Abstract
Two different nematode species were recovered from pomegranate decaying fruit in two localities in Southern Italy: the mycetophagus nematode Sheraphelenchus sucus and a bacterial feeder nematode belonging to the Panagrolaimidae (Rhabditida) family. Morphometrics of the Italian population of S. sucus closely resemble that of the type population, whereas some differences were found when compared with another population from Iran. Molecular characterization of the Italian S. sucus using the 18S rRNA gene, D2–D3 expansion domains of the 28S rDNA, the ITS region, and the partial mitochondrial COI were carried out. Sequences of the 18S rRNA gene, the D2–D3 domains, and the ITS were analyzed using several methods for inferring phylogeny to reconstruct the relationships among Sheraphelenchus and Bursaphelenchus species. The bacterial feeder Panagrellus sp. was characterized at the molecular level only. The D2–D3 expansion domains and ITS sequences of this Italian panagrolaimid were determined. The D2–D3 sequences of the Italian panagrolaimid showed 99% similarity with the corresponding sequence of Panagrellus sp. associated with Rhynchophorus ferrugineus. This is the first report on the tritrophic association of S. sucus and Rhabditida that uses both insects and pomegranate fruit as hosts.
Keywords: 18S rRNA gene, D2–D3 expansion domains, molecular analysis, Panagrolaimidae, phylogenesis, Sheraphelenchus sucus
Among the Mediterranean small fruit producers, Italy is one of the largest pomegranate (Punica granatum) producers. More than 75% of Italian commercial orchards are located in the South of the country where the importance of this crop has largely increased in the last 10 years. The harvesting period of this crop is from the end of September to the middle of November (when it is particularly humid in the circum-Mediterranean countries) and this period corresponds to an increase in disease development upon maturity. The most serious problem with pomegranate is the occurrence of biotic and abiotic diseases causing loss of production and also resulting in fruit splitting on the plant. While leaf drop is commonly considered of limited importance, fruit splitting is one of the major abiotic diseases because it usually occurs just as the fruits begin to mature predisposing them to fungal colonization also during storage. Palou et al. (2013) listed the main fungal species known to cause pomegranate fruit rot worldwide (natural or laboratory infections), including Alternaria spp., Aspergillus niger van Tiegh. and other Aspergillus spp., Botrytis cinerea Pers. Fr., Colletotrichum gloeosporioides (Penz.) Penz. & Sacc., Coniella spp., Nematospora spp., Penicillium spp., Pestalotiopsis versicolor Speg. Steyaert, Pilidiella granati Sacc., Rhizopus spp., and Syncephalastrum racemosum Cohn ex J. Schröt.
Nitidulid beetles on rotten/decaying fruits are frequently observed; the insects can vector the causal agents of fungal infections and mycophagous nematodes, mainly belonging to the family of Aphelenchoididae. One of the nematode species recovered from decaying orange in Hawaii and fig in California (Nickle, 1970) associated with nitidulid beetles was identified as Sheraphelenchus entomophagus Nickle, 1970. The genus includes four nominal species: S. entomophagus Nickle, 1970; Sheraphelenchus brevigulonis (Massey and Hinds, 1970) Hunt, 1993, S. sucus (Kanzaki and Tanaka, 2013); and Sheraphelenchus parabrevigulonis Fang et al. (2015). Then, several entomophagus-like morphospecies have been isolated: S. entomophagus from orange in Italy (Lo Giudice and Lanza, 1973), S. sucus from sap flow in Japan (Kanzaki and Tanaka, 2013), S. entomophagus from pomegranate in Iran (Moosavi et al., 2014), S. sucus from onion bulbs imported from South Korea (Fang et al., 2015), and from pomegranate in Italy (the present study). All these Sheraphelenchus morphospecies are characterized by conserved morphology and identical ribosomal sequences suggesting that they could be conspecific therefore further investigations on type material of S. entomophagus from Hawaii are needed.
During a field survey for fungal diseases on pomegranate orchards in Southern Italy, abundant populations of nematodes and fungi were collected from pomegranate fruit showing rot symptoms. Two different nematode species (a mycetophagus and a bacterial-feeder species) were recovered, belonging to Aphelenchoididae and Panagrolaimidae families, respectively. The mycetophagus nematode population was abundant and identified as S. sucus based on morphological and molecular analyses; by contrast, the number of the bacterial feeder nematode was low, thus it was characterized at molecular level only and identified as Panagrellus sp.
The objectives of the present study were to (i) characterize morphologically and molecularly the Italian population of the mycetophagus nematode species; (ii) infer the phylogenetic relationships with other species and genera of Aphelenchoididae using the ITS, the SSU, and D2–D3 expansion domains; (iii) characterize the bacterial feeder nematodes belonging to Panagrolaimidae associated with decaying pomegranate fruit.
Materials and Methods
Fruit symptoms and nematode extraction:
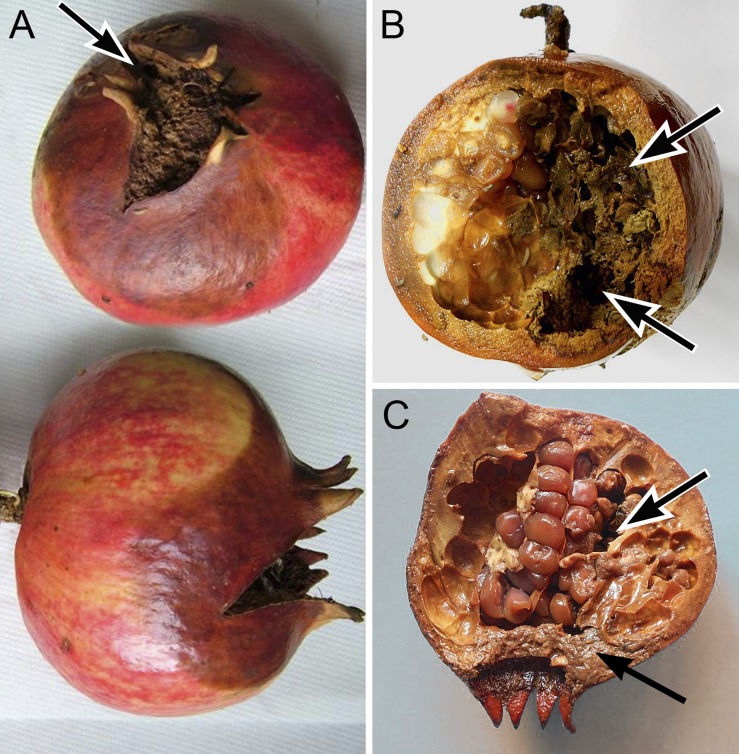
Pomegranate fruit showing rotting symptoms (Fig. 1) were collected at Metaponto (Matera province, Basilicata) and at Fasano (Brindisi province, Apulia), in Southern Italy on October 2015. Three out of twelve sampled localities revealed the occurrence of decaying fruit, still hanging on the tree, with simultaneous presence of fungi, bacterial- and fungal-feeder nematodes. Pomegranate fruit was fragmented into small pieces of 1 × 2 cm which were incubated in water in Petri dishes. After 2–3 hr of incubation, the emerged nematodes were used to conduct morphological and molecular observations.
Fig. 1.
Pomegranate decaying fruit hosting fungi, the nematode mycetophagus (Sheraphelenchus sucus), and bacteriophagus (Panagrolaimus sp.) populations. A. Top (top) and lateral (bottom) view of the entire decaying fruit; B, C. Cross-section of an affected pomegranate showing more or less extensive rotting tissue portions.
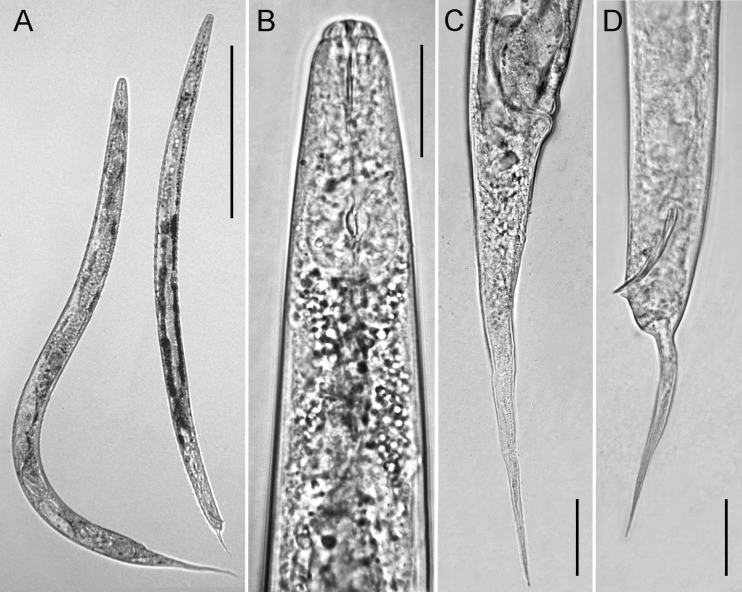
Adult specimens of the mycetophagus population for microscopic observation were handpicked and killed by gentle heat, fixed in a solution of 4% of formaldehyde +1% propionic acid, and processed to glycerol according to Seinhorst’s method (Hooper, 1986). Glycerin-infiltrated specimens were examined by light microscopy for diagnosis. Photographs were taken with a Leica DFC 425 system mounted on a Leitz Wetzlar optical microscope, using specimens mounted on water agar temporary slides (Troccoli, 2002) (Fig. 2), whereas measurements were made at the camera lucida on glycerine infiltrated specimens. Abbreviations and terms used in the morphometric data presentation as defined in Siddiqi (2000).
Fig. 2.
Light microscopy micrographs illustrating morphodiagnostic characters of freshly mounted specimens of the Italian bisexual population of Sheraphelenchus sucus. A. Female (left) and male (right) entire body; B. Female anterior end; C. Female tail; D. Male tail with spicules (Scale bars: A = 250 μm; B–D = 20 μm).
DNA extraction, PCR amplification, and sequencing:
Individual nematodes were crushed with a sterile microspatula under a stereomicroscope. Genomic DNA was extracted from fifteen and ten individual mycetophagus and bacterial nematodes, respectively, as described by De Luca et al. (2004). The crude DNA isolated from each individual nematode was directly amplified. The ITS1-5.8S-ITS2 regions were amplified using the forward primer TW81 (5′-GTTTCCGTAGGTGAACCTGC-3′) and the reverse primer AB28 (5′-ATATGCTTAAGTTCAGCGGGT-3′) (Joyce et al., 1994); the 18S rDNA was amplified using the 18SnF (5′-TGGATAACTGTGGTAATTCTAGAGC-3′) and 18SnR (5′-TTACGACTTTTGCCCGGTTC-3′) (Kanzaki and Futai, 2002); the D2A-D3B expansion segments of 28S rRNA gene amplified using the primers D2A (5′-ACAAGTACCGTGGGGAAAGTTG-3′) and the D3B (5′-TCGGAAGGAACCAGCTACTA-3′) (Nunn, 1992) and the portion of the mitochondrial cytochrome oxidase c subunit 1 (mtCOI) gene was amplified with this primer set: COI-F1 (5′-CCTACTATGATTGGTGGTTTTGGTAATTG-3′) and COI-R2 (5′-GTAGCAGCAGTAAAATAAGCACG-3′) (Kanzaki and Futai, 2002). PCR cycling conditions used for amplification were an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 50 sec, annealing at 55°C for 50 sec, extension at 72°C for 1 min, and a final step at 72°C for 7 min. The size of the amplification products was determined by comparison with the molecular weight marker ladder 100 (Fermentas, St. Leon-Rot, Germany) after electrophoresis of 10 μl on a 1% agarose gel. PCR products of the ITS containing region, the 18 S gene, the D2–D3 expansion domains, and the mitochondrial COI, from three individual nematodes, were purified using the protocol suggested by the manufacturer (High Pure PCR elution kit; Roche, Germany). Purified DNA fragments were cloned and sent for sequencing, in both directions, at MWG-Eurofins in Germany.
Phylogenetic analysis:
A BLAST (Basic Local Alignment Search Tool) search at NCBI (National Center for Biotechnology Information) was performed to confirm their nematode origins and species (Altschul et al., 1997). The newly obtained sequences for the ITS, 18S rRNA gene, the D2–D3 expansion domains of 28S gene, and mtcoI were aligned using ClustalW with default parameters with the corresponding published gene sequences of Aphelenchoididae. Sequence alignments were manually edited using BioEdit to improve the multialignment. Outgroup taxa for each dataset were chosen based on the results of previously published data. Phylogenetic trees, obtained for the ITS, 18S rRNA gene, and the D2–D3 expansion domains were performed with Neighbour-Joining, Minimum Evolution, Maximum Likelihood (ML), and Maximum Parsimony methods using MEGA version 7 software (Kumar et al., 2016). No significant conflict in branching order and support level among methods is observed and, therefore, only ML tree is shown for each marker. The phylograms were bootstrapped 1,000 times to assess the degree of support for the phylogenetic branching indicated by the optimal tree for each method. The newly ITS and D2–D3 sequences of the bacterial nematodes Panagrellus sp. were aligned with the corresponding sequences of Rhabditida. Rhabditophanes sp. and Halicephalobus gingivalis were used as outgroup taxa. The newly obtained sequences were submitted to GenBank with the following accession numbers: LT903555–LT903556 for the ITS of S. sucus; LT837694 for the 18S gene of S. sucus; LT837695–LT837696 for the D2–D3 domains of 28S gene of S. sucus; LT-837697–LT837698 for the mitochondrial COI of S. sucus; LT837699–LT8837700 for the ITS of Panagrellus sp., and LT908055–LT908056 for the D2–D3 domains of 28S gene of Panagrellus sp.
Results
Identification of Sheraphelenchus population:
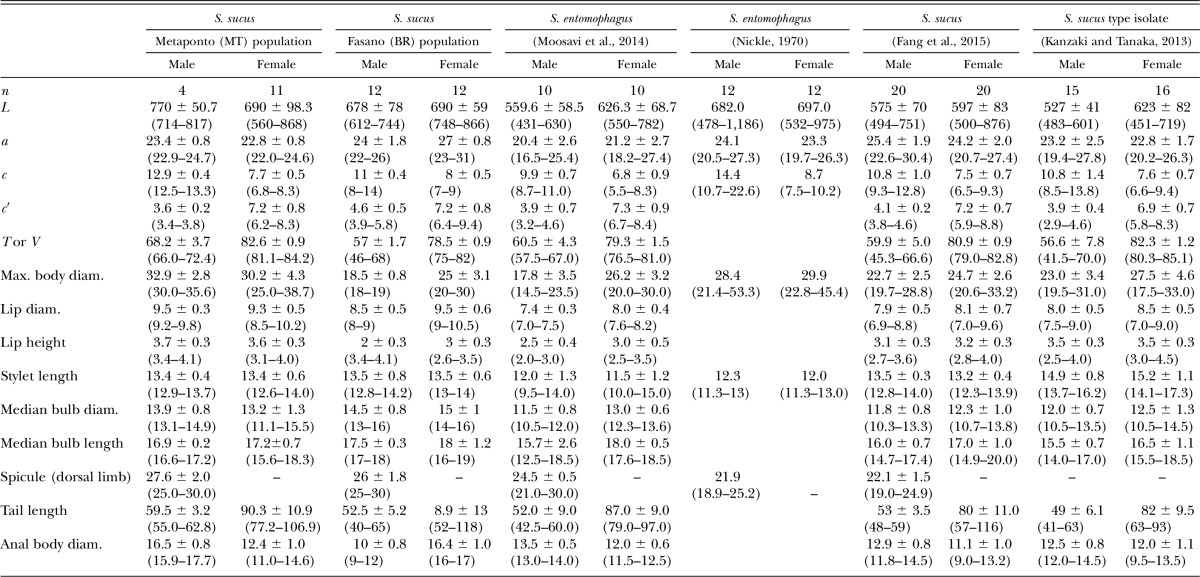
In Table 1, the comparative morphometrics of populations of S. entomophagus (type and Iranian populations) and of S. sucus from the present study and from literature are presented. The morphology of the two populations of S. sucus from Italy closely resembles that of the type population of S. sucus from Japan (Kanzaki and Tanaka, 2013) and that from South Korea (Fang et al., 2015). However, they are also morphologically close to that of the type population of S. entomophagus from Hawaii (Nickle, 1970) as well as to the Iranian S. entomophagus (Moosavi et al., 2014) although with little variation concerning the slightly smaller size of specimens of the latter population. The stylet length, which was originally considered the only diagnostic character useful to separate these species (sensu Kanzaki and Tanaka, 2013), was afterward amended as being underestimated (Kanzaki, 2015) resulting in the close similarity between the two species S. sucus and S. entomophagus.
Table 1.
Morphometric comparison of the Italian populations of Sheraphelenchus sucus with populations of S. entomophagus and S. sucus from the literature. All measurements are in micrometers and in the form: mean ± s.d. (range).
Molecular identification:
Amplification of the ITS, 18S rRNA gene, the D2–D3 expansion segments of the 28S gene, and the mitochondrial COI of Italian S. sucus yielded single fragments of 1,218, 1,610, 779, and 711 bp, respectively.
The ITS sequences of Italian S. sucus from pomegranate showed 99% similarity (8–9 nucleotides different) to S. sucus original population (KC875230 and KC875231). The partial 18S rRNA sequences of S. sucus from pomegranate were identical to S. sucus (AB808720 and KC875229) from the database (99% similarity and 5–8 different nucleotides, respectively) and S. parabrevigulonis (KC875226 and 98% similarity, 30 different nucleotides). The D2–D3 expansion domains of the Italian S. sucus showed a 99% similarity with the corresponding sequences of S. sucus (KC894755 and AB808721; 3 and 8 different nucleotides, respectively) and S. entomophagus from Iran (KF373753 and KF373754; 1 different nucleotide), present in the database. No corresponding sequences were present in the database for the partial mitochondrial COI of the Italian S. sucus population.
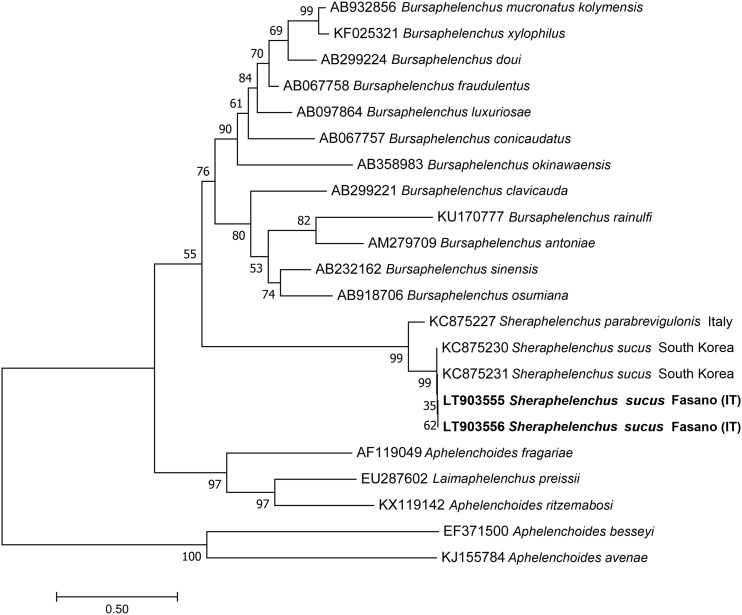
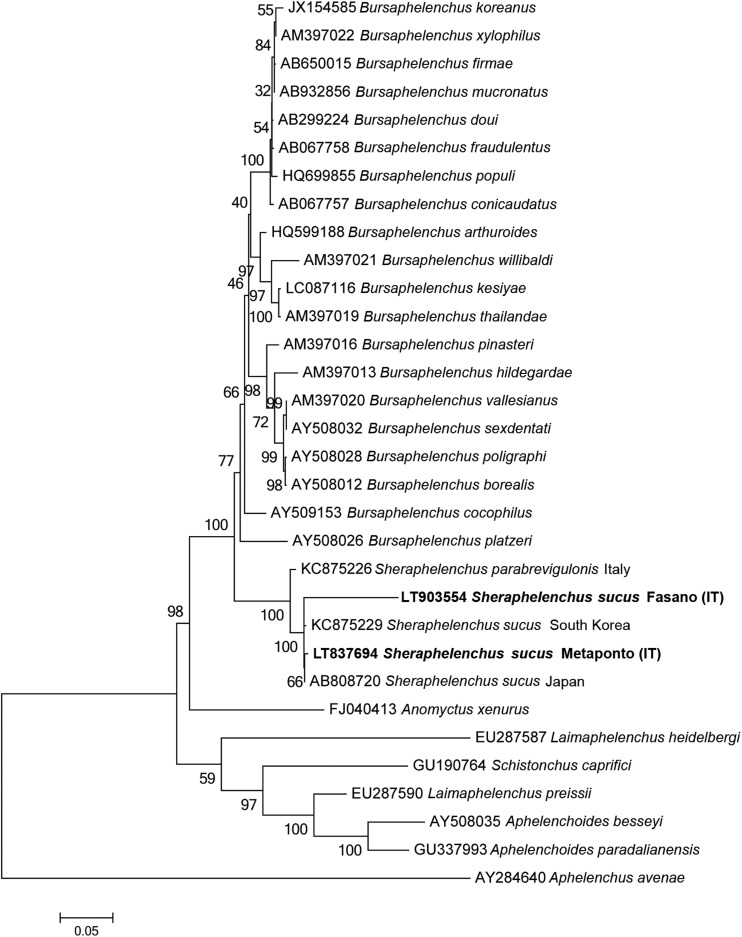
Phylogenetic relationships of the Italian population of S. sucus based on the ITS of a multiple edit alignment including 22 sequences, using ML method with Aphelenchus avenae as outgroup are reported in Fig. 3. All populations of S. sucus clustered together in the same clade with 99% of bootstrap support and revealed sister relationships with S. parabrevigulonis.
Fig. 3.
Phylogenetic tree based on ITS sequences describing the evolutionary relationships among different Sheraphelenchus and Bursaphelenchus species using the Maximum Likely (ML) method. Branch lengths are proportional to the distances as derived from the distance matrix obtained using the General Time Reversible (GTR) method with the invariant site plus gamma options. Numbers at nodes indicate bootstrap values.
Phylogenetic relationships of the Italian population of S. sucus based on 18S rRNA gene of a multiple edited alignment including 31 sequences, using ML method with Aphelenchus avenae as an outgroup is reported in Fig. 4. This ML tree showed that all populations of S. sucus formed a well-supported group with 100% support and resulted sister species with S. parabrevigulonis.
Fig. 4.
Phylogenetic tree based on 18S rRNA gene describing the evolutionary relationships among different Sheraphelenchus and Bursaphelenchus species using the Maximum Likely (ML) method. Branch lengths are proportional to the distances as derived from the distance matrix obtained using the General Time Reversible (GTR) method with the invariant site plus gamma options. Numbers at nodes indicate bootstrap values.
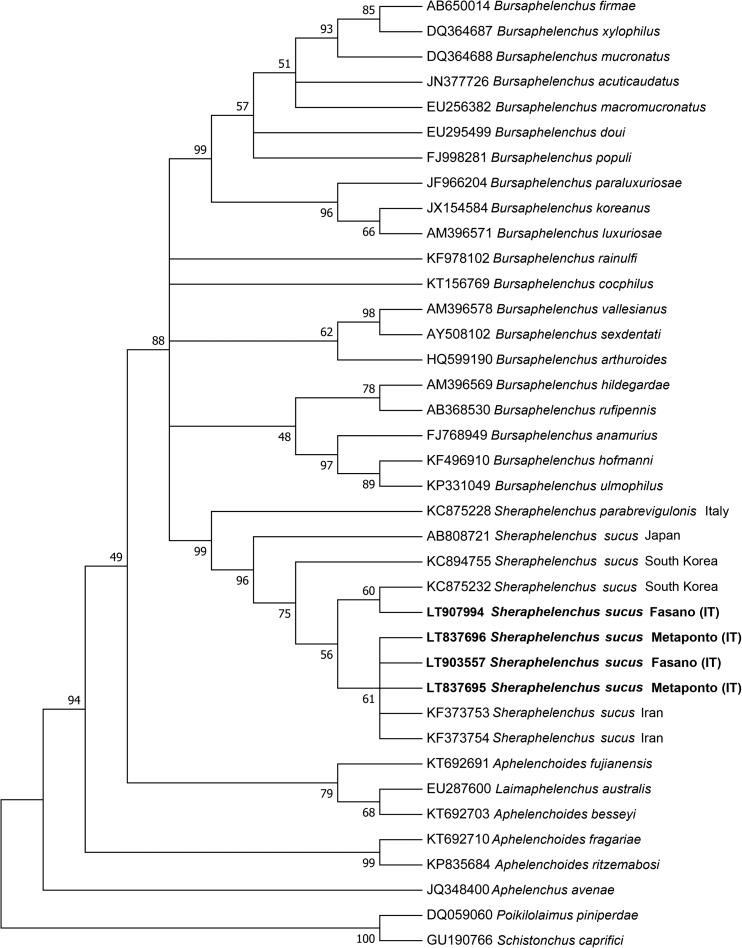
Phylogenetic relationships of S. sucus from Italy based on D2–D3 expansion domains of a multiple edited alignment including 34 sequences were also determined (Fig. 5). Sheraphelenchus sucus populations formed a well-supported grouping with S. entomophagus from Iran, congruent with those obtained by using the 18S rRNA gene and ITS.
Fig. 5.
Phylogenetic tree based on D2–D3 expansion domains of 28S rRNA gene describing the evolutionary relationships among different Sheraphelenchus and Bursaphelenchus species using the Maximum Likely (ML) method. Branch lengths are proportional to the distances as derived from the distance matrix obtained using the General Time Reversible (GTR) method with the invariant site plus gamma options. Numbers at nodes indicate bootstrap values.
The ITS containing region of Panagrellus sp. produced a single fragment of 1,261 bp. No corresponding sequences were present in the database. The D2–D3 sequences of Panagrellus sp. from pomegranate showed 99% similarity (8 nucleotide different) with Panagrellus sp. isolated from aberrant adult specimen of R. ferrugineus (Camerota et al., 2016). Thus, selected Panagrellus and rhabditid sequences were aligned with those of D2–D3 of Panagrellus sp. from pomegranate.
Discussion
In the present study, the molecular identification of the two nematode species occurring in the same rotting pomegranate fruit revealed that the ITS, the partial 18S rRNA gene, the D2–D3 expansion domains, and the partial mitochondrial COI clearly identified S.sucus, whereas the ITS and D2–D3 sequences of the bacterial feeder nematode from pomegranate were identified as Panagrellus sp. The results obtained by sequence and phylogenetic analyses conducted in the present study strongly suggest that S. sucus and S. entomophagus from Iran are conspecific. In addition, morphology and measurements of these two species, including the type population of S. entomophagus from Hawaii fit well with the intraspecific variability of species as reported for other nematode species (Gutiérrez-Gutiérrez et al., 2013; Groza et al., 2017) thus giving strong support for synonymization. However, no molecular data are available for the S. entomophagus type population, suggesting the necessity of reisolation of the S. entomophagus type population from the original locality to organize the species status using morphological characters and molecular barcoding.
Very little is known about the morphological and molecular characterization of Panagrellus species and their interaction with other organisms and corresponding habitats (Stock and Nadler, 2006). Panagrellus species have been already reported as insect associates (Massey, 1974) in bark beetles. In Italy, Smith et al. (1992) described a yeast–nematode–insect association with Drosophila flies in grape sour rot suggesting that Drosophila contributed to the dissemination of these insect associates.
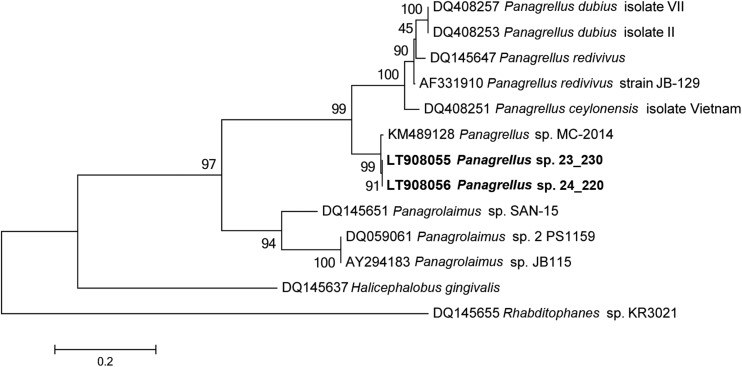
In the present study, phylogenetic relationships of Panagrellus species based on the D2–D3 sequences supported the grouping of Panagrellus sp. from rotting pomegranate with Panagrellus sp. from R. ferrugineus, with 99% overlap (Fig. 6). This finding suggests that the Italian isolates of Panagrellus sp. share the same habitat preference, decaying fruits or rotting materials, and the phoretic relationships occurred casually when they occupy the same habitat of insects (Félix and Duveau, 2012; Camerota et al., 2016; Kanzaki, 2017). These observations strongly suggest that the Italian Panagrellus sp. isolates undergo host switching.
Fig. 6.
Phylogenetic tree based on D2–D3 expansion domains of 28S rRNA gene describing the evolutionary relationships among different Panagrellus species using the Maximum Likely (ML) method. Branch lengths are proportional to the distances as derived from the distance matrix obtained using the General Time Reversible (GTR) method with the invariant site plus gamma options. Numbers at nodes indicate bootstrap values.
The coexistence of the mycetophagus nematode, S. sucus, and the bacterial feeder nematode, Panagrellus sp. in the same rotting pomegranate has never been reported so far. As both nematode species were found in hanging fruit, it is conceivable that they have been carried by insects with which they are phoretically or occasionally associated. More studies are needed to understand the role of phoretic insect–nematode associations in insect ecology and evolution and how they influence reproduction, survival, etc.
Literature Cited
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerota M, Mazza G, Carta LK, Paoli F, Torrini G, Benvenuti C, Carletti B, Francardi V, Roversi PF. Occurrence of Panagrellus (Rhabditida: Panagrolaimidae) nematodes in a morphologically aberrant adult specimen of Rhynchophorus ferrugineus (Coleoptera: Dryophthoridae) Journal of Nematology. 2016;48:1–6. doi: 10.21307/jofnem-2017-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca F, Fanelli E, Di Vito M, Reyes A, De Giorgi C. Comparison of the sequences of the d3 expansion of the 26s ribosomal genes reveals different degrees of heterogeneity in different populations and species of Pratylenchus from the Mediterranean region. European Journal of Plant Pathology. 2004;111:949–957. [Google Scholar]
- Fang Y, Gu J, Wang X, Wang J, Li H. Description of Sheraphelenchus parabrevigulonis n. sp (Nematoda: Aphelenchoidinae) in pine wood packing from Italy and redescription of S. sucus in onion bulbs from S. Korea, isolated at Nigbo, China. Nematology. 2015;17:213–229. [Google Scholar]
- Félix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biology. 2012;10:59–77. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groza M, Lazarova S, De Luca F, Fanelli E, Elshishka M, Radoslavov G, Hristov P, Coman M, Peneva V. The morphological and molecular identity of Longidorus piceicola Lišková, Robbins & Brown, 1997 from Romania (Nematoda, Dorylaimida) Zookeys. 2017;667:1–19. doi: 10.3897/zookeys.667.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Gutiérrez C, Cantalapiedra-Navarrete C, Remesal E, Palomares-Rius JE, Navas-Cortés JA, Castillo P. New insight into the identification and molecular phylogeny of dagger nematodes of the genus Xiphinema (Nematoda: Longidoridae) with description of two new species. Zoological Journal of the Linnean Society. 2013;169:548–579. [Google Scholar]
- Hooper DJ. Handling, fixing, staining and mounting nematodes. In: Southey JF, editor. Laboratory methods for work with plant and soil nematodes. Reference book 402. London, UK: Her Majesty’s Stationery Office; 1986. pp. 59–80. [Google Scholar]
- Hunt DJ. Aphelenchida, Longidoridae and Trichodoridae: Their systematics and bionomics. Wallingford, UK: CAB International; 1993. [Google Scholar]
- Joyce SA, Burnell AM, Powers TO. Characterization of Heterorhabditis isolates by PCR amplification of segments of mtDNA and rDNA genes. Journal of Nematology. 1994;26:260–270. [PMC free article] [PubMed] [Google Scholar]
- Kanzaki N. Comment on the stylet of Sheraphelenchus sucus Kanzaki & Tanaka, 2013. Nematology. 2015;17:499–500. [Google Scholar]
- Kanzaki N. Current status of entomophilic nematode survey in Japan. In: Motokawa M, Kajihara H, editors. Species diversity of animals in Japan. 2017. pp. 285–317. Japan: Springer. DOI 10.1007/978-4-431-56432-4-11. [Google Scholar]
- Kanzaki N, Futai K. A PCR primer set for determination of phylogenetic relationships of Bursaphelenchus species within xylophilus group. Nematology. 2002;4:35–41. [Google Scholar]
- Kanzaki N, Tanaka R. Sheraphelenchus sucus n. sp. (Tylenchina: Aphelenchoididae) isolated from sap flow of Quercus serrata in Japan. Nematology. 2013;15:975–990. [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA 7: Molecular evolutionary genetics analysis version 7 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giudice V, Lanza G. Presenza di Sheraphelenchus entomophagus su frutti di arancio. Informatore Fitopatologico. 1973;3:15–16. [Google Scholar]
- Massey CL. 1974 Biology and taxonomy of nematode parasites and associates of bark beetles in the United States. USDA Forest Service Agriculture Handbook No. 446, Washington, DC: U.S. Government Printing Office. [Google Scholar]
- Massey CL, Hinds TS. Nematodes from aspen cankers in Colorado and New Mexico. Canadian Journal of Zoology. 1970;48:97–108. [Google Scholar]
- Moosavi MR, Liramaji F, Pourjam E, Pedram M. Occurrence of Sheraphelenchus entomophagus Nickle, 1970 (Nematoda: Aphelenchoidinae) in Iran and its molecular phylogenetic analysis, with proposal for Japonema gen. n. International Journal of Nematology. 2014;34:81–86. [Google Scholar]
- Nickle WR. Description of Entaphelenchidae fam. n., Roveaphelenchus jonesi gen. n., sp. n., and Sheraphelenchus entomophagus gen. n., sp. n. (Nematoda: Aphelenchoidea) Proceedings of the Helminthological Society of Washington. 1970;37:105–109. [Google Scholar]
- Nunn GB. Ph.D. thesis. University of Nottingham, UK; 1992. Nematode molecular evolution. . [Google Scholar]
- Palou L, Taberner V, Guardado A, Del Río MÁ, Montesinos-Herero C. Incidence and etiology of postharvest fungal diseases of pomegranate (Punica Granatum cv. Mollar De Elche) in Spain. Phytopathologia Mediterranea. 2013;52:478–489. [Google Scholar]
- Siddiqi MR. Tylenchida. Parasites of plants and insects. London, UK: Commonwealth Agricultural Bureaux; 2000. [Google Scholar]
- Smith M, Shann C, Batenburg Van Der Vegetew C, Schmitt R, Wehrli E, Roeijmans HJ, Van Eijk GW. Botryozyma nematodophila new genus new species Candidaceae. Antonie van Leeuwenhoek. 1992;61:277–284. doi: 10.1007/BF00713936. [DOI] [PubMed] [Google Scholar]
- Stock SP, Nadler SA. Morphological and molecular characterisation of Panagrellus spp. (Cephalobina: Panagrolaimidae): Taxonomic status and phylogenetic relationships. Nematology. 2006;8:921–938. [Google Scholar]
- Troccoli A. Aspetti pratici e comparativi di alcune metodologie tradizionali applicate all’dentificazione dei nematodi da quarantena. Nematologia mediterranea. 2002;30:107–110. [Google Scholar]