicrobial, insecticidal, or cytotxic activity.
Only a few SM have been tested under laboratory conditions with a limited number of targeted pathogens. In particular, more bioassays should be carried out to test the bioactivity of specific SM molecules, specially increasing the range of targeted plant pathogens and parasites. These assays should be followed by pilot greenhouse trials to assess the effectiveness of the studied SM in more natural scenarios. Focus should also be placed on understanding their specificity; biosafety, and environmental impact to nontarget organism. Specifically, studies should be conducted to evaluate their allergenicity and toxinogencity on plants, animals, and humans.
Moving toward the commercial delivery of Photorhabdus SM as pesticides, efforts should be placed on their production at an industrial scale. Considerations should be made toward the evaluation of their shelf life and on their compatibility with additives and/or adjuvants that could potentially improve their application and stability as a final product (Montesinos, 2003).
There is no doubt that Photorhabdus represent an abundant and valuable source of bioactive and chemically novel compounds with potential for exploitation in agriculture. Indeed, we are yet in an infancy stage, but the promise that Photorhabdus SM warrant will continue to lead the path for further discoveries and applications into pest management.
Literature Cited
- Ahn JY, Lee JY, Yang EJ, Lee YJ, Koo KB, Song KS, Lee KY. Mosquitocidal activity of anthraquinones isolated from symbiotic bacteria Photorhabdus of entomopathogenic nematode. Journal of Asia-Pacific Entomology. 2013;16:317–320. [Google Scholar]
- Akhurst RJ. Antibiotic activity of Xenorhabdus ssp. bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. Journal of General Microbiology. 1982;128:3061–3065. doi: 10.1099/00221287-128-12-3061. [DOI] [PubMed] [Google Scholar]
- Akhurst RJ, Dunphy GB. Tripartite interactions between symbiotically associated entomopathogenic bacteria, nematodes, and their insect hosts. In: Beckage N, Thompson S, Federici B, editors. Parasites and pathogens of insects, vol. 2. San Diego: Academic Press; 1993. pp. 1–23. [Google Scholar]
- Baur ME, Kaya HK, Strong DR. Foraging ants as scavengers on entomopathogenic nematode-killed insects. Biological Control. 1998;12:231–236. [Google Scholar]
- Berdy J. Bioactive microbial metabolites. Journal of Antibiotics. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Current Opinion in Chemical Biology. 2009;13:224–230. doi: 10.1016/j.cbpa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- Bode HB. 2011. Insect-associated microorganisms as a source for novel secondary metabolites with therapeutic potential. Pp. 77-93 in Insect biotechnology. The Netherlands: Springer. [Google Scholar]
- Bode HB, Brachmann AO, Jadhav KB, Seyfarth L, Dauth C, Fuchs SW, Kaiser M, Waterfield NR, Sack H, Heinemann SH, Arndt HD. Structure elucidation and activity of kolossin A, the D-/L-pentadecapeptide product of a giant nonribosomal peptide synthetase. Angewandte Chemie International Edition. 2015;54:10352–10355. doi: 10.1002/anie.201502835. [DOI] [PubMed] [Google Scholar]
- Bode HB, Reimer D, Fuchs SW, Kirchner F, Dauth C, Kegler C, Lorenzen W, Brachmann AO, Grün P. Determination of the absolute configuration of peptide natural products by using stable isotope labeling and mass spectrometry. Chemistry: A European Journal. 2012;18:2342–2348. doi: 10.1002/chem.201103479. [DOI] [PubMed] [Google Scholar]
- Bock CH, Shapiro-Ilan DI, Wedge DE, Cantrell CL. Identification of the antifungal compound, trans-cinnamic acid, produced by Photorhabdus luminescens, a potential biopesticide against pecan scab. Journal of Pest Science. 2014;87:155–162. [Google Scholar]
- Brachmann AO, Brameyer S, Kresovic D, Hitkova I, Kopp Y, Manske C, Schubert K, Bode HB, Heermann R. Pyrones as bacterial signaling molecules. Nature Chemical Biology. 2013;9:573–578. doi: 10.1038/nchembio.1295. [DOI] [PubMed] [Google Scholar]
- Brachmann AO, Joyce SA, Jenke-Kodama H, Schwär G, Clarke DJ, Bode HB. A type II polyketide synthase is responsible for anthraquinone biosynthesis in Photorhabdus luminescens. Chembiochemistry. 2007;8:1721–1728. doi: 10.1002/cbic.200700300. [DOI] [PubMed] [Google Scholar]
- Brachmann AO, Kirchner F, Kegler C, Kinski SC, Schmitt I, Bode HB. Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. Journal of Biotechnology. 2012;157:96–99. doi: 10.1016/j.jbiotec.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Burnell AM, Stock SP. Heterorhabditis, Steinernema and their bacterial symbionts – Lethal pathogens of insects. Nematology. 2000;2:31–42. [Google Scholar]
- Chalabaev S, Turlin E, Bay S, Ganneau C, Brito-Fravallo E, Charles JF, Danchin A, Biville F. Cinnamic acid, an autoinducer of its own biosynthesis, is processed via Hca enzymes in Photorhabdus luminescens. Applied and Environmental Microbiology. 2008;74:1717–1725. doi: 10.1128/AEM.02589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challinor VL, Bode HB. Bioactive natural products from novel microbial sources. Annals of the New York Academy of Sciences. 2015;1354:82–97. doi: 10.1111/nyas.12954. [DOI] [PubMed] [Google Scholar]
- Chen G. Antimicrobial activity of the nematode symbionts, Xenorhabdus and Photorhabdus (Enterobacteriaceae) and the discovery of two novel groups of antimicrobial substances, nematophin and xenorides. 1996 Ph.D. thesis, Simon Fraser University, BC, Canada. [Google Scholar]
- Chen ZX, Dickson DW. Review of Pasteuria penetrans: Biology, ecology, and biological control potential. Journal of Nematology. 1998;30:313–340. [PMC free article] [PubMed] [Google Scholar]
- Chen G, Webster JM, Li J, Hu K, Liu W, Zhu J. Anti-inflammatory and psoriasis treatment and protein kinase inhibition by hydroxy stilbenes and novel stilbene derivatives and analogues. 2008 U.S. Patent 7,321,050. [Google Scholar]
- Chitwood DJ. Research on plant-parasitic nematode biology conducted by the United States Department of Agriculture–Agricultural Research Service. Pest Management Science. 2003;59:748–753. doi: 10.1002/ps.684. [DOI] [PubMed] [Google Scholar]
- Ciche TA, Blackburn M, Carney JR, Ensign JC. Photobactin: A catechol siderophore produced by Photorhabdus luminescens, an entomopathogen mutually associated with Heterorhabditis bacteriophora NC1 nematodes. Applied Environmental Microbiology. 2003;69:4706–4713. doi: 10.1128/AEM.69.8.4706-4713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DJ. Photorhabdus: A model for the analysis of pathogenicity and mutualism. Cellular Microbiology. 2008;10:2159–2167. doi: 10.1111/j.1462-5822.2008.01209.x. [DOI] [PubMed] [Google Scholar]
- Clarke DJ. 2016. The regulation of secondary metabolism in Photorhabdus, in R. ffrench-Constant, ed. Current topics in microbiology and immunology, vol. 402. The molecular biology of Photorhabdus bacteria. Cham: Springer. doi: 10.1007/82_2016_21. [Google Scholar]
- Coulthurst SJ, Barnard AM, Salmond GP. Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nature Reviews Microbiology. 2005;3:295–306. doi: 10.1038/nrmicro1128. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Portmann C, Zhang X, Roeffaers MB, Clardy J. Small molecule perimeter defense in entomopathogenic bacteria. Proceedings of the National Academy of Sciences. 2012;109:10821–10826. doi: 10.1073/pnas.1201160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain AL, Fang A. The natural functions of secondary metabolites. In: Fietcher A, editor. Advances in biochemical engineering/biotechnology: History of modern biotechnology I. vol. 69. Berlin: Springer; 2000. pp. 2–39. [DOI] [PubMed] [Google Scholar]
- Derzelle S, Duchaud E, Kunst F, Danchin A, Bertin P. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Applied Environmental Microbiology. 2002;68:3780–3789. doi: 10.1128/AEM.68.8.3780-3789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadio SX, Monciardini P, Alduina R, Mazza P, Chiocchini C, Cavaletti C, Sosio M, Puglia AM. Microbial technologies for the discovery of novel bioactive metabolites. Journal of Biotechnology. 2012;99:187–198. doi: 10.1016/s0168-1656(02)00209-2. [DOI] [PubMed] [Google Scholar]
- Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, Dassa E, Derose R, Derzelle S, Freyssinet G, Gaudriault S, Médigue C, Lanois A, Powell K, Siguier P, Vincent R, Wingate V, Zouine M, Glaser P, Boemare N, Danchin A, Kunst F. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nature Biotechnology. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- Dudnik A, Bigler L, Dudler R. Heterologous expression of a Photorhabdus luminescens syrbactin-like gene cluster results in production of the potent proteasome inhibitor glidobactin A. Microbiological Research. 2013;168:73–76. doi: 10.1016/j.micres.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Easom CA, Clarke DJ. Motility is required for the competitive fitness of entomopathogenic Photorhabdus luminescens during insect infection. BMC Microbiology. 2008;8:168. doi: 10.1186/1471-2180-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D, Holt K. Beginner’s guide to comparative bacterial genome analysis using next-generation sequence data. Microbial Informatics and Experimentation. 2013;3:1–9. doi: 10.1186/2042-5783-3-2. doi: 10.1186/2042-5783-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftherianos I, Boundy S, Joyce SA, Aslam S, Marshall JW, Cox RJ, Simpson TJ, Clarke DJ, Reynolds SE. An antibiotic produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Proceedings of the National Academy of Sciences. 2007;104:2419–2424. doi: 10.1073/pnas.0610525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- fFrench-Constant R, Waterfield N, Daborn P, Joyce S, Bennett H, Au C, Dowling A, Boundy S, Reynolds S, Clarke D. Photorhabdus: Towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiology Reviews. 2003;26:433–456. doi: 10.1111/j.1574-6976.2003.tb00625.x. [DOI] [PubMed] [Google Scholar]
- fFrench-Constant RH, Dowling A, Waterfield NR. Insecticidal toxins from Photorhabdus bacteria and their potential use in agriculture. Toxicon. 2007;49:436–451. doi: 10.1016/j.toxicon.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Foster V, Mourato S. Valuing the multiple impacts of pesticide use in the UK: A contingent ranking approach. Journal of Agricultural Economics. 2000;51:1–21. [Google Scholar]
- Gaugler R. Matching nematodes and insects to achieve optimal field performance. In: Polavarapu S, editor. Optimal use of insecticidal nematodes in pest management. New Brunswick, NJ: Rutgers University; 1999. pp. 9–14. [Google Scholar]
- Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: Two roads to the same destination. Molecular Microbiology. 2007;64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- Grewal PS, Ehlers RU, Shapiro-Ilan D. Nematodes as biocontrol agents. Cambridge, MA: CABI; 2005. [Google Scholar]
- Haas D, Gamper M, Zimmermann A, Galli E, Silver S, Witholt B. Pseudomonas biology and biotechnology. Washington, DC: American Society for Microbiology; 1992. [Google Scholar]
- Han RC, Ehlers RU. Trans-specific nematicidal activity of Photorhabdus luminescens. Nematology. 1999;1:687–693. [Google Scholar]
- Hattori M. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proceedings of the National Academy of Science. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazir S, Shapiro-Ilan D, Bock CH, Hazir C, Leite LG, Hotchkiss MW. Relative potency of culture supernatants of Xenorhabdus and Photorhabdus spp. on growth of some fungal phytopathogens. European Journal of Plant Pathology. 2016;146:369–381. [Google Scholar]
- Hevesi M, Al-Arabi K, Göndör M, Papp J, Honty K, Kasa K, Toth M. 2004 Development of eco-friendly strategies for the control of fire blight in Hungary. Pp. 345–348 in X International Workshop on Fire Blight. vol. 704. [Google Scholar]
- Hillocks RJ. Farming with fewer pesticides: EU pesticide review and resulting challenges for UK agriculture. Crop Protection. 2012;31:85–93. [Google Scholar]
- Hu KJ, Li JX, Wang WJ, Wu HM, Lin H, Webster JM. Comparison of metabolites produced in vitro and in vivo by Photorhabdus luminescens, a bacterial symbiont of the entomopathogenic nematode Heterorhabditis megidis. Canadian Journal of Microbiology. 1998;44:1072–1077. [Google Scholar]
- Hu K, Li J, Webster JM. 3,5-Dihydroxy-4-isopropylstilbene: A selective nematicidal compound from the culture filtrate of Photorhabdus luminescens. Canadian Journal of Plant Pathology. 1996;18:104. (Abstr.). [Google Scholar]
- Hu K, Li J, Webster JM. Nematicidal metabolites produced by Photorhabdus luminescens (Enterobacteriaceae), bacterial symbiont of entomopathogenic nematodes. Nematology. 1999;1:457–69. [Google Scholar]
- Hu KJ, Li JX, Li B, Webster JM, Chen GH. A novel antimicrobial epoxide isolated from larval Galleria mellonella infected by the nematode symbiont, Photorhabdus luminescens (Enterobacteriaceae) Bioorganic and Medicinal Chemistry. 2006;14:4677–4681. doi: 10.1016/j.bmc.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Hu K, Webster JM. Mortality of plant-parasitic nematodes caused by bacterial (Xenorhabdus spp. and Photorhabdus luminescens) culture media. Journal of Nematology. 1995;27:502–503. [Google Scholar]
- Huber J, Donald RG, Lee SH, Jarantow LW, Salvatore MJ, Meng X, Painter R, Onishi RH, Occi J, Dorso K, Young K. Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chemistry and Biology. 2009;16:837–848. doi: 10.1016/j.chembiol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Inman FL, III, Holmes L. Antibacterial screening of secreted compounds produced by the phase I variant of Photorhabdus luminescens. Indian Journal of Microbiology. 2012;5:708–709. doi: 10.1007/s12088-012-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce SA, Brachmann AO, Glazer I, Lango L, Schwär G, Clarke DJ, Bode HB. Bacterial biosynthesis of a multipotent stilbene. Angewandte Chemie International Edition. 2008;47:1942–1945. doi: 10.1002/anie.200705148. [DOI] [PubMed] [Google Scholar]
- Kaya HK, Gaugler R. Entomopathogenic nematodes. Annual Reviews in Entomology. 1993;38:181–206. [Google Scholar]
- Kerry BR. 1998. Progress towards biological control strategies for plant-parasitic nematodes. Pp. 739–746 in Proceedings of the Brighton Crop Protect Conference: Pests and Disease British Crop Protection Council, Farnham, Surrey, UK. [Google Scholar]
- Kontnik R, Crawford JM, Clardy J. Exploiting a global regulator for small molecule discovery in Photorhabdus luminescens. ACS Chemical Biology. 2010;5:659–665. doi: 10.1021/cb100117k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz TO, Winston DJ, Bruckner DA, Martin WJ. Comparative in vitro synergistic activity of new beta-lactam antimicrobial agents and amikacin against Pseudomonas aeruginosa and Serratia marcescens. Antimicrobial Agents and Chemotherapy. 1981;20:239–243. doi: 10.1128/aac.20.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange L, Sanchez Lopez C. Microorganisms as a source of biologically active secondary metabolites. In: Copping LG, editor. Crop protection agents from nature: Natural products analogues. Cambridge, UK: The Royal Society of Chemistry; 1996. pp. 1–26. [Google Scholar]
- Li J, Chen G, Webster JM, Czyzewska E. Antimicrobial metabolites from a bacterial symbiont. Journal of Natural Products. 1995;58:1081–1086. doi: 10.1021/np50121a016. [DOI] [PubMed] [Google Scholar]
- Maxwell PW, Chen G, Webster JM, Dunphy GB. Stability and activities of antibiotics produced during infection of the insect Galleria mellonella by two isolates of Xenorhabdus nematophilus. Applied and Environmental Microbiology. 1994;60:715–721. doi: 10.1128/aem.60.2.715-721.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney BV, Gregson RP, Lacey MJ, Akhurst RJ, Lyons GR, Rhodes SH, Smith DR, Engelhardt LM, White AH. Biologically active metabolites from Xenorhabdus spp., Part 1. Dithiolopyrrolone derivatives with antibiotic activity. Journal of Natural Products. 1991a;54:774–784. doi: 10.1021/np50075a005. [DOI] [PubMed] [Google Scholar]
- McInerney BV, Taylor WC, Lacey MJ, Akhurst RJ, Gregson RP. Biologically active metabolites from Xenorhabdus spp. Part 2. Benzopyrane-1-one derivatives with gastro-protective activity. Journal of Natural Products. 1991b;54:785–795. doi: 10.1021/np50075a006. [DOI] [PubMed] [Google Scholar]
- Montesinos E. Development, registration and commercialization of microbial pesticides for plant protection. International Microbiology. 2003;6:245–252. doi: 10.1007/s10123-003-0144-x. [DOI] [PubMed] [Google Scholar]
- Mukherjee AK, Das K. Correlation between diverse cyclic lipopeptides production and regulation of growth and substrate utilization by Bacillus subtilis strains in a particular habitat. FEMS Microbiology Ecology. 2005;54:479–489. doi: 10.1016/j.femsec.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Nollmann FI, Heinrich AK, Brachmann AO, Morisseau C, Mukherjee K, CasanovaTorres ÁM, Strobl F, Kleinhans D, Kinski S, Schultz K, Beeton ML. A Photorhabdus natural product inhibits insect juvenile hormone epoxide hydrolase. Chemistry and BioChemistry. 2015;16:766–771. doi: 10.1002/cbic.201402650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena M, Jacques P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends in Microbiology. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Orozco RA, Molnár I, Bode H, Stock SP. Bioprospecting for secondary metabolites in the entomopathogenic bacterium Photorhabdus luminescens subsp. sonorensis. Journal of Invertebrate Pathology. 2016;141:45–52. doi: 10.1016/j.jip.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Park HB, Crawford JM. Lumiquinone A, an α-aminomalonate-derived aminobenzoquinone from Photorhabdus luminescens. Journal of Natural Products. 2015;78:1437–1441. doi: 10.1021/np500974f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul VJ, Frautschy S, Fenical W, Nealson KH. Isolation and structure assignment of several new antibacterial compounds from the insect-symbiotic bacteria Xenorhabdus spp. Journal of Chemical Ecology. 1981;7:589–597. doi: 10.1007/BF00987707. [DOI] [PubMed] [Google Scholar]
- Pearce D, Koundouri P. Fertilizer and pesticide taxes for controlling non-point agricultural pollution. Agricultural and Rural Development Department, World Bank Group, ; 2003. www.worldbank.org/rural. [Google Scholar]
- Pimentel H, Acquay M, Biltonen P, Rice M, Silva J, Nelson V, Lipner S, Giordano A, Horowitz M, D’Amore M. Environmental and human costs of pesticide use. Bioscience. 1992;42:750–760. [Google Scholar]
- Pimentel D, Greiner A. Environmental and socio-economic costs of pesticide use. In: Pimentel D, editor. Techniques for reducing pesticide use: Economic and environmental benefits. Chichester: John Wiley and Sons; 1997. pp. 51–78. [Google Scholar]
- Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecological Economics. 2005;52:273–288. [Google Scholar]
- Poinar GO, Thomas G, Haygood M, Nealson KH. Growth and luminescence of the symbolic bacteria associated with the terrestrial nematode, Heterorhabditis bacteriophora. Soil Biology and Biochemistry. 1980;121:5–10. [Google Scholar]
- Richardson WH, Schmidt TM, Nealson KH. Identification of an anthraquinone pigment and a hydroxystilbene antibiotic from Xenorhabdus luminescens. Applied Environmental Microbiology. 1988;54:1602–1605. doi: 10.1128/aem.54.6.1602-1605.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Karunarathna A, Buckley NA, Manuweera G, Rezvi Sheriff M, Eddleston MH. Influence of pesticide regulation on acute poisoning deaths in Sri Lanka. Bulletin WHO. 2003;81:789–798. [PMC free article] [PubMed] [Google Scholar]
- San-Blas E, Carrillo Z, Parra Y. Effect of Xenorhabdus and Photorhabdus bacteria and their exudates on Moniliophthora roreri. Archives of Phytopathology and Plant Protection. 2012;45:1950–1967. [Google Scholar]
- Sansinenea E, Ortiz A. Secondary metabolites of soil Bacillus spp. Biotechnology Letters. 2011;33:1523–1538. doi: 10.1007/s10529-011-0617-5. [DOI] [PubMed] [Google Scholar]
- Shapiro-Ilan DI, Reilly CC, Hotchkiss MW. Suppressive effects of metabolites from Photorhabdus and Xenorhabdus spp. on phytopathogens of peach and pecan. Archives of Phytopathology and Plant Protection. 2009;42:715–728. [Google Scholar]
- Shi H, Zeng H, Yang X, Zhao J, Chen M, Qiu D. An insecticidal protein from Xenorhabdus ehlersii triggers prophenoloxidase activation and hemocyte decrease in Galleria mellonella. Current Microbiology. 2012;64:604–610. doi: 10.1007/s00284-012-0114-7. [DOI] [PubMed] [Google Scholar]
- Shrestha YK, Lee KY. Oral toxicity of Photorhabdus culture media on gene expression of the adult sweetpotato whitefly, Bemisia tabaci. Journal of Invertebrate Pathology. 2012;109:91–96. doi: 10.1016/j.jip.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Sikora RA, Hoffmann-Hergarten S. Biological control of plant-parasitic nematodes with plant-health promoting rhizobacteria. In: Lumsden RD, Vaughn JL, editors. Pest management: Biologically based technologies; Proceedings of Beltsville Symposium XVIII. ; Washington, DC: American Chemical Society; 1993. pp. 166–172. [Google Scholar]
- Siddiqi ZA, Mahmood I. Role of bacteria in the management of plant-parasitic nematodes: A review. Bioresources and Technology. 1999;69:167–179. [Google Scholar]
- Somvanshi VS, Sloup RE, Crawford JM, Martin AR, Heidt AJ, Kim KS, Clardy J, Ciche TA. A single promoter inversion switches Photorhabdus between pathogenic and mutualistic states. Science. 2012;337:88–93. doi: 10.1126/science.1216641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein ML, Beck P, Kaiser M, Dudler R, Becker CF, Groll M. One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor. Proceedings of the National Academy of Sciences. 2012;109:18367–18371. doi: 10.1073/pnas.1211423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickle WE. EPA regulation of inerts under the bush administration: A new departure. In: Mueningroff JC, Vietz AK, Downer RA, editors. Pesticide formulations and application systems. vol. 23. ASTM International; 2003. [Google Scholar]
- Stock SP, Goodrich-Blair H. Entomopathogenic nematodes and their bacterial symbionts: The inside out of a mutualistic association. Symbiosis. 2008;46:65–76. [Google Scholar]
- Sztaricskai F, Dinya Z, Batta G, Szallas E, Szentirmai A, Fodor A. Anthraquinones produced by enterobacters and nematodes. Acta Chimica Hungary. 1992;129:697–707. [Google Scholar]
- Teasdale ME, Liu J, Wallace J, Akhlaghi F, Rowley DC. Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Applied and Environmental Microbiology. 2009;75:567–572. doi: 10.1128/AEM.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore CM, King JB, You J, Cichewicz RH. Production of cytotoxic glidobactins/luminmycins by Photorhabdus asymbiotica in liquid media and live crickets. Journal of natural products. 2012;75:2007–2011. doi: 10.1021/np300623x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson NR, Crow MA, McGowan SJ, Cox A, Salmond GPC. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Molecular Microbiology. 2000;36:539–556. doi: 10.1046/j.1365-2958.2000.01872.x. [DOI] [PubMed] [Google Scholar]
- Tobias NJ, Mishra B, Gupta DK, Sharma R, Thines M, Stinear TP, Bode HB. Genome comparisons provide insights into the role of secondary metabolites in the pathogenic phase of the Photorhabdus life cycle. BMC Genomics. 2016;17:537. doi: 10.1186/s12864-016-2862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uma GP, Prabhuraj A, Patil MB. Antibiotic and antibacterial activity of a symbiotic bacterium, Photorhabdus luminescens. Journal of Biological Control. 2010;24:168–172. [Google Scholar]
- Ullah I, Khan AL, Ali L, Khan AR, Waqas M, Hussain J, Lee IJ, Shin JH. Benzaldehyde as an insecticidal, antimicrobial, and antioxidant compound produced by Photorhabdus temperata M1021. The Journal of Microbiology. 2015;53:127–133. doi: 10.1007/s12275-015-4632-4. [DOI] [PubMed] [Google Scholar]
- Vidaver AK. Uses of antimicrobials in plant agriculture. Clinical Infectious Diseases. 2002;34:S107–S110. doi: 10.1086/340247. [DOI] [PubMed] [Google Scholar]
- Voisard C, Keel C, Haas D, Dèfago G. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. The EMBO Journal. 1989;8(2):351–358. doi: 10.1002/j.1460-2075.1989.tb03384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield NR, Ciche T, Clarke D. Photorhabdus and a host of hosts. Annual Review of Microbiology. 2009;63:557–574. doi: 10.1146/annurev.micro.091208.073507. [DOI] [PubMed] [Google Scholar]
- Webster JM, Chen G, Hu K, Li J. Bacterial metabolites. In: Gaugler R, editor. Entomopathogenic nematology. New York, NY: CABI Publishing; 2002. pp. 99–114. [Google Scholar]