Abstract
The olive fruit fly, Bactrocera oleae, is the single most important pest for the majority of olive plantations. Oxitec’s self-limiting olive fly technology (OX3097D-Bol) offers an alternative management approach to this insect pest. Because of previously reported asynchrony in the mating time of wild and laboratory strains, we have characterized the olive fly circadian clock applying molecular, evolutionary, anatomical and behavioural approaches. Here we demonstrate that the olive fly clock relies on a Drosophila melanogaster-like organization and that OX3097D-Bol carries a functional clock similar to wild-type strains, confirming its suitability for operational use.
Introduction
The fruit fly Bactrocera oleae (Rossi) (Diptera: Tephritidae) is a major pest of cultivated olives. Its presence has been historically reported in Mediterranean and African countries and, more recently, it has spread to Central America and California1. The adult female fly lays approximately 800 eggs during its lifetime, through the skin of olive fruits. The hatching larvae feed while tunneling in the fruit mesocarp causing crop damage and premature drop. Multiple and overlapping generations every year cause enormous loss to olive agriculture and the economy of olive products2. Current control methods against olive fly rely overwhelmingly on the use of chemical insecticides. Several insecticides have been, or are being, phased out due to concerns about their negative impact on the environment or human health. This reduces control options and increases the rate at which the olive fly becomes resistant to the remaining insecticides3.
The Sterile Insect Technique (SIT) is a targeted method of pest control that involves the mass rearing and release of sterilized insects (traditionally by irradiation) into the wild. The introduction of an excess of sterile insects reduces the reproductive potential of the target population through infertile matings, leading to suppression4–6. Due to its high economic burden and the over-use of chemical control, the olive fly was among the first insects to be considered for SIT. However, previous attempts using irradiated mixed-sex insects achieved very limited success, due to the poor quality of the irradiated insects and mating asynchrony between the laboratory-reared and wild olive flies7–9. “Self-limiting” engineered olive fly strains can unlock the SIT potential for the control of this economically important agricultural pest10.
The success of SIT, whether traditional or using novel technologies, is largely driven by the mating behaviour of the insects of concern. The mass-released males must exhibit the same mating behaviour as wild males, or they may be unsuccessful in gaining females11. In most Bactrocera species mating occurs at a species-specific time window during the day, most commonly associated with dusk and less frequently midday12, at which time male sexual activity has to be synchronous with female receptivity13. In Bactrocera, this timing of mating behaviour is known to be modulated by the circadian clock14,15, as it is in Drosophila16. It has been shown that in Bactrocera this can be very sensitive to insect colony adaptation in laboratory conditions17.
Yet, very little is known about the mechanisms of the circadian clock in Bactrocera species (Tephritidae); in contrast to the fruit fly, Drosophila melanogaster (Drosophilids), that has been widely adopted as the primary insect model. D. melanogaster possesses a molecular clock which relies on the interaction between several clock genes and their respective products. Two main transcription/translation feedback loops (TTFLs) drive single cell molecular oscillations which are at the basis of circadian physiology and behaviour18. A first loop involves the clock genes period (per), timeless (tim), Clock (Clk) and cycle (cyc). A second loop involves the clock genes Par-domain-protein-ε (Pdp1ε) and vrille (vri) that interlock with the first loop regulating the expression of Clk. Under light-dark cycles, the molecular oscillation is reset every day via the blue-light photoreceptor cryptochrome (CRY)19,20.
The master pacemaker in Drosophila is located in the Central Nervous System (CNS), where around 75 clock neurons per brain hemisphere drive synchronous oscillations in seven distinct neural cluster (s-LNv, l-LNv, LNd, LPN, DN1, DN2, DN3). The clock network uses neuropeptides like Pigment Dispersing Factor (PDF)21,22 and Ion Transport Peptide (ITP)23,24 to coordinate the oscillations in the different clusters. This complex network supports rhythmicity in locomotor activity under cycles of alternated light and dark (LD) and constant darkness (DD), whereas it is lost under constant light (LL)25.
Circadian rhythms in B. oleae were previously described for several behaviours (i.e. mating26, exodus of larvae from diet27,28 and pheromone emission29). Here we characterize the circadian clock of B. oleae at molecular, evolutionary, anatomical, and behavioural levels, and we show that the olive fly carries a Drosophila-like clockwork in all of these aspects. Furthermore, we demonstrate that the self-limiting olive fly strain OX3097D-Bol possesses a functional circadian clock that does not differ from wild-type genotypes.
Results
Identification of B. oleae clock genes
We identified the whole length transcript sequences of the clock genes per, cyc and cry, as well as partial sequence of Clk (Table 1).
Table 1.
Clock genes identified from Bactrocera oleae (and see Fig. 1).
| Gene | Length | Putative protein coding and other details |
|---|---|---|
| per CDS | 3105 bp | A protein of 1034 aa residues, containing two Nuclear Localization Signals (NLS, aa 72 to 78 and 691 to 718), two PER-ARNT-SIM domains (PAS-A, aa 193 to 242; PAS-B, aa 343 to 395) and a Cytoplasmic Localization Domain (CLD, aa 398 to 457). There is only a single Threonine-Glycine tandem repeat (T-G) in the B. oleae PER sequence (Supplementary Fig. S1) |
| cyc CDS | 1206 bp | A 401 aa polypeptide containing a basic Helix-Loop-Helix motif (bHLH, aa 22 to 74) followed by PAS-A (aa 114 to 157) and PAS-B (aa 306 to 353) domains (Supplementary Fig. S2) |
| cry CDS | 1644 bp | A 547 aa protein, in which the DNA photolyase activity domain (aa 4 to 195) and the FAD binding domain (aa 213 to 516) contains the conserved putative interaction sites for FAD (Flavin Adenine Dinucleotide; 13/14 identity), MTHF (methenyltetrahydrofolate; 7/7 identity) and CPD (cyclobutane pyrimidine dimers; 14/14 identity) (Supplementary Fig. S3) |
| Clk (5′Clk CDS) | 990 bp | A 330 aa truncated peptide that contains most of the functional domains, such as bHLH (aa 13 to 58) and PAS (PAS-A, aa 92 to 140; PAS-B, aa 252 to 297), and has high sequence homology to Clk of D. melanogaster (Supplementary Fig. S4). |
When aligned with their respective Drosophila and Bactrocera counterparts, the predicted amino acid sequences show high identity levels (Supplementary Figs S1–S4), particularly within the characterized functional domains (Fig. 1). However, other Diptera such as mosquitoes and sandflies (Nematocera) carry the mammalian form of cyc, cry and Clk (see Discussion). The overall high similarity between Bactrocera and Drosophila clock genes, especially for cyc, cry and Clk, suggests that the molecular clock of the two genera is very similar, but clearly different from the clock of other insects (Fig. 2).
Figure 1.
Schematic representation of the predicted structure of B. oleae PER, CLK, CYC and CRY. B. oleae sequences (top) are compared to their respective D. melanogaster counterparts (bottom). Coloured parts are the functional domains for which the percentage of identity between the two sequences is indicated. Rectangle length corresponds to sequence length; the entire length of the proteins is indicated at the right end. An identification key for the different domains is present in the figure. Multiple sequence alignment and a more detailed overview of proteins structure can be found in Supplementary Figures S1–S4.
Figure 2.
Phylogenetic analysis of B. oleae PER and CYC. Gene trees of (A) CYC and (B) PER reconstructed from amino acids with Maximum Likelihood using RaxML. Values at the nodes represent bootstrap values, determined with 1000 replicates.
Our phylogenetic analysis also suggests that the evolutionary pace of substitutions for the Brachycera branch was by far higher than expected regarding the remaining tree for the cyc gene (χ2 = 73.24, df = 1, p < 0.001), and slightly for per (χ2 = 5.01, df = 1, p < 0.05). This supports the notion of dramatic changes in the evolutionary history of the clock genes in this group in comparison to other insects.
Temporal expression of clock genes in B. oleae heads
In order to see whether the TTFLs of B. oleae run similar to those of D. melanogaster, we analysed the temporal expression of per, Clk, cyc and cry under light-dark cycles of 12 h:12 h (LD12:12). As reported previously for D. melanogaster30,31, we detected strong and significant circadian oscillations in per and Clk mRNA levels that were in anti-phase to each other, with their respective peaks at Zeitgeber time (ZT) 12 and ZT3 and troughs at ZT0 and ZT12 (Fig. 3). In contrast, the clock gene cyc was constantly expressed as shown in D. melanogaster32. Furthermore, similar to D. melanogaster19, we observed weak circadian oscillations in cry mRNA levels of B. oleae, with a peak around ZT6 and a trough around ZT18 (Fig. 3). For comparison, cyc is known to oscillate in anti-phase to per in other insects and mammals, whereas Clk is constantly expressed (see Discussion). Taken together, our results show that the regulation of the expression of B. oleae clock genes relies on mechanisms common to the D. melanogaster TTFLs.
Figure 3.
Temporal expression of the clock genes per, Clk, cyc and cry in B. oleae heads. In Argov male flies, per and Clk mRNA levels are strongly rhythmic and in antiphase to each other (per: H(7) = 20.9, p = 0.003; CircWave, p < 0.001 – Clk: H(7) = 21.2, p = 0.003; CircWave, p < 0.001). The gene cyc is constantly expressed (H(7) = 4.1, p = 0.771; CircWave, p > 0.05). cry mRNA shows weak oscillation (H(7) = 11.6, p = 0.115; CircWave, p < 0.001).
Most importantly, when the expression levels of these genes were compared among wild-type and genetically engineered strains (Fig. 4), we found no significant differences in phases or in amplitude (Supplementary Table S1), demonstrating that the TTFLs of the strain OX3097D-Bol do not differ from the wild-type strains in their regulation of clock gene expression.
Figure 4.
Comparison of clock genes expression between wild-type and transgenic B. oleae. Oscillations in the expression of per, Clk, cyc and cry under LD12:12 was investigated in three different genotypes: Demokritos, Argov (wild-type) and OX3097D-Bol. No significant interaction among timepoint and genotype was found using 2-way ANOVA (all p-values > 0.05, see Supplemetal Table 1), indicating no differences in amplitudes nor phases among strains.
The clock network in B. oleae brain
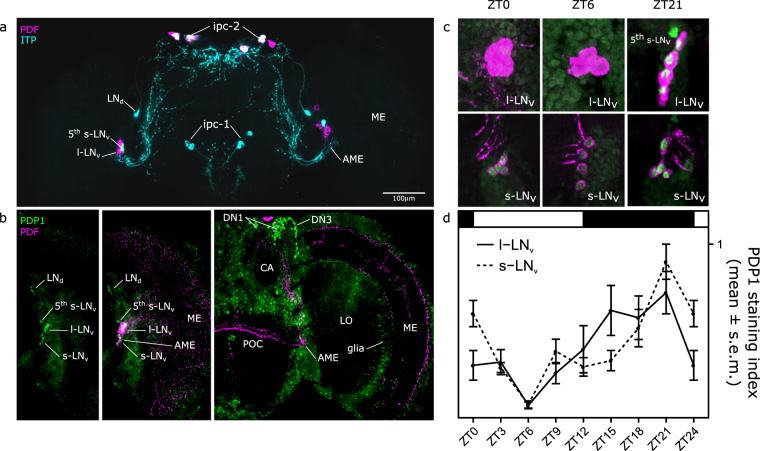
We first characterized the neuroarchitecture of the B. oleae clock network by investigating the expression pattern of the clock neuropeptides PDF, ITP and the core clock protein PDP1 (Fig. 5a–c). We identified four small (s-LNv) and four large (l-LNv) ventro-Lateral-Neurons that express PDF, as well as four neurosecretory cells (putative IPCs, Insulin-Producing-Cells; we call them ipc-2 cells according to the naming in D. melanogaster) in the pars lateralis (PL).
Figure 5.
The clock network of B. oleae. (a) The clock neuropeptide PDF (magenta) is expressed in 4 l-LNv and 4 s-LNv, as well as in 4 putative insulin producing cells (ipc-2) in the PL. ITP (cyan) is expressed in the 5th s-LNv and in one cell of the LNd group, as well as in putative ipc-1 and ipc-2. (b) anti-PDP1 (green) co-localize in the nuclei of PDF positive cells (magenta) in the LNv cluster. Antibody reveals also other putative clock clusters (LNd, DN), and stains many other non-clock cells. (c,d) PDP1 protein level in the nuclei oscillates under LD12:12 in the s-LNv (H(7) = 34.78, p < 0.001) and l-LNv (H(7) = 19.259, p < 0.05). AME: accessory medulla; ME: medulla; CA: calyx; LO: lobula.
The 4 s-LNv project to the dorsal protocerebrum, whereas the 4 l-LNv innervate the medulla and project to the contralateral hemisphere via the posterior optic commissure (POC) (Fig. 5a,b). ITP is expressed in the 5th s-LNv and in one cell of the dorsal Lateral Neurons (LNd), as well as in three of the four PDF positive ipc-2 cells in the PL. ITP is additionally stained in three other putative IPC cells (ipc-1 as they are named in D. melanogaster) in the central brain. When we perfomed double-staining using anti-PDF and anti-PDP1 antibodies together, cytoplasmic PDF signal co-localized with nuclear PDP1 signal in both small and large clusters, confirming the homology with D. melanogaster lateral ventral clock neurons (Fig. 5c). In addition, anti-PDP1 antibody labeled also the other clock clusters (DN1, DN3, LNd) that were located in a similar position as the homologous neurons in different Drosophila species33. In order to see whether PDP1 oscillates in its abundance, we analysed the staining intensity of PDP1 in the LNv (co-labelled with PDF) at several timepoints across the LD12:12 cycle. We found synchronous oscillations in the levels of PDP1 in both s-LNvs (H(7) = 34.78, p < 0.001) and l-LNvs (H(7) = 19.26, p < 0.05) (Fig. 5d), reaching their maxima and minima at ZT21 and ZT6, respectively. These oscillations are similar to the oscillations of PDP1ε described in D. melanogaster34.
Locomotor activity rhythm of B. oleae
In order to determine the endogenous clock properties of wild-type and self-limiting strains, we recorded locomotor activity as a readout of the circadian pacemaker activity. In general, activity recording worked well in olive flies, although they showed a high mortality rate when isolated into the glass tubes (~50% of the flies died within the first week). Furthermore, olive flies showed low activity levels (on average 17 beam crosses/hours, Supplementary Fig. S5). Under LD12:12 cycles, all flies were diurnal and more or less uniformly active throughout the day (Fig. 6). Only OX3097D-Bol males were significantly more active (on average 39 beam crosses/hours; H(5) = 56.17, p < 0.001; Supplementary Fig. S5) and exhibited bimodal activity, with more activity in the morning and evening as described for D. melanogaster (Fig. 6). The amount of activity associated with the evening was significantly different among strains (F(5) = 6.8, p < 0.001), with OX3097D-Bol males showing the highest level (Supplementary Fig. 6).
Figure 6.
Locomotor activity of B. oleae under different conditions. Daily activity profile under LD12:12 (top) and one representative actogram, respectively under DD (middle) and LL (bottom) of males (right) and females (left) of Demokritos, Argov and OX3097D-Bol at 20 °C. Horizontal bars aside the actograms separate LD and DD/LL phases.
After transfer in DD, the majority of flies showed self-sustained rhythmicity in their locomotor activity, with a free-running period shorter than 24 hours in all genotypes tested (Figs 6 and 7 and Table 2). No significant differences were found in the percentage of rhythmic flies (Table 2; χ2 = 4.8, df = 5, p = 0.44) and in the free-running period (Fig. 7, H(5) = 10.92, p = 0.053) between the different strains and sexes. When flies were released in constant light (LL), their locomotor activity became arrhythmic as known to happen in D. melanogaster (Fig. 6 and Table 2).
Figure 7.
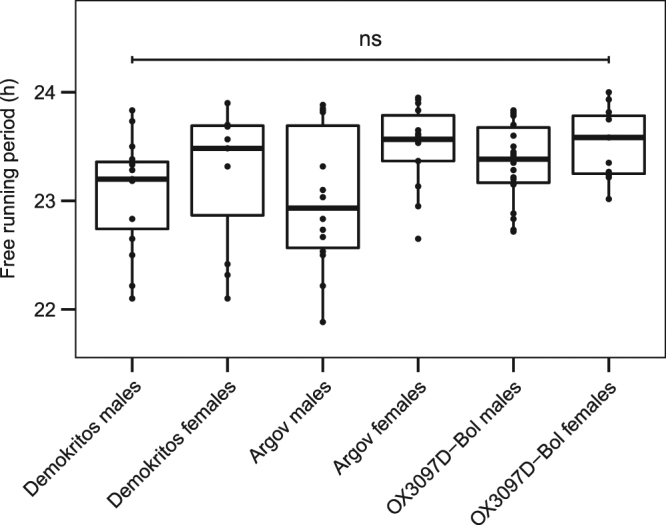
Average free running periods of the different strains of B. oleae. No significant differences in the free-running periods of males and females of Demokritos, Argov and OX3097D-Bol strains are found under DD (H(5) = 10.92, p > 0.05).
Table 2.
Free running period and rhythmicity percentage of olive flies under DD and LL.
| genotype | gender | DD | LL | |
|---|---|---|---|---|
| % rhythmic flies (N) | period ± s.e.m. (h) | % rhythmic flies (N) | ||
| Demokritos | M | 72.7 (22) | 22.99 ± 0.15 | 0 (16) |
| F | 91.7 (12) | 23.24 ± 0.19 | 0 (26) | |
| Argov | M | 73.7 (19) | 23.01 ± 0.17 | 0 (15) |
| F | 93.3 (15) | 23.49 ± 0.1 | 0 (12) | |
| OX3097D-Bol | M | 78.6 (28) | 23.36 ± 0.08 | 0 (14) |
| F | 68.8 (16) | 23.54 ± 0.1 | 0 (20) | |
Taken together, these results demonstrate that locomotor activity of B. oleae is clock-controlled and that the fundamental clock properties in the olive fly are common with D. melanogaster.
Discussion
The molecular clock of B. oleae
Although the fruit fly Drosophila melanogaster has been of exceptional importance in understanding how the molecular clock in an insect model ticks, studies in other insects with partially annotated genomes highlight differences in the underlying molecular mechanisms. Striking differences in the structure and expression of some clock genes are already evident among flies (order Diptera).
For example, Nematocera (lower Diptera, such as mosquitoes or sandflies) have been shown to carry the mammalian form of the gene cyc35, also called Bmal1 (Brain and muscle ARNT-like). This alternative form encodes a peptide that carries an extra domain at the C-terminus (BCTR, BMAL C-terminus region) working as transactivation domain required for binding and activating transcription of the per gene. Most probably because of this, cyc is rhythmically expressed in mosquitoes35,36, whereas in Drosophila and other higher Diptera (=Brachycera) it is constantly expressed32,37,38. Here we report that B. oleae cyc shares the same features as cyc in other Brachycera flies.
In higher Diptera, the CLK protein possesses a transactivation domain (multiple poly-Q sites at the C-terminus) and is responsible of activating per transcription and oscillates in abundance39. The CLK protein of lower Diptera (mosquitoes and sandflies) is considerably shorter than in higher Diptera and neither oscillates in abundance nor binds and activates per35,36. Since we could only isolate the 5′Clk CDS of Bactrocera, we do not know whether Clk possesses the poly-Q sites typical for D. melanogaster Clk. Nevertheless, we found that Clk mRNA cycles in abundance, making it very likely that the transactivation domain is present.
Most likely, the CYC and CLK found in lower Diptera are the ancestral forms that are conserved in most insects and mammals. During evolution, the higher Diptera (Brachycera) seem to have lost the BCTR activation domains at the C-terminus of CYC but gained the poly-Q repeats at the C-terminus of CLK (see38 for discussion). Bactrocera oleae, belonging to the higher Diptera (Brachycera; family Tephritidae), fits perfectly into this picture.
Like cyc and Clk, the gene cryptochrome is also found in two distinct variants in Diptera: cry1 and cry2. Cry1 is the only cryptochrome present in Drosophila; it is light sensitive and acts as the main circadian photoreceptor in Drosophila’s master clock19,20. In contrast, cry2 is the mammalian form of cryptochrome; it is not light-sensitive, and it is instead involved in the core feedback loop40,41. Nematocera, for example mosquitoes, express both types of cryptochrome. In B. oleae we have identified cry1, but not cry2: the sequences annotated in GenBank as Bactrocera cry2 are phylogenetically closer to photolyases rather than to the mammalian-like cryptochrome (see Supplementary Fig. S7). A similar situation occurred already in Musca domestica, where Bazalová and Dolezel42 found that the gene previously annotated as cry2 actually belongs to photolyases. Most importantly, B. oleae cry1 carries all the domains needed for binding the chromophores flavin and MTHF, indicating that it is likely to work as a circadian photoreceptor. This feature is also confirmed by the arrhythmic activity of Bactrocera flies under LL. In addition, our findings fit in with the results obtained from other Bactrocera species, such as B. tryoni, B. neohumeralis and Z. curcubitae43,44.
The per gene has been characterized in many insect species including some tephritid fruit flies (Bactrocera and Ceratitis species)45–48. In all these species, per carries the same structure and functional motifs and exhibits daily rhythmic expression as in D. melanogaster. Here we confirmed that also B. oleae per shares the same features.
In other insect orders, such as the Lepidoptera and Hymenoptera for example, the clock scenario gets more complex. Nevertheless, all investigated species so far express the mammalian form of CYC, CLK and CRY40,49–51. In contrast, the overall picture shows that B. oleae carries clock components that are of the Drosophila type (perhaps better Brachycera type), and their expression under light-dark cycles is highly coherent with the respective Drosophila profiles. Our phylogenetic analysis on B. oleae PER and CYC highlights how the clock of Brachycera species might be exceptional among insects, and that a molecular clockwork different from the one of Drosophila might be the most common model among Hexapoda.
The clock network in the brain
At the anatomical level, several studies have shown that the distribution of certain neuropeptides in the clock network are quite different within closely related species in the Brachycera. For example, Drosophila species that inhabit cold habitats (high latitudes) differ in their PDF expression pattern from D. melanogaster: They lack PDF in the s-LNv33,52,53. Here we show that the olive fly, like D. melanogaster, expresses PDF in both s-LNv and l-LNv. Additionally, we demonstrate that the expression of the other central clock neuropeptide, ITP23,24, is highly conserved.
Anti-PDP1 antibody was previously used to reveal the clock neurons in several Drosophila species33. It worked also in B. oleae and stained neurons in the lateral and dorsal brain that appear homologous to those seen in all Drosophila species tested so far. Furthermore, PDP1 cycled in a circadian fashion in the s-LNv and l-LNv as reported for D. melanogaster34.
We conclude that the clock network in the brain of B. oleae is highly similar to that of D. melanogaster; even more similar than the clock network of Drosophila flies from higher latitudes. B. oleae is prevalent in warm habitats mostly around the Mediterranean area. This underscores our hypothesis that changes in the clock network preferentially happened in species that radiated to the north, but not in flies that remained in subtropical regions53. We are not aware of any study performed on the clock network in the brain of lower Diptera. To further prove this hypothesis, it would be most interesting to investigate the PDF-pattern in lower Diptera, such as mosquitoes, of which species are found at all latitudes ranging from tropical regions to the very north or south.
Behavioural rhythms of B. oleae
Despite the high mortality rate and low activity of the isolated flies, we could clearly show that B. oleae locomotor activity is under the control of the circadian clock: rhythmic locomotor activity pattern persisted in the absence of all Zeitgebers under constant darkness (DD). We suspect that the relative low activity level of olive flies might be due to their natural life style. In the wild, Bactrocera flies (especially males) spend much of the day resting in the canopy of trees54, with resource foraging and mating behaviours at temporally distinct times55. Laboratory mass rearing is known to further decrease the tendency of flies to move56. The lack of natural environmental stimuli in the glass tubes may also lead to the observed low activity. Nevertheless, the activity level of the majority of flies was high enough to reveal a circadian rhythm in locomotor activity under DD conditions. This is consistent with our finding that PDF is present in the s-LNv. The latter has been shown to be essential for robust activity rhythms under constant darkness53. Fly species lacking PDF in the s-LNv show arrhythmic activity under DD conditions52,53,57,58. In contrast to DD conditions, B. oleae flies were arrhythmic under constant light. The same is observed in D. melanogaster and can be explained by the permanent degradation of the clock protein TIM via the light-sensitive CRY159. The same model seems to apply to B. oleae.
Taken together, our results clearly depict, from three different points of view (molecular, anatomical and behavioural), that the general organization of the B. oleae circadian clock highly matches the well known clock of D. melanogaster. For this reason, we propose that the molecular basis of the olive fly clock relies on a Drosophila-like mechanism.
Impact of our study on the application of SIT method against B. oleae
One relevant aspect of our study was the investigation of the self-limiting strain, OX3097D-Bol, which offers an alternative olive fly pest control strategy to chemical interventions. Despite the continued broad use of pesticides to keep insect pest populations under control, reduced or eliminated pesticide use is getting the consensus of growers and consumers as the more sustainable way to manage pest species in agriculture. Among these, SIT is already applied in modern agriculture to fight several Tephritidae species such as the Mediterranean fruit fly (Ceratitis capitata), the Oriental fruit fly (Bactrocera dorsalis), the Melon fly (Bactrocera cucurbitae) and the Mexican fruit fly (Anastrepha ludens)60–63. Unexpectedly, this method has proven to be inefficient in the control of B. oleae in the past. One of the reasons suggested as a cause for the low success was the mating asynchrony reported between wild and laboratory-reared individuals8,9. In these studies, the possible role of the circadian clock was not taken into account, and it was for this reason that we examined the circadian clock of the self-limiting olive fly strain OX3097D-Bol. We now clearly demonstrate that OX3097D-Bol expresses the clock genes in the same temporal manner as wild-type flies. Additionally, our behavioural studies show that the endogenous period of this strain does not differ from the wild-type genotypes. Together, this supports the fact that the circadian clock of OX3097D-Bol is functional and unaltered from the wild-type strains.
Interestingly, OX3097D-Bol showed higher overall locomotor activity, especially in the evening, compared to wild-type flies. These increased evening activity levels overlap with the timing of mating, which is known to occur in the hours prior to scotophase both under natural and artificial conditions26. Whether this may be advantageous to future operational applications is not investigated here. However, in other Bactrocera species, increased male activity at time of mating has been directly linked to increased male mating success64. We thus speculate that increased locomotor activity of OX3097D-Bol males during the time window of female receptivity may increase mating success. In conclusion, our results suggest that the clock structure and expression remain intact in the self-limiting olive fly strain OX3097D-Bol, and together with previously published data10 we conclude that sustained release of OX3097D-Bol males can be a viable stategy for olive fly pest control.
Material and Methods
Fly strains and husbandry
The olive fly strains considered in this article are the following: the wild-type strains Argov65 and Demokritos66 and the self-limiting strain OX3097D-Bol10. The self-limiting strain of B. oleae, OX3097D-Bol, incorporates a tetracycline-off system designed to produce a conditional female-specific self-limiting trait when reared in the absence of a sufficient concentration of tetracycline. The OX3097D-Bol also expresses a fluorescent marker gene to enable detection and monitoring in operational deployment. Flies were reared on standard olive fly larval and adult diet and maintained under cycles of 12 hours of light followed by 12 hours of darkness (LD12:12) at 23°C (±2 °C) and 50% humidity (±10%)10.
Cloning of clock genes
Wild-type flies were collected in dry ice, and RNA was isolated from heads using the Total RNA Purification Kit (Norgen Biotek). cDNA was obtained by reverse transcription using oligo-dT primers and the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). A PCR-based strategy using degenerate oligonucleotide primers designed over highly conserved regions of reported Drosophila and Bactrocera clock genes was used to isolate the olive fly homologous of per, cry, Clk and cyc (see primer sequences in SupplementaryTable S2). The same strategy was initially used for the isolation of timeless, which has also been annotated in the genome of B. oleae (XM_014247668.1). Nevertheless, for technical reasons we decided not to pursue the charachterization of this gene any further. 3′ and 5′ RACE PCRs were used to obtain the full-length cDNA sequence (SMARTer RACE cDNA Amplification Kit, Clontech Laboratories). PCRs were performed in a T3000 Thermal Cycler (Biometra) using Q5 Hot Start Polymerase (NEB). PCR products were purified either directly from the reaction (QIAquick PCR Purification Kit, QIAGEN) or from agarose gel (QIAquick Gel Extraction Kit, QIAGEN), and cloned into pJet1.2/blunt vector (Thermo Fisher Scientific). Positive clones were identified by colony PCR screening and purified plasmids (GeneJET Plasmid Miniprep Kit, Thermo Fisher Scientific) were sent to sequencing (GATC Biotech).
Phylogenetic analysis
A phylogenetic analysis was conducted for each of the genes of interest, to infer information about the evolutionary background of the genes. For this, sequences of cyc and per were respectively aligned using ClustalOmega67 with corresponding reference sequences obtained from GenBank68 from other insect taxa, and as the outgroup for the analysis human and mouse (GenBank identifiers in Supplementary Table S3). Only homologous parts of the sequences in at least 50% of the sequences were kept, the rest masked out using GBlocks69 to avoid too much divergency for phylogenetic interference. This clean alignment for each of the genes was then used for a phylogenetic reconstruction using RaxML v8.0.070 with 100 bootstraps and the BLOSUM62 substitution matrix. To test whether the Brachycera branch of each phylogeny corresponded to the overall substitution rates of the corresponding gene tree or evolved with a different evolutionary speed, PAML71 was used (mammals excluded). For this we estimated the log likelihood (lnL) of the null-model with a fixed rate over all branches and one model with a released rate for the Brachycera branch for each gene tree. The differences between both models were compared with χ2 = 2* (lnLnullmodel − lnLmodel) and the statistical significance inferred with a χ2 table.
Real time PCR and analysis
From the late pupal stage onwards, flies were entrained under LD12:12 (23 °C, 60% humidity) and 3 to 5 days individuals were sampled separately. Total RNA was isolated from heads (4 heads/sample) and cDNA generated as described in the previous paragraph. Experiments were carried out using a thermal cycler (MX3005P, Stratagene) and reverse transcription products were detected using a DNA-binding fluorescent dye (SYBR Green PCR Master Mix, Applied Biosystems) following the manufacturer’s protocol. Thermal cycling consisted of 10 minutes incubation at 95 °C, followed by 40 cycles of denaturation (30 seconds at 95 °C), annealing (1 minute at 60 °C) and extension (1 minute at 72°C) followed by melting curves (55 °C–95 °C). For each time series, reactions were set up in duplicates on one 96-well plate, including negative controls (-cDNA) for each gene. Relative clock gene mRNA abundance was normalized against the constantly expressed reference gene 17 S rRNA transcript and analyzed using the ddCt method.
Immunocytochemistry and staining quantification
Flies were sampled in 4% paraformaldehyde (PFA) in Phosphate Buffer Saline with the addition of Triton-X100 at the concentration of 0.1% (PBST) and fixed for 4 hours at room temperature. PFA was removed by several washes of PBS, and right afterwards brains were manually dissected. Blocking solution (5% Normal Goat Serum (NGS) in PBST) was applied for 4 hours at room temperature and afterwards samples were transferred in primary antibodies solution for at least one day at room temperature. Primary antibodies were diluted in PBST with 5% NGS and 0.02% NaN3. Those used in this work are the following: mouse anti-PDF c7 (1:500, DHSB), rabbit anti-PDF-cricket (1:150072); rabbit anti-ITP (1:1000073); rabbit anti-PDP1 (1:100074). After incubation, primary antibodies were removed from the tissue by washes with PBST and secondary antibodies (anti-mouse or anti-rabbit, Alexa Fluor 488, Alexa 555 or Alexa Fluor 635 conjugated; 1:400 in PBST with 5% NGS) applied overnight at 4 °C. Finally, samples were washed with PBS and mounted on slides using Vectashield medium (Vector Laboratories). For the double labelling with anti-PDP1 and anti-PDF antibodies (Fig. 4), which are both raised in rabbit, the full staining procedure was performed twice. The first anti-PDP1 staining (labelled with anti-rabbit Alexa Fluor 488) was fixed a second time and the protocol repeated using anti-PDF-cricket antibody (labelled with anti-rabbit Alexa Fluor 635). Confocal images were acquired every 2 μm Z-stacks using a Leica TCS SPE (Leica). The quantification of PDP1 levels was performed measuring the staining intensity of PDP1 in the nuclear region of PDF positive cells using the software FIJI75. Thereafter, background fluorescence was substracted from the mean intensity of each cluster and final values were averaged among hemispheres.
Locomotor activity recording and analysis
Adult flies (5 to 10 days after eclosion) were placed in single locomotor activity tubes (10 cm length, 1 cm diameter) where diet was supplied on one side as 4% saccharose and 2% agarose in water. Locomotor activity was recorded using a custom monitoring device (TriKinetics Inc) as the number of times that an infrared light beam crossing the middle of the tube was interrupted by a passing fly76. Monitors were placed in incubators (Panasonic, MIR-154-PE) where temperature was kept constant at 20 °C and flies were entrained to artificial square cycles of 12 hours of light and 12 hours of dark (LD12:12, approximately 200 lux). Constant dark and constant light was applied after entrainment at the same constant temperature.
Activity was recorded every minute; later the dataset was binned in 15 minute-intervals to get rid of noisy and erratic activity bouts in recordings. The daily profile of locomotor activity has been calculated on manually selected days for each single flies.
The total amount of daily activity was calculated as the sum of beam crosses counted during the light phase in each single fly and averaged across all insects recorded under the same condition. The relative amount of evening activity was calculated using the same procedure considering only the last part of the day (from ZT9 to ZT12) and dividing it by the total activity.
Representative double-plotted actograms in this paper show the last four days of entrainment followed by 10 days of constant darkness (DD) or constant light (LL).
The free-running period in DD has been determined on selected consecutive days for each single individual using the Lomb-Scargle periodogram implemented in the ImageJ plugin ActoJ77. Flies were considered rhythmic judged on both visual inspection and significance of the Lomb-Scargle periodogram (p < 0.05).
Statistical analysis
Statistical analysis was performed using R software. Normal distribution of samples was tested using the Shapiro test. Equality of variances among samples was tested using Levene’s test. According to the outcome, 1-way ANOVA, 2-way ANOVA or Kruskal-Wallis H tests was applied. Pairwise comparisons between group levels were corrected for multiple testing using the Bonferroni method. To detect cycling in the daily gene expression profiles, we additionally used the software CircWave78.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on request.
Electronic supplementary material
Acknowledgements
E.B. and C.K. were funded by the INsecTIME (FP7- PEOPLE-2012-ITN, grant no. 316790) and E.B. additionally by the CRC 1047 “Insect timing” (project A1). Authors would like to thank Thea Marubbi and Nuria Morales Puerto for help with husbandry of flies and immunocytochemistry; Justin Blau, Heinrich Dircksen, and Kenji Tomioka for antibodies; Anthony Clarke and David Dolezel for comments on the manuscript.
Author Contributions
E.B. performed the behavioural experiments, identified the clock network in the brain by immunocytochemical stainings, analysed the majority of experiments, compiled the figures and wrote the second draft of the manuscript; C.K. identified the clock genes of B. oleae, studied the temporal expression of clock genes in B. oleae heads and wrote the first draft of the manuscript; P.M. helped with stainings and partly supervised E.B.; A.K. performed the phylogenetic analysis of the clock genes together with E.B., M.K. supervised C.K. and provided funding; C.H.F. supervised E.B., helped with the histological analysis and the figure and provided funding. All authors participated in the study design and worked on the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Enrico Bertolini and Christa Kistenpfennig contributed equally to this work.
A correction to this article is available online at https://doi.org/10.1038/s41598-018-24850-w.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19255-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/30/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
- 1.Daane KM, Johnson MW. Olive fruit fly: managing an ancient pest in modern times. Annu. Rev. Entomol. 2010;55:151–169. doi: 10.1146/annurev.ento.54.110807.090553. [DOI] [PubMed] [Google Scholar]
- 2.Tzanakakis ME. Seasonal development and dormancy of insects and mites feeding on olive: a review. Netherlands J. Zool. 2003;52:87–224. doi: 10.1163/156854203764817670. [DOI] [Google Scholar]
- 3.Vontas JG, et al. Resistance-associated point mutations of organophosphate insensitive acetylcholinesterase, in the olive fruit fly Bactrocera oleae. Insect Mol. Biol. 2002;11:329–336. doi: 10.1046/j.1365-2583.2002.00343.x. [DOI] [PubMed] [Google Scholar]
- 4.Alphey, L. S. Engineering insects for the sterile insect technique. Area-Wide Control Insect Pests From Res. to F. Implement. 51–60 (2007).
- 5.Hendrichs JP. Use of the sterile insect technique against key insect pests. Sustain. Dev. Int. 2000;2:75–79. [Google Scholar]
- 6.Vreysen M, Robinson A. Ionising radiation and area-wide management of insect pests to promote sustainable agriculture. A review. Agron. Sustain. Dev. 2011;31:233–250. doi: 10.1051/agro/2010009. [DOI] [Google Scholar]
- 7.Economopoulos A. Sexual competitiveness of gamma-ray sterilized males of Dacus oleae. Mating frequency of artificially reared and wild females. Environ. Entomol. 1972;1:490–497. doi: 10.1093/ee/1.4.490. [DOI] [Google Scholar]
- 8.Zervas GA, Economopoulos A. Mating frequency in caged populations of wild and artificially reared (normal or gamma-sterilized) olive fruit flies. Environ. Entomol. 1982;11:17–20. doi: 10.1093/ee/11.1.17. [DOI] [Google Scholar]
- 9.Economopoulos, A. & Zervas, G. A. The quality problem in olive flies produced for SIT experiments. in IAEA-SM-255/39 357–368 (1982).
- 10.Ant T, et al. Control of the olive fruit fly using genetics-enhanced sterile insect technique. BMC Biol. 2012;10:1–8. doi: 10.1186/1741-7007-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shelly TE, Whittier TS, Kaneshiro KY. Sterile insect release and the natural mating system of the mediterranean fruit fly, Ceratitis capitata (Diptera: Tephritidae) Ann. Entomol. Soc. Am. 1994;87:470–481. doi: 10.1093/aesa/87.4.470. [DOI] [Google Scholar]
- 12.Fletcher BS. The biology of Dacine fruit flies. Annu. Rev. Entomol. 1987;32:115–144. doi: 10.1146/annurev.en.32.010187.000555. [DOI] [Google Scholar]
- 13.Ekanayake WMTD, Jayasundara MSH, Peek T, Clarke AR, Schutze MK. The mating system of the true fruit fly Bactrocera tryoni and its sister species, Bactrocera neohumeralis. Insect Sci. 2017;24:478–490. doi: 10.1111/1744-7917.12337. [DOI] [PubMed] [Google Scholar]
- 14.Tychsen PH, Fletcher BS. Studies on the rhythm of mating in the Queensland fruit fly. Dacus tryoni. J. Insect Physiol. 1971;17:2139–2156. doi: 10.1016/0022-1910(71)90174-0. [DOI] [Google Scholar]
- 15.Smith PH. Genetic manipulation of the circadian clock’s timing of sexual behaviour in the Queensland fruit flies, Dacus tryoni and Dacus neohumeralis. Physiol. Entomol. 1979;4:71–78. doi: 10.1111/j.1365-3032.1979.tb00179.x. [DOI] [Google Scholar]
- 16.Sakai T, Kitamoto T. Clock, love and memory: Circadian and non-circadian regulation of Drosophila mating behavior by clock genes. Sleep Biol. Rhythms. 2006;4:255–262. doi: 10.1111/j.1479-8425.2006.00224.x. [DOI] [Google Scholar]
- 17.Schutze MK, et al. Effects of laboratory colonization on Bactrocera dorsalis (Diptera, Tephritidae) mating behaviour: ‘what a difference a year makes’. Zookeys. 2015;540:369–383. doi: 10.3897/zookeys.540.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Özkaya Ö, Rosato E. The Circadian Clock of the Fly: A Neurogenetics Journey Through Time. Adv. Genet. 2012;77:79–123. doi: 10.1016/B978-0-12-387687-4.00004-0. [DOI] [PubMed] [Google Scholar]
- 19.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/S0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 20.Stanewsky R, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/S0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 21.Helfrich-Förster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshii T, et al. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J. Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johard HAD, et al. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J. Comp. Neurol. 2009;516:59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- 24.Hermann-Luibl C, Yoshii T, Senthilan PR, Dircksen H, Helfrich-Förster C. The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. J. Neurosci. 2014;34:9522–9536. doi: 10.1523/JNEUROSCI.0111-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konopka RJ, Pittendrigh CS, Orr D. Reciprocal behaviour associated with altered homeostasis and photosensitivity of Drosophila clock mutants. J. Neurogenet. 1989;6:1–10. doi: 10.3109/01677068909107096. [DOI] [PubMed] [Google Scholar]
- 26.Loher WJ, Zervas G. The mating rhythm of the olive fruitfly, Dacus oleae Gmelin. J. Appl. Entomol. 1979;88:425–435. [Google Scholar]
- 27.Tsitsipis, J. A. Exodus of olive fly larvae, Dacus oleae, from the diet for pupation as affected by photoperiod. In Proceedings of the CEC/IOBC ‘ad-hoc meeting’ Hamburg/23 August 1984 89–93 (1986).
- 28.Tsitsipis, J. A. & Loher, W. J. Circadian rhythmical exodus of olive fruit fly larvae from the diet for pupation. In Fruit flies of economic importance 87: proceedings of the CEC/IOBC International Symposium, Rome 7–10, April 1987 203–208 (1989).
- 29.Levi-Zada A, et al. Analyzing diurnal and age-related pheromone emission of the olive fruit fly, Bactrocera oleae by sequential SPME-GCMS analysis. J. Chem. Ecol. 2012;38:1036–1041. doi: 10.1007/s10886-012-0167-x. [DOI] [PubMed] [Google Scholar]
- 30.Hardin PE, Hall J, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 31.Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol. Cell. Biol. 1998;18:6142–6151. doi: 10.1128/MCB.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae K, Lee C, Hardin PE, Edery I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J. Neurosci. 2000;20:1746–1753. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermann C, et al. The circadian clock network in the brain of different Drosophila species. J. Comp. Neurol. 2013;521:367–388. doi: 10.1002/cne.23178. [DOI] [PubMed] [Google Scholar]
- 34.Cyran SA, et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/S0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 35.Meireles-Filho ACA, Amoretty PR, De Souza NA, Kyriacou CP, Peixoto AA. Rhythmic expression of the cycle gene in a hematophagous insect vector. BMC Mol. Biol. 2006;7:1–10. doi: 10.1186/1471-2199-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meuti ME, Stone M, Ikeno T, Denlinger DL. Functional circadian clock genes are essential for the overwintering diapause of the Northern house mosquito, Culex pipiens. J. Exp. Biol. 2015;218:412–422. doi: 10.1242/jeb.113233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Codd V, et al. Circadian rhythm gene regulation in the housefly Musca domestica. Genetics. 2007;177:1539–1551. doi: 10.1534/genetics.107.079160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chahad-Ehlers S, et al. Expanding the view of Clock and cycle gene evolution in Diptera. Insect Mol. Biol. 2017;26:317–331. doi: 10.1111/imb.12296. [DOI] [PubMed] [Google Scholar]
- 39.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/S0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 40.Sandrelli F, Costa R, Kyriacou CP, Rosato E. Comparative analysis of circadian clock genes in insects. Insect Mol. Biol. 2008;17:447–463. doi: 10.1111/j.1365-2583.2008.00832.x. [DOI] [PubMed] [Google Scholar]
- 41.Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 2007;24:948–955. doi: 10.1093/molbev/msm011. [DOI] [PubMed] [Google Scholar]
- 42.Bazalová O, Dolezel D. Daily activity of the housefly, Musca domestica, is influenced by temperature independent of 3’ UTR period gene splicing. G3. 2017;7:2637–2649. doi: 10.1534/g3.117.042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An X, Tebo M, Song S, Frommer M, Raphael KA. The cryptochrome (cry) gene and a mating isolation mechanism in tephritid fruit flies. Genetics. 2004;168:2025–2036. doi: 10.1534/genetics.104.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuchikawa T, et al. The clock gene cryptochrome of Bactrocera cucurbitae (Diptera: Tephritidae) in strains with different mating times. Heredity. 2010;104:387–392. doi: 10.1038/hdy.2009.167. [DOI] [PubMed] [Google Scholar]
- 45.An X, et al. The period gene in two species of tephritid fruit fly differentiated by mating behaviour. Insect Mol. Biol. 2002;11:419–430. doi: 10.1046/j.1365-2583.2002.00351.x. [DOI] [PubMed] [Google Scholar]
- 46.Miyatake T. Pleiotropic effect, clock genes, and reproductive isolation. Popul. Ecol. 2002;44:201–207. doi: 10.1007/s101440200023. [DOI] [Google Scholar]
- 47.Mazzotta GM, et al. The clock gene period in the medfly Ceratitis capitata. Genet. Res. 2005;86:13–30. doi: 10.1017/S0016672305007664. [DOI] [PubMed] [Google Scholar]
- 48.Matsumoto A, et al. Period gene of Bactrocera cucurbitae (Diptera: Tephritidae) among strains with different mating times and sterile insect technique. Ann. Entomol. Soc. Am. 2008;101:1121–1130. doi: 10.1603/0013-8746-101.6.1121. [DOI] [Google Scholar]
- 49.Chang DC, et al. Constructing a feedback loop with circadian clock molecules from the silkmoth, Antheraea pernyi. J. Biol. Chem. 2003;278:38149–38158. doi: 10.1074/jbc.M306937200. [DOI] [PubMed] [Google Scholar]
- 50.Rubin EB, et al. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 2006;16:1352–1365. doi: 10.1101/gr.5094806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomioka K, Matsumoto A. Circadian molecular clockworks in non-model insects. Curr. Opin. Insect Sci. 2015;7:58–64. doi: 10.1016/j.cois.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Bahn JH, Lee G, Park JH. Comparative analysis of pdf-mediated circadian behaviors between Drosophila melanogaster and D. virilis. Genetics. 2009;181:965–975. doi: 10.1534/genetics.108.099069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menegazzi P, et al. Adaptation of circadian neuronal network to photoperiod in high-latitude European Drosophilids. Curr. Biol. 2017;27:1–7. doi: 10.1016/j.cub.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 54.Ero MM, Hamacek E, Clarke AR. Foraging behaviours of Diachasmimorpha kraussii (Fullaway) (Hymenoptera: Braconidae) and its host Bactrocera tryoni (Froggatt) (Diptera: Tephritidae) in a nectarine (Prunus persica (L.) Batsch var. nectarina (Aiton) Maxim) orch. Aust. J. Entomol. 2011;50:234–240. [Google Scholar]
- 55.Raghu S, Clarke AR. Spatial and temporal partitioning of behaviour by adult dacines: direct evidence for methyl eugenol as a mate rendezvous cue for Bactrocera cacuminata. Physiol. Entomol. 2003;28:175–184. doi: 10.1046/j.1365-3032.2003.00328.x. [DOI] [Google Scholar]
- 56.Weldon CW, Prenter J, Taylor PW. Activity patterns of Queensland fruit flies (Bactrocera tryoni) are affected by both mass-rearing and sterilization. Physiol. Entomol. 2010;35:148–153. doi: 10.1111/j.1365-3032.2010.00726.x. [DOI] [Google Scholar]
- 57.Kauranen H, et al. Flies in the north: locomotor behavior and clock neuron organization of Drosophila montana. J. Biol. Rhythms. 2012;27:377–387. doi: 10.1177/0748730412455916. [DOI] [PubMed] [Google Scholar]
- 58.Vaze KM, Helfrich-Förster C. Drosophila ezoana uses an hour-glass or highly damped circadian clock for measuring night length and inducing diapause. Physiol. Entomol. 2016;41:378–389. doi: 10.1111/phen.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ceriani MF, et al. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 60.Aketarawong N, et al. The oriental fruitfly Bactrocera dorsalis s.s. in East Asia: Disentangling the different forces promoting the invasion and shaping the genetic make-up of populations. Genetica. 2014;142:201–213. doi: 10.1007/s10709-014-9767-4. [DOI] [PubMed] [Google Scholar]
- 61.Ito Y, Kakinohana H, Yamagishi M, Kohama T. Eradication of the melon fly, Bactrocera cucurbitae, from Okinawa, Japan, by means of the sterile insect technique, with special emphasis on the role of basic studies. Journal of Asia-Pacific Entomology. 2003;6:119–129. doi: 10.1016/S1226-8615(08)60177-6. [DOI] [Google Scholar]
- 62.Gómez Y, Teal PEA, Pereira R. Enhancing efficacy of Mexican fruit fly SIT programmes by large-scale incorporation of methoprene into pre-release diet. J. Appl. Entomol. 2013;137:252–259. doi: 10.1111/j.1439-0418.2011.01695.x. [DOI] [Google Scholar]
- 63.Hendrichs J, et al. Medfly areawide sterile insect technique programmes for prevention, suppression or eradication: the importance of mating behavior studies. Florida Entomol. 2012;85:1–13. doi: 10.1653/0015-4040(2002)085[0001:MASITP]2.0.CO;2. [DOI] [Google Scholar]
- 64.Kumaran N, Prentis PJ, Mangalam KP, Schutze MK, Clarke AR. Sexual selection in true fruit flies (Diptera: Tephritidae): Transcriptome and experimental evidences for phytochemicals increasing male competitive ability. Mol. Ecol. 2014;23:4645–4657. doi: 10.1111/mec.12880. [DOI] [PubMed] [Google Scholar]
- 65.Argov, Y., Kuslutzky, W. & Hoelmer, K. Biological control of olive fruit fly, Bactrocera oleae, In Israel. in Proceedings of the meeting at Jerusalem (Israel), 15–20 May, 2011 (2011).
- 66.Tzanakakis ME, Economopoulos A, Tsitsipis JA. Improved artificial food media for larvae of Dacus oleae (Gmelin) (Diptera:Tephritidae) ‘Democritus’ Nuclear Research Center of the Greek Atomic Energy Commission, Athens. 1966;59:127–130. [Google Scholar]
- 67.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:1–6. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benson DA, et al. GenBank. Nucleic Acids Res. 2017;45:37–42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 70.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 72.Abdelsalam S, et al. Characterization of PDF-immunoreactive neurons in the optic lobe and cerebral lobe of the cricket. Gryllus bimaculatus. J. Insect Physiol. 2008;54:1205–1212. doi: 10.1016/j.jinsphys.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 73.Ring M, et al. Expression of Schistocerca gregaria ion transport peptide (ITP) and its homologue (ITP-L) in a Baculovirus/Insect Cell System. Insect Biochem. Mol. Biol. 1998;28:51–58. doi: 10.1016/S0965-1748(97)00096-9. [DOI] [PubMed] [Google Scholar]
- 74.Reddy KL, et al. The Drosophila PAR domain protein 1 (Pdp1) gene encodes multiple differentially expressed mRNAs and proteins through the use of multiple enhancers and promoters. Dev. Biol. 2000;224:401–414. doi: 10.1006/dbio.2000.9797. [DOI] [PubMed] [Google Scholar]
- 75.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosato E, Kyriacou CP. Analysis of locomotor activity rhythms in. Drosophila. Nat. Protoc. 2006;1:559–568. doi: 10.1038/nprot.2006.79. [DOI] [PubMed] [Google Scholar]
- 77.Schmid B, Helfrich-Förster C, Yoshii T. A new ImageJ plug-in ‘ActogramJ’ for chronobiological analyses. J. Biol. Rhythms. 2011;26:464–467. doi: 10.1177/0748730411414264. [DOI] [PubMed] [Google Scholar]
- 78.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J. Biol. Rhythms. 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on request.