Summary
Nuclear RNA metabolism is influenced by protein complexes connecting to both RNA-productive and -destructive pathways. The ZC3H18 protein binds the cap-binding complex (CBC), universally present on capped RNAs, while also associating with the nuclear exosome targeting (NEXT) complex, linking to RNA decay. To dissect ZC3H18 function, we conducted interaction screening and mutagenesis of the protein, which revealed a phosphorylation-dependent isoform. Surprisingly, the modified region of ZC3H18 associates with core histone proteins. Further examination of ZC3H18 function, by genome-wide analyses, demonstrated its impact on transcription of a subset of protein-coding genes. This activity requires the CBC-interacting domain of the protein, with some genes being also dependent on the NEXT- and/or histone-interacting domains. Our data shed light on the domain requirements of a protein positioned centrally in nuclear RNA metabolism, and they suggest that post-translational modification may modulate its function.
Keywords: ZC3H18, NEXT, CBC, RNA exosome, histones, transcription, RNA decay
Graphical Abstract
Highlights
-
•
ZC3H18 uses separate domains for binding to CBCA, NEXT, and histones
-
•
ZC3H18 interacts with histones in a phosphorylation-dependent manner
-
•
RNA sequencing reveals a role for ZC3H18 in mRNA production
-
•
CBCA-binding domain is important for ZC3H18’s role in RNA production and decay
The ZC3H18 protein is involved in RNA decay mediated by the CBC-NEXT complex. Winczura et al. identify a phosphorylation-dependent interaction of ZC3H18 with histones, and they find separate CBCA-, NEXT-, and histone-binding domains. They suggest a role for ZC3H18 in mRNA biogenesis, which for some genes is independent of its role in RNA decay.
Introduction
Nuclear RNA metabolism is a major contributor to eukaryotic RNA homeostasis by sorting transcripts for nuclear function, export or decay. This is a demanding task given the fact that eukaryotic genomes are promiscuously transcribed into a heterogeneous mix of RNAs with highly variable cellular duties and half-lives (Derrien et al., 2012, Jensen et al., 2013). Additionally, all transcripts are produced as longer precursors, requiring appropriate nuclear maturation events to yield functional molecules. How this massive genomic output and its requirements for different activities is organized by participating nuclear machineries, and which molecular interactions execute decision processes dictating RNA fate, is a matter of intense investigation.
A protein-interaction-based framework for RNA fate determination is instigated early during nascent RNA synthesis. For RNA polymerase II (Pol II) transcripts, this starts with m7-G capping of the nascent RNA 5′ end emerging from the Pol II exit channel and its subsequent binding by the CBP20 and CBP80 proteins, composing the cap-binding complex (CBC) (Bentley, 2014, Glover-Cutter et al., 2008, Görnemann et al., 2005, Izaurralde et al., 1994). The CBC binds all classes of m7-G-capped transcripts, and it affects nuclear RNAs productively by stimulating the splicing of cap-proximal introns, RNA 3′ end formation, and the packaging of transport-competent ribonucleoprotein (RNP) particles in general (Cheng et al., 2006, Izaurralde et al., 1994, Narita et al., 2007). These activities are aided, respectively, by the association of the CBC with the U4/U6.U5 tri-snRNP (Pabis et al., 2013); the ARS2, NELF-E, and SLBP proteins (Andersen et al., 2013, Hallais et al., 2013, Narita et al., 2007); and the RNA transport factors ALY/REF and PHAX (Cheng et al., 2006, Kuersten et al., 2001, Ohno et al., 2000).
However, contacts from the CBC can also be established to the ribonucleolytic RNA exosome operating at Pol II-transcribed loci to degrade spurious transcripts like promoter upstream transcripts (PROMPTs) and enhancer RNA (eRNAs) (Andersen et al., 2013, Andersson et al., 2014, Ntini et al., 2013), or to participate in RNA processing (Lubas et al., 2015). Specifically, the CBC recruits the ARS2 protein to form the CBC-ARS2 (CBCA) complex, which harbors transcription termination activity (Andersen et al., 2013, Gruber et al., 2012, Hallais et al., 2013, Iasillo et al., 2017). CBCA may then bind the zinc-finger protein ZC3H18, mediating contact to the nuclear exosome targeting (NEXT) complex, to form the CBC-NEXT (CBCN) assembly, and further to the RNA exosome (Andersen et al., 2013, Lubas et al., 2015). Interestingly, formation of the decay-promoting CBCN assembly is mutually exclusive with formation of the so-called CBC-ARS2-PHAX (CBCAP) complex (Giacometti et al., 2017), where PHAX is recruited to the CBCA complex to act as an adaptor between the CBC/RNP complex and the nuclear transport receptor CRM1 (Kuersten et al., 2001, Ohno et al., 2000). Assembly of CBCN relative to CBCAP appears to be balanced by direct competition between ZC3H18 and PHAX for CBCA-bound RNA (Giacometti et al., 2017). Since this competition can yield opposite functional outcomes, it constitutes an interesting principle of how nuclear RNA sorting may occur at the mechanistic level. In addition, it positions ZC3H18 as a central factor capable of bridging the CBC, universal for Pol II-derived products, with the nuclear RNA exosome in competition with RNP-productive factors.
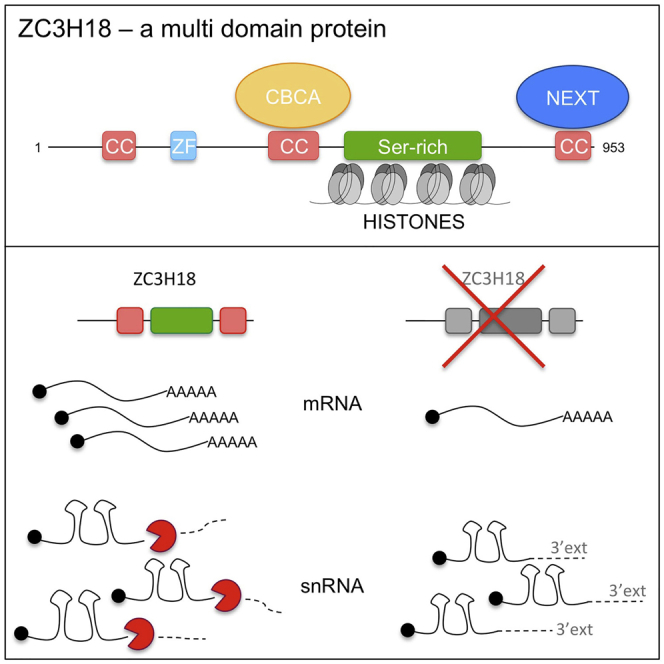
In addition to its affinity for CBC-bound RNA (Andersen et al., 2013, Merz et al., 2007, Pabis et al., 2013) and its presence in the CBCN assembly (Andersen et al., 2013), ZC3H18 was shown to interact with the THOC2 component of the transcription export (TREX) complex, and its depletion resulted in the nuclear accumulation of polyadenylated RNA (Chi et al., 2014). However, whether this latter phenotype reflects an mRNA export function of ZC3H18 or rather its role in targeting the nuclear exosome for RNA decay remains to be determined. Apart from its reported protein-protein interactions, the ZC3H18 protein is completely uncharacterized. It consists of 953 amino acids (aa), yet it is simply predicted to harbor a CCCH zinc-finger domain, three coiled-coil domains, and a few regions enriched for certain aa, namely proline (Pro) and serine (Ser) residues. ZC3H18 can be categorized as an intrinsically disordered protein, some of which are functionally important in RNA biology (Calabretta and Richard, 2015, Järvelin et al., 2016).
To achieve a better understanding of ZC3H18 interaction and function, we conducted an affinity capture (AC) interaction screening followed by mass spectrometry (MS). This allowed a thorough characterization of ZC3H18-containing complexes and the unexpected discovery of its association with core histone proteins. Two ZC3H18 isoforms were revealed, representing phosphorylation-dependent post-translational modification, occurring in the domain of ZC3H18 responsible for histone binding. Further mutagenesis analyses demonstrated separate domains of ZC3H18 engaged in histone, CBCA, and NEXT binding, and functional analysis revealed a role of ZC3H18 in mRNA biogenesis, which for a subset of genes is independent of the RNA decay-promoting activity of the protein. Our data disclose previously unknown physical links of a protein involved in RNA fate decisions, and they imply that post-translational modification is modulating its function.
Results
Interaction Profiling Positions ZC3H18 with Chromatin Factors
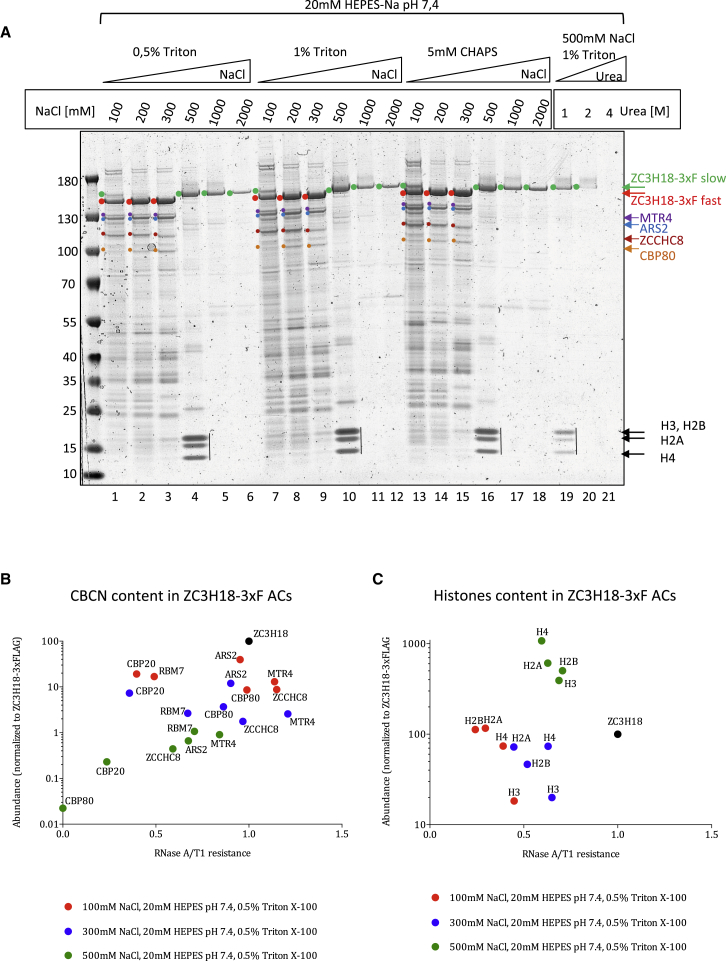
We previously characterized ZC3H18 as a central component of the CBCN complex, facilitating exosome-mediated RNA decay. To further explore its protein-protein interactions, we conducted high-throughput AC screening (Hakhverdyan et al., 2015) of ZC3H18 expressed with a C-terminal 3×FLAG (3xF) tag from a stably integrated and tetracycline-inducible HEK293 Flp-In T-Rex cell line (Andersen et al., 2013). Cell lysates were subjected to anti-FLAG AC across 21 different buffer conditions and visualized by Coomassie staining. The ZC3H18-3xF bait migrated at the expected size, and it was recovered in all conditions except for the harsh 4M Urea buffer incubation (Figure 1A, lane 21). AC eluates in lanes 1–6, 7–12, and 13–18 were from three sets of buffers differing in their content of detergent and each containing increasing concentrations of NaCl. AC profiles of the remaining proteins appeared similar across the three sets, demonstrating a limited impact of detergent choice. Instead, salt concentration-dependent changes were notable: (1) a reduction in the number of proteins recovered across increasing salt concentrations, including components of the CBCN complex, MTR4, ARS2, ZCCHC8, and CBP80; (2) a substantial qualitative shift in ZC3H18-3xF migration between the 300 and 500 mM NaCl buffers (Figure 1A, ZC3H18 forms denoted with red (fast) and green (slow) dots, respectively); and (3) the simultaneous appearance of three strongly stained bands at the bottom of the gel with the transition to the slow-migrating form of ZC3H18 (Figure 1A, lanes 4, 12, and 16). These fast-migrating species formed a pattern characteristic of core nucleosome histones, which was confirmed by MS analysis of relevant excised gel slices (data not shown).
Figure 1.
Interaction Profiling of ZC3H18 Identifies Histones
(A) SDS-PAGE gel of proteins co-precipitating with ZC3H18-3xF at the indicated buffer compositions. Two major isoforms of the ZC3H18-3xF bait protein are indicated with red and green dots for the fast- and slow-migrating forms, respectively. Denoted with color code are also CBCN complex components MTR4, ARS2, ZCCHC8, and CBP80 as well as histones. The identity of the indicated bands was established by MS analysis.
(B and C) Scatterplots presenting MS analysis of co-purified CBCN components (B) and core histones (C) in the ZC3H18-3xF AC experiments performed in 100, 300, and 500 mM NaCl-containing buffers (indicated by red, blue, and green dots, respectively). Exact buffer compositions are shown below the plots. The y axes display protein abundances estimated as the ratio between a protein’s mean peptide intensity from two biological experiments and its molecular weight and normalized to the abundance of ZC3H18-3xF bait protein. The x axes display RNase A/T1 resistance calculated as the ratio between protein abundances in RNase A/T1-treated versus untreated samples.
AC experiments conducted in 100, 300, and 500 mM NaCl, respectively (Figure 1A, lanes 1, 3, and 4), were collected in duplicate and subjected to MS analyses, after which abundances of co-precipitated proteins were calculated based on mean peptide intensities from the two experiments (Table S1). To discriminate any RNA-dependent interactions, bead-bound complexes were treated with RNase A/T1 or with BSA before elution. Using these MS data, we plotted the RNase A/T1 resistance of selected groups of proteins versus their abundance in the ZC3H18 precipitates. The association of ZC3H18 with CBCN complex components decreased gradually with increasing salt concentration (Figure 1B), and its contact with the RNA exosome was practically lost in the 300 mM-NaCl sample (Figure S1A). Consistent with data from Figure 1A, ZC3H18 association with histones was instead strengthened with increasing salt, which was seen for both canonical (Figure 1C) and non-canonical (Figure S1B) histones. Although mostly at quite low abundances, the 500 mM-NaCl sample was further enriched for many chromatin and DNA-related factors, such as (1) components of the facilitate chromatin transcription (FACT) complex, involved in nucleosome reorganization; (2) the poly-ADP polymerase 1 (PARP1), involved in DNA damage repair; and (3) chromobox proteins CBX1, CBX3, and CBX5, involved in heterochromatin organization (Figure S1C). Finally, as previously reported (Chi et al., 2014), ZC3H18 associated with TREX complex components, although only in the low-salt buffer and with ALY/REF as the most prominent interactor (Figure S1D). Generally, the described co-precipitating complexes largely associated with ZC3H18 independently of RNA (RNase A/T1 resistance close to 1), as opposed to bona fide RNA-binding factors (e.g., most heterogeneous nuclear ribonucleoproteins [hnRNPs]), whose association was lost upon RNase A/T1 treatment (Figure S1E).
In conclusion, our AC screening disclosed, in addition to already known ZC3H18 interactors, the co-precipitation of ZC3H18 with core histone proteins and additional factors involved in chromatin biology. These associations appear specific for a slow-migrating isoform of ZC3H18.
ZC3H18 Binds Histones in a Phosphorylation-Dependent Manner
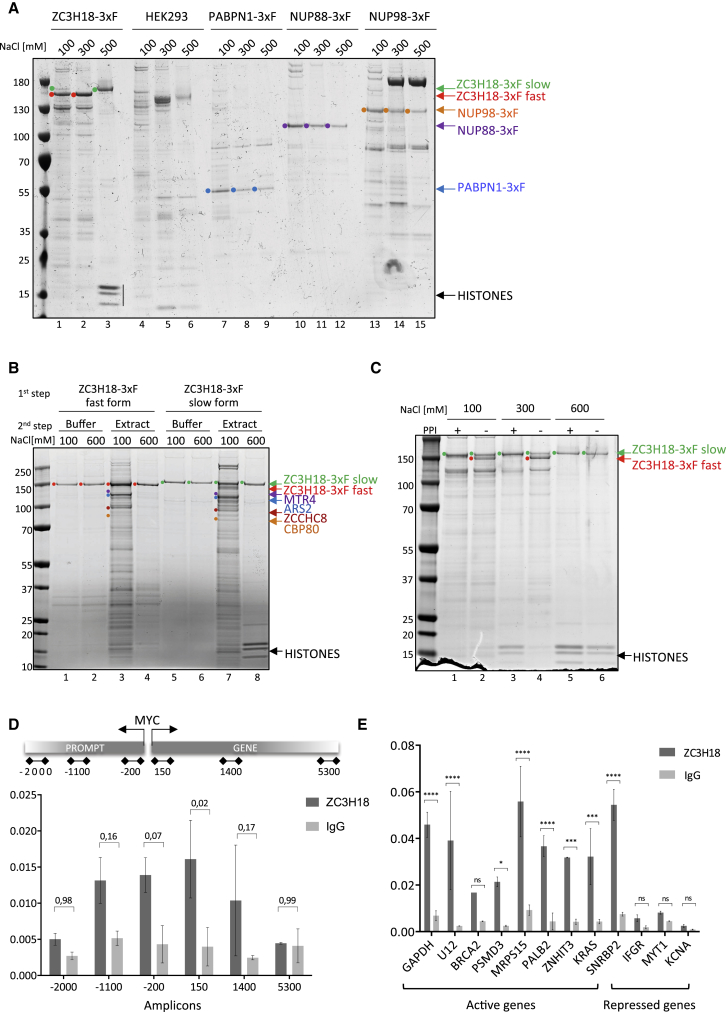
Having established a new link between ZC3H18 and histones, we wanted to characterize the interaction further and to ascertain its specificity. To this end, we first performed anti-FLAG AC in 100, 300, and 500 mM-NaCl conditions of unrelated extracts prepared from additional HEK293 cell lines stably expressing 3xF-tagged fusion proteins, including nuclear poly(A)-binding protein (PABPN1) as well as the two nucleoporins NUP88 and NUP98. For the assessment of background binding, we employed the parental HEK293 cell line devoid of any 3xF fusion construct. Only the ZC3H18-3xF fusion co-precipitated histones in 500 mM NaCl (Figure 2A, lane 3), reinforcing that the ZC3H18-histone interaction is specific.
Figure 2.
ZC3H18 Binding to Histones Is Specific and Phosphorylation Dependent
(A) SDS-PAGE gel of proteins co-precipitating with color-coded ZC3H18, PABPN1, NUP88, and NUP98 proteins expressed with a 3xF tag from HEK293 Flip-In T-Rex cell lines. As a control the HEK293 parental cell line was also used. AC was carried out in three different NaCl buffers (20 mM HEPES [pH 7.4], 0.5% Triton X-100 and 100, 300, or 500 mM NaCl, respectively) as indicated. 3xF-tagged proteins are indicated, based on their expected molecular weights, with color-coded dots and corresponding arrows. Only the ZC3H18-3xF construct co-purified histones in the 500 mM NaCl buffer (indicated by black vertical line in lane 3).
(B) SDS-PAGE gel showing the ZC3H18 isoforms and co-precipitating proteins after a two-step ZC3H18-3xF AC in 100 and 600 mM-NaCl conditions (see main text for details). Migration of ZC3H18-3xF fast- and slow-migrating forms as well as CBCN complex components are indicated as in (A). Note that histones are bound only by the slow-migrating form of ZC3H18-3xF.
(C) SDS-PAGE gel of proteins co-precipitating with ZC3H18-3xF in 100, 300, and 600 mM-NaCl containing buffers supplemented (+) or not (−) with PPI. ZC3H18-3xF isoforms denoted as in (A).
(D) ChIP-PCR analysis of ZC3H18 occupancy along the MYC gene and MYC PROMPT using amplicons from top schematics. IgG ChIP was used to assess background binding. Data are displayed as mean ± SD of two biological replicates. Significance levels between the ZC3H18 and IgG samples were assessed by a two-way ANOVA test with obtained p values presented above the bars.
(E) ChIP-PCR analysis of ZC3H18 occupancy at promoters of transcriptionally active (GAPDH, U12, BRCA1, PSMD3, MRPS15, PALB2, ZNHIT3, KRAS, and SNRPB2) and repressed (IFRG28, MYT1, and KCNA1) genes. IgG ChIP was used to assess background binding. Data are displayed as mean ± SD of two technical replicates of the representative experiment. Significance levels between the ZC3H18 and IgG samples were calculated using a two-way ANOVA test and are denoted with asterisks corresponding to the following p value ranges: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
Next, we explored why histones co-precipitated with ZC3H18-3xF only in the high-salt condition. Different solutions impart different characteristics to protein extracts, influencing the soluble and insoluble fractions, as well as the stabilities of macromolecular complexes. We reasoned that high NaCl concentrations might disturb DNA-histone interaction. To test this notion, we extracted proteins from HEK293 cells in buffers containing 100, 300, 500, or 1,000 mM NaCl, and we examined the soluble and insoluble fractions for each sample by SDS-PAGE. As predicted, increasing the NaCl concentration from 300 to 500 mM led to the transfer of histones into the soluble fraction (Figure S2A). Thus, ZC3H18 might interact with histones at high-salt concentrations, simply because they become available for binding in these conditions and not because of any altered ZC3H18 isoform. To discriminate these possibilities, we performed a two-step AC experiment to examine whether ZC3H18 would also precipitate histones in low salt, given their sufficient availability. First, the fast- and slow-migrating forms of ZC3H18 were purified in 300 and 800 mM NaCl buffers, respectively, followed by washing with 800 mM NaCl buffer, leaving only the respective ZC3H18-3xF isoforms bound to beads (Figure 2B, lanes 1, 2 and 5, 6). Next, protein extracts prepared in 300 or 600 mM NaCl buffers, lacking and containing histones, respectively, were used for AC with the two forms of ZC3H18-3xF. Critically, only the slow-migrating form of ZC3H18 co-purified histones from the 600 mM protein extract (Figure 2B, compare lanes 4 and 8), while both forms bound CBCN complex components when low-salt conditions were employed (Figure 2B, lanes 3 and 7). We conclude that the ZC3H18 isoforms bind differentially to histones and presumably use different domains to interact with CBCN components.
What is then different between the two major isoforms of ZC3H18? As their N and C termini were identical (data from MS and western blotting analyses not shown), we reasoned that their mass difference might be caused by post-translational modification (PTM). ZC3H18 is extensively phosphorylated (Beli et al., 2012, Hornbeck et al., 2015) and contains multiple Ser, threonine (Thr), and tyrosine (Tyr) residues as putative phosphorylation targets. We therefore used protein phosphatase inhibitors (PPIs) during protein extraction to prevent post-lysis dephosphorylation, and we performed anti-FLAG AC. Remarkably, adding PPIs to low-salt protein extracts prevented formation of the fast-migrating ZC3H18 isoform otherwise visible in lanes without the inhibitors (Figure 2C, compare lanes 1 and 2 as well as 3 and 4). Thus, ZC3H18 is post-translationally modified in a phosphorylation-dependent manner. To assess the ratio of modified to unmodified ZC3H18 in living cells, we prepared cell extracts in SDS-containing low-salt buffer, which rapidly denatures all proteins at the time of cell lysis. Western blotting analysis revealed that, upon such extraction, only the slow-migrating form of ZC3H18 was detected (Figure S2B). Furthermore, ZC3H18 AC in 100 mM NaCl buffer, performed after cells had been crosslinked with 1% formaldehyde (FA), preserved the slow-migrating form (Figure S2C, compare lanes 1 and 2). These results suggest that cellular ZC3H18 is predominantly in its modified, slow-migrating form and that capture of the fast-migrating form in low-salt concentrations is caused by post-lysis dephosphorylation.
To this point, our experiments were performed using protein extracts. Hence, to assess any in vivo relevance of the observed ZC3H18-histone association, we turned to chromatin immunoprecipitation (ChIP) assays capable of capturing protein-chromatin associations inside cells. Gratifyingly, the employed antibody, detecting endogenous protein, chromatin immunoprecipitated ZC3H18 over background around the transcription start sites (TSSs) and bodies of the interrogated MYC and MYC-PROMPT transcription units (TUs) (Figure 2D). Moreover, low-resolution ZC3H18 ChIP sequencing (ChIP-seq) analysis (data not shown) pointed to ZC3H18 binding at the promoter regions of a few genes, which we validated by ChIP-qPCR over the active GAPDH, U12, BRCA1, PSMD3, MRPS15, PALB2, ZNHIT3, KRAS, and SNRPB2 promoters (Figure 2E). Only minor signals were detected over three repressed genes, IFRG28, MYT1, and KCNA1. These data confirmed an association of ZC3H18 with chromatin in vivo, and they suggest that binding may preferentially occur around active promoter regions, possibly due to the CBC-associating nature of the protein.
ZC3H18 Interacts with CBCA, NEXT, and Histones through Distinct Domains
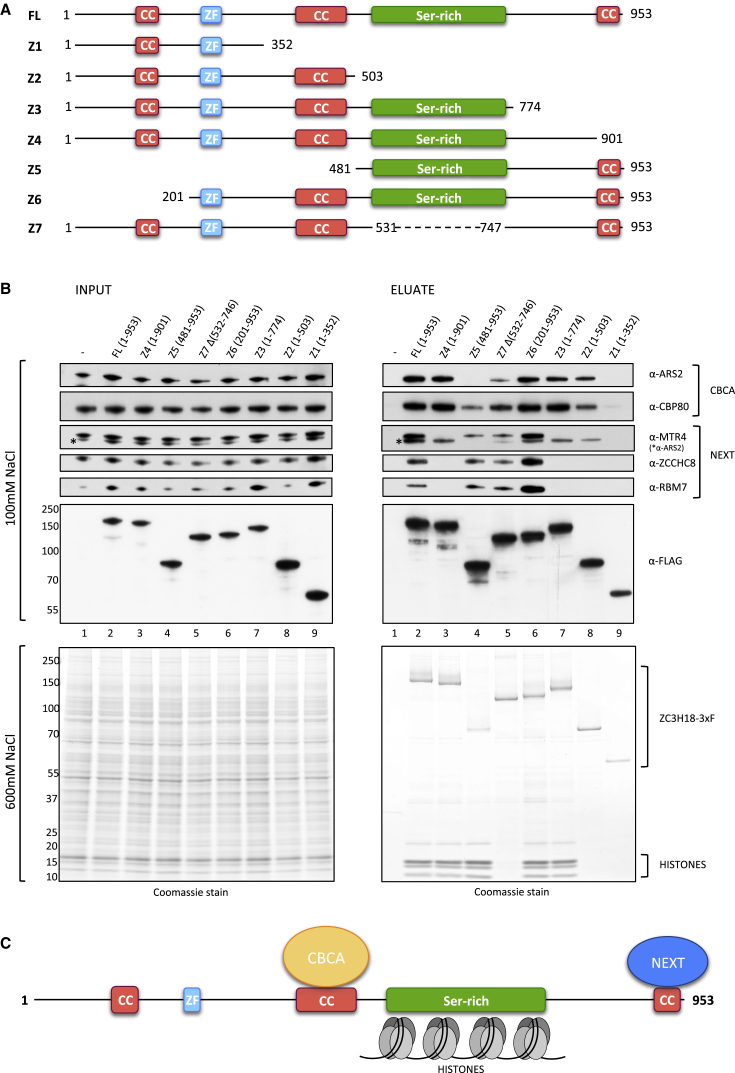
Results obtained so far imply that ZC3H18 engages in at least two sub-complexes, with CBCN and with histones, and that ZC3H18 may utilize distinct domains to form these interactions. To further analyze this issue, we conducted mutational analysis of ZC3H18, whose 953 aa residues consist of a zinc finger, three coiled-coil (cc) domains, and a large region enriched in Ser residues (Figure 3A, top full-length [FL] construct). To interrogate any function of these domains, we designed four C-terminal (Z1–Z4) and two N-terminal (Z5–Z6) ZC3H18 truncations as well as a mutant variant internally deleted for the Ser-rich region (Z7) (Figure 3A). All fragments were C-terminally fused to a 3xF tag, facilitating immunolocalization analyses. Upon transfection into HeLa Kyoto cells, fragments Z4–Z7 all appeared nuclear, like the FL ZC3H18-3xF fusion, while fragments Z1–Z3 localized to both the cytoplasm and the nucleus (Figure S3A). This suggests that a nuclear localization signal (NLS) likely resides in the C terminus of ZC3H18 (Figure S3B).
Figure 3.
Domain Mapping of ZC3H18
(A) Schematic representation of the FL and Z1-Z7 mutant variant ZC3H18 constructs. Dashed line in Z7 construct indicates deletion. CC, coiled coil; ZF, zinc finger; Ser-rich, serine-rich region (UniProt: Q86VM9; www.uniprot.org). All constructs were C-terminally fused to 3xF.
(B) Western blotting analysis of the indicated proteins co-purifying with ZC3H18-3xF FL and mutant variants in 100 mM-NaCl (top panel) or 600 mM-NaCl (bottom panel) AC conditions. In the low-salt condition, input and eluate samples were probed with antibodies against the CBCA components ARS2 and CBP80 and the NEXT components MTR4, ZCCHC8, and RBM7 as well as α-FLAG antibodies against the 3xF-tagged constructs. In the high-salt condition, ZC3H18-3xF variants and histones were visualized using Coomassie staining. Note that the lower band in the α-MTR4 western analysis, denoted with an asterisk, corresponds to ARS2 signal from a previous exposure of the same membrane.
(C) Schematic representation of interaction domains on ZC3H18.
We then performed FLAG AC of the ZC3H18 fragments. To monitor the contents of CBCN and histone components in the resulting eluates, protein extracts were prepared in 100 and 600 mM NaCl buffers, respectively, and CBCN and histone components were detected by western blotting analysis and Coomassie staining. Reassuringly, the FL ZC3H18 construct co-purified all studied interactors (Figure 3B, right panel lane 1). A similar result was obtained employing fragment Z6, indicating that the 200-aa N-terminal part of ZC3H18 is not crucial for these interactions. Fragment Z5 co-purified all three NEXT complex components, but not ARS2 and only low CBP80 amounts, demonstrating that a major binding site for the CBCA complex resides within residues 201–480. Remarkably, fragment Z4, which lacks only 52 residues from the ZC3H18 C terminus, completely lost association with the NEXT complex but preserved binding to CBP80, ARS2, and histones. Thus, the C-terminal coiled-coil domain of ZC3H18 provides a platform for NEXT binding, in agreement with previous results (Giacometti et al., 2017). Consistently, fragments Z1, Z2, and Z3, all lacking this domain, did not precipitate the NEXT complex. Z2 and Z3 bound CBP80 and ARS2 to similar extents as the FL construct, while Z1 did not bind any of the CBCN components. Finally, histones were bound by the FL and Z3–Z6 fragments, and this binding was abolished for Z1, Z2, and Z7, which pinpoints the Ser-rich region as a histone-binding domain. Taken together, these analyses not only identified a distinct histone-binding domain but also revealed that the association of ZC3H18 with the CBCA and NEXT complexes is physically separated (Figure 3C).
The ZC3H18 fragments also allowed a more detailed analysis of domains involved in defining the slow- and fast-migrating isoforms. Hence, we tested the response of the various ZC3H18 mutants to high/low salt extraction and to PPI treatment. Interestingly, fragments lacking the Ser-rich region (Z1, Z2, and Z7) did not display any gel migration differences, regardless of the extraction method or PPI addition (Figure S3C). In contrast, all remaining fragments (Z3–Z6) and the FL construct displayed slow-migrating isoforms when extracted with the high-salt concentration or when the low-salt extraction was supplied with PPI. This strongly suggests that the Ser-rich domain is required for the phosphorylation-dependent modification of ZC3H18 (Figure S3B). Moreover, robust co-purification of histones could be achieved even in low salt, simply provided that PPI was supplied. This further supports the notion that it is primarily the slow-migrating form of ZC3H18 that shows specificity toward these proteins. Whether ZC3H18 isoform switching is driven directly by phosphorylation of the Ser-rich region, which are the phosphorylation sites, and which are the involved kinases remain to be elucidated.
ZC3H18 Depletion Differentially Affects Transcript Levels
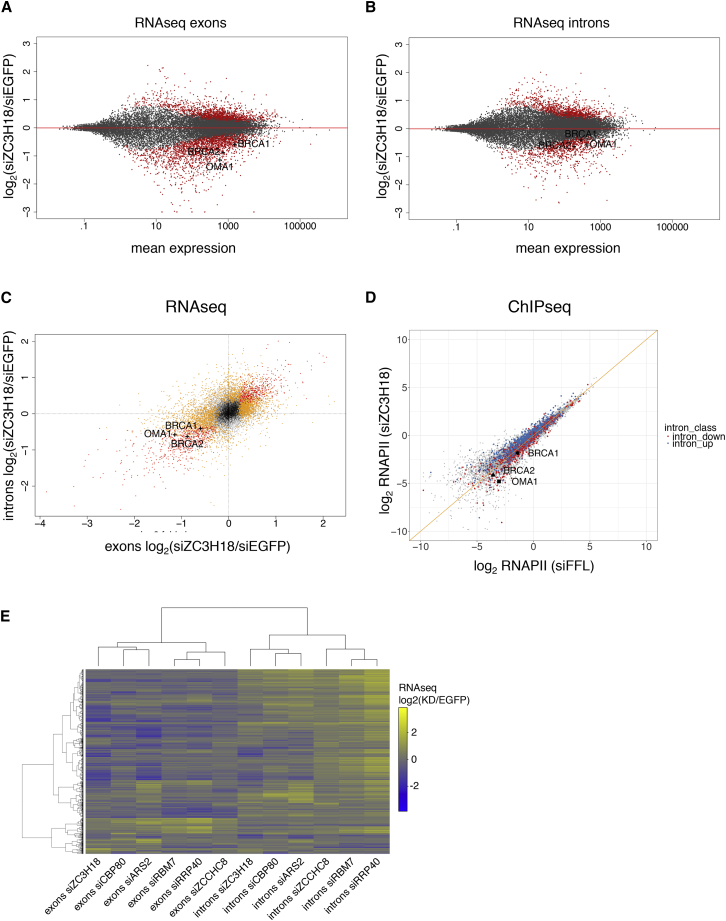
We previously demonstrated that ZC3H18 depletion provokes elevated levels of otherwise labile nuclear transcripts, like eRNAs, PROMPTs, and 3′ extended small nuclear RNAs (snRNAs), consistent with a role of ZC3H18 in CBCN-mediated RNA decay by the exosome (Andersen et al., 2013, Giacometti et al., 2017, Iasillo et al., 2017). To inquire whether any protein-coding (pc) transcripts might also be affected by ZC3H18 depletion, we explored previously published RNA sequencing (RNA-seq) data of total RNA from HeLa Kyoto cells subjected to triplicate ZC3H18 small interfering RNA (siRNA)-mediated depletion or to the addition of EGFP control siRNA (Iasillo et al., 2017). RNA-seq reads filtered to map exclusively to either exonic or intronic regions of annotated pc genes, as approximations for mRNA and pre-mRNA, respectively, were subjected to differential expression analysis.
ZC3H18 depletion resulted in 1,756 and 2,110 significantly up- and downregulated mRNAs, respectively (Figure 4A). Since downregulated transcripts cannot be rationalized by a degradative role of ZC3H18, we speculated that these might reflect a currently undisclosed exosome-independent function of ZC3H18. Interestingly, similar numbers of up- and downregulation were observed when considering intronic reads (Figure 4B), and intron and exon reads were generally correlated (Figure 4C). Highlighted BRCA1, BRCA2, and OMA1 RNAs exemplify transcripts that were significantly downregulated both at intronic and exonic levels and, therefore, selected as test cases for further analysis (Figures S4A–S4C; see below).
Figure 4.
ZC3H18 Depletion Affects Transcription of a Subset of Protein-Coding Genes
(A) RNA-seq-derived differential expression changes of exonic reads between ZC3H18 depletion and control (EGFP) libraries. The y axis displays the log2 fold change of read counts, and the x axis displays the log10-transformed exonic read count per gene. Data were computed by the DESeq2 software (Love et al., 2014). All individual genes are shown in gray and significantly called transcripts (padj = FDR < 0.1) are colored red. Of the n = 18,731 genes shown, n = 1,756 and n = 2,110 were significantly up- and downregulated, respectively. BRCA1, BRCA2, and OMA1 RNAs are highlighted.
(B) As in (A) but based on reads from intronic regions of each gene, scaled using sizing factors from exonic reads. Of the n = 17,872 genes shown, n = 1,277 and n = 905 were significantly up- and downregulated, respectively.
(C) Scatterplot showing RNA-seq log2 fold changes of exonic (from A) versus intronic (from B) regions for individual genes (n = 16,157, Pearson correlation coefficient r2 = 0.56). RNAs that were significantly up- or downregulated (padj = FDR < 0.1) at exon or intronic levels are colored yellow. RNAs that were significantly affected in both exon and intron read counts are colored red. BRCA1, BRCA2, and OMA1 RNAs are highlighted.
(D) Scatterplot showing Pol II ChIP-seq log2 signals in ZC3H18 depletion (y axis) relative to control (siFFL) (x axis) samples. Genes expressing RNAs that were significantly (padj = FDR < 0.1) up- or downregulated in the RNA-seq intronic analysis are colored blue and red, respectively. BRCA1, BRCA2, and OMA1 genes are highlighted.
(E) Heatmap of hierarchical clustering analysis of log2 fold changes of RNA-seq exonic (left part) or intronic (right part) reads from ZC3H18, CBP80, ARS2, RBM7, RRP40, and ZCCHC8 depletion relative to control samples (denoted by siXXXX below the figure). RNAs with padj < 0.01 and log2 fold change >1 or log2 fold change <−1 were included (n = 2,626).
Changes in intronic read content are often taken as proxies for changes in transcription levels (Gaidatzis et al., 2016, Iasillo et al., 2017, Jonkers et al., 2014). We therefore interrogated previously reported Pol II ChIP-seq data from ZC3H18-depleted and control cells (Iasillo et al., 2017). Globally, only a few genes exhibited strong changes in Pol II ChIP-seq signal upon ZC3H18 depletion. However, the data provided a clear stratification of transcription levels, as measured by intronic reads (Figure 4D). Consistently, the BRCA1, BRCA2, and OMA1 genes all showed lower Pol II ChIP-seq signal upon ZC3H18 depletion (Figures 4D and S4D). These results suggest that the downregulation of genes in ZC3H18-depleted cells is due to decreased transcription.
The negative impact of ZC3H18 depletion on select mRNA levels was surprising, and we therefore asked whether this phenomenon depended on known ZC3H18 interaction partners. Hence, we performed a comparative analysis of published RNA-seq data of transcripts from HeLa Kyoto cells individually depleted of CBCN components ZC3H18, CBP80, ARS2, ZCCHC8, and RBM7 as well as the exosome core component RRP40, together with corresponding siEGFP control samples (Iasillo et al., 2017, Meola et al., 2016). We selected significantly up- or downregulated genes based on their exonic or intronic reads (false discovery rate [FDR] < 0.1, log2 fold change >1 or <−1) for each depletion condition, and we observed a highly consistent overlap among all samples (data not shown). We then used the log2 fold change values for hierarchical clustering analysis. For both exonic and intronic reads, this revealed the clustering of the different depletion samples in two major groups: one comprising ZC3H18, ARS2, and CBC80 samples and the other comprising ZCCHC8, RBM7, and RRP40 samples (Figure 4E). These analyses suggest that ZC3H18’s function in mRNA metabolism is primarily linked to the CBCA complex.
Impact of ZC3H18 Interaction Domains on Protein Function
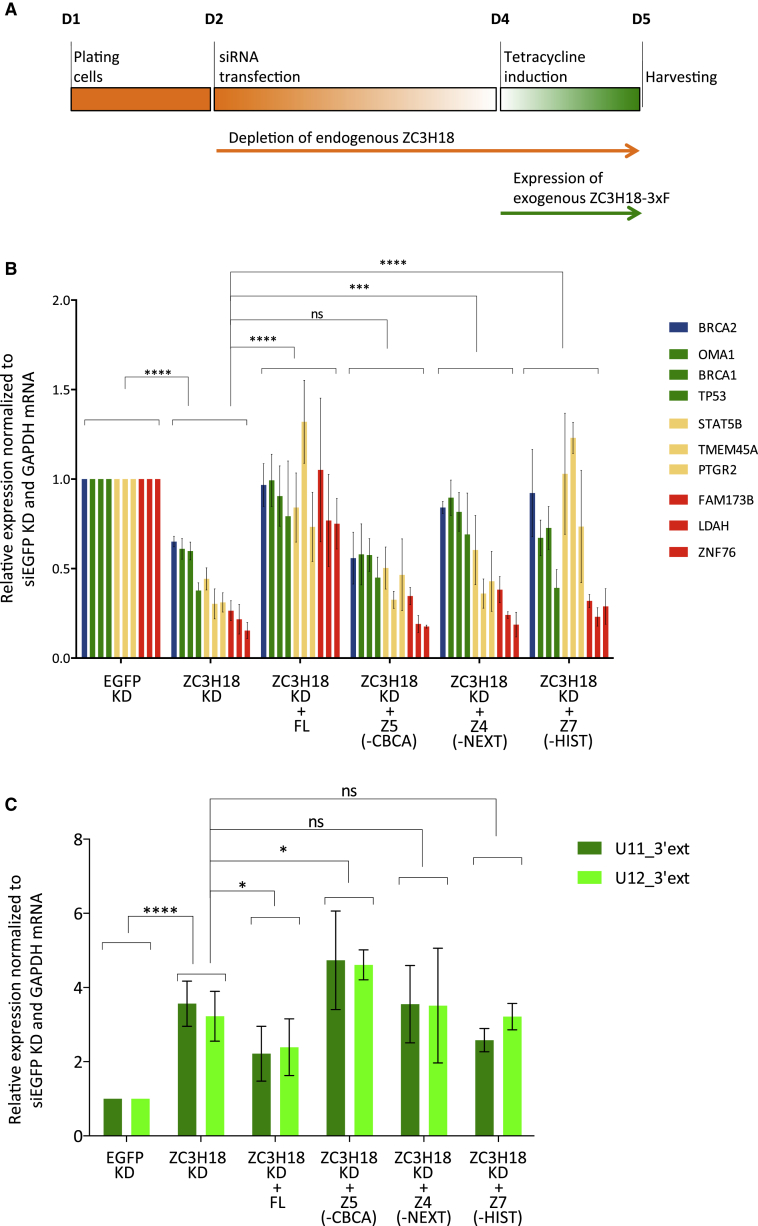
ZC3H18 acts, directly or indirectly, on different RNAs and is functionally and physically linked to the CBCA and NEXT complexes. Having established protein-protein interaction domains of ZC3H18, we were well positioned to inquire how interactions with CBCA, NEXT, and histones contribute to the dual role of ZC3H18 in decay of 3′ extended snRNA and in mRNA regulation. Thus, we depleted endogenous ZC3H18 protein with siRNA targeting the 3′ UTR of the ZC3H18 mRNA, followed by the attempt to rescue the interrogated phenotype by exogenously expressing RNAi-immune FL ZC3H18 and its fragment variants defective in binding to NEXT (Z4), CBCA (Z5), and histones (Z7), respectively (Figure 5A). These ZC3H18 constructs were stably expressed in HEK293 Flp-In T-Rex cell lines under the control of a tetracycline-inducible CMV promoter. Depletion efficiency and levels of exogenous ZC3H18 variant expression was monitored by western blotting analysis (Figures S5A and S5B), which revealed a leaky expression of exogenous ZC3H18 constructs in the absence of tetracycline. This was most visible for fragments Z7 and Z5, which could be clearly distinguished from endogenous ZC3H18 (Figure S5A, lanes 13–15 and 17–19, respectively). Hence, to avoid interference on target RNAs of this expression, we used a parental HEK293 cell line to assess steady-state RNA levels upon endogenous ZC3H18 depletion.
Figure 5.
Functional Impact of ZC3H18 Protein Interaction Domains
(A) Schematic representation of rescue experiments performed in HEK293 cell lines containing stably integrated and inducible ZC3H18-3xF FL and mutant variants. Endogenous ZC3H18 depletion was subjected to possible suppression by exogenous expression of ZC3H18-3xF variants.
(B) qRT-PCR analysis of indicated mRNA levels in total RNA samples from HEK293 cells depleted for endogenous ZC3H18 and expressing the indicated exogenous and ZC3H18-3′ UTR siRNA-resistant variants. Data are displayed as mean ± SD of at least three biological replicates normalized to the control (EGFP) sample and GAPDH mRNA. Significance levels between the mean values of mRNA levels in ZC3H18 depletion versus rescued samples were calculated using a two-way ANOVA test and are denoted with asterisks corresponding to the following p value ranges: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
(C) qRT-PCR analysis as in (B) but using primers detecting U11 and U12 snRNA 3′ extended regions.
Expression levels of 10 mRNAs, BRCA1, BRCA2, OMA1, TP53, STAT5B, TMEM45A, PTGR2, ZNF76, FAM173B, and LDAH, which were downregulated upon ZC3H18 depletion, were all recovered by the expression of FL ZC3H18 (Figure 5B). In contrast, the Z5 CBCA-binding mutant did not rescue expression of any of these transcripts. This is consistent with ZC3H18 and CBCA depletions yielding comparable phenotypes (Figure 4E), and it suggests that CBCA interaction is crucial for ZC3H18 function in mRNA metabolism. The Z4 NEXT-binding and the Z7 histone-binding mutants, on the other hand, demonstrated statistically significant rescue effects for only a subset of the tested mRNAs. Based on the extent and the significance of the rescue (Figures 5B and S5C), we identified targets whose expression depended on both histone- and NEXT-binding domains (Figure 5B, red columns); NEXT, but not histone, interaction (Figure 5B, yellow columns); histone, but not NEXT, interaction (Figure 5B, green columns); or neither of these domains (Figure 5B, blue column). Altogether, these results suggest a complex role of ZC3H18 in the expression of human genes, where the CBCA interaction domain is solely required for function.
In the case of 3′ extended U11 and U12, which were upregulated upon ZC3H18 depletion, expression of the FL ZC3H18 construct only partially rescued the phenotype (Figures 5C and S5C). This made interpretation of the functional effects of the three analyzed ZC3H18 mutants more challenging. However, consistent with the established role of the CBCN complex in the degradation of 3′ extended snRNAs (Lubas et al., 2015, Giacometti et al., 2017), the CBCA- and NEXT-binding mutants failed to rescue the phenotype. The Z7 histone-binding mutant provided partial recovery in the case of U11 and no recovery in the case of U12 snRNA. However, in light of the low statistical strength, this result does not allow us to discriminate whether the histone-binding domain might subtly impact snRNA biogenesis.
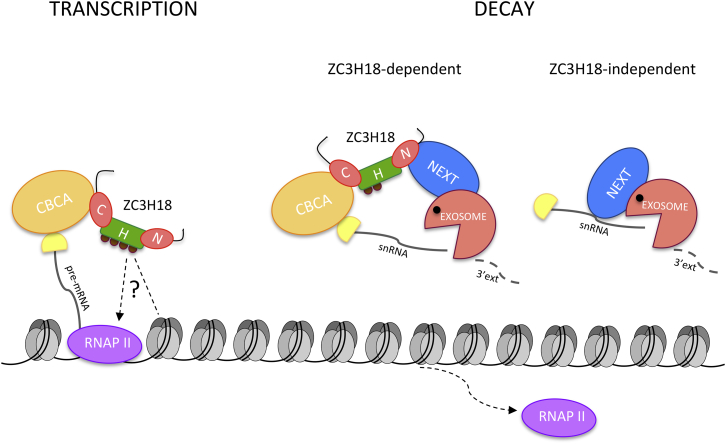
Taken together, the data underscore a fundamental role of the CBC as a platform for interactions affecting the fate of capped RNAs, and they establish ZC3H18 as a central factor engaged in RNA transactions involving both anabolic and catabolic decisions (Figure 6).
Figure 6.
Dual Roles of ZC3H18 in Nuclear RNA Metabolism
Schematic representation of the dual engagement of ZC3H18 in RNA transcription and decay processes. Left: the C terminus of ZC3H18 interacts with the CBCA complex to regulate protein-coding gene transcription directly or indirectly. Right: short non-coding RNAs, such as 3′ extended snRNAs, are targeted ZC3H18 dependently in a process requiring both CBCA and NEXT interaction of ZC3H18. Such RNA decay can also occur ZC3H18 independently via direct NEXT exosome targeting. See the main text for details. C, CBCA-interacting domain; H, histone-interacting domain; N, NEXT-interacting domain. Question mark indicates the elusive role of the histone-interacting domain and its modification in modifying the functionality of ZC3H18.
Discussion
RNA metabolic processes initiate early during nascent transcript synthesis. This is best understood for Pol II-derived RNAs, where the CBC affects the splicing of cap-proximal pre-mRNA introns as well as the 3′ end processing or decay of a variety of short RNAs (Müller-McNicoll and Neugebauer, 2014). The CBC recruits the ARS2 protein to form the CBCA complex, which harbors transcription termination activity and is capable of both linking to RNA-productive and -destructive activities (Andersen et al., 2013, Hallais et al., 2013, Iasillo et al., 2017). This choice is, at least to some extent, decided by a competition for CBCA binding between PHAX and ZC3H18, where the latter protein can exclude PHAX and instead connect the CBCA-bound transcript to the NEXT complex, hereby assembling CBCN, which in turn makes contact to the ribonucleolytic exosome (Giacometti et al., 2017). As indicated in this study, the CBCA-ZC3H18 axis may also participate in the transcription-based production of certain mRNAs, independently of the NEXT complex. Thus, to better comprehend the molecular detail underlying the roles of ZC3H18 as a central contributor to nuclear RNA fate decisions, we set out to map the domain organization of this 953-aa-large and uncharacterized protein. As expected from its CBCN-forming capacity, ZC3H18 utilizes two separate domains to bind the CBCA and NEXT complexes. More surprisingly, an unrelated part of the protein binds histones, establishing three distinct interaction domains of ZC3H18 (Figure 3C). The relevance and interplay of these domains in yielding competence to ZC3H18’s participation in the decay of short RNAs as well the production of selected mRNAs are discussed below.
Any analysis based on affinity isolation of protein complexes is limited by the quality of the extract and the experimental condition employed during complex purification. By applying a multi-well screening procedure (Hakhverdyan et al., 2015), we interrogated proteins co-purifying with FLAG-tagged ZC3H18 across several different buffer conditions. This revealed the robust co-precipitation of ZC3H18 with core nucleosomal histones, an interaction that had gone unnoticed when applying standard low-salt AC conditions (Andersen et al., 2013). Here, histones are apparently insoluble and therefore unavailable for binding to ZC3H18. Our screening approach also identified fast- and slow-migrating isoforms of ZC3H18, specific for low- and high-salt precipitation conditions, respectively. The slow-migrating form appears to dominate in vivo, but it is converted to the fast-migrating form upon subjection of cell extracts to 100–300 mM-NaCl conditions. Most likely, these extracts contain an enzymatic activity capable of demodifying ZC3H18 in low-salt conditions. Indeed, the addition of protein phosphatase inhibitors to the extracts resulted in preservation of the slow-migrating form, strongly suggesting that phosphorylation is participating in the modification of ZC3H18. Although we cannot exclude that ZC3H18 is modified by a phosphorylation-dependent alternative modification event, we suggest that the protein is phosphorylated directly. This is because in silico tools (https://www.phosphosite.org/homeAction.action ; http://www.hprd.org/PhosphoMotif_finder) predict a multitude of phosphorylation sites on ZC3H18 serine, threonine, and tyrosine residues, some of which were detected in high-throughput MS-based phosphoproteome studies (Amanchy et al., 2007, Beli et al., 2012, Hornbeck et al., 2015). In further support, our mutational analyses of ZC3H18 revealed the serine-rich mid-domain of the protein as a major PTM target.
Interestingly, only the slow-migrating form of ZC3H18 can interact with histones. Consistently, this interaction critically depends on the presence of dephosphorylation-incompetent extract as well as on the serine-rich domain, suggesting that histone binding is regulated by phosphorylation. Parts of ZC3H18 contain low-complexity sequences indicative of an intrinsically disordered protein, which may be predisposed to PTMs, likely phosphorylation, to stabilize its structure (Iakoucheva et al., 2004). The resulting negatively charged phosphorylated residues might then provide binding epitopes for positively charged histones. Although our study argues that the serine-rich domain of ZC3H18 is functionally relevant for the proper expression of some mRNAs, it is not clear whether modification of the protein provides a mechanistically relevant regulatory switch possibly controlling histone interaction. Given that the slow-migrating form of ZC3H18 predominates in vivo, such PTM-based regulation would have to occur on only a small population of the protein or with kinetics limiting the amounts of the unmodified protein, which may also be targeted for degradation, explaining its low in vivo levels. It also remains to be determined whether ZC3H18 has a preference for certain specific histone modifications. We show that ZC3H18 is readily chromatin immunoprecipitated to several active gene promoters and further enriched in both upstream and downstream regions around the divergent MYC promoter. Thus, histone configurations affiliated with promoters might attract ZC3H18 to active loci needing the function of this central RNP factor. The robust interaction of ZC3H18 with the cap-bound CBCA complex might then further contribute to efficient nascent RNA targeting. It is also possible that the affinity of ZC3H18 for chromatin is involved in other activities, like the release of ZC3H18 from the RNP in case transport factors like PHAX outcompete its presence or to provide substrate availability if the RNA is subject to decay. Further detailed analysis is clearly justified to delineate which exact role the serine-rich domain, and its modification, plays for ZC3H18 function.
An interpretation of the functions and interplay between the other two established domains of ZC3H18 appears more straightforward. As expected from the dependence on the NEXT complex and the RNA exosome for the decay of many short cryptic RNAs, the NEXT-binding ZC3H18 variant was unable to substitute the FL ZC3H18 construct in degrading the 3′ extended snRNA test substrates. This establishes the C-terminal 52 aa of ZC3H18 as the central binding platform for NEXT, possibly via a direct ZCCHC8-ZC3H18 interaction (Giacometti et al., 2017). The ZC3H18 variant compromised for CBCA binding, though capable of binding the NEXT complex as robustly as FL ZC3H18, did also not facilitate substrate turnover. The simplest interpretation of this result is that CBCA targeting by ZC3H18 is required for productive assembly of the CBCN complex, leading to exosomal decay. It is important here to note that depletion of NEXT or exosome components generally results in more dramatic RNA changes than depletion of ZC3H18 (Andersen et al., 2013, Iasillo et al., 2017), which is likely because the NEXT complex can also be targeted to RNA in ZC3H18-independent manners (Giacometti et al., 2017, Lubas et al., 2015). The extent to which RNA levels increase upon ZC3H18 depletion might, in other words, be fully explained by what can be accounted for by diminished CBC-directed RNA turnover. This notion implies that ZC3H18 exercises most of its functions through its connection with the CBCA complex, which is supported by the similar RNA-binding profiles of CBP20, ARS2, and ZC3H18 in vivo (Giacometti et al., 2017).
A tight functional connection between the CBCA complex and ZC3H18 is also illustrated by the second RNA phenotype we uncovered in the present study, namely the downregulation of certain mRNAs upon ZC3H18 depletion. Again, the ZC3H18 variant compromised for CBCA binding fails to rescue this phenotype. The NEXT-binding mutant, however, offers two distinct effects. For some mRNAs, it is as functionally adept as FL ZC3H18, which argues against a degradation/exosome-mediated mechanism. Consistently, RNA exonic and intronic changes caused by ZC3H18 depletion correlate well with the phenotypes caused by CBP80 and ARS2 depletion but less well with RBM7, ZCCHC8, and RRP40 depletion phenotypes. Thus, ZC3H18 is engaged in regulating RNA levels independently on transcript turnover. For another set of mRNAs, expression of the NEXT-binding mutant provides no rescue of the downregulated transcripts. Indeed, some of these mRNAs are also downregulated after ZCCHC8 and RBM7 depletion (data not shown), implying an indirect role of the NEXT complex in the positive regulation of some mRNAs.
What might be the mechanism of ZC3H18 effects on pre-mRNA/mRNA? Based on the associated decline of intronic RNA-seq reads and Pol II ChIP levels in ZC3H18 depletion conditions, it appears that CBCA-ZC3H18 positively affects, either directly or indirectly, transcription of a subset of protein-coding genes in the HeLa cell genome. Apart from a report demonstrating the involvement of ZC3H18 in nuclear factor κB (NF-κB)-activated transcription (Gewurz et al., 2012), such activity has not previously been described. Although at this point any functional model remains speculative, it is an interesting possibility that transcriptional support mediated by ZC3H18 might occur via its early targeting of the nascent RNA 5′ end via CBCA. Such reverse coupling, where RNA metabolic processes exercise an impact on transcription initiation levels, has been described (Damgaard et al., 2008, Manley, 2002) and constitutes a possible line of future investigation.
The protein-coding genes impacted transcriptionally by ZC3H18 depletion include BRCA1 and BRCA2, whose protein products are involved in DNA damage response and repair processes (Roy et al., 2011). For example, the BRCA1 protein forms a DNA damage-induced complex with BCLAF1 and key components of the splicing machinery to regulate RNA processing from key genes involved in DNA damage response and repair (Savage et al., 2014). Interestingly, ZC3H18 is found to be phosphorylated upon various DNA insults (Beli et al., 2012), providing a possible link between ZC3H18 chromatin-related activities and the cellular stress response. Hence, further examination of the repertoire of ZC3H18 target genes and their response to ZC3H18 depletion during stress might aid our physiological understanding of this large protein, functioning at the crossroad between RNA metabolism and transcription.
Experimental Procedures
Cell Lines and Cell Culture
HeLa Kyoto and HEK293 cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Invitrogen). For transient transfection, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer’s recommendations. ZC3H18-3 × F FL (Andersen et al., 2013) and fragments Z1–Z7 were stably expressed in HEK293 Flp-In T-REx cells. Fusion proteins (see the Supplemental Experimental Procedures and Table S6 for description of their construction) were introduced using a modified pcDNA5/FRT/TO vector that contains the protein of interest followed by a C-terminal 3× FLAG tag (Domanski et al., 2012). Expression of fusion proteins was induced by replacing cell growth media with fresh media, containing 2 ng/mL or 5 ng/mL tetracycline (Sigma) for rescue and AC experiments, respectively.
RNAi
Transfections were performed with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions, with siRNAs at a final concentration of 20 nM. Cells were harvested 3 days after transfection. The siRNA sequences are listed in Table S2.
AC Experiments
AC experiments presented in Figures 1 and 2 were performed with cell powder and in Figure 3 with cell pellets. Cryogenic grinding of human cells and α-FLAG AC were performed as previously described (Domanski et al., 2012, LaCava et al., 2016). Interaction screening was performed as previously described (Hakhverdyan et al., 2015). See the Supplemental Experimental Procedures and Table S3 for details.
MS Analysis
All AC/MS experiments were performed in label-free conditions and in duplicate. The Filter Aided Sample Preparation (FASP) protocol (Wiśniewski et al., 2009) was used to prepare the samples for MS analysis. Samples were analyzed with an LTQ Orbitrap Velos instrument (Thermo Scientific). Data acquisition was performed as described (Andersen et al., 2013). See the Supplemental Experimental Procedures for details.
Western Blotting Analysis
See the Supplemental Experimental Procedures for details on western blotting. A list of primary and secondary antibodies can be found in Table S4.
ChIP-qPCR
The ChIP experiment was performed with α-ZC3H18 antibody (Sigma, HPA040847). See the Supplemental Experimental Procedures for details. Primers used for qPCR are listed in Table S5.
Immunolocalization Analysis
Localization of ZC3H18-3xF FL protein and mutants was obtained with α-FLAG antibody (Sigma, F7425) followed by incubation with α-rabbit Alexa Fluor 488 (Life Technologies, A-11008). See the Supplemental Experimental Procedures for details.
RNA Isolation and Reverse Transcription
RNA was purified with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Random hexamers and anchored oligo-dT primers were used for cDNA synthesis. Reverse transcription was performed using SuperscriptIII (Invitrogen) according to the manufacturer’s instructions. qPCR was performed with the primers listed in Table S7. Data were processed with the ddCt method, with normalization to both GAPDH mRNA levels and EGFP siRNA control samples.
Statistical Analysis
Data are shown as means ± SDs. The significance of differences between different groups was evaluated using two-way ANOVA in the GraphPad Prism software. p values < 0.05 were considered to represent significant differences and are denoted with asterisks corresponding to the following p value ranges: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0,0001.
Bioinformatics Analyses
The details of bioinformatics analyses, including annotations used for RNA-seq and ChIP-seq analysis, RNA-seq libraries, RNA-seq differential expression, clustering analysis, and ChIP-seq analysis, can be found in the Supplemental Experimental Procedures.
Published Datasets
The accession numbers for the RNA-seq data reported in this paper are GEO: GSE99059 (siARS2, siZC3H18 and siCBP80, and siEGFP) and GSE84172 (siRRP40, siZCCHC8, siRBM7, and siEGFP). The accession number for the ChIP-seq data reported in this paper is GEO: GSE99344.
Acknowledgments
We thank Evdoxia Karadoulama for initial processing of the RNA-seq data; Dorthe Caroline Riishøj, Muhammad Ahmad Maqbool, and Jean-Christophe Andrau (IGMM, Montpellier, France) for technical assistance; and Søren Lykke-Andersen and Edouard Bertrand for constructive comments on the manuscript. Work in the T.H.J. laboratory was supported by the ERC (grant 339953), the Danish National Research Council, and the Lundbeck Foundation and the Novo Nordisk Foundation. The work was aided by collaboration with the National Center for Dynamic Interactome Research and the National Resource for the Mass Spectrometric Analysis of Biological Macromolecules, supported in part by NIH grant P41 GM109824 and grant P41 GM103314. This paper is subject to the NIH Public Access Policy.
Author Contributions
K.W. performed the experiments presented in Figures 1, 2, 3, and 5. C.I. performed the experiments presented in Figure 4. K.R.M., L.M.H., and J.S.A. performed MS experiments and assisted with data analysis. M.S. performed computational analyses. J.L. and T.H.J. conceived and supervised the project. K.W., M.S., and T.H.J. wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: January 2, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and seven tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.12.037.
Data and Software Availability
The accession number for the protein interactions reported in this paper is IMEx: IM-26173 (http://www.imexconsortium.org, IMEx consortium through IntAct, Orchard et al., 2014).
Supplemental Information
References
- Amanchy R., Periaswamy B., Mathivanan S., Reddy R., Tattikota S.G., Pandey A. A curated compendium of phosphorylation motifs. Nat. Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- Andersen P.R., Domanski M., Kristiansen M.S., Storvall H., Ntini E., Verheggen C., Schein A., Bunkenborg J., Poser I., Hallais M. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 2013;20:1367–1376. doi: 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beli P., Lukashchuk N., Wagner S.A., Weinert B.T., Olsen J.V., Baskcomb L., Mann M., Jackson S.P., Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta S., Richard S. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem. Sci. 2015;40:662–672. doi: 10.1016/j.tibs.2015.08.012. [DOI] [PubMed] [Google Scholar]
- Cheng H., Dufu K., Lee C.S., Hsu J.L., Dias A., Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Chi B., Wang K., Du Y., Gui B., Chang X., Wang L., Fan J., Chen S., Wu X., Li G., Cheng H. A Sub-Element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucleic Acids Res. 2014;42:7305–7318. doi: 10.1093/nar/gku350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard C.K., Kahns S., Lykke-Andersen S., Nielsen A.L., Jensen T.H., Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanski M., Molloy K., Jiang H., Chait B.T., Rout M.P., Jensen T.H., LaCava J. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. Biotechniques. 2012;0:1–6. doi: 10.2144/000113864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidatzis D., Burger L., Florescu M., Stadler M.B. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol. 2016;34:210. doi: 10.1038/nbt0216-210a. [DOI] [PubMed] [Google Scholar]
- Gewurz B.E., Towfic F., Mar J.C., Shinners N.P., Takasaki K., Zhao B., Cahir-McFarland E.D., Quackenbush J., Xavier R.J., Kieff E. Genome-wide siRNA screen for mediators of NF-κB activation. Proc. Natl. Acad. Sci. USA. 2012;109:2467–2472. doi: 10.1073/pnas.1120542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti S., Benbahouche N.E.H., Domanski M., Robert M.C., Meola N., Lubas M., Bukenborg J., Andersen J.S., Schulze W.M., Verheggen C. Mutually Exclusive CBC-Containing Complexes Contribute to RNA Fate. Cell Rep. 2017;18:2635–2650. doi: 10.1016/j.celrep.2017.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K., Kim S., Espinosa J., Bentley D.L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görnemann J., Kotovic K.M., Hujer K., Neugebauer K.M. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Gruber J.J., Olejniczak S.H., Yong J., La Rocca G., Dreyfuss G., Thompson C.B. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol. Cell. 2012;45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakhverdyan Z., Domanski M., Hough L.E., Oroskar A.A., Oroskar A.R., Keegan S., Dilworth D.J., Molloy K.R., Sherman V., Aitchison J.D. Rapid, optimized interactomic screening. Nat. Methods. 2015;12:553–560. doi: 10.1038/nmeth.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallais M., Pontvianne F., Andersen P.R., Clerici M., Lener D., Benbahouche Nel.H., Gostan T., Vandermoere F., Robert M.C., Cusack S. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 2013;20:1358–1366. doi: 10.1038/nsmb.2720. [DOI] [PubMed] [Google Scholar]
- Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva L.M., Radivojac P., Brown C.J., O’Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iasillo C., Schmid M., Yahia Y., Maqbool M.A., Descostes N., Karadoulama E., Bertrand E., Andrau J.C., Jensen T.H. ARS2 is a general suppressor of pervasive transcription. Nucleic Acids Res. 2017;45:10229–10241. doi: 10.1093/nar/gkx647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I.W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Järvelin A.I., Noerenberg M., Davis I., Castello A. The new (dis)order in RNA regulation. Cell Commun. Signal. 2016;14:9. doi: 10.1186/s12964-016-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T.H., Jacquier A., Libri D. Dealing with pervasive transcription. Mol. Cell. 2013;52:473–484. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- Jonkers I., Kwak H., Lis J.T. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S., Ohno M., Mattaj I.W. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 2001;11:497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- LaCava J., Jiang H., Rout M.P. Protein Complex Affinity Capture from Cryomilled Mammalian Cells. J. Vis. Exp. 2016;(118):54518. doi: 10.3791/54518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M., Andersen P.R., Schein A., Dziembowski A., Kudla G., Jensen T.H. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 2015;10:178–192. doi: 10.1016/j.celrep.2014.12.026. [DOI] [PubMed] [Google Scholar]
- Manley J.L. Nuclear coupling: RNA processing reaches back to transcription. Nat. Struct. Biol. 2002;9:790–791. doi: 10.1038/nsb1102-790. [DOI] [PubMed] [Google Scholar]
- Meola N., Domanski M., Karadoulama E., Chen Y., Gentil C., Pultz D., Vitting-Seerup K., Lykke-Andersen S., Andersen J.S., Sandelin A., Jensen T.H. Identification of a Nuclear Exosome Decay Pathway for Processed Transcripts. Mol. Cell. 2016;64:520–533. doi: 10.1016/j.molcel.2016.09.025. [DOI] [PubMed] [Google Scholar]
- Merz C., Urlaub H., Will C.L., Lührmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-McNicoll M., Neugebauer K.M. Good cap/bad cap: how the cap-binding complex determines RNA fate. Nat. Struct. Mol. Biol. 2014;21:9–12. doi: 10.1038/nsmb.2751. [DOI] [PubMed] [Google Scholar]
- Narita T., Yung T.M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Ntini E., Järvelin A.I., Bornholdt J., Chen Y., Boyd M., Jørgensen M., Andersson R., Hoof I., Schein A., Andersen P.R. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- Ohno M., Segref A., Bachi A., Wilm M., Mattaj I.W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabis M., Neufeld N., Steiner M.C., Bojic T., Shav-Tal Y., Neugebauer K.M. The nuclear cap-binding complex interacts with the U4/U6·U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA. 2013;19:1054–1063. doi: 10.1261/rna.037069.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Chun J., Powell S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage K.I., Gorski J.J., Barros E.M., Irwin G.W., Manti L., Powell A.J., Pellagatti A., Lukashchuk N., McCance D.J., McCluggage W.G. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell. 2014;54:445–459. doi: 10.1016/j.molcel.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.