Abstract
A wide range of miRNAs have been identified as phloem-mobile molecules that play important roles in coordinating plant development and physiology. Phytoplasmas are associated with hundreds of plant diseases, and the pathogenesis involved in the interactions between phytoplasmas and plants is still poorly understood. To analyse the molecular mechanisms of phytoplasma pathogenicity, the miRNAs profiles in mulberry phloem saps were examined in response to phytoplasma infection. A total of 86 conserved miRNAs and 19 novel miRNAs were identified, and 30 conserved miRNAs and 13 novel miRNAs were differentially expressed upon infection with phytoplasmas. The target genes of the differentially expressed miRNAs are involved in diverse signalling pathways showing the complex interactions between mulberry and phytoplasma. Interestingly, we found that mul-miR482a-5p was up-regulated in the infected phloem saps, and grafting experiments showed that it can be transported from scions to rootstock. Based on the results, the complexity and roles of the miRNAs in phloem sap and the potential molecular mechanisms of their changes were discussed. It is likely that the phytoplasma-responsive miRNAs in the phloem sap modulate multiple pathways and work cooperatively in response to phytoplasma infection, and their expression changes may be responsible for some symptoms in the infected plants.
Introduction
Mulberry trees that have long been cultivated for sericulture are susceptible to many diseases, among which yellow dwarf disease caused by phytoplasma is one of the most devastating1. Phytoplasmas are wall-less, obligate intracellular plant pathogens in the class Mollicutes2 that infect several hundred economically important plants and cause devastating losses in agriculture and forestry3. The inability to culture phytoplasmas in vitro makes it difficult to characterize the plant pathogens at the molecular level, and the underlying molecular mechanisms of their pathogenicity are still poorly understood4.
When subjected to pathogen infection, the host plant activates sophisticated response mechanisms to reprogramme the expression of genes, proteins and metabolites5. The gene, protein and metabolite profiles in some host plants challenged with phytoplasmas have been investigated by differential methods6–16. MiRNAs functioning as negative regulators of gene expression are involved in the control of plant development and immunity17. Increasing evidence showed that miRNAs serve as an important mechanism for mediating gene expression during plant-pathogen interactions, and many miRNAs have been linked to resistance responses in plants18–21. A group of bacteria-responsive miRNAs and their target genes have been identified, and their regulatory functions have been extensively characterized in model plant species21–29. However, to our knowledge, only three studies have explored phytoplasma-responsive miRNAs in Mexican lime (Citrus aurantifolia L.), mulberry (Morus multicaulis Perr.) and Paulownia fortunei30–32. Although some miRNA families are conserved among various plant species, every species will have their own specific miRNAs, and the functions of these miRNAs may also be species-specific33,34. Moreover, different miRNAs, in addition to individual miRNAs in the same family, may be expressed differentially in various tissues and have different functions in response to the same pathogen infection35. Therefore, phytoplasma-responsive miRNAs have not been fully explored, and their mediating mechanisms for gene expression in response to phytoplasma are largely unknown.
Phloem is not only the major route for the translocation and distribution of organic metabolites but also an important mediator of whole-plant communication involved in whole plant events, including stress responses and long-distance signalling36. Recently, many miRNAs have been identified from the phloem exudates from Brassica napus37, Malus domestica (apple)38, and Lupinus albus39. Although the role of phloem miRNAs is not yet clear, some miRNAs in the phloem have been demonstrated to use long-distance signalling and have a role in mediating plant developmental patterning and stress responses38,40–48. In plants, phytoplasmas are restricted to the sieve elements of phloem tissues49. Therefore, it is reasonable to assume that the phloem is the site where the host immediate defence response against phytoplasma occurs and is involved in the coordination of the defence response at the whole plant level. Identification and characterization of phytoplasma-responsive miRNAs in the phloem sap promises to enhance our understanding of the molecular mechanisms involved in yellow dwarf disease symptom development.
In the present study, based on transcriptome information for mulberry, we employed high-throughput sequencing to profile the miRNAs in the phloem sap during the response of mulberry to phytoplasma infection. Phytoplasma-responsive miRNAs were identified, and their potential target genes were predicted. In addition, the translocation and functions of the mul-miR482a-5p involved in the response of mulberry to phytoplasma infection were discussed. The results reported here may facilitate our understanding of phytoplasma pathogenicity.
Results
Purity assessment of phloem sap
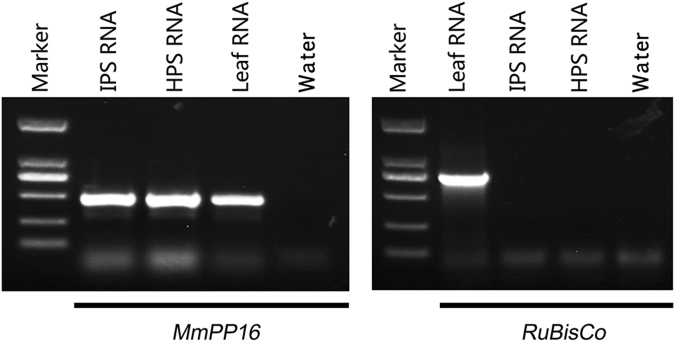
To identify phloem-enriched sRNAs and ensure that the sRNAs identified in phloem sap did not result from contamination during sampling, the frequency of the ribulose bisphosphate carboxylase oxygenase (RuBisCo) large subunit gene that would be expected to in leaves, but not the sieve element–companion cell complex, was determined to assess the purity of the phloem sap sampled. The results detected no RuBisCo mRNA in the phloem sap samples, but this mRNA was clearly present in leaf tissue. Meanwhile, phloem-specific MmPP16 mRNA was detected in phloem sap samples (Fig. 1). This indicates that contamination from surrounding tissues in the collected phloem sap samples was very low.
Figure 1.
Total RNAs extracted from mulberry leaves and phloem saps were analysed by RT-PCR for the presence of RuBisCo and MmPP16 mRNAs. IPS, phloem sap sampled from infected trees. HPS, phloem sap sampled from healthy trees. The gels used were cropped from different gels showed in the Supplementary Figure 1.
Overview of small RNA in phloem saps
To examine the phytoplasma-responsive miRNAs in phloem sap, the sRNA libraries were constructed from phytoplasma-infected and healthy mulberry phloem saps and subjected to Solexa deep sequencing. After removing the rRNA, tRNA, and degradation products from the matching sequences, the remaining clean small RNA sequences were aligned with the miRNA precursors/mature miRNAs in the miRBase database v21.0, and 86 known miRNAs members belonging to 78 families were identified (Table 1). All non-annotated sRNA sequences were mapped to our mulberry transcriptome database, and 19 sequences were found to perfectly match the transcriptome sequences. The mapped RNAs were able to fold into hairpin structures and had negative folding free energies (from −21.0 to −95.65 kcal mol−1 with an average of about −49.05 kcal mol−1) according to Mfold; these values were lower than the folding free energies of rRNA (−33 kcal mol−1) and tRNA (−27.5 kcal mol−1)50. Therefore, these mapped RNAs were identified as candidate novel miRNAs (Table 2). Interestingly, some miRNA-3p sequences, such as mul-miR160b-3p, mul-miR166h-3p and mul-miR169p-3p, were identified, but the corresponding miRNA-5p sequences were not detected. This may indicate that these miRNA-3ps may be the authentic miRNAs. The size distribution of the small RNA sequences identified in the two libraries showed that more than 80% of the mapped small RNAs were 20–24 nt long, with 24 nt and 21 nt as the major size groups (Fig. 2).
Table 1.
Profiles of conserved miRNAs in mulberry phloem saps.
| MiRNA-name | Sequence (5′-3′) | Normalized value | Fold-change (log2 IPS/HPS) | P-value | Significance lable | |
|---|---|---|---|---|---|---|
| IPS | HPS | |||||
| mul-miR1134 | CAGAACGAAGAAGAAGAAGAAGA | 22.10 | 22.34 | −0.02 | 0.90042163 | |
| mul-miR1223e | UUGAGAUGUCAUGCACCACUCUG | 1.58 | 5.11 | −1.69 | 1.18E-06 | ** |
| mul-miR1310 | GAGGCAUCGGGGGCGCAACG | 44.79 | 24.47 | 0.87 | 5.19E-18 | |
| mul-miR1511 | ACUAUGCUCUGAUACCAUGUUAA | 1.25 | 5.35 | −2.10 | 9.19E-09 | ** |
| mul-miR1520m | AAUUCAAACUGAGAUGUGACAUU | 132.28 | 124.54 | 0.09 | 0.08960825 | |
| mul-miR156a-5p | UGACAGAAGAGAGUGAGCAC | 20.68 | 7.55 | 1.45 | 8.14E-19 | ** |
| mul-miR157a-5p | UUGACAGAAGAUAGAGAGCAC | 881.9 | 44.69 | 4.30 | 0 | ** |
| mul-miR159a-3p | UUUGGAUUGAAGGGAGCUCU | 84.07 | 158.84 | −0.92 | 1.63E-64 | |
| mul-miR160a-5p | UGCCUGGCUCCCUGUAUGCCA | 1.97 | 0.75 | 1.39 | 0.00994309 | ** |
| mul-miR160b-3p | GCGUAUGAGGAGCCAUGCAUA | 22.44 | 15.66 | 0.52 | 0.00010904 | |
| mul-miR164a-5p | UGGAGAAGCAGGGCACGUGCA | 3.54 | 8.42 | −1.25 | 5.02E-07 | ** |
| mul-miR165b-5p | GAAGUGUUCGGAUCGAGGC | 9181.72 | 2752.65 | 1.74 | 0 | ** |
| mul-miR166a-3p | UCGGACCAGGCUUCAUUCCCC | 2068.07 | 1700.73 | 0.28 | 2.47E-98 | |
| mul-miR166h-3p | UCGGACCAGGCUUCAUUCCC | 1899.18 | 1541.57 | 0.30 | 4.99E-102 | |
| mul-miR167d-5p | UGAAGCUGCCAGCAUGAUCUG | 2.25 | 1.81 | 0.32 | 0.44192478 | |
| mul-miR168a-5p | UCGCUUGGUGCAGGUCGGGAA | 161.64 | 163.80 | −0.02 | 0.67470362 | |
| mul-miR168a-3p | CCCGCCUUGCAUCAACUGAAU | 1.75 | 2.44 | −0.48 | 0.24497579 | |
| mul-miR169b | CAGCCAAGGAUGACUUGCCGG | 1.75 | 2.67 | −0.61 | 0.12748684 | |
| mul-miR169p-3p | GGCAUAUGAUCAUCUUGGGGCUAG | 3.84 | 10.86 | −1.50 | 6.76E-11 | ** |
| mul-miR171g-5p | UUUUGGAUGGCUCAACACG | 20.10 | 39.26 | −0.97 | 1.48E-18 | |
| mul-miR172a-3p | AGAAUCUUGAUGAUGCUGCAU | 21.85 | 38.16 | −0.80 | 1.15E-13 | |
| mul-miR172e-3p | GAAUCUUGAUGAUGCUGCAU | 21.85 | 38.94 | −0.83 | 1.04E-14 | |
| mul-miR1856 | UACGUAGAGGCGGAUUCGUA | 88.82 | 43.58 | 1.03 | 2.35E-44 | ** |
| mul-miR1858a | GAGCGGAGGACUGUAGUGGGUGC | 37.11 | 93.62 | −1.33 | 2.27E-69 | ** |
| mul-miR2108b | GUUAGAUGUGAUUGUUUGUGAG | 329.36 | 282.59 | 0.22 | 2.99E-11 | |
| mul-miR2118-5p | GUCGAUGGAACAAUGUAGGCAAGG | 562.31 | 157.74 | 1.83 | 0 | ** |
| mul-miR2199 | UGAUAACUCGACGGAUCGC | 61232.99 | 54719.33 | 0.16 | 0 | |
| mul-miR2670f | GGGUCUGUUUGGUUGGGGGA | 89.83 | 49.64 | 0.86 | 1.89E-33 | |
| mul-miR2867-3p | CCAGGACGGUGGUCAUGGA | 89.58 | 77.81 | 0.20 | 0.00138453 | |
| mul-miR2873b | AUUGGCUGGAGAUAUUGGUAUG | 30.36 | 26.98 | 0.17 | 0.11724709 | |
| mul-miR2911 | GCCGGGGGACGGACTGGGAA | 153874.88 | 140579.96 | 0.13 | 0 | |
| mul-miR2916 | GUUGGGGGCUCGAAGACGAUCAGA | 6604.64 | 5708.74 | 0.21 | 3.21E-177 | |
| mul-miR319a | UUGGACUGAAGGGAGCUCCC | 1.33 | 7.63 | −2.52 | 1.56E-14 | ** |
| mul-miR3630-3p | GGGAAUCUCUCUGAUGCA | 0.50 | 1.65 | −1.72 | 0.00597169 | ** |
| mul-miR3630-5p | GCAAGUGAUGAUAAACAGACA | 2.09 | 3.46 | −0.73 | 0.04145473 | |
| mul-miR390a-5p | AAGCUCAGGAGGGAUAGCGCC | 25.19 | 10.31 | 1.29 | 4.65E-19 | ** |
| mul-miR390a-3p | CGCUAUCUAUCCUGAGUUUCA | 3.17 | 1.97 | 0.69 | 0.06255943 | |
| mul-miR393a-5p | UCCAAAGGGAUCGCAUUGA | 1.50 | 2.12 | −0.50 | 0.25832055 | |
| mul-miR396a | UUCCACAGCUUUCUUGAACUG | 0.92 | 1.73 | −0.92 | 0.08385227 | |
| mul-miR396b-3p | GCUCAAGAAAGCUGUGGGAGA | 4.50 | 6.53 | −0.54 | 0.03282992 | |
| mul-miR397a-5p | UCAUUGAGUGCAGCGUUGAUG | 1.33 | 0.16 | 3.08 | 0.00046347 | ** |
| mul-miR398a-5p | GGCGUGACCCCUGAGAACACAAG | 1.08 | 2.75 | −1.34 | 0.00276708 | ** |
| mul-miR408b-5p | CAGGGAACGGACAGAGCAUGG | 61.30 | 63.57 | −0.05 | 0.47719536 | |
| mul-miR4403 | ACGGCACAAACACGACACGAGCAC | 3.75 | 2.52 | 0.58 | 0.08327133 | |
| mul-miR4414a-3p | AUCCAACGAUGCAGGAGCUAGCC | 3.67 | 7.32 | −0.99 | 0.00010668 | |
| mul-miR4414a-5p | AGCUGCUGACUCGUUGGUUCA | 47.54 | 75.37 | −0.66 | 9.44E-19 | |
| mul-miR447b-5p | ACUCUCACUCAAGGGCUUCA | 1.67 | 0.79 | 1.08 | 0.04888033 | * |
| mul-miR472b-3p | UUUUCCCAACACCACCCAUACC | 22.77 | 19.51 | 0.22 | 0.07812493 | |
| mul-miR473a-5p | ACUCUCCCCCUUAAGGCUUCCA | 70.81 | 115.18 | −0.70 | 1.95E-30 | |
| mul-miR477c | CUCUCCCCCUUAAGGCUUCC | 77.90 | 139.01 | −0.84 | 1.32E-48 | |
| mul-miR482a-3p | UUCCCAAGGCCGCCCAUUCCGA | 217.77 | 160.57 | 0.44 | 3.91E-25 | |
| mul-miR482a-5p | GGAAUGGGCUGUUUGGGAAGA | 2140.30 | 964.91 | 1.15 | 0 | ** |
| mul-miR5021 | GAGGGAGAAGAAGAAGAAGA | 47.04 | 54.12 | −0.20 | 0.01340026 | |
| mul-miR5039 | CCCUAUUUUUAAUCGUUGGA | 0.92 | 1.42 | −0.63 | 0.26283984 | |
| mul-miR5054 | GUGCCCCACGGUGGGCGCCA | 16.99 | 1.25 | 3.76 | 1.51E-44 | ** |
| mul-miR5059 | CGGGCCUGGCGCACCCCA | 940.30 | 1455.27 | −0.63 | 9.97E-301 | |
| mul-miR5072 | GUUCCCCAGUGGAGUCGCCA | 72.69 | 2.92 | 4.64 | 3.41E-214 | ** |
| mul-miR5077 | UUCACGUCGGGUUCACCA | 12.51 | 28.32 | −1.18 | 1.67E-18 | ** |
| mul-miR5085 | AAGGACAUUUGGUUGUGGCUC | 129.36 | 187.63 | −0.54 | 1.08E-30 | |
| mul-miR5139 | GUAACCUGGCUCUGAUACCA | 2.25 | 1.10 | 1.03 | 0.02729391 | * |
| mul-miR5224a | UUGAUGGACAUGAAGACGUUAU | 4.84 | 4.09 | 0.24 | 0.38021350 | |
| mul-miR5266 | CGGGGGACGGACUGGGGC | 25.35 | 36.27 | −0.52 | 1.02E-06 | |
| mul-miR5279 | GGAACCUCGGAUGAUCGGUUA | 6.17 | 9.91 | −0.68 | 0.00105543 | |
| mul-miR5293 | GGAGGAAGUGAGAAGAAGAAGA | 9.34 | 10.78 | −0.21 | 0.2625989 | |
| mul-miR529-3p | GCUGUACCCCCUCUCUUCUC | 1.83 | 0.55 | 1.74 | 0.00316862 | ** |
| mul-miR529b | AGAAGAGAGAGAGUACAGCUU | 21.35 | 5.35 | 2.00 | 1.85E-29 | ** |
| mul-miR5368 | GGGACAGUCUCAGGUAGACAGUU | 1.17 | 0.63 | 0.89 | 0.16322128 | |
| mul-miR5386 | CGUCAGCUGUCGGCGGACUG | 33.70 | 47.28 | −0.49 | 1.11E-07 | |
| mul-miR5568f-3p | GUCUGGUAAUUGGAAUGAG | 447.29 | 559.12 | −0.32 | 3.04E-35 | |
| mul-miR5641 | UGGAACGAACAGAGAUAGAAUUA | 3.17 | 2.60 | 0.29 | 0.40238294 | |
| mul-miR5813 | ACAGCAGGACGGUGGUCAUGGA | 29487.21 | 24982.87 | 0.24 | 0 | |
| mul-miR6030 | UCCCCCAACCAAACAGACCCU | 77.98 | 114.39 | −0.55 | 2.53E-20 | |
| mul-miR6150 | AGUUUGUUUGAUGGUACUUGC | 764.73 | 1088.74 | −0.51 | 1.54E-154 | |
| mul-miR6180 | AGGGUCGGAGGAAAGAGGGCC | 2.00 | 0.63 | 1.67 | 0.00267402 | ** |
| mul-miR6191 | AUAAUUUGUCUGGUUAUGAA | 19.77 | 22.34 | −0.18 | 0.16412556 | |
| mul-miR6196 | GAGGACAGGAGUAGAGAGGA | 5.59 | 3.07 | 0.86 | 0.00251337 | |
| mul-miR6214 | CACGACACGAGCUGACGACA | 5.09 | 0.01 | 8.99 | 6.91E-20 | ** |
| mul-miR6235-5p | UGUGAGAGAAAAUACUGUAGCGA | 115.68 | 110.14 | 0.07 | 0.19506665 | |
| mul-miR6300 | GUCGUUGUAGUAUAGUGGU | 5283.44 | 18100.50 | −1.78 | 0 | ** |
| mul-miR6478 | CCGACCUUAGCUCAGUUGG | 41.53 | 9.20 | 2.17 | 4.78E-62 | ** |
| mul-miR845a | CGGCUCUGAUACCAACUGUGACG | 6.92 | 6.37 | 0.12 | 0.59522396 | |
| mul-miR854a | GGAUGGGAUGGAGGAGGAG | 30.19 | 33.36 | −0.14 | 0.16388474 | |
| mul-miR894 | GUUUCACGUCGGGUUCAC | 46.96 | 97.32 | −1.051 | 6.61E-50 | ** |
| mul-miR952b | AACGAGGAUCCAUUGGAG | 911.10 | 888.21 | 0.04 | 0.05791621 | |
| mul-miRn10-3p | AGGUGCAGAUGCAGAUGCAGG | 6.6723 | 0.01 | 9.38 | 7.50E-26 | ** |
| mul-miRn12-3p | UCUUGCCGAGACCUCCCAUA | 33.6116 | 28.2432 | 0.25 | 0.016399105 | |
IPS, phloem sap sampled from infected trees. HPS, phloem sap sampled from healthy trees.
Table 2.
Novel miRNAs in mulberry phloem saps by Illumina sequencing.
| MiRNA-name | Sequence (5′-3′) | Precursor sequence (5′-3′) | Energy (kcal mol-1) |
|---|---|---|---|
| mul-miRn21-3p | GAGCAGUGCGGAGUAGCUGAG | GGUUCCGGCUGUUGCGUUGGAACUGAAUGUCUUAUUUAUUCACUUCAUCAUUUAUUUAUUUAACGCUUUCUUAUUUAUUUAUAAAGUUAUUUCAAAUUAUAUCCUACUGGACCUUUUACUCACGUUUUAUUGUUUUAAAAAUGUUAACCCCUCUCUUGAGACUCUAAGAGCAGUGCGGAGUAGCUGAGUUG | −33.60 |
| mul-miRn22-5p | CAGCGAACUAAACGGGCCCU | AUCAGCGAACUAAACGGGCCCUUAAACUUUCGUUUUUUCACCUCAUCAUUCUGUACUUGUUAACUUCUGUAGAAAUCUUAAAAAAUUCAAUUAAACACUCAUUUUGUCCCCACUCAGAGCUGAUAAUUUAAAAGUUUAAAAAUUCUAGCCAUACUCACUUAGGGCGUGUUUGGUUCGGGGGA | −35.84 |
| mul-miRn23-5p | UGAGGAUGUAUCAGAAGAUAG | AAAGGGUCCCUGAGGAUGUAUCAGAAGAUAGUGCAGAUAUUUGGUUUUGAUAGGCAUUCUUGUUCUGGAGUAUGCUUUUGCUUAUCCUUCCCCCACUACAUUCGCAUUGCUGGAUCUGCUUGAGGAAACUAAAACCAACAAUAAUUUUAGUUAAAGUAUUCUGCUGUUGUGGAAUAUUGGUUGACACAUGAGAUAUCUCCCUUUCUUUCACCGUCAAAGGCAGAACUCUCUUGUUCCCACUUUCCUCUAACACUUCAGCUCCAGCUGUAGAAGAGAAAGUAUCUCGCUUAGCCACACCAAUGUAUUUUGACUUGCCUCUGACAGAGGUGUUUUUGAUAGCAUCUCUCAAAACAUACAUUUUAGUGGUAUUCC | −95.65 |
| mul-miRn24-5p | AAGCUAGCUGUGUGGAUGAUA | UAUGCAACAAAAGCUAGCUGUGUGGAUGAUAUUAAUUGUCGUUUUAGCGGCAGCGUGUUUCGUUGUCGC | −21.80 |
| mul-miRn25-3p | UUCCAAAUCCACCCAUGCCCAC | UUUGAGCUUUAUGAAGUUGUCGGGCCUGGGAGGUUUGGUAGGAGUAAUAAGUAAUUACCAUUUAGUUUUUUGUUCACUUAAUUGAUAUUAUAAUUGUAUGUUUUAAUUUAGUUCUCCUUCCAAAUCCACCCAUGCCCACAAUUUCCUCAGGCUUCUCUC | −51.80 |
| mul-miRn26-5p | GCUUCCUCGGAGACGGCGCACG | AGAAUAACAACAAAUCGGCAGCUUCCUCGGAGACGGCGCACGACAGCAAGAAGGGUGGGUCGUCCUCGGGAAGCGGAGAGCAGGCGGCGGCGCCG | −37.00 |
| mul-miRn27-5p | CAGACAUUGAGUGGGGGAGG | UGAUAUUUUCAGCCUGUAAUCAGACAUUGAGUGGGGGAGGAAGAGAAGAUCUCGUUACCGGCGGGUCGACCCGGAUAAACCGCUGGAAAAUGACGGUUUUGUUCCUUGCCACGCGGCGGCCGGCGAUCGGUGCCAGCCAUGGUAGCCGCGGUCUCAUCUCUCUCCCGCGCUUUGUCUGAGAAACGGCCAGUCUGAGCCC | −82.7 |
| mul-miRn28-3p | GGACUUUAUGGACCCGUCGGUG | CGGGCAAUGCUGUGAACGGUCAGAUCCCGCCGGCCGACCGGUCUGUGAGUCCUUUGUUACCGGACUUUAUGGACCCGUCGGUGUGCUAUGUCC | −37.9 |
| mul-miRn29-5p | GUGGAUCAAGAACUGGAGGC | UUUGCAACAUGUGGAUCAAGAACUGGAGGCAAAGGUUACUGCUUCAUUUGCACCACAGAGAUCUCAAGUGGCACAACCUCCUGCAACCAAAGGUUCUUAUUCACAGGUUGGAUU | −35.1 |
| mul-miRn30-3p | UCCAGAAGCAAUCGUACGGGA | CGUGGAGCACCCUGUUCCUGUAGUUGCUCCUGGAUCUGCUAAGAAUCUCUCUGAAGUCAAAAUUAAUCCAGAAGCAAUCGUACGGGAAGGACACACU | −27.9 |
| mul-miRn31-3p | AUGCACUGCCUCUUCCCUGGC | AGAGGGGGUCAAAAAGCAGAAUAAGGCAGGGAACGGACAGAGCAUGGAUGGAGCCUUCAACAGAAGAAGGAAUGCUGUUGUGGCUCUACUCAUGCACUGCCUCUUCCCUGGCUGUGCCUCUC | −56.7 |
| mul-miRn32-5p | GGAAUGUUGUCUGGCUCGAGG | AAUCCCGCUAAGAAGUCUUUGUUUAAGAGCUAUAGACUAUAAAGUAAGGGAAUGGGACCCCGACGGGAAUGUAUCCCAAGCAGCGGAGUAAGUUCACUCUUUGGUAAGCUUAGGCGCCUAAAUGUCUGAGUUCGGAAAGCAGAGUAAGCGGCGAUC | −45.0 |
| mul-miRn33-3p | GGGAGAAAGAGGAAAAUAGGC | UUUGUUUUUGUUUUUUUUUUUAAUAUCACAAGAUUCACAAGUCACAACCCUGCUUGUUAGAGCACAGACACAAGCAACUGUAGGUGGACUAAGUAGCCAUGGGCCAGGCUGAGAGCGGUAUCAUGAGGCCUCGCAGAGUAGCUUCAAUGCAUUGGAUGCAUUUCCAACGUAAAAAAUUUCACUUUUAAGCGUGUAAUUUUUAAAUAAUUUUUAAGAAUUUUUUUGGGAGAAAGAGGAAAAUAGGC | −55.6 |
| mul-miRn34-5p | GGGAGCUGAGUUGAUGAGCA | GGACUGCUAAACAAGCUCGUCUCUCGCUCCCUCUCCGUCGCUGGAAAAUGGCAGCAGCAACAGCUCCGCCGUCUUAACAUCCAUGAAUAUCAGGGAGCUGAGUUGAUGAGCAAAUGUGGGAU | −43.6 |
| mul-miRn35-5p | GCAGAAGAGUCAGAGCUUUGA | AGGUAGUUUUGCAGAAGAGUCAGAGCUUUGAUUUGAAUCUCAGAAAAAAUAAAAACCGAGAAAGAAAAAAGAAAUGGCGAGCCCGAAAUCCGGCAGCCAAAACGACGGCGUUCCGUGCGACUUCUGCAGCGAGCAAACGGCGGUGCUGUACUGCAGAGCCGACUCGGCGAAGCUCUGCCUCUUCUGCGACCAGCACGUCCA | −75.2 |
| mul-miRn36-3p | GCUGAAGCUGGGGUGGGGCC | GCGGCGGCUCUGGUGCUCCACACCUUCUUCAGCUGGCUGCUGAUGCUGAAGCUGGGGUGGGGCCUUGCCGGCGG | −41 |
| mul-miRn37-3p | GGACGGCAUCGAUCGGAGCUC | CGGCGAGGGAGCUCCGACCGAAGCUUCCUCUUGGCGAUGGACGGCAUCGAUCGGAGCUCUUGCUUCGCU | −37.2 |
| mul-miRn38-3p | GACUGAAAGCGGACCUGGUGGUG | GAAGAUUUGUAUUUGGCCGCCAGGUCCACCUUCAGUCUUCUUCAAAGACCUUCGUUGCUGCCACACAGCCAGCUUUGGUUUCAAGGACUGAAAGCGGACCUGGUGGUGAUAAUUCAAA | −49.5 |
| mul-miRn39-5p | UCGACCAGCCGAGUAGAAGUA | AUAUUGGAUCUCGACCAGCCGAGUAGAAGUAAGUAUCUCAUUUCCCUCGAUUGUUACUUCUACUCGGCUGGUAGAGAUUCAAUGUU | −50.6 |
Figure 2.

Length distribution of small RNA in mulberry phloem sap sRNA libraries. IPS, phloem sap sampled from infected trees. HPS, phloem sap sampled from healthy trees.
Expression profiling of miRNAs in response to phytoplasma-infection
The profiles of some miRNAs differed between the healthy and infected phloem sap libraries. A total of 30 known miRNAs and 13 novel miRNAs were found to be differentially expressed between phytoplasma-infected and healthy mulberry phloem sap libraries (Tables 1, 3). These differentially expressed miRNAs were considered phytoplasma-responsive miRNAs, among which 15 miRNAs decreased and 28 miRNAs increased significantly in the infected phloem saps (P < 0.05, fold 2.0). These phytoplasma-response miRNAs consist of not only highly expressed miRNAs such as mul-miR166a, mul-miR166h-3p, mul-miR2199, and mul-miR5813 but also low-abundance miRNAs such as mul-miR160a, mul-miR167d-5p, mul-miR3630-3p, mul-miR3630-5p, and mul-miR396a. Although both the miRNA-5p and miRNA-3p of some miRNAs were detected, only miRNA-5p or miRNA-3p changed during phytoplasma-infection. This strongly suggested that single-strand miRNAs, and not miRNA-5p/miRNA-3p duplexes, are the phytoplasma infection-relevant molecular species. To validate the miRNA expression differences revealed by the sequencing experiments, we performed RT-qPCR analysis for 12 known miRNAs and 10 novel miRNAs covering different expression patterns. The results obtained by RT-qPCR showed a very strong correlation with read frequencies, demonstrating that our sequencing data are quantitative and reliable (Fig. 3).
Table 3.
Expression profiling of novel miRNAs in mulberry phloem saps.
| No. | MiRNA-name | Normalized value | Fold-change (log2 IPS/HPS) | P-value | Significance lable | |
|---|---|---|---|---|---|---|
| IPS | HPS | |||||
| 1 | mul-miRn21-3p | 0.01 | 1.8095 | −7.50 | 2.38E-07 | ** |
| 2 | mul-miRn22-5p | 0.01 | 4.7203 | −8.88 | 5.02E-18 | ** |
| 3 | mul-miRn23-5p | 1.6681 | 0.01 | 7.38 | 5.12E-07 | ** |
| 4 | mul-miRn24-5p | 2.3353 | 0.01 | 7.87 | 1.58E-09 | ** |
| 5 | mul-miRn25-3p | 5.7548 | 5.507 | 0.063 | 0.793593809 | |
| 6 | mul-miRn26-5p | 1.7515 | 0.9441 | 0.89 | 0.085551202 | |
| 7 | mul-miRn27-5p | 20.0168 | 12.5088 | 0.68 | 3.39E-06 | |
| 8 | mul-miRn28-3p | 9.7582 | 0.01 | 9.93 | 1.82E-37 | ** |
| 9 | mul-miRn29-5p | 2.0017 | 0.01 | 7.65 | 2.84E-08 | ** |
| 10 | mul-miRn30-3p | 2.4187 | 0.01 | 7.92 | 7.66E-10 | ** |
| 11 | mul-miRn31-3p | 0.9174 | 1.8881 | −1.04 | 0.044254454 | * |
| 12 | mul-miRn32-5p | 15.2628 | 34.9303 | −1.19 | 6.62E-23 | ** |
| 13 | mul-miRn33-3p | 4.8374 | 0.01 | 8.92 | 6.04E-19 | ** |
| 14 | mul-miRn34-5p | 1.0842 | 1.6521 | −0.61 | 0.237320172 | |
| 15 | mul-miRn35-5p | 1.7515 | 0.7867 | 1.15 | 0.03366659 | * |
| 16 | mul-miRn36-3p | 1.7515 | 1.3374 | 0.39 | 0.410468979 | |
| 17 | mul-miRn37-3p | 1.8349 | 0.01 | 7.52 | 1.21E-07 | ** |
| 18 | mul-miRn38-3p | 1.7515 | 1.4948 | −0.22863852 | 0.617178011 | |
| 19 | mul-miRn39-5p | 65.5551 | 38.6279 | −0.76306477 | 1.07E-20 | |
IPS, phloem sap sampled from infected trees. HPS, phloem sap sampled from healthy trees.
Figure 3.

Verification of selected miRNAs from deep sequencing by RT-qPCR. (A) Conserved miRNA abundance analysis by RT-qPCR. (B) Novel miRNA abundance analysis by RT-qPCR. Relative miRNA abundance was evaluated using comparative Ct method with U6 as the reference. Log2 values of the ratio of phytoplasma-infected samples to healthy samples are plotted. Values are given as the mean ± SD of three experiments per group. IPS, phloem sap from infected trees. HPS, phloem sap from healthy trees.
Phytoplasma-responsive miRNAs related to diverse biologic processes
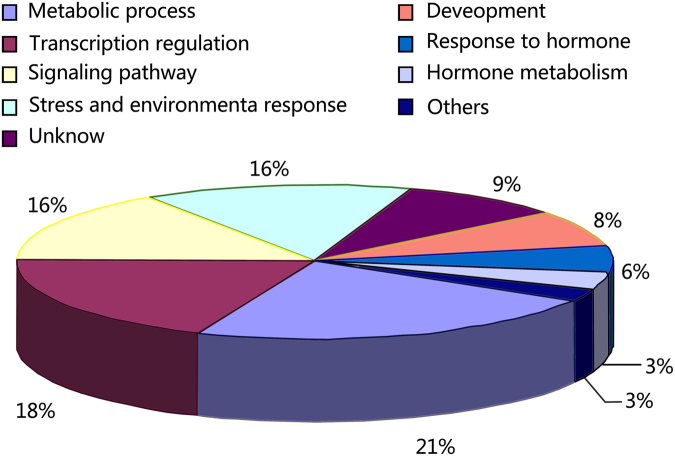
To understand the biological functions of phytoplasma-responsive miRNAs reported here, the target genes of these miRNAs were predicted and subjected to Gene Ontology (GO) analysis. Together, 131 target genes of 36 phytoplasma-responsive miRNAs were predicted and classified into nine categories according to their ontologies in Arabidopsis, based on KEGG functional annotations (Tables 4, 5; Fig. 4). The categories included metabolic process, transcription regulation, signalling pathway, stress and environmental response, development, response to hormones and hormone metabolism. Interestingly, 16% of target genes were found to be associated with signalling pathways. This indicated that many miRNAs in the phloem sap were associated with signal transduction and that diverse signalling pathways were involved in phytoplasma infection in the infected plants. In addition, many genes homologous to the sequences of unknown functions were predicted as targets of phytoplasma-responsive miRNAs. Further analyses of these genes and miRNAs may reveal new biological functions for phloem. Thus, these phytoplasma-responsive miRNAs in the phloem saps might be related to diverse biologic processes, and the regulatory networks involved in the response to phytoplasma-infection in mulberry were intricate.
Table 4.
Predicted targets of the differential conserved miRNAs.
| MiRNA-name | Putative GO_process | Predicted target annotations in mulberry transcriptome data |
|---|---|---|
| mul-miR1223e | Hormone metabolism | O-fucosyltransferase |
| Metabolic process | Glutaredoxin | |
| Metabolic process | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | |
| mul-miR1511 | Unknow | Transposable |
| Stress response | Trichome | |
| mul-miR156a | Transcription regulation | Squamosa promoter binding protein-like 7 |
| Transcription regulation | Squamosa promoter binding protein-like 9 | |
| Transcription regulation | Squamosa promoter binding protein-like 10 | |
| Signaling pathway | Protein kinase superfamily protein | |
| Signaling pathway | Cysteine/Histidine-rich C1 domain family protein | |
| Metabolic process | Putative pyridine nucleotide-disulphide oxidoreductase | |
| Metabolic process | UDP-glycosyltransferase-like protein | |
| mul-miR157a | Transcription regulation | Squamosa promoter binding protein-like 7 |
| Transcription regulation | Squamosa promoter binding protein-like 10 | |
| Transcription regulation | LIM domain-containing protein | |
| Unknown | Unknown protein | |
| Hormone metabolism | Galactose oxidase/kelch repeat superfamily protein | |
| Signaling pathway; Development | F-box family protein | |
| Stress response | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| Metabolic process | Dioxygenase-like protein | |
| mul-miR160a | Auxin signaling; Transcription regulation | Auxin response factor 10 |
| Auxin signaling; Transcription regulation | Auxin response factor 16 | |
| Auxin signaling; Transcription regulation | Auxin response factor 18 | |
| Transcription regulation | NAC domain containing protein 1 | |
| Transcription regulation | NAC domain containing protein 6 | |
| Unknown | Unknown protein | |
| Signaling pathway | CBL-interacting protein kinase | |
| mul-miR165b-5p | Metabolic process | Methyl esterase 17 |
| Stress response | Leucine-rich repeat receptor-like protein kinase | |
| Development | Embryo defective 1379 protein | |
| mul-miR1856 | Metabolic process | Glucan synthase-like 3 |
| mul-miR1858a | Metabolic process | Transmembrane amino acid transporter family protein |
| Stress response | Major facilitator superfamily protein | |
| mul-miR2118-5p | Secondary metabolitic process; Environmental responses | Cytochrome P450 like_TBP |
| mul-miR319a | Transcription regulation | R2R3-MYB transcription factor |
| Transcription regulation | Transcription factor MYB811 | |
| Defense response | Disease resistance protein | |
| mul-miR3630-3p | Signal transduction | Leucine-rich receptor-like protein kinase |
| Signal transduction | Leucine-rich repeat transmembrane protein kinase-like protein | |
| Transcription regulation | Smg-4/UPF3-like protein | |
| Secondary metabolitic process | Lycopene beta-cyclase | |
| RNA process | Endoribonuclease | |
| Unknown | Uncharacterized protein | |
| mul-miR390a | Auxin signaling; Development | TAS3/TASIR-ARF (TRANS-ACTING SIRNA3) |
| Signal transduction | Protein kinase superfamily protein | |
| Signal transduction | Leucine-rich repeat protein kinase-like protein | |
| mul-miR397a | Metabolic process; Stress response | Laccase 2 |
| Metabolic process | Laccase 11 | |
| Transcription regulation | CLP protease proteolytic subunit 3 | |
| Response to stress | TRICHOME BIREFRINGENCE-LIKE 14 | |
| Signal transduction | Protein kinase superfamily protein | |
| mul-miR398a-5p | Metabolic process; Transcription regulation | Purple acid phosphatase 14 |
| mul-miR447 | Metabolic process | P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| Microtubule-based process | ATP binding microtubule motor family protein | |
| mul-miR482a-5p | Transcription regulation | Regulator of chromosome condensation family protein |
| Metabolic process | Trehalose 6-phosphate synthase | |
| Metabolic process | Inositol 1,3,4-trisphosphate 5/6-kinase family protein | |
| mul-miR5072 | Metabolic process | Heteroglycan glucosidase 1 |
| mul-miR5077 | Signal transduction | Calponin domain-containing protein |
| Metabolic process | Slpha-rhamnosidase-like protein | |
| Metabolic process | ATP-sulfurylase precursor | |
| Defense response | Guanylate-binding-like protein | |
| Unknown | Uncharacterized protein | |
| mul-miR5139 | Unknown | Transposable element gene |
| Transcription regulation | RNA-dependent RNA polymerase family protein | |
| mul-miR529b | Transcription regulation | SPL domain class transcription factor |
| Transcription regulation | Squamosa promoter binding protein-like 9 | |
| Development and environmental responses | Pentatricopeptide repeat (PPR) superfamily protein | |
| Responses to biotic stress | Glycine/proline-rich protein | |
| Response to jasmonic acid and wounding | Inosine-uridine preferring nucleoside hydrolase family protein | |
| Responses to viral infection | Cysteine-rich repeat secretory protein 60 | |
| Signal transduction | Leucine-rich repeat protein kinase family protein | |
| Metabolic process; Environmental responses | 3-ketoacyl-CoA synthase 19 | |
| Development, response to auxin | SAUR-like auxin-responsive protein family | |
| Unknown | Hypothetical protein | |
| mul-miR529-3p | Response to stress | ARM repeat-containing protein-like protein |
| Response to stress | Sensitive to freezing 6 | |
| Development | Growth-regulating factor 2 | |
| Metabolic process | UDP-glucosyl transferase 75B2 | |
| Metabolic process | RING/U-box superfamily protei | |
| Signaling pathway | Shikimate kinase 1 | |
| mul-miR6180 | Metabolic process | Wax synthase isoform 1 |
| Response to stress | Class III peroxidase | |
| Metabolic process | Acyl-CoA sterol acyl transferase 1 | |
| mul-miR6300 | Response to biotic and abiotic stresses, | Glycosyl hydrolase family 1 protein |
| Hormone-mediated signaling pathway | Leucine-rich repeat receptor-like protein kinase | |
| Unknown | Predicted protein | |
| mul-miR894 | Response to auxin and ethylene; Transcription regulation | Auxin and ethylene responsive GH3-like protein |
| Calcium-mediated signalling | C2 domain-containing protein | |
| Unknown | Coiled-coil domain-containing protein 55 | |
| mul-miRn10-3p | Auxin signaling; Transcription regulation | Auxin response factor 19 |
| Transcription regulation; Development | Zinc finger family protein | |
| Transcription regulation | Squamosa promoter binding protein-like 14 | |
| Transcription regulation | RNA polymerase II transcription mediators | |
| Signal transduction; Development | Leucine-rich receptor-like protein kinase family protein | |
| Signaling pathway | Protein kinase superfamily protein | |
| Metabolic process | S-formylglutathione hydrolase | |
| Metabolic process; | Esterase/lipase/thioesterase family protein | |
| Metabolic process; Response to stress | Aconitase 3 | |
| Response to stress | AWPM-19-like family protein | |
| Unknown | Uncharacterized protein |
Table 5.
Predicted targets for the differential novel miRNAs.
| MiRNA-name | Putative GO process | Predicted target annotations in mulberry transcriptome data |
|---|---|---|
| mul-miRn21-3p | Signal transduction | Transducin/WD40 repeat-like superfamily protein |
| RNA processing | Tetratricopeptide repeat (TPR)-like superfamily protein | |
| mul-miRn22-5p | Metabolic process | 2-oxoglutarate dehydrogenase |
| mul-miRn23-5p | Cytokinin metabolic; Development | SOB five-like 2 |
| Development; Auxin homeostasis; Gibberellic acid mediated signaling pathway | Lateral root primordium (LRP) protein-related | |
| Transcription; Development; Gibberellin biosynthetic process | Integrase-type DNA-binding superfamily protein | |
| Response to auxin stimulus | SAUR-like auxin-responsive protein family | |
| Defense response | Disease resistance protein (TIR-NBS-LRR class) family | |
| Unknown | Calcium-dependent lipid-binding family protein | |
| Metabolic process | Cellulose synthase family protein | |
| Unknown | TPR-like superfamily protein | |
| Unknown | Transposable element gene | |
| mul-miRn24-5p | Metabolic process | ATP-citrate lyase A-3 |
| Membrane transport | Oligopeptide transporter 1 | |
| mul-miRn28-3p | Defence response | ADR1-like 1 |
| mul-miRn29-5p | Ehylene mediated signaling pathway; Transcription regulation | Integrase-type DNA-binding superfamily protein |
| Secondary metabolitic process; Environmental responses | Cytochrome P450, family 96, subfamily A, polypeptide 5 | |
| Transcription regulation | Tudor/PWWP/MBT domain-containing protein | |
| Defence response; Flavonoid biosynthetic process | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | |
| Unknown | Transposable element gene | |
| Defence response | Disease resistance protein | |
| mul-miRn30-3p | Development; Environmental responses | PPR repeat-containing protein, |
| Signaling pathway | Serine/threonine-protein kinase-like protein CCR2 | |
| Metabolic process | Oxidoreductase family protein | |
| Transcription regulation | SAP domain-containing protein | |
| mul-miRn31-3p | Metabolic process; Development | ARPN | plantacyanin |
| Transcription regulation | SAC3/GANP/Nin1/mts3/eIF-3 p25-family protein | |
| mul-miRn32-5p | Response to stress; Signaling pathway | U-box domain-containing protein kinase family protein |
| Signaling pathway | Leucine-rich repeat protein kinase family protein | |
| Signaling pathway | Protein kinase superfamily protein |
Figure 4.
Percentage distributions of predicted target genes for differentially expressed phloem sap miRNAs in various categories.
Mul-miR482a-5p accumulated in phloem sap under phytoplasma-infection is mobile
It was reported that some miRNAs accumulated in the phloem sap are translocatable51. Our data showed that the mul-miR482a-5p accumulated strongly in the mulberry phloem sap under phytoplasma infection, so it is reasonable to suspect that it was mobile in the phloem sap. To investigate whether mul-miR482a-5p was mobile in the phloem, we performed grafting experiments using mul-miR482a-5p-overexpression and hen1-1 mutant Arabidopsis thaliana. After the establishment of graft unions, different parts of the successful grafts were used to analyse the mul-miR482a-5p abundance by RT-qPCR. As expected, the translocation of mul-miR482a-5p from overexpressing scions to hen1-1 rootstocks was observed in various independently grafted plants both with and without scions suffering Pseudomonas syringae pv. tomato DC3000 (Pst. DC3000) infection. However, little mul-miR482a-5p was detected in the hen1-1 scions grafted with mul-miR482a-5p overexpressing rootstock for scions with or without Pst. DC3000 infection (Fig. 5). This result indicates that mul-miR482a-5p can be transported efficiently across the graft junction from scions to rootstock under both infective and uninfective conditions. However, mul-miR482a-5p can scarcely be transported in the opposite direction.
Figure 5.
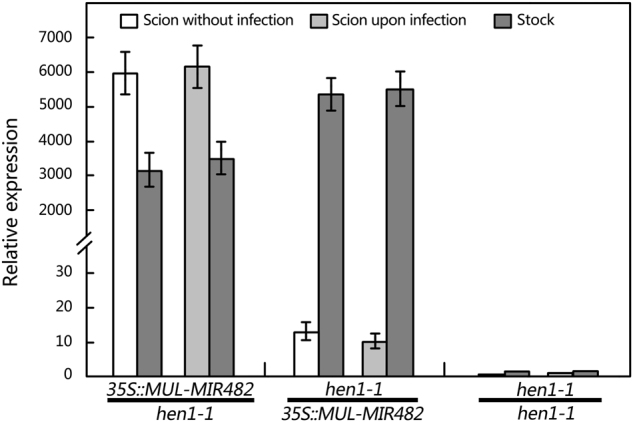
Measurement of mul-miR482a-5p in scions and rootstocks of grafted plants. Infection experiments were performed by spraying Pst. DC3000 suspensions at 108 CFU mL−1 in 10 mM MgCl2 with 0.04% (v/v) Silwet L-77 onto leaves of scions. Mul-miR482a-5p abundance was detected by RT-qPCR. The relative miRNA abundance was evaluated using comparative Ct method taking U6 as a reference. Values are given as the mean ± SD of three experiments in each group.
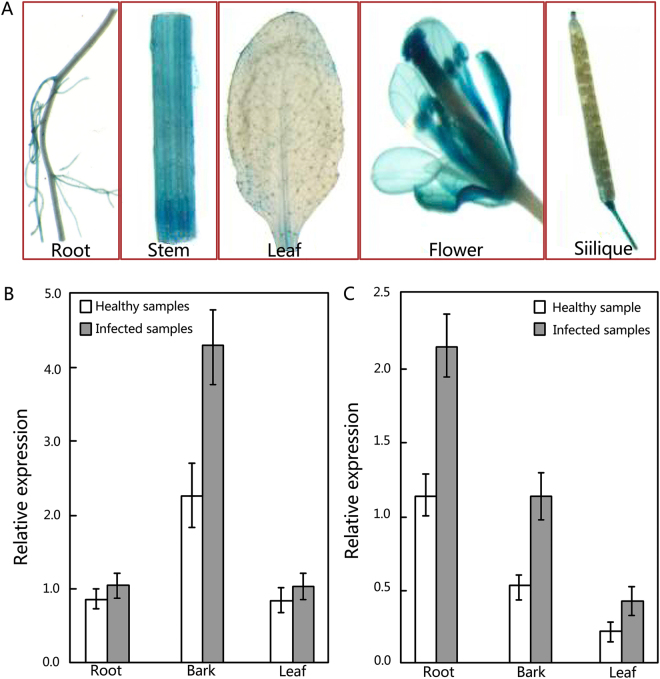
To provide further evidence that the mul-miR482a-5p is mobile, the promoter of MUL-MIR482A-5p was cloned and fused to the reporter gene encoding β-glucuronidase (GUS) to analyse the expression pattern of mul-miR482a-5p in various tissues. Staining results showed that GUS activity was predominantly observed in stems and flowers, and the reporter signal in roots was very low, with no signal detected in the leaves and siliques (Fig. 6A). Corresponding to the staining results, when the pri-mul-miR482a transcript was examined in mulberry, more pri-mul-miR482a transcript was detected in the stem than in the root (Fig. 6B). However, this did not result in higher accumulation of mature mul-miR482a-5p in the stem than the root. In contrast, more mature mul-miR482a-5p accumulated in the root than the stem (Fig. 6C). The opposite accumulation of pri-mul-miR482a and mature mul-miR482a-5p in stems and roots implies that MUL-MIR482A was highly expressed in stem and mature mul-miR482a-5p was transported to roots. However, the abundance of mature mul-miR482a-5p increased in the infected roots compared to healthy roots (Fig. 6C), and the abundance of mul-miR482a-5p primary transcripts did not increase in the infected roots (Fig. 6B). Therefore, mul-miR482a-5p, but not its precursors, is mobile within the phloem. Although the level of mul-miR482a-5p increased in the infected stem barks, the abundance of mul-miR482a-5p did not increase in the infected leaves (Fig. 6C). This may confirm that mul-miR482a-5p can be transported from upper to lower parts but can scarcely be transported in the opposite direction.
Figure 6.
Tissue localization of MUL-MIR482A and measurement of pri-mul-miR482a and mul-miR482a-5p. (A) GUS staining in MUL-MIR482 promoter::reporter transgenic plants. (B and C) Measurement of pri-mul-miR482a (B) and mature mul-miR482a-5p (C) in various tissues of mulberry, respectively. Pri-mul-miR482a and mul-miR482a-5p abundance were detected by RT-qPCR, and relative abundance was evaluated using comparative Ct method using actin (Accession No. DQ785808) and U6 as the reference, respectively. Values are given as the mean ± SD of three experiments in each group.
Mul-miR482a-5p accumulated in phloem sap has physiological functions
The regulator of chromosome condensation family protein (RCC1) gene, trehalose 6-phosphate synthase gene and inositol 1,3,4-trisphosphate 5/6-kinase family protein gene were predicted as targets of mul-miR482a-5p and were experimentally verified by 5′-RLM RACE analyses (Fig. 7). To examine whether the translocation of mul-miR482a-5p in the phloem had physiological functions, the expression levels of its target genes in leaves, phloem saps, and roots were analysed by RT-qPCR (Fig. 8). The results showed that the expression levels of the three target genes have no significant change between infected and healthy leaves. This was consistent with the level of mul-miR482a-5p, which did not differ between infected and healthy leaves. The expression levels of the three target genes showed no significant change between infected and healthy phloem sap, although the level of mul-miR482a-5p was significantly increased in the infected phloem sap. This may be because there was no RNase, which was necessary for the cleavage of target mRNAs. Therefore, mul-miR482a-5p may have no physiological function that directs cleavage of its target mRNAs in the phloem sap. However, the expression of all three target genes was down regulated in the infected roots compared to healthy roots, and this coincided with the changes in mul-miR482a-5p in the roots. Thus, the translocation of mul-miR482a-5p in the phloem might have physiological cleavage functions for its target genes in the roots.
Figure 7.

Validation of predicted target genes of mul-miR482a-5p using 5′ RLM-RACE. The mul-miR482a-5p cleavage sites on its target genes were highlighted with an arrow. The number is the frequency of accurate clones when validating cleavage sites of target mRNAs. RCC1, regulator of chromosome condensation family protein gene. T6PS, trehalose 6-phosphate synthase gene. ITPK, inositol 1,3,4-trisphosphate 5/6-kinase family protein gene.
Figure 8.
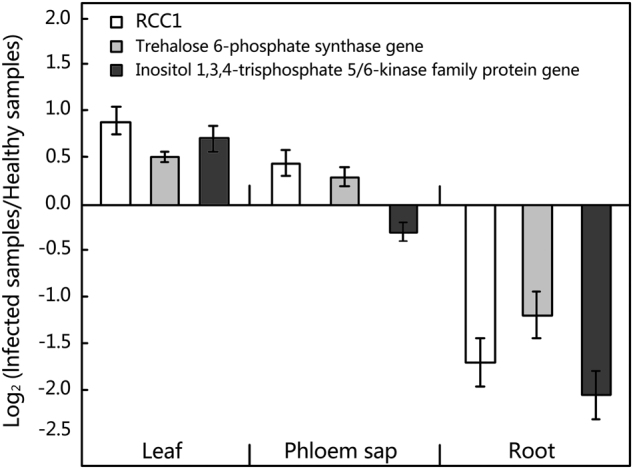
Abundance analysis of predicted target genes of mul-miR482a-5p by RT-qPCR. Relative gene expression was evaluated using comparative Ct method with actin (Accession No. DQ785808) as the reference gene. Log2 values of the ratio of phytoplasma-infected samples to healthy samples are plotted. Values are given as the mean ± SD of three experiments per group.
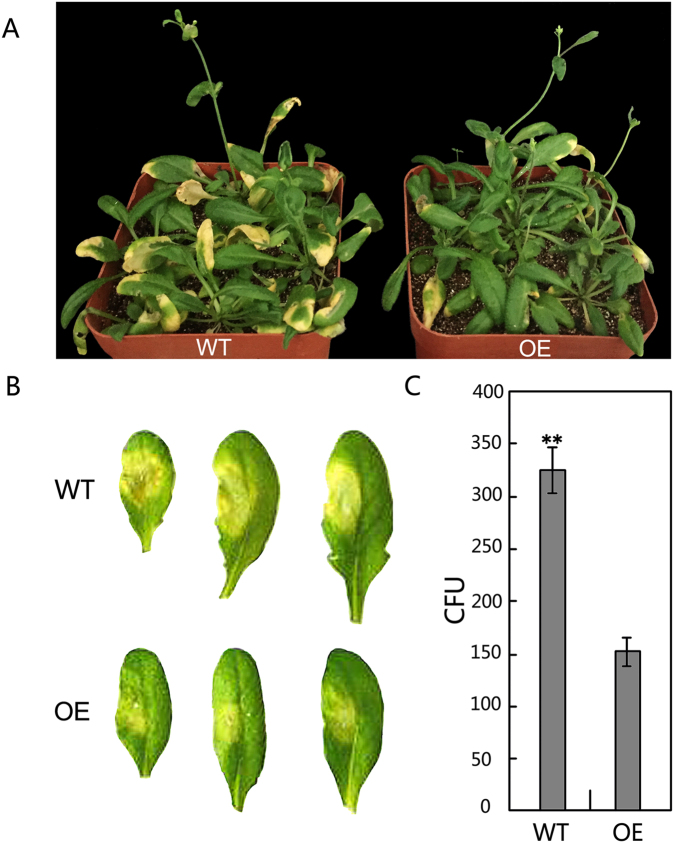
To further explore the physiological functions of mul-miR482a-5p, one of the target genes, RCC1, was also cloned and transformed into Arabidopsis thaliana, and RCC1-overexpressing and wild type Arabidopsis thaliana were inoculated with Pst. DC3000. The results showed that the RCC1 transgenic Arabidopsis plants showed stronger resistance to Pst. DC3000 than wild-type plants, suggesting that the RCC1 gene may be a positive regulator of defence responses (Fig. 9). Therefore, mul-miR482a-5p may repress the expression of RCC1 in infected mulberry and reduce host resistance to biotic stress.
Figure 9.
Analysis of resistance of transgenic Arabidopsis plants to Pst. DC3000. (A) Phenotypes of plants spray-inoculated with Pst. DC3000. (B) Phenotypes of leaves vacuum-infiltrated with Pst. DC3000; disease symptoms were recorded using a camera 3 days after inoculation; (C) Colony-forming units (CFU) of Pst. DC3000 in infected Arabidopsis leaves. Bacterial numbers were calculated at 3 days after inoculation and represented as CFU per gram leaf tissue, and CFU of Pst. DC3000 in infected leaves was counted in a 1/1000-fold bacterium solution. Bioassays were performed three times, each with three replicates, and each value is mean ± SD of three experiments. Asterisks indicate significant difference based on Student’s t-test (**P < 0.01). WT, Wild type Arabidopsis Col-0; OE, Transgenic RCC1 Arabidopsis plants.
Discussion
Component complexity of miRNAs in phloem sap
Although a number of miRNAs have been previously identified in the phloem sap of several herbaceous plants, to the best of our knowledge, no data on phloem miRNAs is available for woody perennials except for apple. Our study demonstrates that mulberry phloem sap contains small RNAs that are major contributors to the phloem sap RNA population. Some highly expressed miRNAs such as miR166, miR167, and miR172 identified in mulberry phloem sap were also detected in the phloem sap of B. napus37, apple (M. domestica “Royal Gala”)39, and pumpkin (Cucurbita maxima)40, suggesting that some miRNAs were conserved across plant species. However, some miRNAs, such as miR171, which were detected in mulberry, B. napus37 and pumpkin40 phloem sap were not detected in apple phloem sap. Meanwhile, miR403 and miR162 were detected in B. napus37, apple39, and pumpkin40 phloem sap but were not present in mulberry. In addition, some novel miRNA candidates were identified in our data. Therefore, the compositions of phloem sap miRNAs differed between herbaceous and woody plants and even among woody species. Furthermore, our data showed that 43 miRNAs were differentially expressed in mulberry phloem sap in response to phytoplasma-infection. It was also reported that the composition of phloem sap miRNAs differed under different nutrient stresses. Thus, the miRNA composition of phloem sap is complex, and identification and characterization of the phloem miRNAs of mulberry may enhance current knowledge of the miRNA composition of phloem sap and help to discover new candidates that have significant action on phloem functions.
MiRNAs in phloem sap and leaves are distinct in complement and expression pattern in response to phytoplasma infection
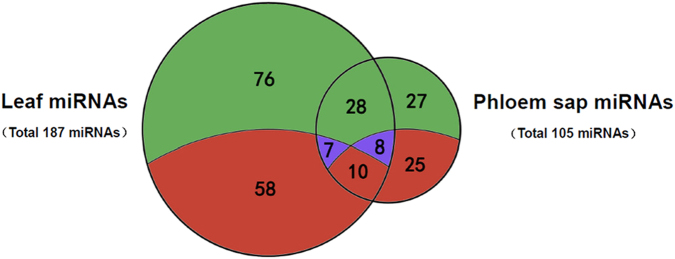
Plant miRNAs often show differential expression among various tissues39, and many miRNAs present in the phloem sap of mulberry were identified in this study. When the identified miRNAs were compared to previously published collections of miRNAs in the leaves of mulberry in the response to phytoplasma infection31, there were 52 miRNAs identified in the phloem sap but not the leaves, and 134 miRNAs were identified in the leaves but not the phloem sap (Fig. 10). Among the 53 miRNAs common to the leaves and phloem sap, the relative levels of expression of some miRNAs, such as mul-miR2199, mul-miR2916, mul-miR5813 and mul-miR6300, were high in the phloem sap but low in the leaves. This demonstrates that phloem sap contains a specific set of miRNAs distinct from leaves and that a set of phloem-enriched sRNAs exists. Moreover, among the 43 phytoplasma-responsive miRNAs identified in phloem sap, only 10 miRNAs were expressed differently in healthy and infected leaves. This may be because not all miRNAs in phloem sap can be translocated, and mobile miRNAs might be translocated in different directions. Therefore, the expression pattern of miRNAs in phloem sap was distinct from leaves in response to phytoplasma infection, and different miRNAs might have distinct localizations and functions. Interestingly, the 10 common phytoplasma-responsive miRNAs between phloem sap and leaves showed the same expression pattern. These miRNAs could potentially act as a long-distance information transmitters in response to phytoplasma infection in mulberry. Further experiments are required to uncover the translocatability and functions of these miRNAs in modulating the response of mulberry to phytoplasmas.
Figure 10.
Venn diagram indicating miRNA identification profiles in phloem sap and leaves from mulberry. Values in green and red sections represent numbers of undifferentially and differentially expressed miRNAs. Values in blue sections represent numbers of miRNAs differentially expressed specifically in phloem sap or leaves.
Role of miRNAs in phloem sap
Although many miRNAs have been detected in many plant phloem saps, only a few miRNAs have been shown to translocate between cells and over long distances42–44,47. It is not clear whether all differentially expressed miRNAs in the phloem sap are mobile. Since phytoplasmas are restricted to sieve elements of phloem tissues, the sieve elements may experience phytoplasma infection earlier than other tissues. Thus, the translocated phytoplasma-responsive miRNAs in phloem sap might have roles in long-distance signalling in response to phytoplasma infection and serve to coordinate physiological responses with other plant parts that are not yet infected. Even if some differentially expressed miRNAs in the phloem sap were not mobile, it was reported that the miRNAs in the phloem sap may act to prevent translation and movement of their target mRNAs39. Although the putative target genes were bioinformatically predicted, the role of differentially expressed miRNAs to prevent translation and movement of their target mRNAs, which may be involved in signalling in response to phytoplasma infection, remains to be studied. The elucidation of the roles of these phytoplasma-responsive miRNAs in the phloem sap may reveal the mechanisms underlying the interactions of phytoplasma and mulberry.
The most typical symptoms of phytoplasma diseases indicate perturbations in plant hormonal balance52–55. In this study, we also found expression changes for several phloem sap miRNAs involved in auxin signalling and auxin metabolism, e.g., differentially expressed mul-miR319a was predicted to target the MYB transcription factor, which redirects auxin signal transduction by interacting with ARFs and plays a role in plant hormone responses27. Differential mul-miR1223e was predicted to target genes coding for O-fucosyltransferase family protein and SAUR-like auxin-responsive protein, which are associated with auxin metabolism (Table 4). In addition to the auxin signalling pathway, mul-miR1223e was found to target the genes involved in salicylic acid signalling, and mul-miR157a was predicted to target the gene coding galactose oxidase/kelch repeat superfamily protein associated with brassinosteroid biosynthesis. Meanwhile, mul-miR529b was found to target the gene involved in abscisic acid and jasmonic acid signalling pathways. In addition, mul-miR894 and mul-miRn25-5p were found to be associated with the ethylene signalling pathway, and mul-miRn23-5p was predicted to target the lateral root primordium (LRP) protein-related gene and SOB five-like 1 gene, which was related to the gibberellic acid-mediated signalling pathway and cytokinin metabolic processes. These data are consistent with earlier reports that show phytoplasma infection-induced alteration in hormonal signalling leading to symptoms in infected plants.
Symptoms induced in infected plants suggest that phytoplasma infection may modulate developmental processes within the plant host56. Our data showed that some phytoplasma-responsive miRNAs target the transcription factors involved in development. For example, mul-miR156a was predicted to target the Squamosa Promoter-Binding Protein-Like (SPL) family, which plays important roles in flower and fruit development, plant architecture and phase transition in plants57,58. Meanwhile, mul-miR319a was predicted to target the MYB transcription factor, which was crucial to the control of proliferation and differentiation in a number of cell types and key factors in regulatory networks controlling development59. In addition, our results showed differentially expressed miRNAs—such as mul-miR1223e, mul-miR165b-5p, mul-miR529, mul-miRn23-5p—that target the genes associated with modulating plant development through various pathways. The changes in these miRNAs in the phloem sap may disorder the expression of many genes involved in diverse development processes, causing symptoms of phytoplasma disease in the infected plants.
As intracellular parasites, phytoplasmas have lost many metabolic genes and must obtain essential metabolites from their hosts60, which has a great impact on the metabolome of infected plants7,14,52. Our results showed that phytoplasma infection alters the profiles of a number of miRNAs involved in metabolism in phloem sap. These differentially expressed miRNAs target genes associated with protein metabolism, CHO metabolism and lipid metabolism. Furthermore, some miRNAs targeting the genes associated with secondary metabolism were also differentially expressed. For example, mul-miR156a and mul-miR529-3p were predicted to target UDP-glucosyltransferase genes, which were involved in the flavonoid biosynthesis pathway61. Therefore, differentially expressed miRNAs in the phloem sap may disturb many metabolic processes and have a significant effect on the response against phytoplasmas.
Some phytoplasma-responsive miRNAs modulate overlapping signalling of biotic and abiotic stresses
Plants have evolved sophisticated mechanisms to sense and respond to diverse biotic stresses62. Our miRNA expression analysis showed many differentially expressed miRNAs targeting genes associated with defence response genes. The changes in these miRNAs may down-regulate or up-regulate their target gene expression and alter plant resistance to phytoplasma. Interestingly, we also found that some differentially expressed miRNAs were responsive to abiotic stresses. For example, mul-miR397a were predicted to target the gene casein kinase II beta chain 3, which has roles in response to light stimulus response63, and mul-miR529b, which was predicted to target the 3-ketoacyl-CoA synthase 19 gene involved in response to cold stress64. In addition, mul-miR529-3p was predicted to target the ARM repeat superfamily protein associated with salt stress and shoot gravitropism65. Meanwhile, several phytoplasma-responsive miRNAs were reported to be differentially expressed under various abiotic stresses in other plant species. For instance, miR156, miR157, and miR390 were reported to be responsive to salt, drought, cold, and heat stress in many other plants29,62 and were detected to be differentially expressed in phloem sap in response to phytoplasma infection in this study. This may be because phytoplasma infection had a great influence on the growth and development of mulberry, and these miRNAs may contribute to modulation of the necessary growth and developmental adjustments to adapt to phytoplasma-infected conditions. In conclusion, our results suggested that phytoplasma infection may cause both biotic and abiotic stress in the mulberry, and some phytoplasma-responsive miRNAs involved in both biotic and abiotic stress signalling converge upstream of phytoplasma infection. The infected plant can modulate protective responses by controlling the abundance of these miRNAs via overlapping signalling. However, further experiments are required to uncover signalling in the phloem sap that modulates the response of mulberry to phytoplasmas.
Mul-miR482a-5p might negatively regulate mulberry resistance to phytoplasma-infection
The miR482 superfamily is a group of plant-specific miRNAs targeting the NBS-LRR gene, and several reports have demonstrated that the miR482-NBS-LRR regulatory loop is part of the immune response induced by pathogens66–69. Moreover, miR482 was also found to participate in guidance for the biosynthesis of secondary phasiRNAs, which are involved in controlling immune-response genes66,70. To date, miR482 has been confirmed to be distributed in more than 20 species, and approximately sixty primary transcripts of miR482 have been identified. Among the primary transcripts identified, there are approximately 20 primary transcripts that can be processed to generate both miR482-5p and miR482-3p (http://www.mirbase.org). To date, there are many reports of miR482-3p, but the function of miR482-5p is still not well understood. Plants infected with pathogens showed a reduced level of miR482 and an increased level of miR482 target mRNAs, suggesting that the miR482-mediated silencing cascade is suppressed by pathogen attack and may be a defence response of plants71. However, the level of mul-miR482a-5p, not mul-miR482a-3p, was changed significantly in the phloem sap infected by phytoplasma (Table 1). Mul-miR482a-5p was predicted to target the RCC1 gene, which is the guanine nucleotide exchange factor for the nuclear GTP binding protein Ran, and is probably involved in various biological processes, but the role of this gene under stress is currently not clear72. Our results showed that the RCC1 gene may be a positive regulator of defence responses (Fig. 9). Therefore, the up-regulation of mul-miR482a-5p may repress the expression of RCC1 in the infected plant and reduce host resistance to phytoplasma. This is consistent with the report that RCC1 family proteins were down-regulated in the incompatible interaction between soybean and Phytophthora sojae73. It was suggested that nucleocytoplasmic trafficking plays an essential role in the expression of disease resistance74, and nuclear localization of some disease resistance (R) proteins, such as members of CC-NB-LRR and TIR-NB-LRR proteins, is essential for their resistance function75,76. As RCC1 plays a major role in nucleocytoplasmic transport72, silencing of RCC1 may result in partial impairment of nucleocytoplasmic trafficking and loss of resistance function of some disease resistance (R) proteins. Thus, when mulberry plants were infected by phytoplasma, the mul-miR482a-3p did not decrease, suggesting that the miR482-NBS-LRR regulatory loop was not induced. Moreover, increased mul-miR482a-5p might repress the resistance to phytoplasma mediated by RCC1. Therefore, phytoplasma-derived suppression of RNA silencing may repress the whole host immune system during infection and potentially enhance phytoplasma colonization and amplification. However, further research is required to elucidate the regulatory mechanisms of the RCC1 gene.
In conclusion, the characterization of miRNA-Seq-based expression profiling of miRNAs allowed for the identification of many phytoplasma-responsive miRNAs in mulberry phloem sap. Future investigation will explore the functions and regulatory networks of these miRNAs. The information provided here will be particularly useful for a complete understanding of the function of miRNAs in phloem sap and will lay the foundation to reveal the mechanisms underlying phytoplasma pathogenicity.
Methods
Plant material
One-year-old cutting seedlings collected from Husang 32 (Morus multicaulis Perr.) were used as rootstock and grafted to scions from healthy or phytoplasma-infected mulberry trees (Husang 32). All establishment graft unions were incubated in a greenhouse, and plants showing Witches’ broom disease symptoms were confirmed by PCR assay with an amplified fragment of the phytoplasma 16S rRNA gene (GenBank Accession No. EF532410) using the primers (P16mF: 5′-TAAAAG ACCTAGCAATAGG-3′ and P16mR: 5′-CAATCCGAACTGAGACTGT-3′) as previously described53.
Phloem saps collection and purity assessing
Phloem sap was collected from infected and healthy mulberry plants using the shoot exudation method77. First, shoots were excised with a sterile razor blade, the first droplets were discarded, and the cut surface was blotted with sterile filter paper several times to avoid contamination. Exuding phloem sap was collected into an Eppendorf tube containing TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Leaf and phloem sap RNAs were isolated using a TRIzol kit (Invitrogen) following the manufacturer’s instructions. To detect RuBisCO and MmPP16 transcripts, cDNA was synthesized using oligo (dT)18 primer (GACTCTAGACGACATCGA(T)15) and ReverTra Ace M-MLV RTase (TaKaRa, Dalian, China). RT-PCR assays were performed in 25-µl reaction volumes containing 20 ng cDNA and 150 nM forward and reverse primers. The primers used for amplification of RuBisCO and MmPP16 genes are shown in Supplementary Table 1.
Small RNA library construction and high-throughput sequencing
Isolated phloem sap RNAs were used to prepare a small RNA library according to the protocol of the TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA). Single-end sequencing (36 bp) was performed on an Illumina Hiseq2500 instrument following standard protocols. Three independent libraries each (biological replicates) were analysed for infected and healthy phloem saps.
MiRNAs identification
The raw sequences tags obtained from HiSeq sequencing were cleaned to remove adapter dimers, junk, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA) and repeats, and the length distribution of the clean tags was summarized. The trimmed reads longer than 18 nt were annotated into different categories, and the sequences of 18–25 nucleotides were compared to a miRBase database v21.0 (http://www.mirbase.org/). The sequences with identical or related sequences from other plants were regarded as conserved miRNAs. All remaining unmapped sequences were BLASTed against our mulberry transcriptome database, and the hairpin RNA structures containing sequences were predicted using RNAfold software and used to predict novel miRNAs using Mireap (http://sourceforge.net/projects/mireap/).
Differential expression analyses of miRNAs
The frequency of miRNA was normalized by the total number of miRNAs in every sample, where normalized expression = (Actual miRNAs sequencing reads count/Total clean reads count) × 1,000,000. The fold change between infected (IPS) and healthy phloem sap (HPS) was calculated as follows: fold-change = log2(IPS/HPS). Statistical analysis was performed according to Poisson distribution, and the P value was calculated based on the formula:
N1 and N2 represent the total count of clean reads of a given miRNA in the sRNA library of infected and healthy phloem sap, respectively. The x and y represent normalized expression levels of a given miRNA in the sRNA library of infected and healthy phloem saps, respectively.
A fold-change ≥ 2 and P ≤ 0.05 were used as criteria to identify differentially expressed miRNAs, and an miRNA was designated as significantly differentially expressed if its expression value varied more than two-fold and P ≤ 0.05 between infected and healthy phloem saps.
Target prediction of differential miRNAs
The target genes of the differentially expressed miRNAs were predicted using the software psRNATarget (http://plantgrn.noble.org/psRNATarget/) by submitting miRNA sequences to a search against our in-house mulberry transcriptome data following the criteria of (i) maximum expectation less than 3.0; (ii) multiplicity of target sites 2; (iii) range of central mismatch for translational inhibition 9–11 nt; and iv) maximum mismatches at the complementary site ≦ 4 without any gaps. All predicted target genes were aligned with the reference Arabidopsis thaliana database downloaded from TAIR (http://www.arabidopsis.org/; version TAIR10) to annotate their functions, and the GO terms of these targets were also annotated based on their TAIR GO categories.
Quantitative real-time PCR analysis for miRNAs and mRNAs
RNA was extracted using the TRIzol® reagent following the manufacturer’s recommendations (Invitrogen, Carlsbad, CA, USA) and digested with DNase I. Real-time PCR analyses for miRNAs and mRNAs were performed using the PrimeScriptTM miRNA qPCR Starter Kit Ver.2.0 (TaKaRa, Dalian, China) and the SYBR Premix Ex TaqTM kit (TaKaRa, Dalian, China) on the Rotor-Gene 3000A system (Bio-Rad, Munich, Germany), respectively, according to the manufacturer’s protocol for the Rotor-Gene 3000A system. The U6 and actin genes were used as reference genes for miRNA and mRNA normalization, respectively. The U6 gene was amplified using the primer (5′-ATGGCCCCTGCGTAAGGATG-3′), and actin was amplified using primer pair (F: 5′-CAGTGCTTCTCACTGAGGCTC-3′ and R: 5′-GGAAGAGGACTTCTGGGCATC-3′). The primers (Supplementary Tables 1, 2) used to amplify the genes and miRNAs were designed based on our available mulberry transcriptome data. The relative expression levels of miRNA and mRNA were evaluated using the Comparative cycle threshold (Ct) method78. All samples were assayed in triplicate.
Target validation
For miRNA target validation, a modified gene-specific 5′ RNA ligase-mediated rapid amplification of cDNA ends (5′ RLM-RACE) was performed using the GeneRacer Kit (Invitrogen, Carlsbad, CA, USA). Total RNA was isolated from mulberry seedlings using the TRIzol kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions, ligated to the RNA oligo adapter (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′) and reverse transcribed with SuperScript III reverse transcriptase using oligo(dT) primer. The resulting cDNA was PCR-amplified with GeneRacer 5′ primer (5′-CGACTGGAGCACGAGGACACTGA-3′) and each respective gene-specific outer primer (shown in Supplementary Table 3). The PCR product was further amplified by nested PCR using GeneRacer 5′ nested primer (5′-GGACACTGACATGGACTGAAGGAGTA-3′) and each respective gene-specific inner primer (shown in Supplementary Table 3). The final PCR product was gel-purified and cloned into a pMD18-T vector (Invitrogen, Carlsbad, CA, USA) for sequencing.
Micrografting experiments
Four-day-old seedlings of hen1-1 mutant and transgenic Arabidopsis thaliana were used for micrografting experiments. The seedlings were cut transversely using a sterile razor blade and combined inside silicon tubing (0.3 mm internal diameter). The graft unions were cultured on 1.5% (w/v) agar plates with half-strength MS medium for 9 days and were hydroponically cultured for 10 d. Grafted plants without adventitious roots were selected, and the scions and stocks of the selected grafted plants were harvested for RNA isolation. Then, the RNAs were used for RT-qPCR of the miRNA. To investigate whether the miRNA moves during infective conditions, the leaves of the scions of graft unions were spray-inoculated with Pst. DC3000 suspensions at 108 CFU mL−1 in 10 mM MgCl2 with 0.04% (v/v) Silwet L-77. Three days after inoculation, the RNAs of the scions and stocks of the grafted plants were isolated and used for RT-qPCR of the miRNA.
Promoter activity analysis
The promoter was cloned using a Tail-PCR method and ligated into the vector pBI121 to replace the cauliflower mosaic virus (CaMV) 35S promoter and fused to the GUS (β-glucuronidase) reporter gene to create the promoter expression vector pMIR482::GUS. The derived construct vector was introduced into Agrobacterium tumefaciens strain GV3101, and the WT Arabidopsis plants were transformed with a floral dipping method. Histochemical staining for GUS activity was performed referring to the previously described method79.
Detection of resistance against Pst. DC3000
The RCC1 gene coding sequence was cloned and ligated into the vector pBI121, and the derived construct vector was introduced into A. tumefaciens strain GV3101 under the control of 35S. The WT Arabidopsis plants were transformed with floral dipping method. After transformation, the transformed plants were selected. Four-week-old transgenic and wild-type Arabidopsis seedlings were spray-inoculated with Pst. DC3000 suspensions at 108 CFU mL−1 in 10 mM MgCl2 with 0.04% (v/v) Silwet L-77 or vacuum-infiltrated with bacterial suspensions at 107 CFU mL−1 with a syringe. Three days after inoculation, disease symptoms were recorded using a camera, and bacterial numbers were calculated and represented as colony-forming units (CFU) per gram leaf tissue in a 1/1000-fold bacterium solution. Bioassays were performed three times, with three replicates each.
Data Availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. 30972366, 31070573, and 31100478), Natural Science Foundation of Shandong Province (No. ZR2015CM008 and ZR2015CM019) and Modern Agricultural Technology System of Shandong Province (No. SDAIT-18-04).
Author Contributions
Y.P. and X.L. were responsible for design, sample preparations, and contributed to write the manuscript. H.N. and Y.N. were responsible for high-throughput deep sequencing analysis. B.S. and S.S. were responsible for PCR analysis. S. and F.Y. were responsible for mul-miR482-5p analysis. All authors edited the manuscript and reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Ying-Ping Gai and Huai-Ning Zhao contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19210-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jiang H, et al. Distribution patterns of mulberry dwarf phytoplasma in reproductive organs, winter buds, and roots of mulberry trees. J. Gen. Plant Pathol. 2004;70:168–173. doi: 10.1007/s10327-004-0103-1. [DOI] [Google Scholar]
- 2.Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA. 2011;108:E1254–E1263. doi: 10.1073/pnas.1105664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weintraub PG, Beanland L. Insect vectors of phytoplasmas. Annu. Rev. Entomol. 2006;51:91–111. doi: 10.1146/annurev.ento.51.110104.151039. [DOI] [PubMed] [Google Scholar]
- 4.Wei W, Davis RE, Nuss DL, Zhao Y. Phytoplasmal infection derails genetically preprogrammed meristem fate and alters plant architecture. Proc. Natl. Acad. Sci. USA. 2013;110:19149–19154. doi: 10.1073/pnas.1318489110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheel D. Resistance response physiology and signal transduction. Curr. Opin. Plant Biol. 1998;1:305–310. doi: 10.1016/1369-5266(88)80051-7. [DOI] [PubMed] [Google Scholar]
- 6.Carginale V, et al. Identification of genes expressed in response to phytoplasma infection in leaves of Prunus armeniaca by messenger RNA differential display. Gene. 2004;332:29–34. doi: 10.1016/j.gene.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 7.Choi YH, et al. Metabolic discrimination of Catharanthus roseus leaves infected by phytoplasma using 1H-NMR spectroscopy and multivariate data analysis. Plant Physiol. 2004;135:2398–2410. doi: 10.1104/pp.104.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hren M, et al. ‘Bois noir’ phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genomics. 2009;10:460. doi: 10.1186/1471-2164-10-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margaria P, Palmano S. Response of the Vitis vinifera L. cv. ‘Nebbiolo’ proteome to Flavescence dorée phytoplasma infection. Proteomics. 2011;11:212–224. doi: 10.1002/pmic.201000409. [DOI] [PubMed] [Google Scholar]
- 10.Taheri F, et al. Proteomic analysis of the Mexican lime tree response to “Candidatus Phytoplasma aurantifolia” infection. Mol. Biosyst. 2011;11:3028–3035. doi: 10.1039/c1mb05268c. [DOI] [PubMed] [Google Scholar]
- 11.Zamharir MG, Mardi M, Alavi SM, Hasanzadeh N, Nekouei MK. Identification of genes differentially expressed during interaction of Mexican lime tree infected with Candidatus Phytoplasma aurantifolia. BMC Microbiol. 2011;11:1. doi: 10.1186/1471-2180-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margaria P, Abbà S, Palmano S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phospho-proteomic approach with data integration into functional networks. BMC Genomics. 2013;14:38. doi: 10.1186/1471-2164-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monavarfeshani A, et al. Shotgun proteomic analysis of the Mexican Lime tree infected with “CandidatusPhytoplasma aurantifolia”. J. Proteome Res. 2013;12:785–795. doi: 10.1021/pr300865t. [DOI] [PubMed] [Google Scholar]
- 14.Mou HQ, et al. Transcriptomic analysis of Paulownia infected by Paulownia witches’-broom Phytoplasma. PLoS One. 2013;8:e77217. doi: 10.1371/journal.pone.0077217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luge T, et al. Transcriptomics assisted proteomic analysis of Nicotiana occidentalis infected by Candidatus Phytoplasma mali strain AT. Proteomics. 2014;14:1882–1889. doi: 10.1002/pmic.201300551. [DOI] [PubMed] [Google Scholar]
- 16.Mollayi S, Zadali R, Farzaneh M, Ghassempour A. Metabolite profiling of Mexican lime (Citrus aurantifolia) leaves during the progression of witches’ broom disease. Phytochem. Lett. 2015;13:290–296. doi: 10.1016/j.phytol.2015.07.010. [DOI] [Google Scholar]
- 17.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 19.Katiyar-Agarwal S, Jin H. Role of small RNAs in host microbe interactions. Annu Rev Phytopathol. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao JP, Jiang XL, Zhang BY, Su XH. Involvement of microRNA-mediated gene expression regulation in the pathological development of stem canker disease in Populus trichocarpa. PLoS One. 2012;7:e44968. doi: 10.1371/journal.pone.0044968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldrich P, et al. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 2015;12:847–863. doi: 10.1080/15476286.2015.1050577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 23.He XF, Fang YY, Feng L, Guo HS. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 2008;2:445–2452. doi: 10.1016/j.febslet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Jagadeeswaran G, Saini A, Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta. 2009;9:009–1014. doi: 10.1007/s00425-009-0889-3. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Deng Y, Wu T, Subramanian S, Yu O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 2010;153:1759–1770. doi: 10.1104/pp.110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;2:222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 2011;75:93–105. doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baldrich P, San Segundo B. MicroRNAs in rice innate immunity. Rice (N Y) 2016;9:6. doi: 10.1186/s12284-016-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Ehya F, et al. Phytoplasma-responsive microRNAs modulate hormonal, nutritional, and stress signalling pathways in Mexican lime trees. PLoS One. 2013;8:e66372. doi: 10.1371/journal.pone.0066372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gai YP, et al. Analysis of phytoplasma-responsive sRNAs provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 2014;4:5378. doi: 10.1038/srep05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan G, et al. Identification of microRNAs and their targets in Paulownia fortunei plants free from phytoplasma pathogen after methyl methane sulfonate treatment. Biochimie. 2016;127:271–280. doi: 10.1016/j.biochi.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenz D, May P, Walther D. Comparative analysis of miRNAs and their targets across four plant species. BMC Res. Notes. 2011;4:483. doi: 10.1186/1756-0500-4-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong DH, et al. Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiol. 2013;162:1225–1245. doi: 10.1104/pp.113.219873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kehr J. Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 2006;57:767–774. doi: 10.1093/jxb/erj087. [DOI] [PubMed] [Google Scholar]
- 37.Buhtz A, Springer F, Chappell L, Baulcombe D, Kehr J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 2008;53:739–749. doi: 10.1111/j.1365-313X.2007.03368.x. [DOI] [PubMed] [Google Scholar]
- 38.Varkonyi-Gasic E, Gould N, Sandanayaka M, Sutherland P, MacDiarmid RM. Characterisation of microRNAs from apple (Malus domestica “Royal Gala”) vascular tissue and phloem sap. BMC Plant Biol. 2010;10:159–173. doi: 10.1186/1471-2229-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Medina C, Atkins CA, Mann AJ, Jordan ME, Smith PM. Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.) BMC Biol. 2011;11:36. doi: 10.1186/1471-213X-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo BC, et al. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin SI, et al. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 2008;147:732–746. doi: 10.1104/pp.108.116269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008;53:731–738. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buhtz A, Pieritz J, Springer F, Kehr J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 2010;10:64. doi: 10.1186/1471-2229-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasai A, Kanehira A, Harada T. MiR172 can move long distances in Nicotiana benthamiana. Open Plant Sci. J. 2010;4:1–6. doi: 10.2174/1874294701004010001. [DOI] [Google Scholar]
- 45.Marín-González E, Suárez-López P. “And yet it moves”: cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci. 2012;196:18–30. doi: 10.1016/j.plantsci.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Turnbull CG, Lopez-Cobollo RM. Heavy traffic in the fast lane: long-distance signalling by macromolecules. New Phytol. 2013;198:33–51. doi: 10.1111/nph.12167. [DOI] [PubMed] [Google Scholar]
- 47.Bhogale S, et al. MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014;164:1011–1027. doi: 10.1104/pp.113.230714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Notaguchi M, Okamoto S. Dynamics of long-distance signaling via plant vascular tissues. Front Plant Sci. 2015;6:161. doi: 10.3389/fpls.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertaccini A, Duduk B. Phytoplasma and phytoplasma diseases: a review of recent research. Phytopathol. Mediterr. 2009;48:355–378. [Google Scholar]
- 50.Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 51.Kehr J. Systemic regulation of mineral homeostasis by microRNAs. Front Plant Sci. 2013;4:145. doi: 10.3389/fpls.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gai YP, et al. Metabolomic analysis reveals the potential metabolites and pathogenesis involved in mulberry yellow dwarf disease. Plant Cell Environ. 2014;37:1474–1490. doi: 10.1111/pce.12255. [DOI] [PubMed] [Google Scholar]
- 53.Ji X, Gai Y, Zheng C, Mu Z. Comparative proteomic analysis provides new insights into mulberry dwarf responses in mulberry (Morus alba L.) Proteomics. 2009;9:5328–5339. doi: 10.1002/pmic.200900012. [DOI] [PubMed] [Google Scholar]
- 54.Weintraub, P. G. & Jones, P. Phytoplasmas: Genomes, Plant Hosts and Vectors; CABI: Wallingford, UK (2010).
- 55.Omar AF, Dewir YH, El-Mahrouk ME. Molecular identification of phytoplasmas in fascinated cacti and succulent species and associated hormonal perturbation. J. Plant Interact. 2014;9:632–639. doi: 10.1080/17429145.2014.882421. [DOI] [Google Scholar]
- 56.Sugio A, Hogenhout SA. The genome biology of phytoplasma: modulators of plants and insects. Curr. Opin. Microbiol. 2012;15:247–254. doi: 10.1016/j.mib.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009;50:2133–2145. doi: 10.1093/pcp/pcp148. [DOI] [PubMed] [Google Scholar]
- 58.Chen XB, et al. SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 2010;52:946–951. doi: 10.1111/j.1744-7909.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- 59.Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol. Biol. Plants. 2013;19:307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oshima K, Maejima K, Namba S. Genomic and evolutionary aspects of phytoplasmas. Front Microbiol. 2013;4:230. doi: 10.3389/fmicb.2013.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falcone Ferreyra ML, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khraiwesh B, Zhu JK, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulekar JJ, Bu Q, Chen F, Huq E. Casein kinase II α subunits affect multiple developmental and stress-responsive pathways in Arabidopsis. Plant J. 2012;69:343–354. doi: 10.1111/j.1365-313X.2011.04794.x. [DOI] [PubMed] [Google Scholar]
- 64.Peng X, Teng L, Yan X, Zhao M, Shen S. The cold responsive mechanism of the paper mulberry: decreased photosynthesis capacity and increased starch accumulation. BMC Genomics. 2015;16:898. doi: 10.1186/s12864-015-2047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samuel MA, Salt JN, Shiu SH, Goring DR. Multifunctional arm repeat domains in plants. Int. Rev. Cytol. 2006;253:1–26. doi: 10.1016/S0074-7696(06)53001-3. [DOI] [PubMed] [Google Scholar]
- 66.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li F, et al. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shivaprasad PV, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu Q, et al. MiR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS One. 2013;8:e84390. doi: 10.1371/journal.pone.0084390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson C, et al. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19:1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eckardt NA. A microRNA cascade in plant defense. Plant Cell. 2012;24:840. doi: 10.1105/tpc.112.240311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/S0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 73.Narayanan NN, et al. Identification of candidate signaling genes including regulators of chromosome condensation 1 protein family differentially expressed in the soybean-Phytophthora sojae interaction. Theor Appl Genet. 2009;118:399–412. doi: 10.1007/s00122-008-0895-z. [DOI] [PubMed] [Google Scholar]
- 74.Deslandes L, Rivas S. The plant cell nucleus: A true arena for the fight between plants and pathogens. Plant Signal Behav. 2011;6:42–48. doi: 10.4161/psb.6.1.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meier I, Somers DE. Regulation of nucleocytoplasmic trafficking in plants. Curr. Opin. Plant Biol. 2011;14:538–546. doi: 10.1016/j.pbi.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 76.Bai S, et al. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012;8:e1002752. doi: 10.1371/journal.ppat.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 79.Jefferson R, Kavanagh T, Bevan M. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.