Abstract
Respiratory infections caused by Pseudomonas aeruginosa and Staphylococcus aureus are the leading cause of morbidity and mortality in cystic fibrosis (CF) patients. The authors aimed to identify volatile biomarkers from bronchoalveolar lavage (BAL) samples that can guide breath biomarker development for pathogen identification. BAL samples (n = 154) from CF patients were analyzed using two-dimensional gas chromatography time-of-flight mass spectrometry. Random Forest was used to select suites of volatiles for identifying P. aeruginosa-positive and S. aureus-positive samples using multiple infection scenarios and validated using test sets. Using nine volatile molecules, we differentiated P. aeruginosa-positive (n = 7) from P. aeruginosa-negative (n = 53) samples with an area under the receiver operating characteristic curve (AUROC) of 0.86 (95% CI 0.71–1.00) and with positive and negative predictive values of 0.67 (95% CI 0.38–0.75) and 0.92 (95% CI 0.88–1.00), respectively. We were also able to discriminate S. aureus-positive (n = 15) from S. aureus-negative (n = 45) samples with an AUROC of 0.88 (95% CI 0.79-1.00) using eight volatiles and with positive and negative predictive values of 0.86 (95% CI 0.61–0.96) and 0.70 (95% CI 0.61–0.75), respectively. Prospective validation of identified biomarkers as screening tools in patient breath may lead to clinical application.
Introduction
The leading cause of morbidity and mortality for patients with cystic fibrosis (CF) is respiratory failure associated with chronic bacterial infections and inflammation of the airways1. Prevalence of microorganisms varies with age, with Staphylococcus aureus and Pseudomonas aeruginosa being the most common bacterial species found in the airways of children and adults, respectively2. Accurate microbiological profiling of the lower airways and, consequently, prompt antibiotic treatment, are crucial for delaying chronic infections3. This is particularly true for P. aeruginosa, where eradication therapy is initiated whenever it is detected4. Early identification of S. aureus, both methicillin-sensitive (MSSA) and methicillin-resistant (MRSA), is also important, as MRSA is associated with increased risk of death, irrespective of CF disease severity5.
In expectorating patients, sputum is cultured for detection of bacterial pathogens. Most children as well as individuals with mild CF disease do not spontaneously expectorate sputum6,7. In non-expectorating patients, bronchoalveolar lavage (BAL) may be performed to diagnose lower airway infection. However, it is an invasive procedure that requires sedation and cannot be performed frequently. Hence, oropharyngeal (OP) swab cultures are used as surrogate specimens for species identification and antibiotic sensitivity determination of lower airway infections8. The current literature is divided on the utility of OP swabs as a reliable surrogate for BAL. Most studies on children report variable positive predictive values (PPV) ranging from 0.44 to 0.83 for P. aeruginosa and 0.33 to 0.64 for S. aureus9–13.
The measurement of volatile molecules from respiratory specimens, including BAL fluid, sputum and breath has been proposed as a minimally-invasive approach for differentiating P. aeruginosa-positive from P. aeruginosa-negative CF patients14–22. Diagnostic volatile molecular ‘suites’ from adult and pediatric patient breath have shown the greatest promise for detecting P. aeruginosa, with sensitivities and specificities approaching 1.00 and 0.88, respectively, in studies ranging from 16–233 subjects17,23. For S. aureus identification, Neerincx et al. showed that volatile molecules from patient breath (age ≥ 6 years) could be used to discriminate between S. aureus-infected CF patients (n = 13) from non-infected CF patients (n = 5) with a sensitivity of 1.00 and a specificity of 0.8024.
To set the groundwork for a breath study targeting biomarker evaluation of both P. aeruginosa and S. aureus, we accessed a heterogeneous set of ex-vivo BAL fluid samples (n = 154) obtained from 13 CF centers in the United States (US), via the Cystic Fibrosis Foundation Therapeutics (CFFT) biorepository. P. aeruginosa and S. aureus, the two most prevalent pathogens associated with CF, were detected by culture in 19% and 32% of samples, respectively. Validation sets were used to establish the sensitivity, specificity, PPV, and negative predictive value (NPV) for each set of putative volatile biomarkers.
Materials and Methods
Study subjects and design
BAL fluid samples stored at the Cystic Fibrosis Foundation Therapeutics (CFFT) Biorepository were accessed after approval from the Committee for the Protection of Human Subjects (CPHS) (STUDY00028597) at Dartmouth College. The stored samples were originally collected from subjects (age range: 2 months –50 years, n = 154) with a confirmed diagnosis of CF from 13 CF centers in the US at the time of clinically-indicated bronchoscopy with BAL fluid collection. Subjects with remnant BAL fluid after clinical testing were eligible to participate. The study was approved by the Institutional Review Board at each site. Written informed consent and HIPAA Authorization were obtained from all subjects ≥ 18 years or from parents or legal guardians of subjects < 18 years. Assent was obtained from subjects between 10–17 years. Clinical data (e.g., age, gender, lung function, body-mass index (BMI), and comorbidities) at the time of bronchoscopy were entered into a secure, web-based electronic database (REDCap).
Specimen collection and processing
Bronchoscopy and BAL fluid collection were performed following each site’s standard clinical procedure with most samples collected via laryngeal mask airway or endotracheal tube, limiting upper airway contamination. Standard BAL fluid culture was performed by the local clinical microbiology laboratory in accordance with Cystic Fibrosis Foundation (CFF) guidelines25 and results recorded. Remnant BAL fluid was frozen neat in 1 mL aliquots within one hour of collection at −70 °C. Research samples collected at participating sites were batch-shipped overnight on dry ice to Children’s Hospital Colorado, United States for storage in the CFFT biorepository specimen bank. For volatile metabolomics analysis, cryovials were shipped overnight on dry ice to Dartmouth College, US and stored at −80 °C. Accompanying de-identified clinical and microbiologic data was shared electronically using encryption.
Chemical measurements and data collection
Five hundred microliters of thawed BAL fluid were transferred to sterile 10 mL glass headspace vials containing a magnetic stir bar, and sealed with a polytetrafluoroethylene/silicone screw cap. Samples were stored at 4 °C prior to analysis by GC × GC-TOFMS. A total of 154 GC × GC chromatograms were analyzed, resulting in the detection of 973 volatile molecules across all samples (see Supplementary Information for details).
Statistical analysis
The aim of the study was to investigate whether suites of volatile molecules discriminate between BAL fluid samples in each of the following comparisons: (1) P. aeruginosa-positive (Pa+) versus no cultured microorganism (NCM), (2) S. aureus-positive (Sa+) versus NCM, (3) culture-positive (Pa+/Sa+) versus NCM, (4) Pa+ versus Sa+, (5) Pa+ versus P. aeruginosa-negative (Pa−), and (6) Sa+ versus S. aureus-negative (Sa−). Clinical microbiology results obtained at the time of bronchoscopy were used as the gold standard. Samples labelled as NCM contained no reported pathogens but could be positive for respiratory flora. Descriptive statistics were calculated for age, gender, genotype, BMI, comorbidities (pancreatic insufficiency and cystic fibrosis-related diabetes (CFRD)), forced vital capacity (FVC) percent predicted, and forced expiratory volume in 1 s (FEV1) percent predicted. To assess confounding demographic variables between study groups, the Mann-Whitney U test26 with Benjamini-Hochberg (BH) correction27 was used to compare continuous variables (age, BMI, FEV1 percent predicted, FVC percent predicted) and Mantel-Haenszel chi-square test28 with BH correction was used to compare categorical variables (gender, genotype, pancreatic insufficiency, CFRD).
Preprocessing of chromatographic data decreases influence of artefacts and allows for biological variation to be visualized. Data were log-transformed and normalised using probabilistic quotient normalization29. Samples were randomly assigned to training sets for all six models. The Random Forest (RF) classification algorithm was performed on the training sets to identify suites of discriminatory volatile biomarkers30. RF creates many de-correlated decision trees from a randomly selected subset of volatile compounds and predicts the sample class assignment. Two-thirds of the samples in the training sets were randomly selected with replacement for each decision tree (with an equal number of samples selected per class) and the remaining one-third were used to calculate the performance of the RF classification model. A total of 1000 trees were built for each round of RF. Discriminatory volatile molecules were ranked based on their mean decrease in accuracy over a 100 independent rounds of RF. Volatile molecules that contributed to decrease in accuracy of >30% in at least 90/100 rounds were chosen as the most discriminatory features. A test set was used to validate the accuracy of selected discriminatory panel of volatile molecules for all models.
For visualization purposes, principal component analysis (PCA) score plot was used to demonstrate the relatedness between all samples in the data. In a PCA score plot, each single point is represented by a sample. Points that lie close to each other have similar volatile molecule profiles, while points that are distant have different properties. ROC curves were plotted to represent performance of the predictive models made by RF. The area under the ROC curve (AUROC) is used to assess predictive performance: a value close to 1 indicates high predictive power of the model, whereas an AUC close to 0.5 means that the model has no predictive power31. Confidence intervals (CIs) for sensitivity and specificity are exact Clopper-Pearson CIs. Confidence intervals for PPV and NPV, were calculated using standard logit CIs. CIs for AUROC were calculated using DeLong method. All statistical analyses were performed in R 3.3.2.
Multivariate analysis of variance (MANOVA) was performed to test the influence of confounders on the profiles of putative discriminatory volatile biomarkers. Of the 38 volatiles identified from all models, the Mann-Whitney U-test with BH correction was used to assess differences in the mean concentration between sample classes.
Putative identification for volatile molecules
In total, 553, 400, and 200 chromatographic peaks were identified in at least one chromatogram of Pa+, Sa+ and NCM, respectively. Putative chemical class and chemical identification for volatile molecules that were present in at least 80% of one study group was based on mass spectral similarity score to a compound in the NIST library of at least 850/1000 and having a molecular weight of at least 60.0 amu. Volatile molecules that passed the above criteria and have been reported in the literature were assigned putative names. Experimentally-determined retention indices (RI) that are consistent with the volatile molecules assigned putative names on the mid-polar Rxi-624Sil stationary phase, were quantified using published median RIs for non-polar and polar columns, and the following equation:
Retention indices less than 600 (corresponding to C6) or greater than 1600 (corresponding to C16) were not extrapolated.
Pathway identification
For each putative volatile molecule assigned a name, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was searched for pathways identified in bacteria32.
Data availability
The datasets generated during and/or analysed in the current study are available from the corresponding author on reasonable request.
Results
BAL fluid samples from subjects with cystic fibrosis are polymicrobial and heterogeneous
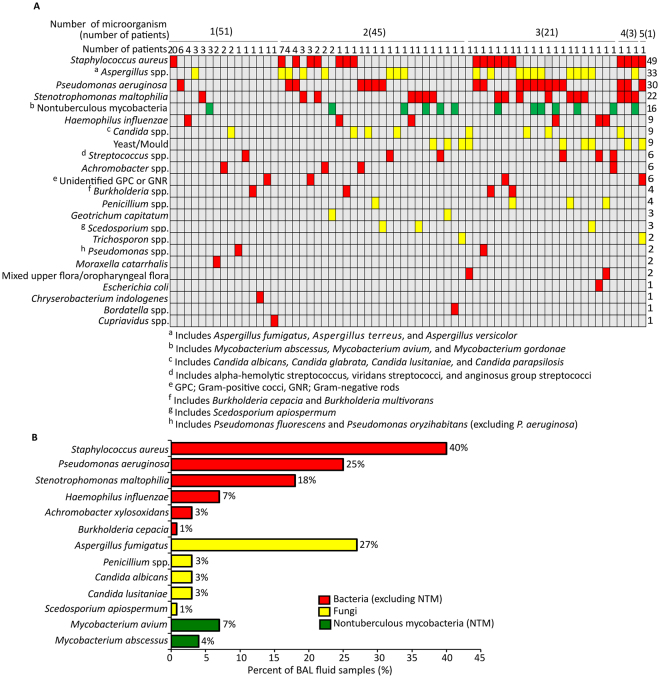
BAL fluid samples from subjects with CF (n = 154) were cultured for the identification of bacterial and fungal pathogens; 79% (n = 121) were identified as culture-positive and the remaining 21% (n = 33) culture-negative. The largest proportion of samples had only one culturable microorganism (n = 51), while 45, 21, 3, and 1 BAL fluid samples cultured two, three, four, and five microorganisms, respectively (Fig. 1A). Of the 66 unique microbiological profiles identified, 74% (n = 49) were specific to a single patient. Organisms cultured from at least 20% of BAL fluid samples were S. aureus (40%), followed by Aspergillus fumigatus (27%) and P. aeruginosa (25%) (Fig. 1B).
Figure 1.
Microbiological results from BAL fluid samples of patients with cystic fibrosis (n = 121; samples with no cultured microorganisms (NCM) excluded for clarity). (A) Colour matrix of the microbiological profiles observed among the samples; (red) bacteria (excluding nontuberculous mycobacteria (NTM)), (yellow) fungi, (green) NTM. (B) Bar plot of the most prevalent microorganisms in the samples (for organisms present in ≥1% samples).
S. aureus and P. aeruginosa represent the two most prevalent pathogens in the setting of CF, therefore BAL fluid samples that cultured either S. aureus (Sa+) or P. aeruginosa (Pa+) were selected for further analysis. BAL fluid samples containing no cultured microorganism (NCM), P. aeruginosa-negative (Pa−) and S. aureus-negative (Sa−) were used as comparison groups. For statistical analysis, we excluded data from samples that cultured both S. aureus and P. aeruginosa (n = 9) as well as 19 samples with incomplete clinical or demographic information. Age and BMI were significantly different in our Pa+ versus NCM and Pa+ versus Pa− model and could therefore confound our analysis (Table 1). We did not include medication in our analysis because the combination of drugs used by patients in our study groups exceeds the number of subjects in each group.
Table 1.
Demographic and clinical characteristics of subjects in the study groups.
| Study groups¥ | p-value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pa+ | Sa+ | NCM | Pa− | Sa− | P+ vs NCM | Sa+ vs NCM | Pa+/Sa+vs NCM | Pa+ vs Sa+ | Pa+ vs Pa− | Sa+ vs Sa− | |
| n | 19 | 40 | 32 | 114 | 93 | — | — | — | — | — | — |
| Age | 17.6 (±8.6) | 13.3 (±5.2) | 9.8 (±6.9) | 10.5 (±6.2) | 11.4 (±7.5) | *0.03 | 0.74 | 0.25 | 0.12 | **0.003 | 0.72 |
| Gender (M/F) | 9/10 | 25/15 | 15/17 | 55/59 | 39/54 | 1.00 | 0.74 | 0.51 | 0.66 | 1.00 | 0.19 |
| Genotype (F508del/F508del / F508del/other) | 6/7 | 15/14 | 13/7 | 66/41 | 53/34 | 0.64 | 0.82 | 0.51 | 1.00 | 0.60 | 0.72 |
| BMI, kg/m2 | 20.0 (±2.8) | 18.2 (±3.7) | 17.6 (±2.7) | 17.9 (±3.2) | 18.2 (±2.9) | *0.03 | 0.82 | 0.25 | 0.12 | **0.01 | 0.94 |
| Comorbidities (Y/N) Pancreatic insufficiency CF-related diabetes | 17/2 | 33/7 | 21/11 | 110/4 | 90/3 | 0.19 | 0.74 | 0.25 | 0.95 | 0.60 | 0.10 |
| 4/15 | 6/34 | 7/25 | 18/96 | 14/79 | 1.00 | 0.82 | 0.77 | 0.95 | 0.93 | 1.00 | |
| FVC % predicted Age ≥6 years | 82.0 (±16.7) n = 18 | 94.6 (±15.6) n = 35 | 94.2 (±19.7) n = 22 | 91.5 (±18.8) n = 87 | 88.5 (±20.1) n = 69 | 0.07 | 0.82 | 0.51 | 0.12 | 0.08 | 0.72 |
| FEV1% predicted Age ≥6 years | 74.1 (±20.5) n = 18 | 87.1 (±17.9) n = 35 | 85.5 (±22.3) n = 22 | 83.4 (±20.7) n = 87 | 78.4 (±21.7) n = 69 | 0.19 | 0.82 | 0.50 | 0.13 | 0.16 | 0.22 |
¥Samples can belong to more than one study group.
Data shown as mean (standard deviation) except where indicated.
p-value <0.05*, 0.01** considered statistically significant after Benjamini-Hochberg correction.
M, male; F, female, Y, yes; N, no; FVC, forced vital capacity; FEV1, forced expiratory volume in 1 s.
Volatile molecular profiles of study groups
In total, 973 peaks were identified in the headspace of the BAL fluid samples (Supplementary Figure S1). After removal of chromatographic artefacts and known contaminants (e.g., siloxanes, phthalates, atmospheric gasses), 805 peaks were selected for further analysis and putative chemical class identifications were assigned to 60 molecules covering a variety of chemical classes. Hydrocarbons were the most abundant chemical class in each group (Pa+ = 11, Sa+ = 9, NCM = 9), followed by ketones (Pa+ = 7, Sa+ = 6, NCM = 2) and alcohols (Pa+ = 5, Sa+ = 4, NCM = 3) (Fig. 2).
Figure 2.
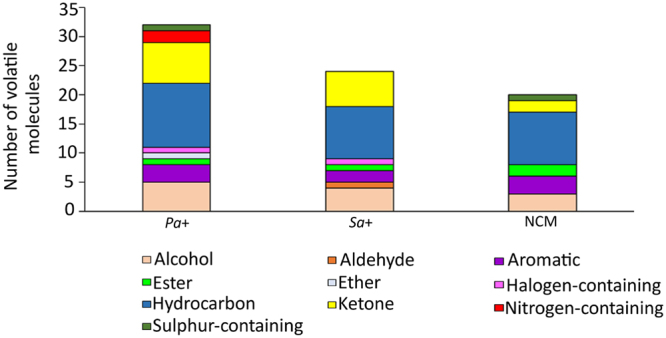
Putative chemical class identifications of volatile molecules present in BAL fluid samples of Pa+, Sa+, and NCM groups.
Identifying P. aeruginosa and S. aureus from BAL fluid samples using suites of volatile biomarkers
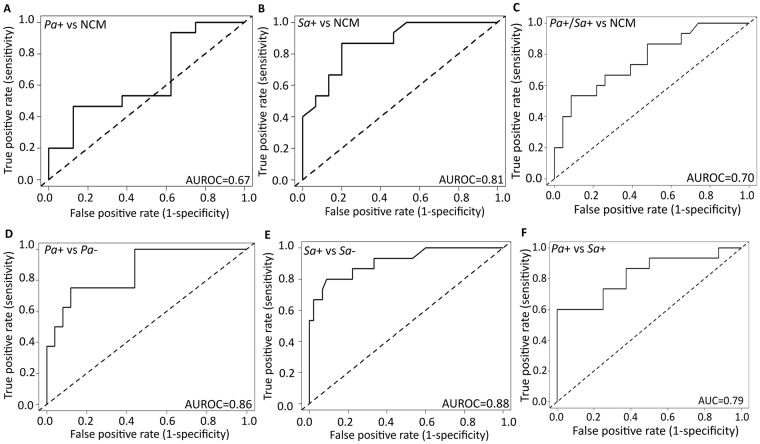
Starting from a basis of 805 peaks, the RF classification algorithm30 was used to identify suites of discriminatory volatile molecules for all models. Each model was validated using a test set, and sensitivity, specificity, PPV, NPV and AUROC values were calculated (Table 2). First, we evaluated the diagnostic accuracy of volatile molecules in discriminating culture-positive from culture-negative samples. Using a set of six, seven, and nine discriminatory volatile molecules selected from training sets for Pa+ versus NCM, Sa+ versus NCM, and Pa+/Sa+ versus NCM, respectively, AUROC values of 0.67 (95% CI 0.39–0.89), 0.81 (95% CI 0.74–0.99), and 0.70 (95% CI 0.62–0.89) were obtained on respective test set samples (Fig. 3A–E). Our Pa+/Sa+ versus NCM model had a sensitivity of 0.79 and PPV of 0.81 for the presence of either P. aeruginosa or S. aureus. In addition, we tested models for Pa+ versus Pa−, which yielded an AUROC of 0.86 (95% CI 0.71–1.00) and NPV of 0.92 using nine volatile molecules, and models for Sa+ vs Sa−, which yielded an AUROC of 0.88 (0.79–1.00) and a PPV of 0.86 using eight volatile molecules.
Table 2.
Model error rate, sensitivity, specificity, PPV, NPV, AUROC (95% CI) for volatile molecules from BAL fluid as diagnostic for Pa+ versus NCM, Sa+ versus NCM, Pa+/Sa+ versus NCM, Pa+ versus Sa+, Sa+ versus Pa+, Pa+ versus Pa− and Sa+ versus Sa−.
| n | Model error rate | Sensitivity | Specificity | PPV | NPV | AUROC | |
|---|---|---|---|---|---|---|---|
| Pa+ vs NCM (n = 53), volatile molecules used for test set = 6 | |||||||
| Training set | 30 | 0.30 | 0.35 (0.05–0.76) | 0.70 (0.40–0.90) | 0.30 (0.10–0.62) | 0.72 (0.55–0.88) | 0.70 (0.45–0.91) |
| Test set | 23 | 0.40 | 0.29 (0.04–0.71) | 0.63 (0.35–0.85) | 0.25 (0.08–0.56) | 0.66 (0.52–0.79) | 0.67 (0.39–0.89) |
| Sa + vs NCM (n = 73), volatile molecules used for test set = 7 | |||||||
| Training set | 43 | 0.32 | 0.58 (0.30–0.80) | 0.60 (0.27–0.85) | 0.62 (0.45–0.81) | 0.51 (0.33–0.66) | 0.87 (0.77–1.00) |
| Test set | 30 | 0.46 | 0.53 (0.27–0.77) | 0.54 (0.25–0.81) | 0.60 (0.42–0.76) | 0.45 (0.30–0.64) | 0.81 (0.74–0.99) |
| Pa +/ Sa + vs NCM (n = 91), volatile molecules used for test set = 9 | |||||||
| Training set | 53 | 0.30 | 0.82 (0.66–0.95) | 0.72 (0.39–0.90) | 0.86 (0.71–0.95) | 0.72 (0.46–0.89) | 0.80 (0.69–0.97) |
| Test set | 38 | 0.40 | 0.79 (0.63–0.90) | 0.60 (0.33–0.82) | 0.81 (0.70–0.89) | 0.56 (0.38–0.72) | 0.70 (0.62–0.89) |
| Pa + vs Sa + (n = 59), volatile molecules used for test set = 11 * | |||||||
| Training set | 36 | 0.25 | 0.70 (0.35–0.93) | 0.92 (0.63–1.00) | 0.88 (0.51–0.98) | 0.80 (0.61–0.91) | 0.84 (0.71–0.98) |
| Test set | 23 | 0.30 | 0.73 (0.46–0.87) | 0.81 (0.60–0.94) | 0.67 (0.43–0.85) | 0.72 (0.59–0.82) | 0.79 (0.67–0.95) |
| Sa + vs Pa + (n = 59), volatile molecules used for test set = 11 * | |||||||
| Training set | 36 | 0.25 | 0.92 (0.63–1.00) | 0.70 (0.35–0.93) | 0.80 (0.61–0.91) | 0.88 (0.51–0.98) | 0.84 (0.71–0.98) |
| Test set | 23 | 0.30 | 0.81 (0.60–0.94) | 0.73 (0.46–0.87) | 0.72 (0.59–0.82) | 0.67 (0.43–0.85) | 0.79 (0.67–0.95) |
| Pa + vs Pa− (n = 133), volatile molecules used for test set = 9 | |||||||
| Training set | 73 | 0.20 | 0.77 (0.60–0.90) | 0.91 (0.48–0.94) | 0.71 (0.37–0.80) | 0.95 (0.91–1.00) | 0.90 (0.82–1.00) |
| Test set | 60 | 0.25 | 0.75 (0.63–1.00) | 0.88 (0.48–0.90) | 0.67 (0.38–0.75) | 0.92 (0.88–1.00) | 0.86 (0.71–1.00) |
| Sa+ vs Sa− (n = 133), volatile molecules used for test set = 8 | |||||||
| Training set | 73 | 0.30 | 0.85 (0.80–0.99) | 0.56 (0.36–0.79) | 0.90 (0.81–0.95) | 0.73 (0.65–0.80) | 0.89 (0.80–1.00) |
| Test set | 60 | 0.37 | 0.80 (0.78–0.92) | 0.52 (0.31–0.72) | 0.86 (0.61–0.96) | 0.70 (0.61–0.75) | 0.88 (0.79–1.00) |
*Volatile molecules are the same for both models.
Figure 3.
Receiver operating characteristic (ROC) curves on test set samples of (A) Pa+ (n = 8) versus NCM (n = 15) using a panel of six volatile molecules, (B) Sa+ (n = 15) versus NCM (n = 15) using seven volatile molecules, (C) Pa+/Sa+ (n = 23) versus NCM (n = 15) using nine volatile molecules, (D) Pa+ (n = 7) versus Pa− (n = 53) using nine volatile molecules, (E) Sa+ (n = 15) versus Sa− (n = 45) using eight volatile molecules and (F) Pa+ (n = 8) versus Sa+ (n = 15) using 11 volatile molecules. (Dotted line indicates random classification).
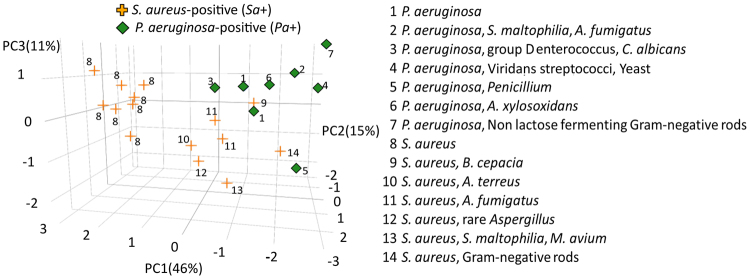
To determine whether volatile molecules can differentiate between polymicrobial P. aeruginosa-positive and S. aureus-positive samples, the Pa+ versus Sa+ model was built and validated on a test group of samples using a set of 11 discriminatory volatile molecules. The model had a specificity and NPV of 0.81 and 0.72, respectively for detecting P. aeruginosa, sensitivity and PPV of 0.67 and 0.66, respectively for detecting S. aureus and an AUROC of 0.79 (95% CI 0.67–0.95) (Fig. 3F). PCA plot was used to visualize the clustering of test set samples using discriminatory volatiles for the Pa+ versus Sa+ model, with labels indicating the complete clinical microbiology profiles (Fig. 4). Samples that cultured only S. aureus (profile #8, n = 8) clustered together, while samples that cultured S. aureus with additional Gram-negative microorganisms (profiles #9, #14, n = 2), or S. aureus along with Aspergillus spp. (profiles #10, #11, #12, n = 4) clustered closer to P. aeruginosa samples.
Figure 4.
Three-dimensional principal component scores plot on test set samples of Pa+ (diamond, n = 8) versus Sa+ (plus, n = 15) with complete microbiology profiles.
In pediatric patients, S. aureus pre-colonization has been shown to be a risk factor for initial P. aeruginosa airway infection33. Hence, it is important to assess the utility of diagnostic biomarkers in S. aureus and P. aeruginosa co-infected samples. Using the panel of our 28 Pa+ biomarkers identified from all our models above, we tested whether samples that were co-infected with P. aeruginosa and S. aureus (n = 9) could be identified as Pa+. We added these samples in our test set of Pa+ versus Pa− analysis and achieved a sensitivity and specificity of 0.74 and 0.89, suggesting that our reported suite has the potential to rule in P. aeruginosa in polymicrobial samples.
Putative identifications and relative abundance of the 38 discriminatory volatile molecules from all six models were determined using mass spectrometric and chromatographic data (Table 3, Supplementary Figures S2–S7). MANOVA was used to ensure that the volatile profile was not confounded by clinical and demographic characteristics of CF patients in Table 1. None of the characteristics significantly influenced the models (Supplementary Table S1). Six discriminatory volatile molecules had been previously reported and were given putative names; 2-butanone, 2-methyl-2-butanol, 1-butanol, ethyl acetate, 2-butanol and 3-methyl-2-butanone. Two out of the six volatile molecules were associated with metabolic pathways in the KEGG database; 2-butanone and 1-butanol (Table 4).
Table 3.
Putative identification of 38 discriminatory volatile molecules for all six models.
| Peak # | Putative identification | Putative molecular formula | Pa+ vs NCM | Sa+ vs NCM | Pa+/Sa+ vs NCM | Pa+ vs Sa+ | Pa+ vs Pa− | Sa+ vs Sa− | CAS# | RI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Unknown | Unknown | — | — | ↑ | — | — | — | — | — |
| 2 | Ketone | C6H12O2 | — | — | — | ↓ | — | — | — | — |
| 3 | 2-butanone | C 4 H 8 O | ↑** | ↑** | ↑* | ↓ | ↑* | — | 78-93-3 | 634 |
| 4 | Ether | C4H6O | — | — | — | ↑ | — | — | — | 635 |
| 5 | 2-methyl-2-butanol | C 5 H 12 O | — | — | — | ↑ | — | — | 75-85-4 | 686 |
| 6 | 1-butanol | C 4 H 10 O | — | — | — | ↓ | — | — | 71-36-3 | 717 |
| 7 | Aromatic | C6H8O | ↓ | ↑ | ↑ | — | — | ↑ | — | 734 |
| 8 | Ether | C6H12O2 | — | — | — | ↑ | — | — | — | 744 |
| 9 | Ketone | C6H12O | — | — | — | ↑ | — | — | — | 743 |
| 10 | Sulfur Containing | C20H42O2S | — | — | — | ↑ | — | — | — | 788 |
| 11 | Hydrocarbon | C15H30 | ↑* | — | ↑ | — | — | — | — | 795 |
| 12 | Ketone | C9H16O3 | — | — | ↑ | — | — | — | — | 826 |
| 13 | Alcohol | C11H22O2 | — | — | ↑ | — | — | — | — | 841 |
| 14 | Ester | C5H8O4 | — | ↑* | — | ↓ | — | ↑ | — | — |
| 15 | Hydrocarbon | C10H20 | ↑* | — | ↑* | — | — | — | — | — |
| 16 | Nitrogen Containing | C12H25NO2 | — | — | — | ↓ | — | — | — | — |
| 17 | Carboxylic Acid | C4H6O4 | — | — | ↑* | — | — | — | — | — |
| 18 | Ketone | C12H14O | — | — | ↓ | — | — | — | — | — |
| 19 | Alcohol | C4H10O3 | — | — | — | ↑ | — | — | — | — |
| 20 | Hydrocarbon | C4H8 | ↑ | — | — | — | ↑** | — | — | — |
| 21 | Ester | C4H8O2 | ↑ | — | — | — | — | — | — | — |
| 22 | Hydrocarbon | C8H18 | — | ↑ | — | — | — | — | — | 734 |
| 23 | Alcohol | C5H12O | — | ↑** | — | — | — | — | — | 768 |
| 24 | Alcohol | C16H14O | — | ↓** | — | — | — | — | — | 789 |
| 25 | Hydrocarbon | C9H20 | — | ↑** | — | — | — | — | — | — |
| 26 | Ethyl acetate | C 4 H 8 O 2 | — | — | — | — | ↓** | — | 141-78-6 | 638 |
| 27 | 2-butanol | C 4 H 10 O | — | — | — | — | ↓* | — | 78-92-2 | 686 |
| 28 | 3-methyl-2-butanone | C 5 H 10 O | — | — | — | — | ↑* | — | 563-80-4 | 700 |
| 29 | Ester | C6H12O2 | — | — | — | — | ↑* | — | — | 784 |
| 30 | Hydrocarbon | C4H10 | — | — | — | — | ↑* | — | — | — |
| 31 | Aromatic | C8H10 | — | — | — | — | ↑* | — | — | — |
| 32 | Nitrogen containing | C10H15N | — | — | — | — | ↓** | — | — | — |
| 33 | Ether | C4H6O | — | — | — | — | — | ↑ | — | — |
| 34 | Hydrocarbon | C7H16 | — | — | — | — | — | ↑ | — | — |
| 35 | Ketone | C6H12O | — | — | — | — | — | ↑ | — | — |
| 36 | Ketone | C8H16O | — | — | — | — | — | ↑ | — | — |
| 37 | Aromatic | C14H22 | — | — | — | — | — | ↑ | — | — |
| 38 | Ketone | C7H14O | — | — | — | — | — | ↑** | — | — |
Volatile molecules in bold have been previously reported in the literature in P. aeruginosa and/or S. aureus volatile metabolomics studies. RI; experimentally determined retention-index, Up; ↑, Down; ↓, based on mean peak area. p-value <0.05*, 0.01** considered statistically significant after Benjamini-Hochberg correction.
Table 4.
Previously reported P. aeruginosa– and S. aureus–-associated volatile molecules.
| Volatile molecule | KEGG pathway ID | Pathway name | Reference | ||
|---|---|---|---|---|---|
| Pa | Sa | Both | |||
| 2-butanone | 00460 | Cyanoamino acid metabolism | 23,34,35,49–52 | 24 | 44 |
| 01110 | Biosynthesis of secondary metabolites | ||||
| 2-methyl-2-butanol | — | — | 51 | — | — |
| 1-butanol | 00650 | Butanoate metabolism | 17,18,49–51 | — | 44,53,54 |
| 01120 | Microbial metabolism in diverse environments | ||||
| 01220 | Degradation of aromatic compounds | ||||
| 2-butanol | — | — | 44 | — | 50 |
| Ethyl acetate | — | — | 51 | — | 44,53 |
| 3-methyl-2-butanone | — | — | 35 | — | — |
Discussion
This is the first study to examine the diagnostic potential of a suite of volatile molecules in identifying the presence of two critical lung pathogens for patients with cystic fibrosis, P. aeruginosa and S. aureus. The molecules identified in the headspace of BAL fluid were similar in chemical class composition to previously reported culture and exhaled breath studies34–38. The range of models presented for the detection of each pathogen were validated using test sets and yield a set of putative discriminatory biomarkers for translation and evaluation in future breath studies.
In non-expectorating subjects, clinicians use OP swab culture as a surrogate for BAL fluid. Jung et al. reported a PPV and NPV of 0.83 and 0.74 respectively, for detecting P. aeruginosa using OP swab culture and a PPV and NPV of 1.00 and 0.94, respectively, using sputum culture in 38 patients13. Seidler et al. reported that in expectorating adults (n = 20), OP swab and sputum cultures both had a PPV of 1.00 for detecting P. aeruginosa and NPV of 0.50 and 0.60, respectively39. In this work, 28 putative volatile biomarkers were selected across all our models for the identification of P. aeruginosa in BAL fluid samples yielding a PPV range of 0.25–0.81 and an NPV range of 0.56–0.92. Our Pa+ versus Pa− model generated a specificity of 0.88 and an NPV of 0.92, suggesting that volatile molecules from the headspace of BAL fluid could be useful to screen patients and ‘rule in’ P. aeruginosa infections. Clinical best practice, as defined by the CF Foundation Pulmonary Guidelines, is to routinely surveil CF patients for new or recurring P. aeruginosa infections and initiate antibiotic eradication therapy at first detection, in an effort to prevent chronic infection and preserve lung function4. Ruling in P. aeruginosa is important as presence of the bacterium in the lung is significantly correlated with lower FEV1% predicted, increased serum C-reactive protein, and increased neutrophil elastase in sputum40.
Identification of S. aureus is also important, as methicillin-resistant S. aureus (MRSA) is associated with worse survival in CF patients5. Seidler et al. reported an NPV of 1.00 for detecting S. aureus in OP swab and sputum cultures versus BAL fluid and a PPV of 0.41 for OP swab and 0.57 for sputum versus BAL fluid39. Neericnx et al. demonstrated that a combination of nine volatile molecules in breath were able to discriminate between CF patients infected with S. aureus (n = 13) and uninfected CF patients (n = 5) with a sensitivity of 1.00 and a specificity of 0.8024. We identified 30 volatile molecules across all of our S. aureus models which yielded a PPV range of 0.60–0.86 and an NPV range of 0.37–0.70, respectively. Our Sa+ versus Sa− model generated a sensitivity of 0.80 and a PPV of 0.86, indicating that volatile molecules from BAL fluid samples could be used to ‘rule out’ S. aureus. The determination of S. aureus infection via volatile molecule panels is a promising avenue of research, however, the development of reliable exhaled breath diagnostics for S. aureus will require the use of paired representative samples from the lower and upper airways of subjects with CF to rule out false positives, as S. aureus is a colonizer of the upper airways of persons with CF41.
Six of the 38 discriminatory volatile molecules have been previously identified as bacterially-derived in volatile metabolomics studies (Table 4). Of these, the KEGG database identified microbial pathways associated with the metabolism of 2-butanone and 1-butanol. Of interest was the release of 2-butanone as a secondary metabolite during a reaction that also results in the formation of hydrogen cyanide (HCN). Hydrogen cyanide is an extensively studied biomarker for P. aeruginosa identification19–21 and more recently also shown to be produced by S. aureus42. We do not report on HCN (27.0 amu) as we did not collect data on ions with a m/z < 30. 2-butanone has also been reported as a biomarker for S. aureus detection in the breath of CF patients24, indicating the potential translation of BAL fluid volatile biomarkers to patient breath. KEGG pathways associated with 1-butanol indicate that it is produced as a by-product of the reversible oxidation of butane and butanal, a metabolic pathway that has been reported in P. butanavora43. 1-butanol has also been reported as a putative biomarker in the breath of CF patients with P. aeruginosa infection17. In addition, production of 1-butanol via the glycolytic fermentation of pyruvate has been reported for S. aureus44,45. These findings point towards possible microbial origins of 2-butanone and 1-butanol in BAL fluid samples.
Although culture remains the gold standard for clinical diagnosis, molecular-based approaches can identify additional microbial species that were not cultured, some of which may affect clinical outcomes in CF patients46–48. To the best of our knowledge, no volatile metabolomics study to-date has compared culture-dependent and culture-independent techniques for the purpose of organism identification. For our Pa+ vs Sa+ model, we compared the bacterial classifications based on 16 S ribosomal RNA (rRNA) sequencing (see supplementary information for details) and culture data. Our results show that samples with concordant microbiological results (average predicted probabilities; Pa+ = 0.70, Sa+ = 0.75) showed higher predicted probabilities than samples that had discordant microbiological findings (average predicted probabilities; Pa+ = 0.35, Sa+ = 0.30) (Supplementary Figure S8). Although the predicted probability of samples with concordant microbiology did not change, indicating the model was robust, the choice of reference sample for model generation in the field of volatile metabolomics is an unexplored issue. Future studies for P. aeruginosa and S. aureus volatile biomarkers should consider incorporating 16 S rRNA or quantitative polymerase chain reaction (qPCR) in order to determine sample status for predictive modeling.
CF lung infections are polymicrobial and our data shows that suites of volatile molecules from BAL fluid produced a predictive signal. We recognize that we have not fully investigated the impact of multiple pathogens on our reported volatile biomarker panel. This was beyond the scope of the study and shall be explored as a future avenue. As a first pass to address this question, we used our 28 Pa+ biomarkers to test whether the samples that were co-infected with P. aeruginosa and S. aureus could be identified as Pa+. Our sensitivity and specificity were 0.74 and 0.89 when these samples were included in the test set of Pa+ versus Pa− model, demonstrating the utility of the volatile biomarkers to rule in P. aeruginosa in polymicrobial samples. In addition, we acknowledge that using the BAL technique to obtain diagnostic samples is invasive, expensive, and thus cannot be performed frequently in a clinical setting. We do not propose the use of volatile molecule suites from BAL fluid as a rapid or non-invasive diagnostic in clinical labs. Furthermore, the PPV and NPV values for our study might not be reflective of the actual prevalence of P. aeruginosa and S. aureus in all CF populations. However, the proposed suite of biomarkers in this study can now serve as a basis for designing well-powered breath studies in the CF patient population, an avenue we are currently exploring. Furthermore, we expect that complementing the culture results with culture-independent data might have the potential to identify additional, clinically relevant microbial species, some of which may affect model generation and interpretation46–48. We conclude that volatile molecules from BAL fluid can provide discriminatory power for ruling in P. aeruginosa and ruling out S. aureus and plan to extend this work to exhaled breath in CF patients.
Electronic supplementary material
Acknowledgements
We are grateful to have received all the BAL fluid samples from Cystic Fibrosis Foundation Therapeutics, Inc. The authors thank the investigators: Richard C. Ahrens, James F. Chmiel, John P. Clancy, Ronald L. Gibson, William T. Harris, Geoffrey Kurland, Theresa A. Laguna, Susanna A. McColley, Karen McCoy, George Retsch-Bogart, Kurtis T. Sobush, and Pamela L. Zeitlin; Research Coordinators, patients and families who participated in the Multicenter Biorepository Study for BALF from Cystic Fibrosis Patients. M.N. and C.A.R. were supported by the Burroughs Wellcome Fund institutional program grant unifying population and laboratory based sciences to Dartmouth College (Grant#1014106). H.D.B. and J.E.H. were supported by grants from CFF (Grant#HILL16I0) and National Institute of Health (NIH) (Grant#UL1TR001086). E.T.Z. was supported by grants from CFFT (Grant#ZEMANI11A0) and NIH/NHLBI (Grant#K23HL114883). A.S. was supported by a grant from Niels Stensen Fellowship.
Author Contributions
E.T.Z. collected the BAL samples. E.T.Z., H.D.B. and J.E.H. outlined the study design. M.N. and C.A.R. analyzed the samples for chromatographic analyses. M.N. and H.D.B. preprocessed the chromatographic data. M.N. and A.S. conducted the statistical analyses. M.N. prepared all figures and tables. M.N., H.D.B. and J.E.H. wrote the manuscript text. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18491-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 2.Patient Registry Annual Data Report. (Cystic Fibrosis Foundation, 2015).
- 3.Padman R, et al. Infant care patterns at epidemiologic study of cystic fibrosis sites that achieve superior childhood lung function. Pediatrics. 2007;119:E531–E537. doi: 10.1542/peds.2006-1414. [DOI] [PubMed] [Google Scholar]
- 4.Mogayzel PJ, Jr, et al. Cystic Fibrosis Foundation pulmonary guideline. pharmacologic approaches to prevention and eradication of initial Pseudomonas aeruginosa infection. Ann Am Thorac Soc. 2014;11:1640–1650. doi: 10.1513/AnnalsATS.201404-166OC. [DOI] [PubMed] [Google Scholar]
- 5.Dasenbrook EC, et al. Association between respiratory tract methicillin-resistant Staphylococcus aureus and survival in cystic fibrosis. JAMA. 2010;303:2386–2392. doi: 10.1001/jama.2010.791. [DOI] [PubMed] [Google Scholar]
- 6.Radhakrishnan DK, Corey M, Dell SD. Realities of expectorated sputum collection in the pediatric cystic fibrosis clinic. Arch Pediatr Adolesc Med. 2007;161:603–606. doi: 10.1001/archpedi.161.6.603. [DOI] [PubMed] [Google Scholar]
- 7.Sagel SD, Kapsner R, Osberg I, Sontag MK, Accurso FJ. Airway inflammation in children with cystic fibrosis and healthy children assessed by sputum induction. Am J Respir Crit Care Med. 2001;164:1425–1431. doi: 10.1164/ajrccm.164.8.2104075. [DOI] [PubMed] [Google Scholar]
- 8.Saiman, L., Siegel, J. & Cystic Fibrosis Foundation Consensus Conference on Infection Control, P. Infection control recommendations for patients with cystic fibrosis: Microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Am J Infect Control31, S1–62 (2003). [PubMed]
- 9.Armstrong DS, et al. Bronchoalveolar lavage or oropharyngeal cultures to identify lower respiratory pathogens in infants with cystic fibrosis. Pediatr Pulmonol. 1996;21:267–275. doi: 10.1002/(SICI)1099-0496(199605)21:5<267::AID-PPUL1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 10.Rosenfeld, M. et al. Diagnostic accuracy of oropharyngeal cultures in infants and young children with cystic fibrosis. Pediatric Pulmonology28, 3.0.co;2-v (1999). [DOI] [PubMed]
- 11.Ramsey BW, et al. Predictive value of oropharyngeal cultures for identifying lower airway bacteria in cystic fibrosis patients. Am Rev Respir Dis. 1991;144:331–337. doi: 10.1164/ajrccm/144.2.331. [DOI] [PubMed] [Google Scholar]
- 12.Burns JL, et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 13.Jung A, et al. Sequential genotyping of Pseudomonas aeruginosa from upper and lower airways of cystic fibrosis patients. Eur Respir J. 2002;20:1457–1463. doi: 10.1183/09031936.02.00268002. [DOI] [PubMed] [Google Scholar]
- 14.Robroeks CM, et al. Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatr Res. 2010;68:75–80. doi: 10.1203/PDR.0b013e3181df4ea0. [DOI] [PubMed] [Google Scholar]
- 15.Kramer R, et al. A rapid method for breath analysis in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 2015;34:745–751. doi: 10.1007/s10096-014-2286-5. [DOI] [PubMed] [Google Scholar]
- 16.Scott-Thomas AJ, et al. 2-Aminoacetophenone as a potential breath biomarker for Pseudomonas aeruginosa in the cystic fibrosis lung. BMC Pulm Med. 2010;10:56. doi: 10.1186/1471-2466-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pabary R, et al. Does mass spectrometric breath analysis detect Pseudomonas aeruginosa in cystic fibrosis? Eur Respir J. 2016;47:994–997. doi: 10.1183/13993003.00944-2015. [DOI] [PubMed] [Google Scholar]
- 18.Savelev SU, et al. Volatile biomarkers of Pseudomonas aeruginosa in cystic fibrosis and noncystic fibrosis bronchiectasis. Lett Appl Microbiol. 2011;52:610–613. doi: 10.1111/j.1472-765X.2011.03049.x. [DOI] [PubMed] [Google Scholar]
- 19.Gilchrist, F. J. et al. Exhaled breath hydrogen cyanide as a marker of early Pseudomonas aeruginosa infection in children with cystic fibrosis. ERJ Open Research1 (2015). [DOI] [PMC free article] [PubMed]
- 20.Enderby B, Smith D, Carroll W, Lenney W. Hydrogen cyanide as a biomarker for Pseudomonas aeruginosa in the breath of children with cystic fibrosis. Pediatr Pulmonol. 2009;44:142–147. doi: 10.1002/ppul.20963. [DOI] [PubMed] [Google Scholar]
- 21.Gilchrist FJ, et al. Hydrogen cyanide concentrations in the breath of adult cystic fibrosis patients with and without Pseudomonas aeruginosa infection. J Breath Res. 2013;7:026010. doi: 10.1088/1752-7155/7/2/026010. [DOI] [PubMed] [Google Scholar]
- 22.Stutz MD, et al. Cyanide in bronchoalveolar lavage is not diagnostic for <em>Pseudomonas aeruginosa</em> in children with cystic fibrosis. European Respiratory Journal. 2011;37:553–558. doi: 10.1183/09031936.00024210. [DOI] [PubMed] [Google Scholar]
- 23.Shestivska V, et al. Variability in the concentrations of volatile metabolites emitted by genotypically different strains of Pseudomonas aeruginosa. J Appl Microbiol. 2012;113:701–713. doi: 10.1111/j.1365-2672.2012.05370.x. [DOI] [PubMed] [Google Scholar]
- 24.Neerincx AH, et al. Detection of Staphylococcus aureus in cystic fibrosis patients using breath VOC profiles. J Breath Res. 2016;10:046014. doi: 10.1088/1752-7155/10/4/046014. [DOI] [PubMed] [Google Scholar]
- 25.Burns JL, et al. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis. 1998;27:158–163. doi: 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- 26.Mann HB, Whitney DR. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Statist. 1947;18:50–60. doi: 10.1214/aoms/1177730491. [DOI] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 28.Cochran WG. Some Methods for Strengthening the Common x2 Tests. Biometrics. 1954;10:417–451. doi: 10.2307/3001616. [DOI] [Google Scholar]
- 29.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in H-1 NMR metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 30.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 31.Hanley JA, Mcneil BJ. The Meaning and Use of the Area under a Receiver Operating Characteristic (Roc) Curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maselli JH, et al. Risk factors for initial acquisition of Pseudomonas aeruginosa in children with cystic fibrosis identified by newborn screening. Pediatr Pulmonol. 2003;35:257–262. doi: 10.1002/ppul.10230. [DOI] [PubMed] [Google Scholar]
- 34.Bean HD, Dimandja JM, Hill JE. Bacterial volatile discovery using solid phase microextraction and comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;901:41–46. doi: 10.1016/j.jchromb.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bean HD, Rees CA, Hill JE. Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates. J Breath Res. 2016;10:047102. doi: 10.1088/1752-7155/10/4/047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nizio KD, et al. In vitro volatile organic compound profiling using GCxGC-TOFMS to differentiate bacteria associated with lung infections: a proof-of-concept study. J Breath Res. 2016;10:026008. doi: 10.1088/1752-7155/10/2/026008. [DOI] [PubMed] [Google Scholar]
- 37.Scholler C, Molin S, Wilkins K. Volatile metabolites from some gram-negative bacteria. Chemosphere. 1997;35:1487–1495. doi: 10.1016/S0045-6535(97)00209-9. [DOI] [PubMed] [Google Scholar]
- 38.Bos LD, Sterk PJ, Schultz MJ. Volatile metabolites of pathogens: a systematic review. PLoS Pathog. 2013;9:e1003311. doi: 10.1371/journal.ppat.1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seidler D, Griffin M, Nymon A, Koeppen K, Ashare A. Throat Swabs and Sputum Culture as Predictors of P. aeruginosa or S. aureus Lung Colonization in Adult Cystic Fibrosis Patients. PLoS One. 2016;11:e0164232. doi: 10.1371/journal.pone.0164232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zemanick ET, et al. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS One. 2013;8:e62917. doi: 10.1371/journal.pone.0062917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neerincx AH, et al. Hydrogen cyanide emission in the lung by Staphylococcus aureus. Eur Respir J. 2016;48:577–579. doi: 10.1183/13993003.02093-2015. [DOI] [PubMed] [Google Scholar]
- 43.Vangnai AS, Sayavedra-Soto LA, Arp DJ. Roles for the two 1-butanol dehydrogenases of Pseudomonas butanovora in butane and 1-butanol metabolism. J Bacteriol. 2002;184:4343–4350. doi: 10.1128/JB.184.16.4343-4350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filipiak W, et al. Molecular analysis of volatile metabolites released specifically by Staphylococcus aureus and Pseudomonas aeruginosa. BMC Microbiol. 2012;12:113. doi: 10.1186/1471-2180-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schomburg, D. & Michal, G. Biochemical pathways: an atlas of biochemistry and molecular biology. 2nd edn, (John Wiley & Sons, 2012).
- 46.LiPuma JJ. Assessing Airway Microbiota in Cystic Fibrosis: What More Should Be Done? J Clin Microbiol. 2015;53:2006–2007. doi: 10.1128/JCM.01218-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zemanick ET, et al. Pulmonary exacerbations in cystic fibrosis with negative bacterial cultures. Pediatr Pulmonol. 2010;45:569–577. doi: 10.1002/ppul.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burns JL, Rolain JM. Culture-based diagnostic microbiology in cystic fibrosis: can we simplify the complexity? J Cyst Fibros. 2014;13:1–9. doi: 10.1016/j.jcf.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Labows, J., McGinley, K., Webster, G. & Leyden, J. Headspace analysis of volatile metabolites of Pseudomonas aeruginosa and related species by gas chromatography-mass spectrometry. Journal of Clinical Microbiology12 (1980). [DOI] [PMC free article] [PubMed]
- 50.Zechman JM, Labows JN., Jr. Volatiles of Pseudomonas aeruginosa and related species by automated headspace concentration–gas chromatography. Can J Microbiol. 1985;31:232–237. doi: 10.1139/m85-045. [DOI] [PubMed] [Google Scholar]
- 51.Goeminne PC, et al. Detection of Pseudomonas aeruginosa in sputum headspace through volatile organic compound analysis. Respir Res. 2012;13:87. doi: 10.1186/1465-9921-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rees, C., Burklund, A., Stefanuto, P. H., Schwartzman, J. D. & Hill, J. E. Comprehensive volatile metabolic fingerprinting of bacterial and fungal pathogen groups. J Breath Res, 10.1088/1752-7163/aa8f7f (2017). [DOI] [PMC free article] [PubMed]
- 53.Thorn RMS, Reynolds DM, Greenman J. Multivariate analysis of bacterial volatile compound profiles for discrimination between selected species and strains in vitro. Journal of Microbiological Methods. 2011;84:258–264. doi: 10.1016/j.mimet.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Zhu J, Bean HD, Kuo YM, Hill JE. Fast detection of volatile organic compounds from bacterial cultures by secondary electrospray ionization-mass spectrometry. J Clin Microbiol. 2010;48:4426–4431. doi: 10.1128/JCM.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed in the current study are available from the corresponding author on reasonable request.