Abstract
The ability of human enteric pathogens to colonize plants and use them as alternate hosts is now well established. Salmonella, similarly to phytobacteria, appears to be capable of producing the plant hormone auxin via an indole-3-pyruvate decarboxylase (IpdC), a key enzyme of the IPyA pathway. A deletion of the Salmonella ipdC significantly reduced auxin synthesis in laboratory culture. The Salmonella ipdC gene was expressed on root surfaces of Medicago truncatula. M. truncatula auxin-responsive GH3::GUS reporter was activated by the wild type Salmonella, and not but the ipdC mutant, implying that the bacterially produced IAA (Indole Acetic Acid) was detected by the seedlings. Seedling infections with the wild type Salmonella caused an increase in secondary root formation, which was not observed in the ipdC mutant. The wild type Salmonella cells were detected as aggregates at the sites of lateral root emergence, whereas the ipdC mutant cells were evenly distributed in the rhizosphere. However, both strains appeared to colonize seedlings well in growth pouch experiments. The ipdC mutant was also less virulent in a murine model of infection. When mice were infected by oral gavage, the ipdC mutant was as proficient as the wild type strain in colonization of the intestine, but it was defective in the ability to cross the intestinal barrier. Fewer cells of the ipdC mutant, compared with the wild type strain, were detected in Peyer's patches, spleen and in the liver. Orthologs of ipdC are found in all Salmonella genomes and are distributed among many animal pathogens and plant-associated bacteria of the Enterobacteriaceae, suggesting a broad ecological role of the IpdC-catalyzed pathway.
Keywords: indole acetic acid, auxin, RIVET, sprouts, produce safety, Medicago truncatula, enteric-plant interactions, tryptophan
Introduction
Recurrent outbreaks of gastroenteritis caused by non-typhoidal Salmonella and shigatoxigenic E. coli and linked to the consumption of sprouts, vegetables, fruits and nuts led to the hypothesis that these enteric pathogens use plants as alternate hosts (Brandl, 2006; Brandl et al., 2013; Hernández-Reyes and Schikora, 2013; Walsh et al., 2014). Both Salmonella and E. coli can persist in manure, soil and water for at least several weeks or months (Brandl, 2006; Brandl et al., 2013; Martínez-Vaz et al., 2014; Wiedemann et al., 2014; Ongeng et al., 2015), from which, they can get established on plants. Salmonella and E. coli colonized seedlings from contaminated manure, and following the feeding of the infected plants to snails and mice, these bacteria were detected within animals and shed in their feces (Franz et al., 2008; Semenov et al., 2010). While these studies establish that enteric pathogens can use plants as alternate hosts or vectors, less is known about the mechanisms by which they interact with plants, despite the significant progress in the characterization of interactions between enteric pathogens and plants (Klerks et al., 2007a,b; Hernández-Reyes and Schikora, 2013; Wiedemann et al., 2014; Han and Micallef, 2016; de Moraes et al., 2017). Plants are colonized with diverse assemblages of epiphytes and endophytes, which interact with each other and their plant hosts in a variety of ways. Epiphytes often congregate at preferred sites, such as hydathodes, stomata, lesions, or breaks in the epidermis caused by lateral root emergence, which offer more nutrients and potential routes for internalization (Leveau and Lindow, 2001; Lindow and Brandl, 2003). Salmonella has been shown to seek similar niches during plant colonization, as it can establish colonies on plant surfaces and may enter internal tissues under conducive conditions (Brandl and Mandrell, 2002; Dong et al., 2003; Klerks et al., 2007a; Golberg et al., 2011; Gu et al., 2013a). However, Salmonella likely faces considerable competition for the colonization of these preferred sites and may benefit from mechanisms that allow it to manipulate plant hosts to increase the availability of growth-conducive sites.
Production of plant hormones, and especially indole-3-acetic acid (IAA), is one of the mechanisms by which plant-associated bacteria and fungi manipulate plant growth and development. Microbial production of IAA alters root architecture to increase availability of nutrients to microbes capable of producing this plant hormone. Microbially derived auxins also contribute to the formation of new organs (such as nodules and galls), suppress plant defenses and regulate virulence in phytobacteria (Lindow and Brandl, 2003). The recognition of the roles auxin signaling in plant-microbial interactions led to the hypothesis that it functions as a reciprocal signal in these relationships (Duca et al., 2014).
The synthesis of auxin is widespread among phytobacteria (Duca et al., 2014; Ludwig-Müller, 2015). At least five bacterial IAA biosynthesic pathways exist. Of these IAA biosynthetic pathways, those proceeding via indole-3-acetamide (IAM) and indole-3-pyruvate (IPyA) are the two most widespread and best characterized. Tryptophan is the general precursor of IAA biosynthesis (Patten et al., 2013). The IAM pathway is a two-step process, conserved in three Kingdoms of Life. In the IAM pathway, tryptophan is first converted by tryptophan-2-monooxygenase (encoded by iaaM) into IAM, and then to IAA by IAM hydrolase (encoded by iaaH) (Lehmann et al., 2010). The IPyA pathway in bacteria is a three-step process mediated by the conversion of indole-3-pyruvate into indole-3-acetaldehyde by indole-3-pyruvate decarboxylase (encoded by ipdC) (Brandl and Lindow, 1996; Brandl et al., 1996). Indole-3-acetaldehyde is then converted into IAA. Production of IAA via IpyA in the common bacterial plant colonist Pantoea agglomerans contributes to its epiphytic fitness (Brandl and Lindow, 1996, 1998). A given phytobacterial species may commonly possess more than one pathway, suggesting a high degree of redundancy, which is likely indicative of an important phenotype (Patten et al., 2013). For example, the plant pathogen E. herbicola pv. gypsophilae harbors both pathways, which appear to function in controlling divergent behaviors. The deletion of ipdC reduced the epiphytic fitness of this pathogen during active growth on bean leaves, whereas a mutation in iaaH was linked to decreased virulence in Gypsophila paniculata (Manulis et al., 1998).
Genomic analysis demonstrates that all salmonellae have a copy of the ipdC gene, similar to that found in other γ-proteobacteria. Although the presence of the ipdC gene in Salmonella genomes has been previously detected (Spaepen et al., 2007), its function in Salmonella remains unclear. The production of IAA by Salmonella, therefore, could enable epiphytic survival in a manner similar to other phytobacteria and could represent a conserved strategy for colonization of plants as a typical part of its life cycle. In this study, we tested the hypothesis that S. enterica produces IAA through the IPyA pathway and that the ipdC-mediated IAA production impacts the outcomes of the interactions of Salmonella with a model plant, Medicago truncatula.
Materials and methods
Culture conditions and strain construction
All strains used in this study are listed in Table 1, and primers used for the strain construction are in Table 2. Unless otherwise specified, strains were routinely grown in LB medium (Miller, 1972). Minimal A medium was prepared as in Ludwig-Müller (2015). A kanamycin marked ipdC mutant was constructed using Datsenko and Wanner mutagenesis (Datsenko and Wanner, 2000) with primers CEC207 and CEC209. The deletion was confirmed with primers BA505 and CEC135. An unmarked mutant was constructed by removing the FRT-kanR-FRT cassette with plasmid pCP20 as in Datsenko and Wanner (2000). The deletion was confirmed with primers CEC134 and CEC135. RIVET reporters were constructed as in Merighi et al. (2005) by placing tnpR immediately downstream of the ipdC stop codon or by placing tnpR immediately downstream of the promoter region in a stream where ipdC has been removed between the start and stop codons. Constructs were confirmed with primers CEC134 and BA184 or CEC210 and BA184. Activation of the RIVET reporter is scored as the loss of the tetracycline resistance marker in the recovered cells (this occurs because activation of the promoter of interest leads to the expression of the TnpR recombinase and the resulting excision of the tetracycline-resistance marker encoded within “res” sites that are substrates for TnpR; Merighi et al., 2005). A plasmid carrying constitutively expressed gfp reporter driven by the Salmonella dppA promoter (Noel et al., 2010) was introduced into strains as needed via electroporation. To ascertain that the genetic manipulations did not result in a growth deficiency, strains were grown in LB broth shake cultures at 22°C and 37°C for 10–24 h and OD600 measurements were taken periodically, at 30 min or 2 h intervals.
Table 1.
Strains used in this study.
| Strain | Relevant genotype | Source |
|---|---|---|
| 14028 | Wild-type S. enterica serovar Typhimurium | American type culture collection |
| JS246 | 14028 yjeP8103::res1-tetAR-res1 | Merighi et al., 2005 |
| CEC1002 | 14028 ΔipdC::FRT-kanR-FRT | This Study |
| CEC2002 | 14028 ΔipdC | This Study |
| CEC5002 | JS246 PipdC-tnpR-lacZY ΔipdC | This Study |
| CEC8002 | JS246 ipdC-tnpR-lacZY | This Study |
| Plasmids | Relevant feature(s) | Source |
| pGFP-ON | pGFP dppA-GFP | Noel et al., 2010 |
| pKD4 | oriR6K bla rgnB FRT-kanR-FRT (kanR) | Datsenko and Wanner, 2000 |
| pKD46 | repA101ts oriR101 araC ParaB- λRed(γ-β-exo)-tL3) (ampR) | Datsenko and Wanner, 2000 |
| pCP20 | repA101ts λpR-Flp ci857 (ampR, kanR) | Cherepanov and Wackernagel, 1995 |
| pCE70 | oriR6K FRT-promoterless tnpR-lacZYα (kanR) | Merighi et al., 2005 |
| pCE71 | oriR6K FRT-promoterless tnpR-lacZYα (kanR) | Merighi et al., 2005 |
Table 2.
Primers used in this study.
| Primer | Sequence | Use |
|---|---|---|
| BA184 | CAAAAAGTCGCATAAAAATTTATCC | RIVET confirmation |
| K2 | CGGTGCCCTGAATGAACTGC | FRT-kanR-FRT confirmation Datsenko and Wanner, 2000 |
| CEC134 | TCCCCCTGTGGCGTGAAT | ipdC confirmation |
| CEC135 | CCTGGCTATTGCTGGCGG | ipdC confirmation |
| CEC207 | GCATTCCTTAATACTCAACATAATATCAACGTCAGAAGGAAAGCTGTCtgaggctggagctgctt | ΔipdC construction |
| CEC208 | TTACTGCGTACCGTGACCCGGGCGCTGGAAGCCCGCAACGGGGGATAAtgtaggctggagctgctt | ipdC-tnpR-lacZY construction |
| CEC209 | TGGCCCCCGCTGCGCCGGATTAGGGTTCGTGACGGTTGGCGGCCAGCAcatatgaatatcctccttag | ipdC_LR |
| CEC210 | GGACAGCCAGTGCGGATT | ipdC confrimation |
Detection of IAA using salkowski reagent
Salmonella enterica sv Typhimurium (S. Typhimurium thereafter) cultures were grown overnight from glycerol stocks at 37°C. 1 mL was washed 3x in PBS and was diluted 1,000x in 5 mL Minimal A medium with or without 1 mM tryptophan. Cultures were incubated at 22°C for 72 h. Supernatant was harvested from 1 mL of culture by centrifugation for 15 min at 7,000 × g. The supernatant was transferred to a 12 mm × 75 mm glass tube and 2 mL of fresh Salkowski reagent R1 (Glickmann and Dessaux, 1995) was added and vortexed to mix. The tubes were incubated for 30 min at 30°C in the dark, aliquots were then withdrawn and A540 was measured using the Shimadzu biospec-mini.
IAA extraction, purification, and identification
Hundred millliter of Salmonella culture filtrates were extracted twice with 150 and 100 mL of ethyl acetate. The extracts were rotary evaporated over a water bath at 40°C. The resulting yellow oily residue was brought up in 600 μl of methanol. The extracts and standards (IAA, tryptophol and IPyA) were subject to reverse phase (C18) liquid chromatography using Waters Atlantis dC18-5 μM column using isocratic elution (72 water 7.2% acetic acid 20.8% methanol), as before (Brandl and Lindow, 1996; Brandl et al., 1996). Eluting substances were detected with a UV/VIS detector set to 230 and 280 nm (Brandl and Lindow, 1996; Brandl et al., 1996). Low Resolution Electrospray Ionization LCMS was carried in a Thermo Scientific LTQ LC-MS instrument using a Grace Vydac 218TP C18 (5 μ, 7.5 cm, 2.1 mm ID) column. LCMS was run using a gradient of 10% acetonitrile (with 0.1% formic acid) and 90% water (with 0.1% formic acid) to 100% acetonitrile (with 0.1% formic acid) from 0 to 15 min and then 100% acetonitrile with formic acid was continued till 21 min.
Plant infections
Seeds of M. truncatula A17 (wild type) and transgenic line containing the GH3::GUS reporter (van Noorden et al., 2007) were surface sterilized in ethanol and then in a diluted chlorox bleach solution as before (Mathesius et al., 2003) and stratified overnight at 4°C. Imbibed seeds were transferred into individual growth pouches and were left to germinate at room temperature overnight. Once radicles reached ~1 cm, six seedlings (per treatment) were inoculated with 100 μl of the suspension containing ~5,000–10,000 CFU of either CEC1000 (S. Typhimurium 14028 marked with a kanamycin cassette in a neutral site) or CEC1002. Plants were watered as needed with the N-free Hoagland solution. To detect activation of the GH3::GUS reporter, seedlings were withdrawn after 48 hrs and 6 days past infection, and stained with X-gluc (10 mg/ml) in 0.2 mM Phosphate buffer (pH 7.2) overnight. They were then washed 3 times with cold phosphate buffer to remove excess substrate and the seedlings were kept in 50% ethanol/phosphate buffer at 4C. They were imaged under a dissecting microscope Olympus MVX10 fit with a MicroFIRE camera (Optronics).
For the reporter assays, washed RIVET reporter suspensions (containing ~10,000–100,000 CFU) were inoculated onto M. truncatula seedlings as described in Supplemental Information. At indicated time intervals, seedlings were excised from pouches, blended with a glass mortar and pestle and dilution plated onto XLD with kanamycin. Bacteria were patched from these onto LB with tetracycline.
To detect changes in lateral root formation, seeds of M. truncatula A17 were surface sterilized, imbibed and then transferred into growth pouches. Seedlings were inoculated with 100 μl of the washed bacterial suspension containing ~5,000–10,000 CFU of either CEC1000 (S. Typhimurium 14028 marked with a kanamycin cassette in a neutral site) or CEC1002 (ipdC::kan).
Mouse infections
BALB/c mice were inoculated by oral gavage, which results in a systemic infection. Prior to the infections, S. enterica sv. Typhimurium 14028 and CEC1002 were streaked on LB agar and incubated at 37°C overnight, colonies were harvested and re-suspended in 1 ml of sterile PBS. Each suspension was adjusted to a final dilution of ~108 CFUs/ml. Groups (n = 3) of female BALB/c mice received three doses of Salmonella in 0.2 ml of PBS. Inoculum doses were confirmed by serial titration, and were found to be 106, 104, and 102 CFU per injection dose for the wild type, and 2.2 × 106, 2.2 × 104, and 2.2 × 102 CFU per injection for the ipdC mutant. Animals were observed daily for 7 days after infection and distressed animals presenting signs of morbidity were euthanized, their organs were not sampled. The experiment was terminated at 7 days, and all surviving animals were sacrificed. Liver, spleen, large intestine, and Peyer's patches were harvested. Tissues were weighed, and homogenized in sterile PBS in TissueLyser II (Qiagen). Bacterial homogenates were serially diluted in PBS and plated onto Xylose-Lysine Deoxycholate (XLD) agar (Beckton, Dickinson and Company) plates, followed by incubation at 37°C overnight for bacterial CFU counts. All animal care and procedures were in accordance with institutional policies for animal health and well-being and approved by the University of Florida Institutional Animal Care and Use Committee (IACUC).
Data analysis
Synteny analysis was carried out using default parameters in the SyntTax web server (Oberto, 2013). Sequences were imported from GenBank. UV peak integration was conducted with proprietary Waters software, and analysis of mass spectral data was carried out using Thermo Scientific proprietary software. Means and standard error of the mean were calculated in Microsoft Excel v. 14.0.0.
Results
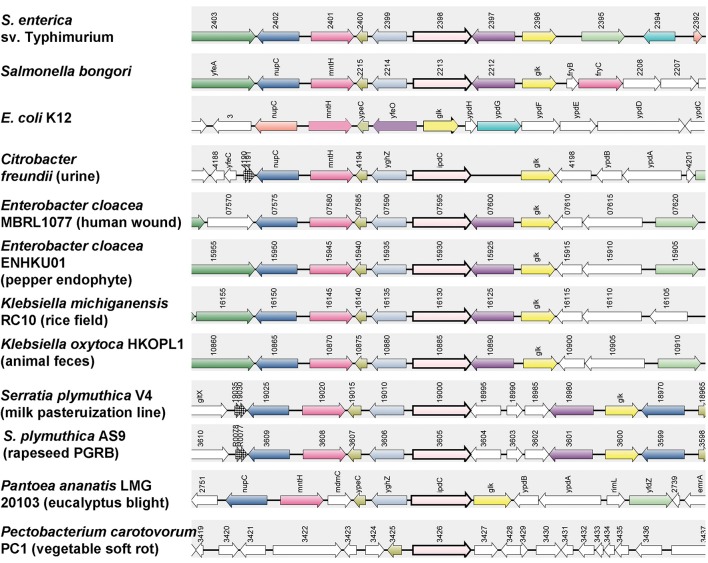
ipdC is conserved in salmonellae. ipdC, the gene encoding indole-3-pyruvate decarboxylase, a key enzyme in the IPyA pathway, is common in many members of Enterobacteriaceae; including organisms that are commonly associated with animals as well as with plants. Genomic analysis showed that all available Salmonella genomes harbor ipdC, but that it is absent in E. coli, Shigella, or Erwinia genomes and in some strains of Enterobacter. The presence or absence from the enterobacterial genomes does not seem to correlate with the source of the isolates, and ipdC was as likely to be found in the genomes of animal as of plant isolates of this genus. Synteny analysis revealed a distribution of orthologs in the genomes of many strains belonging to the Enterobacteriaceae where it is generally found within similar genomic context (Figure 1). In Salmonella, ipdC is located downstream of an ortholog of the glucokinase glk, and a putative ion channel protein known as YfeO. glk was conserved at this location in all genomes except in Pectobacterium, in which it is present at another site. Similarly, YfeO is present at different sites in Citrobacter, Panteoa and Pectobacterium genomes. Immediately downstream of ipdC in the Salmonella genome are an ortholog of the L-glyceraldehyde 3-phosphate reductase gpr, and an uncharacterized periplasmic protein YpeC. Although its function is currently unknown, ypeC was present at a similar location in all genomes. gpr (yghZ) was present between ipdC and ypeC except in E. coli and Pectobacterium. These results suggest that the Salmonella ipdC may encode an ortholog of a well-characterized indolepyruvate decarboxylase, responsible for IAA synthesis in other bacteria. Therefore, we tested whether ipdC is capable of directing IAA production in S. enterica sv Typhimurium.
Figure 1.
Synteny analysis of ipdC in Enterobacteriaceae. Genes homologous to the Salmonella ipdC were retrieved from NCBI. Synteny analysis was carried out using SyntTax (Oberto, 2013). Orthologs of ipdC are shown as pink arrows with a thick black border.
IAA production by Salmonella via ipdC
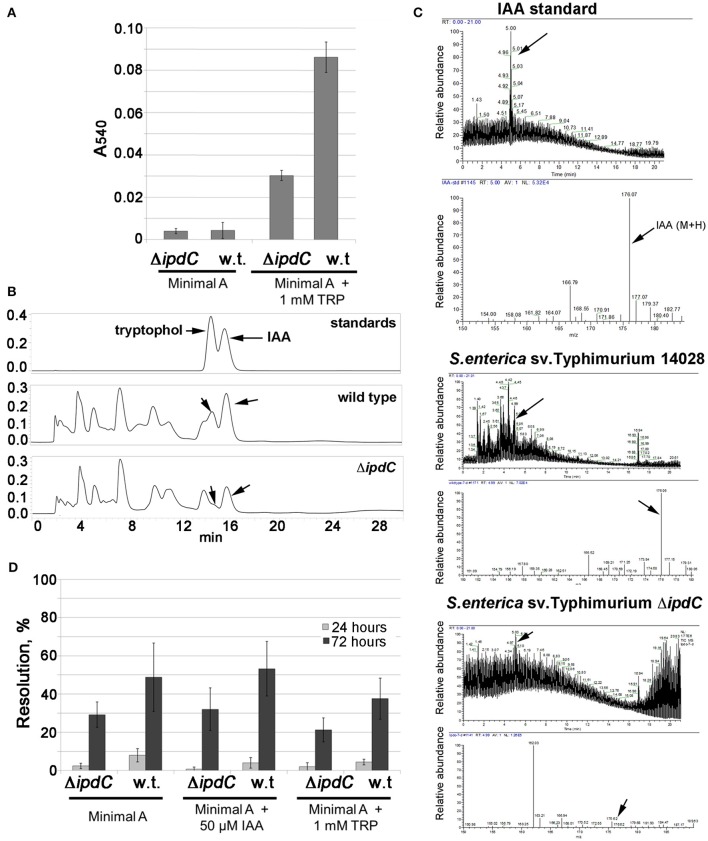
The ability of Salmonella to produce IAA was first tested in culture in Minimal A medium using a colorimetric assay based on the Salkowski reagent. While there was no auxin production by either the wild type or the isogenic ipdC mutant grown in unammended Minimal A medium, production of IAA was detected in the medium supplemented with 1 mM L-tryptophan, the precursor for the IPyA pathway (Figure 2A). The intensity of absorbance at 540 nm after addition of the Salkowski reagent, which correlates with the presence of indole compounds, including IAA, was significantly lower in the culture filtrate extract of the ipdC mutant than that of the wild type strain (Figure 2A). When culture filtrate extracts were separated using HPLC, UV peaks corresponding to IAA and tryptophol were observed for the wild type, but were strongly reduced for the ipdC mutant (Figure 2B). Tryptophol is a product of a reduction of indole-3-acetaldehyde, the product of the reaction catalyzed by ipdC. The production of IAA by the wild type S. enterica sv. Typhimurium 14028 was confirmed by LC-MS/MS. Production of IAA was significantly reduced (but not completely eliminated) in the isogenic strain lacking ipdC (Figure 2C). In laboratory cultures, expression of the ipdC RIVET (recombinase-based in vivo expression technology) reporter was low (less than 10%) after 24 h of incubation, however, it increased by 72 h (Figure 2D). Consistent with the production of IAA in laboratory media, a recombinase in vivo expression technology (RIVET) reporter in ipdC was expressed in a laboratory medium at 45–55% (Figure 2D). Expression of the reporter in the ipdC background was considerably lower than in the wild type, suggesting a feedback regulation. However, supplementation of the medium with 50 μM IAA did not significantly affect expression of the ipdC RIVET reporter in the wild type or in the ipdC mutant. Despite clear differences in the amount of IAA produced in a minimal medium with and without tryptophan, the ipdC RIVET reporter was expressed at similar levels in the minimal medium with and without tryptophan (Figure 2D), suggesting that the availability of the substrate is critical for the function of the IpdC enzyme (as indirectly evidenced by the accumulation of IAA in culture) but not for transcription of the encoding gene.
Figure 2.
Production of IAA in ipdC-dependent manner by Salmonella enterica sv. Typhimuirum 14028. (A) Detection of IAA by the Salkowski reagent. Results are from replicates from three independent experiments, averages are shown. Error bars are standard deviations. (B) HPLC detection of IAA and tryptophol in spent cultures of the wild type and the ipdC mutant. Hydrophobic fractions of Salmonella culture filtrates were subjected to reverse phase (C18) liquid chromatography, eluted isocratically with acidified water/methanol and eluting substances were detected with a UV/VIS detector set to 230 and 280 nm. (C) Low Resolution Electrospray Ionization LC-MS of the hydrophobic fraction of Salmonella culture filtrates. LTQ LC-MS was carried out using a Grace Vydac 218TP C18 and acetonitrile/water gradient. (D) Expression of the ipdC RIVET reporter in the wild type and ΔipdC backgrounds was measured in Minimal A medium with and without synthetic IAA or tryptophan in cultures incubated at 22°C for 72 h. Samples were streaked to xylose lysine deoxycholate (XLD) agar with kanamycin at 24 and 72 h. Plates were incubated at 37°C overnight and colonies were patched to LB agar with tetracycline to quantify resolution. Experiments were repeated five times (without technical replications), averages from the five experiments are shown. Error bars are standard deviations.
Plant responses to IAA from Salmonella
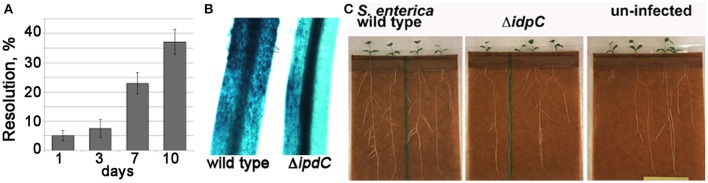
To determine whether Salmonella produces auxin on plant surfaces and whether this bacterially-produced plant hormone has a function during colonization of plant hosts, ipdC expression was assessed in the M. truncatula rhizosphere. The ipdC RIVET reporter was resolved at ~5–8% during the first 3 days of root colonization, reaching 22 and 37% at seven and 10 days post-inoculation, respectively (Figure 3A). When compared with the expression of the same reporter in Minimal A medium (Figure 2D), it does not appear that ipdC is expressed stronger in the rhizosphere, nevertheless, it is clear that ipdC is expressed in S. Typhimurium on roots during colonization. Next, we tested the consequences of the ipdC mutation using a GH3::GUS reporter in M. truncatula. Genes belonging to the GH3 (Gretchen Hagen 3) family are among the regulators that control the dynamic process of endogenous auxin homeostasis (Yang et al., 2015). This GH3::GUS reporter is normally expressed in the stele of the root, while expression in the cortical cells results from exposure to exogenous IAA, including that produced by bacteria such as by the symbiotic Sinorhizobium meliloti (Mathesius et al., 1998, 2000; van Noorden et al., 2007). As shown in Figure 3B, the GH3::GUS reporter was broadly induced in the cortical tissue of M. trucatula roots after inoculation with the S. Typhimurium wild type strain, but only at discrete locations in the cortex after that with the ipdC mutant. Taken together, the activation of the plant GH3::GUS reporter by Salmonella and the expression of the Salmonella ipdC gene in the rhizosphere suggest that this human pathogen synthesizes auxin via IpdC during plant colonization and in quantities that are detectable by the plant cells and sufficient to cause changes in plant gene expression.
Figure 3.
M. truncatula responds to the activation of ipdC in the rhizosphere. (A) Resolution of the Salmonella ipdC RIVET reporter was measured on the seedlings grown in pouches from surface-sterilized M. truncatula Jemalong A17 seeds. (B) Activation of the M. truncatula auxin-responsive GH3::GUS reporter was measured as in Mathesius et al. (2003). Black arrows point to representative sites of GH3::GUS expression in the cortex, likely a result of the IAA production by the wild type Salmonella. Six seedlings were inoculated per treatment, and results of a representative experiment (6 days post- infection) are shown. Care was taken to excise root segments at the same developmental stage (C) Inoculation with the wild type Salmonella Typhimurium 14028 increases lateral root formation in M. truncatula Jemalong A17.
Production of IAA creates a niche for Salmonella
To determine how Salmonella may benefit from the production of IAA, we first tested whether perception of the bacterially-produced IAA led to observable phenotypic changes in the inoculated plants. Secondary root initiation is a phenotype known to be regulated by IAA (Spaepen et al., 2014; Bensmihen, 2015). Therefore, we evaluated formation of secondary roots in the seedlings inoculated with the wild type Salmonella or the isogenic ipdC mutant. As shown in Figure 3C, few secondary roots had emerged in 2-week old M. truncatula seedlings that were not inoculated with Salmonella and treated with the nutrient solution only (control). Seedlings inoculated with the S. Typhimurium wild type strain showed 5–12 secondary roots, while seedlings inoculated with the ipdC mutant had 2–7 secondary roots per plant. When the wild type and the ipdC mutant were tagged with a plasmid constitutively expressing GFP, the ipdC mutant was found to be evenly distributed throughout the root surface and on root hairs, while the wild type tended to form large aggregates at the sites of the lateral root emergence (Figure 4). However, we did not observe differences in the total number of bacteria recovered from the seedlings (data not shown). In laboratory LB shake cultures, the wild type and the ipdC mutant grew with the same kinetics and to the same final optical densities at 22 and 37°C.
Figure 4.
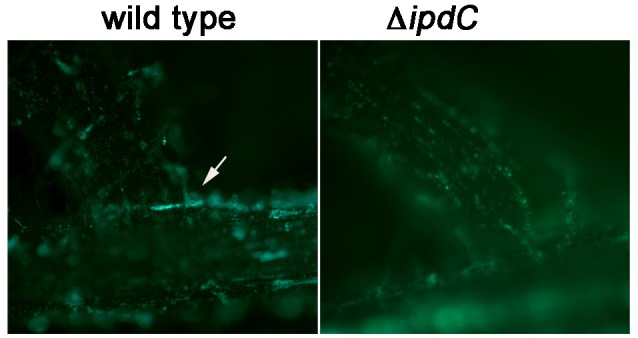
Colonization of sites of lateral root emergence by Salmonella wild type and ΔipdC. M. truncatula seedlings were inoculated with either CEC1000 or CEC1002 labeled with pGFP-ON. Fluorescence of the bacteria on roots of seedlings was documented 12 days after inoculation using Olympus BX41 epifluorescent microscope with 3.3 MPX Olympus camera. The white arrow points to the microcolony of the wild type Salmonella at the base of the lateral root.
Deletion of ipdC reduces virulence in the mouse model
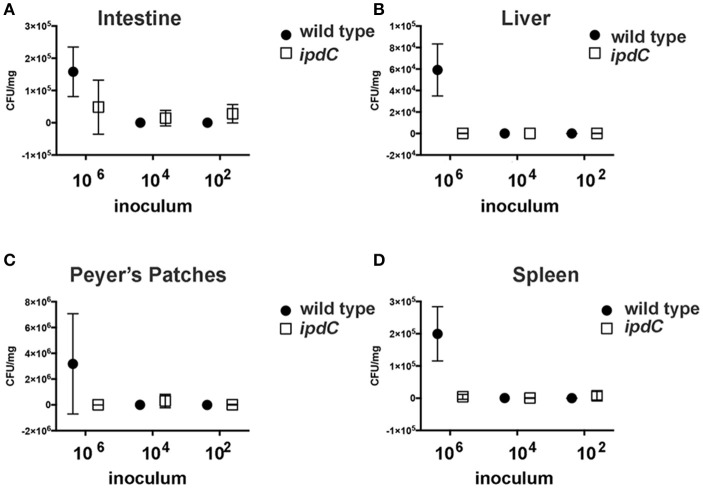
We hypothesized that because IAA is a plant hormone, the function of ipdC may be confined to the interaction of Salmonella with its plant hosts. To test this hypothesis, the comparative ability of the S. Typhimurium wild type and ipdC mutant to colonize mice following an oral gavage was determined. The hypothesis that the mutation in idpC impacts virulence in mice was proven null (Figure 5). While the ipdC mutant did not differ from the wild type in its ability to establish within the intestine, it was deficient in the colonization of the liver and the spleen (Figure 5). However, due to the small number of animals infected per dose, robust statistical analyses of these data were not possible.
Figure 5.
Recovery of Salmonella from mouse organs. Three female BALB/c mice per three infectious doses were infected with the wild type S. Typhimurium 14028 or an isogenic ipdC mutant by oral gavage. After 7 days, surviving animals were euthanized, and organs harvested for the determination of bacterial load. Recovered CFU of the mutant (empty square) and the wild type (filled circle) are shown. Error bars are standard deviations. When error bars are not shown, this indicates that only a single animal was remaining upon completion of the experiment. Salmonella recovery from intestine (A), liver (B), Peyer's patches (C) and spleen (D) is shown.
Discussion
The presence of ipdC in Salmonella was first noted by Spaepen et al. (2007) while constructing a phylogenic analysis to support the differentiation of the ipdC product of Azospirillum brasilense as a phenylpyruvate decarboxylase in contrast to the classical indole-3-pyruvate decarboxylase product of ipdC in Enterobacter cloacae (Spaepen et al., 2007). Of the 33 species with genomes containing putative ipdC homologs considered in their analysis, ipdC of S. Typhimurium LT2 grouped independently but was most closely related to E. cloacae and Pseudomonas putida, the strains most distant from A. brasilense, supporting the hypothetical function of Salmonella ipdC in synthesis of auxin. Our analysis of genomes in the currently available databases, revealed that ipdC is present in all Salmonella genomes, including that of the obligate human pathogen Salmonella Typhi. The synteny analysis presented here confirms also the common presence of ipdC in the genomes of both human and plant pathogenic Enterobacteriaceae.
The results of this work suggest that the Salmonella ipdC contributes to the production of IAA in laboratory cultures, that this gene is expressed in laboratory media and during root colonization, and that the product of the reaction involving IpdC activates plant auxin-responsive promoter. Inoculation of M. truncatula seedlings with the wild type Salmonella, but not that with the ipdC mutant resulted in increased production of lateral roots. Although the wild type and the ipdC mutant were recovered from the seedlings in similar numbers, the wild type cells tended to cluster at the sites of lateral root emergence, where high tryptophan availability to bacterial colonizers was reported with a whole-cell biosensor (Jaeger et al., 1999), whereas cells lacking ipdC were evenly distributed in the rhizosphere. This indicates that Salmonella cells located at sites where tryptophan is likely abundant benefit from IAA production to achieve greater cell densities. (Brandl and Lindow, 1998) suggested that IAA production by bacterial plant colonists may increase their fitness by enhancing nutrient leakage from plant cells by cell wall remodeling (Brandl and Lindow, 1998). Collectively, these results point at the ability of Salmonella to manipulate their plant hosts through IAA production although the entire suite of plant functions subject to this manipulation is not yet known. Plant pathogens, epiphytes, symbionts and endophytes employ auxin to thwart plant defenses, create new organs (nodules and galls), alter plant tissue development, and induce leakage of nutrients (Lindow et al., 1998; Lindow and Brandl, 2003; Duca et al., 2014; Talboys et al., 2014; Ludwig-Müller, 2015). While we present evidence that Salmonella expresses ipdC in the plant rhizosphere and that plants sense and respond to the resulting presence of this exogenous auxin, the broader implications of our observations for the ecology of Salmonella on plants remain unknown. In interpreting the results of this study, it is important to recognize that they were conducted in growth pouches that are largely devoid of native phytomicrobiota, which can strongly influence the outcomes of interactions between human enteric pathogens and plants (Klerks et al., 2007a; Teplitski et al., 2011; Brandl et al., 2013; Gu et al., 2013b). It is not clear whether production of IAA by Salmonella is of consequence during interactions under the field conditions, in natural soils where Salmonella must compete with other rhizosphere microbes, many of which produce IAA as well. In natural environments, effects of IAA on plants are concentration-dependent: low concentrations could induce formation of lateral roots, suppress host defense responses, while at higher doses it can inhibit plant growth (Kazan and Manners, 2009; Duca et al., 2014; Ludwig-Müller, 2015). It is clear that the production of IAA is dependent on the availability of tryptophan. In the rhizosphere, bacteria that produce more tryptophan, rather than depend on exogenous sources, can produce more IAA and therefore tend to have an advantage (Duca et al., 2014). At least initially, in a gnotobiotic rhizosphere Salmonella appears to utilize tryptophan that is exuded from M. truncatula seedlings. It is not certain that Salmonella can scavenge sufficient tryptophan or synthesize it in situ when it competes with the native rhizosphere microorganisms. Therefore, the ecological role of Salmonella-synthesized IAA in natural interactions with plants remains to be elucidated.
The role for ipdC during Salmonella mouse infection was unexpected. We initially hypothesized that the production of IAA must represent a unique adaptation to the plant-associated lifestyle. Upon further reflection, it should come as no surprise that the same molecule may have beneficial functions in a number of environments, where successful opportunists (such as non-typhoidal Salmonella) have evolved to thrive. Studies suggest that in some bacteria, IAA aids in stress adaptation and protection against stressors such as UV, salt and acidity (Duca et al., 2014). It is possible that these functions of IAA explain the mouse phenotype of the mutant. It is also possible that the phenotype of the ipdC mutant in the mouse may be—at least partially—independent of IAA and rather dictated by other functions of IpdC. For example, while indolpyruvate decarboxylases are generally known to have high affinity for indolpyruvate, they can also use phenylpyruvate, pyruvate and benzoylformate as substrates. It is, therefore, possible that IAA production is not the main, but rather incidental, role for these enzymes in some organisms (Duca et al., 2014). The conservation of ipdC in many bacterial animal pathogens that behave as opportunistic plant colonists and, reversely, in epiphytic bacteria that opportunistically colonize animal tissue, provides new incentives to gain insight into the function of this plant hormone in a larger biological context.
Ethics statement
Results of mouse experiments are described. Animal protocols were reviewed and approved by the University of Florida IACUC.
Author contributions
CC, MB, and MT conceived the project and designed the experiments, CC and MT carried out experiments and data analyses in Figures 1, 2A,B,D, 3A, 4. SG carried out chemical identification of IAA (Figures 2B,C). MdM designed and conducted mouse experiments.
Conflict of interest statement
The reviewer AHCVB declared a shared affiliation, with no collaboration, with several of the authors (CC, MdM, and MT) to the handling Editor. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank M. Farias for assistance with plant colonization experiments (Figure 3C), M. Gao for conducting GUS assays (Figure 3B), and C. Hoffman for animal infections. We are grateful to Dr. U. Mathesius for sharing seeds of GH3::GUS reporters.
Footnotes
Funding. This work was supported by a USDA NIFA Post-doctoral Fellowship provided funding to CC under grant number 2012-67012-005175, and by U.S. Department of Agriculture, Agricultural Research Service CRIS project 2030-42000-050-00D.
References
- Bensmihen S. (2015). Hormonal control of lateral root and nodule development in legumes. Plants 4, 523–547. 10.3390/plants4030523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M. T. (2006). Fitness of human enteric pathogens on plants and implications for food safety. Ann. Rev. Phytopath. 44, 367–392. 10.1146/annurev.phyto.44.070505.143359 [DOI] [PubMed] [Google Scholar]
- Brandl M. T., Clark E. M., Lindow S. E. (1996). Characterization of the indole-3-acetic acid (IAA) biosynthetic pathway in an epiphytic strain of Erwinia herbicola and IAA production in vitro. Can. J. Microbiol. 42, 586–592. 10.1139/m96-079 [DOI] [Google Scholar]
- Brandl M. T., Cox C. E., Teplitski M. (2013). Salmonella interactions with plants and their associated microbiota. Phytopathology 103, 316–325. 10.1094/PHYTO-11-12-0295-RVW [DOI] [PubMed] [Google Scholar]
- Brandl M. T., Lindow S. E. (1996). Cloning and characterization of a locus encoding an indolepyruvate decarboxylase involved in indole-3-acetic acid synthesis in Erwinia herbicola. Appl. Environ. Microbiol. 62, 4121–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M. T., Lindow S. E. (1998). Contribution of indole-3-acetic acid production to the epiphytic fitness of Erwinia herbicola. Appl. Environ. Microbiol. 64, 3256–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M. T., Mandrell R. E. (2002). Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68, 3614–3621. 10.1128/AEM.68.7.3614-3621.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. P., Wackernagel W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. 10.1016/0378-1119(95)00193-A [DOI] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moraes M. H., Desai P., Porwollik S., Canals R., Perez D. R., Chu W., et al. (2017). Salmonella persistence in tomatoes requires a distinct set of metabolic functions identified by transposon insertion sequencing. Appl. Environ. Microbiol. 83:e03028-16. 10.1128/AEM.03028-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Iniguez A. L., Ahmer B. M., Triplett E. W. (2003). Kinetics and strain specificity of rhizosphere and endophytic colonization by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl. Environ. Microbiol. 69, 1783–1790. 10.1128/AEM.69.3.1783-1790.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca D., Lorv J., Patten C. L., Rose D., Glick B. R. (2014). Indole-3-acetic acid in plant-microbe interactions. Anton. Leeuw Int. J. G. 106, 85–125. 10.1007/s10482-013-0095-y [DOI] [PubMed] [Google Scholar]
- Franz E., Semenov A. V., van Bruggen A. H. (2008). Modelling the contamination of lettuce with Escherichia coli O157:H7 from manure-amended soil and the effect of intervention strategies. J. Appl. Microbiol. 105, 1569–1584. 10.1111/j.1365-2672.2008.03915.x [DOI] [PubMed] [Google Scholar]
- Glickmann E., Dessaux Y. (1995). A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microb. 61, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golberg D., Kroupitski Y., Belausov E., Pinto R., Sela S. (2011). Salmonella Typhimurium internalization is variable in leafy vegetables and fresh herbs. Int. J. Food Microbiol. 145, 250–257. 10.1016/j.ijfoodmicro.2010.12.031 [DOI] [PubMed] [Google Scholar]
- Gu G., Cevallos-Cevallos J. M., Vallad G. E., van Bruggen A. H. (2013b). Organically managed soils reduce internal colonization of tomato plants by Salmonella enterica serovar Typhimurium. Phytopathology 103, 381–388. 10.1094/PHYTO-04-12-0072-FI [DOI] [PubMed] [Google Scholar]
- Gu G., Cevallos-Cevallos J. M., van Bruggen A. H. (2013a). Ingress of Salmonella enterica Typhimurium into tomato leaves through hydathodes. PLoS ONE 8:e53470. 10.1371/journal.pone.0053470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Micallef S. A. (2016). Environmental metabolomics of the plant surface provides insights on Salmonella enterica colonization of tomato. Appl. Environ. Microbiol. 82, 3131–3142. 10.1128/AEM.00435-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Reyes C., Schikora A. (2013). Salmonella, a cross-kingdom pathogen infecting humans and plants. FEMS Microbiol. Lett. 343, 1–7. 10.1111/1574-6968.12127 [DOI] [PubMed] [Google Scholar]
- Jaeger C. H., III., Lindow S. E., Miller W., Clark E., Firestone M. K. (1999). Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65, 2685–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2009). Linking development to defense: auxin in plant-pathogen interactions. Trend. Plant Sci. 14, 373–382. 10.1016/j.tplants.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Klerks M. M., Franz E., van Gent-Pelzer M., Zijlstra C., van Bruggen A. H. (2007a). Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 1, 620–631. 10.1038/ismej.2007.82 [DOI] [PubMed] [Google Scholar]
- Klerks M. M., van Gent-Pelzer M., Franz E., Zijlstra C., van Bruggen A. H. (2007b). Physiological and molecular responses of Lactuca sativa to colonization by Salmonella enterica serovar Dublin. Appl. Environ. Microbiol. 73, 4905–4914. 10.1128/AEM.02522-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T., Hoffmann M., Hentrich M., Pollmann S. (2010). Indole-3-acetamide-dependent auxin biosynthesis: a widely distributed way of indole-3-acetic acid production? Eur. J. Cell Biol. 89, 895–905. 10.1016/j.ejcb.2010.06.021 [DOI] [PubMed] [Google Scholar]
- Leveau J. H., Lindow S. E. (2001). Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. U.S.A. 98, 3446–3453. 10.1073/pnas.061629598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E., Brandl M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. 10.1128/AEM.69.4.1875-1883.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E., Desurmont C., Elkins R., McGourty G., Clark E., Brandl M. T. (1998). Occurrence of indole-3-acetic Acid-producing bacteria on pear trees and their association with fruit russet. Phytopathology 88, 1149–1157. 10.1094/PHYTO.1998.88.11.1149 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. (2015). Bacteria and fungi controlling plant growth by manipulating auxin: balance between development and defense. J. Plant Physiol. 172, 4–12. 10.1016/j.jplph.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Manulis S., Haviv-Chesner A., Brandl M. T., Lindow S. E., Barash I. (1998). Differential involvement of indole-3-acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae. Mol. Plant Microbe Inter. 11, 634–642. 10.1094/MPMI.1998.11.7.634 [DOI] [PubMed] [Google Scholar]
- Martínez-Vaz B. M., Fink R. C., Diez-Gonzalez F., Sadowsky M. J. (2014). Enteric pathogen-plant interactions: molecular connections leading to colonization and growth and implications for food safety. Microb. Environ. 29, 123–135. 10.1264/jsme2.ME13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U., Mulders S., Gao M., Teplitski M., Caetano-Anolles G., Rolfe B. G., et al. (2003). Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. U.S.A. 100, 1444–1449. 10.1073/pnas.262672599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U., Schlaman H. R. M., Spaink H. P., Sautter C., Rolfe B. G., Djordjevic M. A. (1998). Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 14, 23–34. 10.1046/j.1365-313X.1998.00090.x [DOI] [PubMed] [Google Scholar]
- Mathesius U., Weinman J. J., Rolfe B. G., Djordjevic M. A. (2000). Rhizobia can induce nodules in white clover by “hijacking” mature cortical cells activated during lateral root development. Mol. Plant Microbe Interac. 13, 170–182. 10.1094/MPMI.2000.13.2.170 [DOI] [PubMed] [Google Scholar]
- Merighi M., Ellermeier C. D., Slauch J. M., Gunn J. S. (2005). Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J. Bacteriol. 187, 7407–7416. 10.1128/JB.187.21.7407-7416.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics, Cold Spring Harbor Laboratory. New York, NY: Cold Spring Harbor. [Google Scholar]
- Noel J. T., Arrach N., Alagely A., McClelland M., Teplitski M. (2010). Specific responses of Salmonella enterica to tomato varieties and fruit ripeness identified by in vivo expression technology. PLoS ONE 5:e12406. 10.1371/journal.pone.0012406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberto J. (2013). SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics 14:4. 10.1186/1471-2105-14-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongeng D., Geeraerd A. H., Springael D., Ryckeboer J., Muyanja C., Mauriello G. (2015). Fate of Escherichia coli O157:H7 and Salmonella enterica in the manure-amended soil-plant ecosystem of fresh vegetable crops: a review. Crit. Rev. Microbiol. 41, 273–294. 10.3109/1040841X.2013.829415 [DOI] [PubMed] [Google Scholar]
- Patten C. L., Blakney A. J. C., Coulson T. J. D. (2013). Activity, distribution and function of indole-3-acetic acid biosynthetic pathways in bacteria. Crit. Rev. Microbiol. 39, 395–415. 10.3109/1040841X.2012.716819 [DOI] [PubMed] [Google Scholar]
- Semenov A. M., Kuprianov A. A., van Bruggen A. H. (2010). Transfer of enteric pathogens to successive habitats as part of microbial cycles. Microb. Ecol. 60, 239–249. 10.1007/s00248-010-9663-0 [DOI] [PubMed] [Google Scholar]
- Spaepen S., Bossuyt S., Engelen K., Marchal K., Vanderleyden J. (2014). Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 201, 850–861. 10.1111/nph.12590 [DOI] [PubMed] [Google Scholar]
- Spaepen S., Versées W., Gocke D., Pohl M., Steyaert J., Vanderleyden J. (2007). Characterization of phenylpyruvate decarboxylase, involved in auxin production of Azospirillum brasilense. J. Bacteriol. 189, 7626–7633. 10.1128/JB.00830-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talboys P. J., Owen D. W., Healey J. R., Withers P. J., Jones D. L. (2014). Auxin secretion by Bacillus amyloliquefaciens FZB42 both stimulates root exudation and limits phosphorus uptake in Triticum aestivium. BMC Plant Biol. 14:51. 10.1186/1471-2229-14-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M., Warriner K., Bartz J., Schneider K. R. (2011). Untangling metabolic and communication networks: interactions of enterics with phytobacteria and their implications in produce safety. Trends Microbiol. 19, 121–127. 10.1016/j.tim.2010.11.007 [DOI] [PubMed] [Google Scholar]
- van Noorden G. E., Kerim T., Goffard N., Wiblin R., Pellerone F. I., Rolfe B. G., et al. (2007). Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti. Plant Physiol. 144, 1115–1131. 10.1104/pp.107.099978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K. A., Bennett S. D., Mahovic M., Gould L. H. (2014). Outbreaks associated with cantaloupe, watermelon, and honeydew in the United States, 1973-2011. Foodborn Path Dis. 11, 945–952. 10.1089/fpd.2014.1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann A., Virlogeux-Payant I., Chaussé A. M., Schikora A., Velge P. (2014). Interactions of Salmonella with animals and plants. Front. Microbiol. 5:791. 10.3389/fmicb.2014.00791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yue R., Sun T., Zhang L., Chen W., Zeng H., et al. (2015). Genome-wide identification, expression analysis of GH3 family genes in Medicago truncatula under stress-related hormones and Sinorhizobium meliloti infection. Appl. Microbiol. Biotechnol. 99, 841–854. 10.1007/s00253-014-6311-5 [DOI] [PubMed] [Google Scholar]