Abstract
ACKR1, located on chromosome 1q23.2, is the gene that encodes a glycoprotein expressing the Duffy blood group antigens. This gene is transcribed in two mRNA variants yielding two isoforms, encoding proteins with 338 and 336 amino acids. This review provides a general overview of the Duffy blood group to characterise and elucidate the genetic basis of this system. The Fya and Fyb antigens are encoded by co-dominant FY*A (FY*01) and FY*B (FY*02) alleles, which differ by c.125G>A (rs12075), defining the Fy(a+b−), Fy(a−b+) and Fy(a+b+) phenotypes. The Fy(a−b−) phenotype that occurs in Africans provides an explanation for the apparent absence of Plasmodium vivax in this region: this phenotype arises from homozygosity for the FY*B allele carrying a point mutation c.1-67T>C (rs2814778), which prevents Fyb antigen expression only in red blood cells. The same mutation has also been found on the FY*A allele, but it is very rare. The Fy(a−b−) phenotype in Europeans and Asians arises from mutations in the coding region of the FY*A or FY*B allele, preventing Duffy antigen expression on any cell in the body and thus are true Duffy null phenotypes. According to the International Society for Blood Transfusion, ten alleles are associated with the null expression of the Fy antigens. Furthermore, different allelic forms of FY*B modify Fyb antigen expression, which may result in very weak or equivocal serology results. The mostly common found variants, c.265C>T (rs34599082) and c.298G>A (rs13962) -previously defined in combination only with the FY*B allele - have already been observed in the FY*A allele. Thus, six alleles have been recognised and associated with weak expression of the Fy antigens. Considering the importance of the Duffy blood group system in clinical medicine, additional studies via molecular biology approaches must be performed to resolve and clarify the discrepant results that are present in the erythrocyte phenotyping.
Keywords: Duffy blood group system, ACKR1 gene, DARC, allele variants
Introduction
The blood group systems are characterised by the presence or absence of antigens on the erythrocyte membrane, and these antigens are often polymorphic with respect to sequence and function1. Currently, according to the International Society for Blood Transfusion (ISBT), there are 346 erythrocyte antigens, dispersed over 36 different blood group systems2.
The erythrocyte antigens are genetically inherited and defined by specific sequences of amino acids, constituting proteins connected to lipids or carbohydrates3. These antigens are important to transfusion medicine because their absence from the red blood cells of an individual can result in alloimmunisation after a transfusion with the respective antigen1. Among the various consequences of alloimmunisation, the following stand out: an increased risk of transfusion reactions, reduction of the number of compatible blood bags, destruction of allogeneic erythrocytes, as well as of autologous and foetal erythrocytes, in addition to damage to transplanted tissues4.
In order to minimise the chances of an individual generating erythrocyte alloantibodies, transfusions must be phenotypically compatible to the most immunogenic antigens5. Although phenotyping is essential for the confirmation of the presence of alloantibodies and also for the detection of blood group antigens6–8, phenotyping suffers from certain technical limitations because it is a subjective test, many antibodies are not commercially available and it is a labour-intensive test, so a relatively small number of donors can be typed for a relatively small number of antigens. There are also certain clinical limitations, including the difficulty of phenotyping recently transfused patients as well as red blood cells coated with IgG, and it can be challenging to distinguish an alloantibody from an autoantibody in antigen-positive people9. In these situations, blood group genotyping has proven to be an excellent alternative to phenotypying10,11.
The main indications for performing such a molecular test in immunohaematology are the identification of erythrocyte antigens in recently transfused patients, in patients with a positive direct antiglobulin test (DAT+) and in situations in which there is a risk of developing haemolytic disease of the foetus and newborn (HDFN)7,12,13. Molecular techniques may also be used to identify the presence of variation in genes that encode blood group antigens that are expressed weakly in the membrane, thereby contributing to the prevention of possible haemolytic transfusion reactions6.
DNA-based genotyping of the Duffy blood group system can be an important adjunct to traditional phenotyping, especially in clinical situations in which the risk of HDFN is a concern and for locating matched blood for alloimmunised patients. Accordingly, this review provides a general overview of the Duffy blood group to characterise and clarify the genetic basis of the allelic variants of this system.
Duffy blood group system
The Duffy blood group was initially reported by Cutbush in 1950, who described the reactivity of an antibody found in a male, multitransfused, haemophilic patient who had an alloantibody against an antigen, then denoted as Fya. This antibody was named anti-Fya, in honour of the patient in question14. A year later, an antibody was described in the serum of a multiparous woman which defined its antithetical pair, called anti-Fyb 15. The Duffy antigens reside in an acidic glycoprotein that spans the membrane seven times. The N-terminal portion forms the extracellular domain and the C-terminal portion forms the intracellular domain.
Biological functions
The Duffy glycoprotein, also known as the Duffy antigen receptor for chemokines (DARC), is a promiscuous receptor that binds chemokines of the C-X-C and C-C classes16–18. Examples of C-X-C chemokines are interleukin-8 (IL-8) and melanoma growth stimulatory activity (MGSA), while the C-C chemokines include regulated on activation, normal T expressed and secreted (RANTES) and monocyte chemotactic protein-1 (MCP-1)16–18. The main normal function described for DARC is that it effectively sustains homeostatic levels of circulating chemokines and modulates chemokine gradients between tissues and the blood to mediate the influx of neutrophils and monocytes from blood vessels into tissues during immune responses19,20. Although the specific mechanisms underlying its functions remain uncertain, there is interest in DARC as an explanatory variable for population-specific differences in disease susceptibility21, as demonstrated by ongoing research into its role in inflammation-associated pathology and malignancy21,22, and by the recent, though highly controversial23, surge in interest around the antigen’s role in human immunodeficiency virus infection24.
Much of the research into this blood group has been concerned with elucidating the characteristic expression patterns among different populations25. Interest in the Duffy blood group rose substantially with the recognition of its role as a portal of entry for malarial parasites into human red blood cells26. While Plasmodium falciparum uses a series of receptors on the surface of human erythrocytes to invade them, Plasmodium vivax and Plasmodium knowlesi depend on an interaction with the Duffy antigen, meaning that red blood cells lacking the antigen are refractory to invasion by these merozoites26–29. The proportion of individuals in African populations who do not express the DARC protein in their erythrocytes is high. The gap in distribution of P. vivax in Africa is, therefore, viewed as the consequence of the lack of this protein on red blood cells - suggesting either an adaptive response to the disease or a selective pressure acting on the parasite30,31. Furthermore, a genotype-dosage effect on expression of the DARC protein has been described and the level of DARC expression is associated with susceptibility and resistance to infection32–35. These results imply that the red cells of heterozygotes for the silent allele bind substantially less P. vivax Duffy-binding protein than those of individuals with two active FY alleles, indicating that Duffy-negative heterozygosity confers significant protection and may have a selective advantage in areas where P. vivax is endemic36. However, since 2006 there have been reports of Duffy-negative individuals infected with P. vivax, both throughout Africa (Kenya, Madagascar, Mauritania, Cameroon, Angola, Equatorial Guinea, Ethiopia, and Sudan) and in Brazilian Amazon37–46. The mechanisms involved in this invasion remain to be elucidated: the hypotheses postulated include expansions of the copy number of gene Duffy-binding protein 1 of the P. vivax47 and the use of alternative ligand-receptor pairs30,47.
Genetic basis
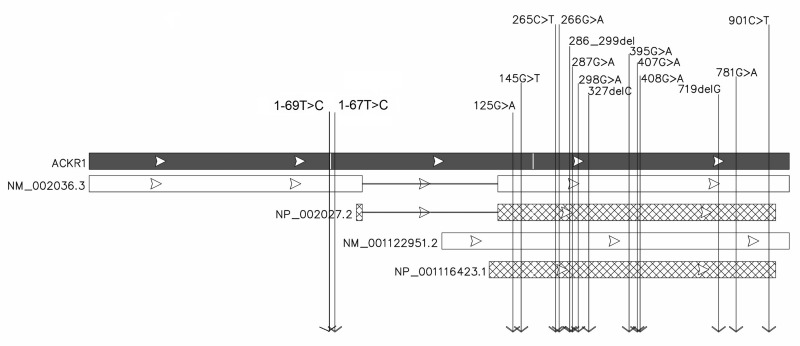
ACKR1, also known as DARC or FY (NCBI), is the gene that encodes a transmembranous glycoprotein expressing the Duffy blood group antigens. Its genetic locus was reported to be on chromosome 1, located formerly in the region 1q21–25 by linkage analysis48. Later, the position of the gene was refined to 1q23.2. The gene (sequence NC_000001.11, region: 159204013–159206500) is transcribed as two mRNA variants. Chaudhuri et al. reported that the first mRNA variant has one exon49. Subsequently, Iwamoto et al. demonstrated the existence of a spliced mRNA variant that has two exons with the intron encompassing sequences in the initial part of the first mRNA variant50. Despite encoding for a shorter protein, the second mRNA variant has a longer transcript than the first because of a longer 5′ untranslated region. These two distinct transcript isoforms are expressed from separate promoters, yielding distinct protein products. The major transcript is derived from exon 1 and exon 2 of ACKR1; the minor product is a transcript initiated at the beginning of exon 2. The minor and major transcripts generate, respectively, isoform A (NM_001122951.2/ NP_001116423.1), encoding a protein of 338 amino acids and isoform B (NM_002036.3/NP_002027.2), which encodes a protein of 336 amino acids51 (Figure 1).
Figure 1.
ACKR1 gene structure and proteins and mRNA isoforms.
Viewing the figure from top to bottom: the black box represents the ACKR1 gene, the white box shows the mRNA of isoform B (NM_002036.3) and the dashed box shows the isoform B protein (NP_002027.2); the next white box represents the mRNA of isoform A (NM_001122951.2) and the dashed box shows the protein of isoform A (NP_001116423.1). The arrows indicate the positions of the main genetic variants already described
The nucleotide and amino acid sequences of ACKR1 were renumbered after the discovery that the spliced mRNA is the major product of the gene. It was proposed that the first nucleotide of the translation initiation codon of the major spliced mRNA be numbered nucleotide 1. This numbering convention avoids inconsistencies created by differing lengths of the 5-prime untranslated region arising from alternative transcription initiation sites52. This isoform B has been chosen as the “canonical” sequence that is known to be relevant for blood group genotyping because is expressed in erythroid lineage cells. All positional information in this review refers to isoform B.
The antithetical antigens, Fya and Fyb, are encoded by co-dominant FY*A (FY*01) and FY*B (FY*02) alleles, which differ by a single nucleotide polymorphism c.125G>A (rs12075)49,50. On the FY*A allele, the base is guanine (G), and on the FY*B allele the base is adenine (A). This missense mutation produces a codon for glycine in the FY*A allele and a codon for aspartic acid in the FY*B allele at position 42 of the major product (p.Gly42Asp)53–55, defining the Fy(a+b−), Fy(a−b+) and Fy(a+b+) phenotypes. Additionally, a number of variants have been identified that cause weak (+w or *W) and null (0 or *N) expression of Duffy Fya or Fyb antigens (Table I). According to the ISBT, there are two mutations associated with weak expression of Fya and five mutations associated with weak expression of Fyb. Seven mutations that cause the null expression of Fya have already been observed and three such mutations for Fyb have been found. Despite the high genetic variability related to Duffy antigen production, some of these variants are more commonly associated with null or weak expression of these antigens than others, and they are described below.
Table I.
Variants of the Duffy blood group system.
| Fy expression | Allele name | Nucleotide | Region | Amino acid | dbSNP |
|---|---|---|---|---|---|
| Fy(a+) | FY*01 | c.125G | Exon 2 | p.Gly42 | rs12075 |
| Fy(b+) | FY*02 | c.125G>A | Exon 2 | p.Gly42Asp | rs12075 |
| Null alleles | |||||
| Fy(a) null | |||||
| Fy(a−) erythroid cells only | FY*01N.01 | c.1-67T>C | 5′UTR | p.0 | rs2814778 |
| Fy(a−) | FY*01N.02 | c.286_299del | Exon 2 | p.Trp96Thrfs | rs587776507 |
| Fy(a−) | FY*01N.03 | c.408G>A | Exon 2 | p.Trp136Ter | – |
| Fy(a−) | FY*01N.04 | c.287G>A | Exon 2 | p.Trp96Ter | rs750052723 |
| Fy(a−) | FY*01N.05 | c.327delC | Exon 2 | p.Phe109fs | – |
| Fy(a−) | FY*01N.06 | c.395G>A | Exon 2 | p.Gly132Asp | rs530992295 |
| Fy(a−) | FY*01N.07 | c.719delG | Exon 2 | p.Gly240fs | – |
| Fy(a−) | – | c.1-69T>C | 5′UTR | p.0 | – |
| Fy(b) null | |||||
| Fy(b−) erythroid cells only, Fyes | FY*02N.01 | c.1-67T>C | 5′UTR | p.0 | rs2814778 |
| Fy(b−) | FY*02N.02 | c.407G>A | Exon 2 | p.Trp136Ter | rs76819093 |
| Fy(b−) | FY*02N.03 | c.781G>A | Exon 2 | p.Gly261Arg | – |
| Weak alleles | |||||
| Fy(a) weak | |||||
| Fy(a+w) | FY*01W.01 | c.265C>T | Exon 2 | p.Arg89Cys | rs34599082 |
| Fy(a+w) | FY*01W.02 | c.265C>T, 298G>A | Exon 2 | p.Arg89Cys, Ala100Thr | rs34599082, rs13962 |
| Fy(b) weak | |||||
| Fy(b+w), Fyx | FY*02W.01 | c.265C>T, 298G>A | Exon 2 | p.Arg89Cys, Ala100Thr | rs34599082, rs13962 |
| Fy(b+w), Fyx | FY*02W.02 | c.145G>T, c.265C>T, 298G>A | Exon 2 | p.Ala49Ser, Arg89Cys, Ala100Thr | – rs34599082, rs13962 |
| Fy(b+w) | FY*02W.03 | c.266G>A | Exon 2 | p.Arg89His | rs371909350 |
| Fy(b+w) | FY*02W.04 | c.901C>T | Exon 2 | p.Pro301Ser | rs753831902 |
The nucleotide position is based on NCBI data. fs: frameshift; UTR: untranslated region.
The Fy(a−b−) phenotype, also known as “erythrocyte silent” (Fyes), occurs in African lineages and, depending on the region, has a prevalence of nearly 100% (e.g. in West Africa) and is also found at greater than 80% frequency in African Americans26,56. This phenotype arises from homozygosity for an FY*B allele carrying a point mutation c.1-67T>C (rs2814778) in the 5′ untranslated region57. This mutation gives rise to the FY*BES (FY*02N.01) allele, which impairs promoter activity in erythroid cells by disrupting a binding site for the GATA-1 erythroid transcription factor57. This mutation prevents Fyb antigen expression only in red blood cells but not in other tissues53,58. As a result, Africans with the Fy(a−b−) phenotype rarely make anti-Fyb50,59,60. The same mutation (previously described as c.-33T>C and c.-46T>C) has been found on the FY*AES allele (FY*01N.01), but only in a heterozygous state in inhabitants of Papua New Guinea and Sudan; it is very rare35,61. Písacka et al. reported a novel mutation at position c.1-69 in the FY promoter that also disrupts the GATA motif and correlates with silencing of the FY*A allele, causing a Fy null phenotype62.
The few documented cases of the Fy(a−b−) phenotype in Europeans and Asians arise from mutations in the coding region of the FY*A or FY*B allele55,63–65. These mutations, when present in the homozygous state, prevent Duffy antigen expression on any cell in the body and thus are true Duffy null phenotypes. Consequently, these individuals are at risk of being alloimmunised when exposed to red blood cells expressing Fy antigens66.
Different allelic forms of the Duffy blood group gene modify the antigen’s expression level, which may cause problems in blood group phenotyping, leading, specifically, to very weak or equivocal serology typing results66–69. The most common variants, c.265C>T (rs34599082) and c.298G>A (rs13962), cause a substitution of arginine to cysteine at position 89 (p.Arg89Cys) and an alanine to threonine substitution at amino acid 100 (p.Ala100Thr) of glycoprotein Duffy, respectively. The aforementioned two variants usually result in the weak expression of Fyb (FY*02W.01 allele, also referred to as Fyx antigen). This allele has been described mainly among Europeans, with a frequency varying from 2 to 3,5%; but it has not been found in Africans69–73. The c.298G>A variant alone does not result in reduced Fyb expression70. Another mutation linked to weak expression of the antigen Fyb, c.145G>T, changes the amino acid alanine to serine at position 49 (p.Ala49Ser), generating the FY*02W.02 allele69.
Although less common, weak serological reactivity of the Fya antigen has already been observed and Lopez et al. recently investigated this phenotype. They identified two variants of the FY*A allele, 265T and 298A, which were consistently present in the donor. Prior to this study, these variants had only been defined in combination with the FY*B allele. Reflecting these facts, FY*01W.02 was the provisional name given to the new allele by the ISBT Working Party on Red Cell Immunogenetics and Blood Group Terminology74. One potential explanation for such complexity among Fy antigens could be the co-expression of alternative ACKR1 gene product isoforms and distinct post-translational modifications between the isoforms acting as immunogens51. The phenotypic and genotypic correlations of the main polymorphisms are presented in Table II.
Table II.
Phenotype and genotype correlations of the main polymorphisms.
| Phenotype | c.125G>A1 | c.265C>T2 | c.298G>A3 | c.1-67T>C4 | Predicted genotype | Predicted antigen |
|---|---|---|---|---|---|---|
| Fy(a+b−) | G/G | C/C | G/G | T/T | FY*A/FY*A or FY*01/FY*01 | Fya |
| G/A | C/C | G/G | T/C | FY*01/FY*02N.01 | Fya/Fyes | |
|
| ||||||
| Fy(a+b+) | G/A | C/C | G/G | T/T | FY*A/FY*B or FY*01/FY*02 | Fya/Fyb |
| G/A | C/T | G/A | T/T | FY*01/FY*02W.01 | Fya/Fyx | |
|
| ||||||
| Fy(a−b+) | A/A | C/C | G/G | T/T | FY*B/FY*B or FY*02/FY*02 | Fyb |
| A/A | C/T | G/A | T/T | FY*02/FY*02W.01 | Fyb/Fyx | |
| A/A | T/T | A/A | T/T | FY*02W.01/FY*02W.01 | Fyx | |
| A/A | C/T | G/A | T/C | FY*02W.01/FY*02N.01 | Fyx/Fyes | |
| A/A | C/C | G/G | T/C | FY*02/FY*02N.01 | Fyb/Fyes | |
|
| ||||||
| Fy(a−b−) | A/A | C/C | G/G | C/C | FY*02N.01/FY*02N.01 | Fyes/Fyes |
rs12075 predicts the expression of the Fya and Fyb antigens;
rs34599082 determines weak expression of the Fyb antigen;
rs13962 determines weak expression of the Fyb antigen;
rs2814778 prevents expression of Fyb antigen in red blood cells.
Geographic distribution
It is characteristic of this blood group system that there is a great diversity of distributions of the Duffy antigenic determinants among different ethnic groups. Some of these variants were described in the 1000 Genomes Project database. According to these data, the rs12075*A single nucleotide polymorphism, which determines the FY*B ancestral allele (0.541), is more prevalent globally than the rs12075*G variant, which determines the FY*A allele (0.459). The FY*B allele is common in Africans (0.981), but not in East Asians (0.077). On the other hand, the FY*A allele is dominant in East Asians (0.923), but is infrequent in Africans (0.019). Finally, the allele frequencies of the variants that determine the Fyx and Fyes antigens are highest in Europeans and Africans, respectively (Table III).
Table III.
Allele frequencies according to the 1000 Genomes Project.
| dbSNP allele | AFR1 | AMR2 | EAS3 | SAS4 | EUR5 |
|---|---|---|---|---|---|
| rs12075*G Fy(a+) |
0.019 | 0.461 | 0.923 | 0.640 | 0.398 |
| rs12075*A Fy(b+) |
0.981 | 0.539 | 0.077 | 0.360 | 0.602 |
| rs34599082*T Fy(b+w) or Fy(a+w) |
0.000 | 0.007 | 0.001 | 0.004 | 0.013 |
| rs13962*A Fy(b+w) or Fy(a+w) |
0.005 | 0.094 | 0.000 | 0.091 | 0.184 |
| rs530992295*A Fy(anull) |
0.000 | 0.000 | 0.000 | 0.002 | 0.000 |
| rs2814778*C Fy(bnull) or Fy(anull) |
0.964 | 0.078 | 0.000 | 0.000 | 0.006 |
AFR: African (from Yoruba in Ibadan, Nigeria; Luhya in Webuye, Kenya; Gambian in Western Divisions in the Gambia; Mende in Sierra Leone; Esan in Nigeria; Americans of African ancestry in the SW USA; African Caribbeans in Barbados);
AMR: Admixed Americans (Mexican ancestry from Los Angeles, USA; Puerto Ricans from Puerto Rico; Colombians from Medellin, Colombia; Peruvians from Lima, Peru;
EAS: East Asian (Han Chinese in Beijing, China; Japanese in Tokyo, Japan; Southern Han Chinese; Chinese Dai in Xishuangbanna, China; Kinh in Ho Chi Minh City, Vietnam);
SAS: South Asian (Gujarati Indians from Houston, Texas, USA; Punjabi from Lahore, Pakistan; Bengali from Bangladesh; Sri Lankan Tamils from the UK; Indian Telugu from the UK);
EUR: European (Utah residents with northern and western European ancestry from the Centre d’Etude du Polymorphisme Humain; Tuscany in Italy; Finnish in Finland; British in England and Scotland; Iberian population in Spain).
The Duffy system is considered one of the most attractive chromosomal loci for evaluating the impact of natural selection in different geographical regions75,76. Because the mutation that confers protection from infection by P. vivax prevents the expression of the DARC protein only in erythrocytes, it is possible to observe differences in phenotypic and genotypic frequencies of the Fyb antigen in Africans. Although the most common genotype is FY*B/FY*B, almost all of the samples type serologically as Fy(a−b−)77. Table IV presents possible phenotypes among different populations.
Table IV.
Typical Duffy phenotype frequencies.
| Phenotype | Frequencies (%) | ||
|---|---|---|---|
| Europeans | Africans | Asians | |
| Fy(a+b−) | 20 | 10 | 89,2 |
| Fy(a−b+) | 32 | 20 | 1,8 |
| Fy(a+b+) | 48 | 3 | 9,0 |
| Fy(a−b−) | Rare | 67 | 0 |
Clinical significance
The anti-Fya antibody is found mainly following transfusion and, less frequently, as a result of pregnancy; it is almost never naturally occuring80,81.The anti-Fyb is about 20 times less common than the anti-Fya and is generally present in sera in combination with other antibodies82.These antibodies are predominantly of the IgG1 type subclass, with lesser contributions from other subclasses, for example, IgG2 (18%) and IgM (25%)83,84. Both antibodies cause immediate and delayed haemolytic transfusion reactions1,85.
The Fya and Fyb antigens are expressed in erythroid and non-erythroid cells, such as endothelial cells, and also in epithelial cells in various organs, including the brain, kidneys, spleen, heart, lungs, pancreas and placenta86 -lending this system an important role in the inflammatory response, in allograft rejection and, possibly, in histocompatibility. The Duffy blood group is thus a polymorphic system that poses a major challenge for researchers at phenotypic, genotypic and tissue levels87.
Conclusions
In view of the importance of the Duffy blood group system in clinical medicine, further studies utilising molecular biology approaches must be developed for the purpose of elucidating and characterising new sequence variants. Such molecular typing can help resolve and clarify the equivocal and discrepant results that arise in erythrocyte phenotyping performed by haemagglutination assays. These techniques, used together, contribute to the optimal use of blood units and, therefore, to the quality of transfusion practice.
Footnotes
The Authors declare no conflicts of interest.
References
- 1.Issitt PD, Anstee DJ. Applied Blood Group Serology. 4th edition. Durham, NC: Montgomery Scientific Publications; 1998. [Google Scholar]
- 2.Storry JR, Castilho L, Chen Q, et al. International Society of Blood Transfusion Working Party on red cell immunogenetics and terminology: report of the Seoul and London meetings. ISBT Science Series. 2016;11:118–22. doi: 10.1111/voxs.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid ME, Yazdanbakhsh K. Molecular insights into blood groups and implications for blood transfusion. Curr Opin Hematol. 1998;5:93–102. doi: 10.1097/00062752-199803000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Poole J, Daniels G. Blood group antibodies and their significance in transfusion medicine. Transfus Med Rev. 2007;21:58–71. doi: 10.1016/j.tmrv.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Coles SM, Klein HG, Holland PV. Alloimmunization in two multitransfused patient populations. Transfusion. 1981;21:462–6. doi: 10.1046/j.1537-2995.1981.21481276005.x. [DOI] [PubMed] [Google Scholar]
- 6.Reid ME. Overview of molecular methods in immunohematology. Transfusion. 2007;47:10S–6S. doi: 10.1111/j.1537-2995.2007.01304.x. [DOI] [PubMed] [Google Scholar]
- 7.Reid ME, Lomas-Francis C. Molecular approaches to blood group identification. Curr Opin Hematol. 2002;9:152–9. doi: 10.1097/00062752-200203000-00012. [DOI] [PubMed] [Google Scholar]
- 8.Strauss D, Reid ME. Value of DNA-based assays for donor screening and regulatory issues. Immunohematology. 2008;24:175–9. [PubMed] [Google Scholar]
- 9.Reid ME. Transfusion in the age of molecular diagnostics. Hematology Am Soc Hematol Educ Program. 2009:171–7. doi: 10.1182/asheducation-2009.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castilho L, Pellegrino J., Júnior Blood group genotyping. ev Bras Hematol Hemoter. 2004;26:135–40. [Google Scholar]
- 11.Castilho L, Rios M, Bianco C, et al. DNA-based typing of blood groups for the management of multiply-transfused sickle cell disease patients. Transfusion. 2002;42:232–8. doi: 10.1046/j.1537-2995.2002.00029.x. [DOI] [PubMed] [Google Scholar]
- 12.Flegel WA, Wagner FF, Muller TH, Gassner C. Rh phenotype prediction by DNA typing and its application to practice. Transfus Med. 1998;8:281–302. doi: 10.1046/j.1365-3148.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 13.Van der Schoot CE, Tax GH, Rijnders RJ, et al. Prenatal typing of Rh and Kell blood group system antigens: the edge of a watershed. Transfus Med Rev. 2003;17:31–44. doi: 10.1053/tmrv.2003.50001. [DOI] [PubMed] [Google Scholar]
- 14.Cutbush M, Mollison PL. The Duffy blood group system. Heredity (Edinb) 1950;4:383–9. doi: 10.1038/hdy.1950.31. [DOI] [PubMed] [Google Scholar]
- 15.Ikin EW, Mourant AE, Pettenkofer HJ, Blumenthal G. Discovery of the expected haemagglutinin, anti-Fyb. Nature. 1951;168:1077–8. doi: 10.1038/1681077b0. [DOI] [PubMed] [Google Scholar]
- 16.Darbonne WC, Rice GC, Mohler MA, et al. Red blood cells are a sink for interleukin 8, a leukocyte chemotaxin. J Clin Invest. 1991;88:1362–9. doi: 10.1172/JCI115442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neote K, Darbonne W, Ogez J, et al. Identification of a promiscuous inflammatory peptide receptor on the surface of red blood cells. J Biol Chem. 1993;268:12247–9. [PubMed] [Google Scholar]
- 18.Horuk R, Colby TJ, Darbonne WC, et al. The human erythrocyte inflammatory peptide (chemokine) receptor, biochemical characterization, solubilization, and development of a binding assay for the soluble receptor. Biochemistry. 1993;32:5733–8. doi: 10.1021/bi00073a002. [DOI] [PubMed] [Google Scholar]
- 19.Mayr FB, Spiel AO, Leitner JM, et al. Duffy antigen modifies the chemokine response in human endotoxemia. Crit Care Med. 2008;36:159–65. doi: 10.1097/01.CCM.0000297875.55969.DB. [DOI] [PubMed] [Google Scholar]
- 20.Reich D, Nalls MA, Kao WH, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anstee DJ. The relationship between blood groups and disease. Blood. 2010;115:4635–43. doi: 10.1182/blood-2010-01-261859. [DOI] [PubMed] [Google Scholar]
- 22.Horne K, Woolley IJ. Shedding light on DARC: the role of the Duffy antigen/receptor for chemokines in inflammation, infection and malignancy. Inflamm Res. 2009;58:431–5. doi: 10.1007/s00011-009-0023-9. [DOI] [PubMed] [Google Scholar]
- 23.Walley NM, Julg B, Dickson SP, et al. The Duffy antigen receptor for chemokines null promoter variant does not influence HIV-1 acquisition or disease progression. Cell Host Microbe. 2009;5:408–10. doi: 10.1016/j.chom.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He W, Neil S, Kulkarni H, et al. Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe. 2008;4:52–62. doi: 10.1016/j.chom.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howes RE, Patil AP, Piel FB, et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 27.Barnwell JW, Nichols ME, Rubinstein P. In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax. J Exp Med. 1989;169:1795–802. doi: 10.1084/jem.169.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wertheimer SP, Barnwell JW. Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol. 1989;69:340–50. doi: 10.1016/0014-4894(89)90083-0. [DOI] [PubMed] [Google Scholar]
- 29.Miller LH, Mason SJ, Dvorak JA, et al. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–3. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 30.Mercereau-Puijalon O, Menard D. Plasmodium vivax and the Duffy antigen: a paradigm revisited. Transfus Clin Biol. 2010;17:176–83. doi: 10.1016/j.tracli.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Hedrick PW. Population genetics of malaria resistance in humans. Heredity (Edinb) 2011;107:283–304. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasehagen LJ, Mueller I, Kiniboro B, et al. Reduced Plasmodium vivax erythrocyte infection in PNG Duffy-negative heterozygotes. PLoS One. 2007;2:e336. doi: 10.1371/journal.pone.0000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavasini CE, de Mattos LC, Couto AA, et al. Duffy blood group gene polymorphisms among malaria vivax patients in four areas of the Brazilian Amazon region. Malar J. 2007;6:167. doi: 10.1186/1475-2875-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasehagen LJ, Mueller I, McNamara DT, et al. Changing patterns of Plasmodium blood-stage infections in the wosera region of papua new Guinea monitored by light microscopy and high throughput PCR diagnosis. Am J Trop Med Hyg. 2006;75:588–96. [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmerman PA, Woolley I, Masinde GL, et al. Emergence of FY*A(null) in a Plasmodium vivax-endemic region of Papua New Guinea. Proc Natl Acad Sci USA. 1999;96:13973–7. doi: 10.1073/pnas.96.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michon P, Woolley I, Wood EM, et al. Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P. vivax ligand required for Blood stage infection. FEBS Lett. 2001;495:111–4. doi: 10.1016/s0014-5793(01)02370-5. [DOI] [PubMed] [Google Scholar]
- 37.Ryan JR, Stoute JA, Amon J, et al. Evidence for transmission of Plasmodium vivax among a Duffy antigen negative population in Western Kenya. Am J Hum Genet. 2006;75:575–81. [PubMed] [Google Scholar]
- 38.Cavasini CE, Mattos LC, Couto AA, et al. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans R Soc Trop Med Hyg. 2007;101:1042–44. doi: 10.1016/j.trstmh.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Ménard D, Barnadas C, Bouchier C, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA. 2010;107:5967–71. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woldearegai TG, Kremsner PG, Kun JF, Mordmüller B. Plasmodium vivax malaria in Duffy-negative individuals from Ethiopia. Trans R Soc Trop Med Hyg. 2013;107:328–31. doi: 10.1093/trstmh/trt016. [DOI] [PubMed] [Google Scholar]
- 41.Mendes C, Dias F, Figueiredo J, et al. Duffy negative antigen is no longer a barrier to Plasmodium vivax: Molecular evidences from the African West Coast (Angola and Equatorial Guinea) PLoS Negl Trop Dis. 2011;5:e1192. doi: 10.1371/journal.pntd.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wurtz N, Mint Lekweiry K, Bogreau H, et al. Vivax malaria in Mauritania includes infection of a Duffy-negative individual. Malar J. 2011;10:336. doi: 10.1186/1475-2875-10-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho TA, Queiroz MG, Cardoso GL, et al. Plasmodium vivax infection in Anajas, State of Para: No differential resistance profile among Duffy-negative and Duffy-positive individuals. Malar J. 2012;11:430. doi: 10.1186/1475-2875-11-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fru-Cho J, Bumah VV, Safeukui I, et al. Molecular typing reveals substantial Plasmodium vivax infection in asymptomatic adults in a rural area of Cameroon. Malar J. 2014;13:170. doi: 10.1186/1475-2875-13-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ngassa Mbenda HG, Das A. Molecular evidence of Plasmodium vivax mono and mixed malaria parasite infections in Duffy-negative native Cameroonians. PLoS One. 2014;9:e103262. doi: 10.1371/journal.pone.0103262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdelraheem MH, Albsheer MM, Mohamed HS, et al. Transmission of Plasmodium vivax in Duffy-negative individuals in central Sudan. Trans R Soc Trop Med Hyg. 2016;110:258–60. doi: 10.1093/trstmh/trw014. [DOI] [PubMed] [Google Scholar]
- 47.Gunalan K, Lo E, Hostetler JB. Role of Plasmodium vivax Duffy-binding protein 1 in invasion of Duffy-null Africans. Proc Natl Acad Sci USA. 2016;113:6271–6. doi: 10.1073/pnas.1606113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dracopoli NC, O’Connell P, Elsner TI, et al. The CEPH consortium linkage map of human chromosome 1. Genomics. 1991;9:686–700. doi: 10.1016/0888-7543(91)90362-i. [DOI] [PubMed] [Google Scholar]
- 49.Chaudhuri A, Polyakova J, Zbrzezna V, et al. Cloning of glycoprotein D cDNA, which encodes the major subunit of the Duffy blood group system and the receptor for the Plasmodium vivax malaria parasite. Proc Natl Acad Sci USA. 1993;90:10793–7. doi: 10.1073/pnas.90.22.10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwamoto S, Li J, Omi T, et al. Identification of a novel exon and spliced form of Duffy mRNA that is the predominant transcript in both erythroid and postcapillary venule endothelium. Blood. 1996;87:378–85. [PubMed] [Google Scholar]
- 51.Davis MB, Walens A, Hire R, et al. Distinct transcript isoforms of the atypical chemokine receptor 1 (ACKR1)/Duffy antigen receptor for chemokines (DARC) gene are expressed in lymphoblasts and altered isoform levels are associated with genetic ancestry and the Duffy-null allele. PLoS One. 2015;10:e0140098. doi: 10.1371/journal.pone.0140098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pogo AO, Chaudhuri A. The Duffy protein: a malarial and chemokine receptor. Semin Hematol. 2000;37:122–9. doi: 10.1016/s0037-1963(00)90037-4. [DOI] [PubMed] [Google Scholar]
- 53.Chaudhuri A, Polyakova J, Zbrzezna V, Pogo AO. The coding sequence of Duffy blood group gene in humans and simians: restriction fragment length polymorphism, antibody and malarial parasite specificities, and expression in nonerythroid tissues in Duffy-negative individuals. Blood. 1995;85:615–21. [PubMed] [Google Scholar]
- 54.Iwamoto S, Omi T, Kajii E, Ikemoto S. Genomic organization of the glycoprotein D gene: Duffy blood group Fya/Fyb alloantigen system is associated with a polymorphism at the 44-amino acid residue. Blood. 1995;85:622–6. [PubMed] [Google Scholar]
- 55.Mallinson G, Soo KS, Schall TJ, et al. Mutations in the erythrocyte chemokine receptor (Duffy) gene: the molecular basis of the Fya/Fyb antigens and identification of a deletion in the Duffy gene of an apparently healthy individual with the Fy(a−b−) phenotype. Br J Haematol. 1995;90:823–9. doi: 10.1111/j.1365-2141.1995.tb05202.x. [DOI] [PubMed] [Google Scholar]
- 56.Schmid P, Ravenell KR, Sheldon SL, Flegel WA. DARC alleles and Duffy phenotypes in African Americans. Transfusion. 2012;52:1260–7. doi: 10.1111/j.1537-2995.2011.03431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat genet. 1995;10:224–8. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 58.Peiper SC, Wang ZX, Neote K, et al. The Duffy antigen/ receptor for chemokines (DARC) is expressed in endothelial cells of Duffy negative individuals who lack the erythrocyte receptor. J Exp Med. 1995;181:1311–7. doi: 10.1084/jem.181.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oberdorfer CE, Kahn B, Moore V, et al. A second example of anti-Fy3 in the Duffy blood group system. Transfusion. 1974;14:608–11. doi: 10.1111/j.1537-2995.1974.tb04588.x. [DOI] [PubMed] [Google Scholar]
- 60.Kosinski KS, Molthan L, White L. Three examples of anti-Fy3 produced in Negroes. Rev Fr Transfus Hemobiol. 1984;27:619–24. doi: 10.1016/s0338-4535(84)80083-5. [DOI] [PubMed] [Google Scholar]
- 61.Kempinska-Podhorodecka A, Knap O, Drozd A, et al. Analysis for genotyping Duffy blood group in inhabitants of Sudan, the fourth cataract of the Nile. Malar J. 2012;11:115. doi: 10.1186/1475-2875-11-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Písacka M, Marinov I, Králová M, et al. FY*A silencing by the GATA-motif variant FY*A(−69C) in a Caucasian family. Transfusion. 2015;55:2616–9. doi: 10.1111/trf.13221. [DOI] [PubMed] [Google Scholar]
- 63.Albrey JA, Vincent EE, Hutchinson J, et al. A new antibody, anti-Fy3, in the Duffy blood group system. Vox Sang. 1971;20:29–35. doi: 10.1111/j.1423-0410.1971.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 64.Rios M, Chaudhuri A, Mallinson G, et al. New genotypes in Fy(a−b−) individuals: nonsense mutations (Trp to stop) in the coding sequence of either FY A or FY B. Br J Haematol. 2000;108:448–54. doi: 10.1046/j.1365-2141.2000.01882.x. [DOI] [PubMed] [Google Scholar]
- 65.Vege S, Hue-Roye K, Velliquette RW, et al. A new Duffy allele, FY*A 395G>A (p.Gly132Asp), associated with silencing Fya expression. [SP278 Abstract] Transfusion. 2013;53(Suppl 2):164A. [Google Scholar]
- 66.Lopez GH, Morrison J, Condon JA, et al. Duffy blood group phenotype-genotype correlations using high-resolution melting analysis PCR and microarray reveal complex cases including a new null FY*A allele: the role for sequencing in genotyping algorithms. Vox Sang. 2015;109:296–303. doi: 10.1111/vox.12273. [DOI] [PubMed] [Google Scholar]
- 67.Chown B, Lewis M, Kaita H. The Duffy blood group system in Caucasians: evidence for a new allele. Am J Hum Genet. 1965;17:384–9. [PMC free article] [PubMed] [Google Scholar]
- 68.Murphy MT, Templeton LJ, Fleming J, et al. Comparison of Fy(b) status as determined serologically and genetically. Transfus Med. 1997;7:135–41. doi: 10.1046/j.1365-3148.1997.d01-16.x. [DOI] [PubMed] [Google Scholar]
- 69.Castilho L, Rios M, Pellegrino J, et al. A novel FY allele in Brazilians. Vox Sang. 2004;87:190–5. doi: 10.1111/j.1423-0410.2004.00554.x. [DOI] [PubMed] [Google Scholar]
- 70.Olsson ML, Smythe JS, Hansson C, et al. The Fy(x) phenotype is associated with a missense mutation in the Fy(b) allele predicting Arg89Cys in the Duffy glycoprotein. Br J Haematol. 1998;103:1184–91. doi: 10.1046/j.1365-2141.1998.01083.x. [DOI] [PubMed] [Google Scholar]
- 71.Tournamille C, Le Van Kim C, Gane P, et al. Arg89Cys substitution results in very low membrane expression of the Duffy antigen/receptor for chemokines in Fy(x) individuals. Blood. 1998;92:2147–56. [PubMed] [Google Scholar]
- 72.Yazdanbakhsh K, Rios M, Storry JR, et al. Molecular mechanisms that lead to reduced expression of Duffy antigens. Transfusion. 2000;40:310–20. doi: 10.1046/j.1537-2995.2000.40030310.x. [DOI] [PubMed] [Google Scholar]
- 73.Parasol N, Reid M, Rios M, et al. A novel mutation in the coding sequence of the FY*B allele of the Duffy chemokine receptor gene is associated with an altered erythrocyte phenotype. Blood. 1998;92:2237–43. [PubMed] [Google Scholar]
- 74.Lopez G, Condon J, Wilson B, et al. A novel FY* A allele with the 265T and 298A SNPs formerly associated exclusively with the FY* B allele and weak Fyb antigen expression: implication for genotyping interpretative algorithms. Vox Sang. 2015;108:52–7. doi: 10.1111/vox.12185. [DOI] [PubMed] [Google Scholar]
- 75.Hamblin MT, Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet. 2002;70:284. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamblin MT, Thompson EE, Di Rienzo A. Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet. 2002;70:369–83. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moulds JM, Hayes S, Wells TD. DNA analysis of Duffy genes in American blacks. Vox Sang. 1998;74:248–52. [PubMed] [Google Scholar]
- 78.Mourant AE, Kopec AC, Domaniewska-Sobczak K. Distribution of Human Blood Groups and Other Polymorphisms. 2th edition. London: Oxford University Press; 1976. [Google Scholar]
- 79.De Silva JR, Lau YL, Fong MY. Genotyping of the Duffy blood group among Plasmodium knowlesi-infected patients in Malaysia. PLoS One. 2014;9:e108951. doi: 10.1371/journal.pone.0108951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosenfield RE, Vogel P, Race RR. [A new case of anti-Fya in human serum]. Rev Hémat. 1950;5:315–7. [In French] [PubMed] [Google Scholar]
- 81.Algora M, Barbolla L, Contreras M. Naturally occurring anti-D, anti-K, anti-Fya, and anti-Leab. Vox Sang. 1991;61:141. doi: 10.1111/j.1423-0410.1991.tb00262.x. [DOI] [PubMed] [Google Scholar]
- 82.Marsh WL. Present status of the Duffy blood group system. CRC Crit Rev Clin Lab Sci. 1975;5:387–412. doi: 10.3109/10408367509107049. [DOI] [PubMed] [Google Scholar]
- 83.Hardman JT, Beck ML. Hemagglutination in capillaries: correlation with blood group specificity and IgG subclass. Transfusion. 1981;21:343–6. doi: 10.1046/j.1537-2995.1981.21381201810.x. [DOI] [PubMed] [Google Scholar]
- 84.Szymanski IO, Huff SR, Delsignore R. An autoanalyzer test to determine immunoglobulin class and IgG subclass of blood group antibodies. Transfusion. 1982;22:90–5. doi: 10.1046/j.1537-2995.1982.22282177133.x. [DOI] [PubMed] [Google Scholar]
- 85.Daniels G, Poole J, de Silva M, et al. The clinical significance of blood group antibodies. Transfus Med. 2002;12:287–95. doi: 10.1046/j.1365-3148.2002.00399.x. [DOI] [PubMed] [Google Scholar]
- 86.Girello AL, Kühn TIB. Fundamentos da Imuno-Hematologia Eritrocitária. 2th ed. São Paulo, SP: Senac; 2007. [Google Scholar]
- 87.Mattos LC. [Duffy: a considerably complex blood group system]. Rev Bras Hematol Hemoter. 2005;27:79–80. [In Portuguese] [Google Scholar]