ABSTRACT
Ubiquitin-mediated protein degradation plays essential roles in proteostasis and is involved in the pathogenesis of neurodegenerative diseases in which ubiquitin-positive aberrant proteins accumulate. However, how such aberrant proteins are processed inside cells has not been fully explored. Here, we show that the product of CG5445, a previously uncharacterized Drosophila gene, prevents the accumulation of aggregate-prone ubiquitinated proteins. We found that ubiquitin conjugates were associated with CG5445, the knockdown of which caused the accumulation of detergent-insoluble ubiquitinated proteins. Furthermore, CG5445 rescued eye degeneration caused by the amyotrophic lateral sclerosis (ALS)-linked mutant TAR DNA-binding protein of 43 kDa (TDP-43), which often forms ubiquitin-positive aggregates in cells through the capacity of CG5445 to bind to ubiquitin chains. Biochemically, CG5445 inhibited the accumulation of insoluble forms and promoted their clearance. Our results demonstrate a new possible mechanism by which cells maintain ubiquitinated aggregation-prone proteins in a soluble form to decrease their cytotoxicity until they are degraded.
KEYWORDS: protein homeostasis, ubiquitin
INTRODUCTION
The ubiquitin-proteasome system (UPS) is the principal pathway for protein degradation in eukaryotic cells (1). In the UPS, proteins to be eliminated are tagged with ubiquitin chains and degraded by the 26S proteasome (2). The UPS plays essential roles in various biological events; therefore, its dysfunction leads to various diseases, including neurodegeneration (3–6). Ubiquitin-positive aggregates are pathological hallmarks of human neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and polyglutamine repeat (polyQ) diseases (7). However, it remains unclear how ubiquitinated proteins are processed inside cells before being degraded by the proteasome.
The most well-studied organism concerning the UPS and its modifiers is the unicellular eukaryote Saccharomyces cerevisiae. Considering that dysfunction of the UPS is involved in various human diseases (6), exploring genes functionally associated with the UPS in multicellular animals with multiple organ systems should provide new findings not yet revealed in yeast. Here, we performed a genetic screen using Drosophila melanogaster to identify genes that are involved in UPS function and identified a previously uncharacterized gene, CG5445, as a possible modifier of the UPS. CG5445 interacted with ubiquitin conjugates via its ubiquitin-associated (UBA) domain. Knockdown of CG5445 in fly compound eyes caused the disorganization and accumulation of ubiquitinated proteins in a detergent-insoluble state. Furthermore, CG5445 mitigated eye degeneration caused by the ALS-linked mutant TAR DNA-binding protein of 43 kDa (TDP-43) in Drosophila, through the inhibition of the accumulation of mutant TDP-43 (8, 9). Our results imply a new possible mechanism by which ubiquitinated proteins are kept soluble by CG5445 and consequently degraded by the proteasome.
RESULTS
Identification of CG5445 as a modifier of the UPS in Drosophila.
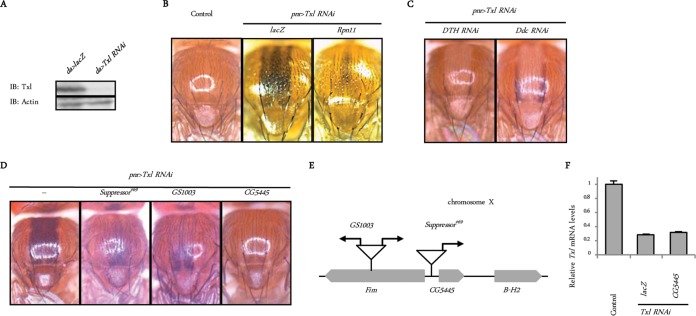
To identify novel genes that positively regulate UPS function, we designed a genetic screen using Drosophila melanogaster. Thioredoxin-like (Txl) is a proteasome-associated disulfide reductase that has been suggested to have a role in proteasome-mediated protein degradation in mammals and fission yeast (10, 11). Using a upstream activating sequence (UAS)-Txl RNA interference (RNAi) strain that efficiently decreased the amount of endogenous Txl (Fig. 1A), we found that Txl knockdown in the dorsal midline under the control of the pnr-Gal4 driver (pnr>Txl RNAi) caused abnormal pigmentation in flies (Fig. 1B). This phenotype was suppressed by the additional knockdown of DTH or Ddc, key enzymes in melanin synthesis (12) (Fig. 1C), indicating that the knockdown of Txl induces melanization. In addition, the upregulation of proteasome function by the ectopic expression of the proteasome subunit Rpn11 (13) suppressed this phenotype, indicating that melanization induced by Txl knockdown is associated with dysfunction in the UPS (Fig. 1B).
FIG 1.
Identification of CG5445 as a possible modifier of the UPS. (A) Confirmation of Txl knockdown. Txl was depleted in the whole body under the control of da-Gal4. Third-instar larvae were homogenized and subjected to immunoblot (IB) analysis with the indicated antibodies. Genotypes are UAS-lacZ/+; da-Gal4/+ and UAS-Txl RNAi/+; da-Gal4/+. (B) Appearance of Drosophila notum. Genotypes (from the left) are pnr-GAL4/+ (control), UAS-Txl RNAi/UAS-lacZ; pnr-GAL4/+, and UAS-Txl RNAi/UAS-Rpn11; pnr-GAL4/+. (C) Effect of downregulation of melanin synthesis on Txl knockdown-induced pigmentation. Genotypes are UAS-Txl RNAi/UAS-DTH RNAi; pnr-GAL4/+ and UAS-Txl RNAi/UAS-Ddc RNAi; pnr-GAL4/+. (D) GS lines and a CG5445 transgenic fly that suppress Txl knockdown-induced melanization. Genotypes (from the left) are Suppressor#69/+; UAS-Txl RNAi/+; pnr-GAL4/+, GS1003/+; UAS-Txl RNAi/+; pnr-GAL4/+, and UAS-Txl RNAi/UAS-CG5445; pnr-GAL4/+. (E) Schematic representation of the genomic locus on chromosome X where the GS vectors are inserted. Arrows represent directions of UAS sequences. (F) Txl was depleted under the control of the da-Gal4 driver. Since flies with Txl knockdown in the whole body exhibited developmental lethality during the pupal stage, flies were raised at 18°C until eclosion to repress GAL4 by GAL80ts, a temperature-sensitive repressor of GAL4, and then incubated at 29°C for 10 days to repress GAL80ts. Total RNAs of each genotype were analyzed by qRT-PCR. Genotypes (from the left) are tub-Gal80ts/UAS-lacZ; da-Gal4/+ (control), tub-Gal80ts UAS-Txl RNAi/UAS-lacZ; da-Gal4/+, and tub-Gal80ts UAS-Txl RNAi/UAS-CG5445; da-Gal4/+. Data are means and standard deviations of results from three experiments.
To find suppressors of melanization, we combined the P-element-based Gene Search (GS) system with the “local hop” technique and performed a gain-of-function screen (13–15). We crossed pnr>Txl RNAi flies with more than 6,000 GS fly lines in which UAS enhancers were randomly inserted into the fly genome and induced the overexpression of downstream genes. We found eight lines that suppressed the melanization of pnr>Txl RNAi flies. One of the identified suppressor GS fly lines, the Suppressor#69 line, partially attenuated melanization (Fig. 1D) and contained a UAS in the upstream region of CG5445 (Fig. 1E). Another GS fly line with GS1003, in which bidirectional UAS enhancers were inserted into the upstream region of CG5445, also reduced melanization (Fig. 1D and E). These results suggest that CG5445 may be responsible for suppression.
To confirm this, we generated CG5445 transgenic flies. The expression of CG5445 in the dorsal midline of pnr>Txl RNAi flies attenuated melanization without affecting Txl mRNA levels (Fig. 1D and F), demonstrating that CG5445 is a suppressor of Txl deficiency.
CG5445 interacts with and affects the solubility of ubiquitinated proteins.
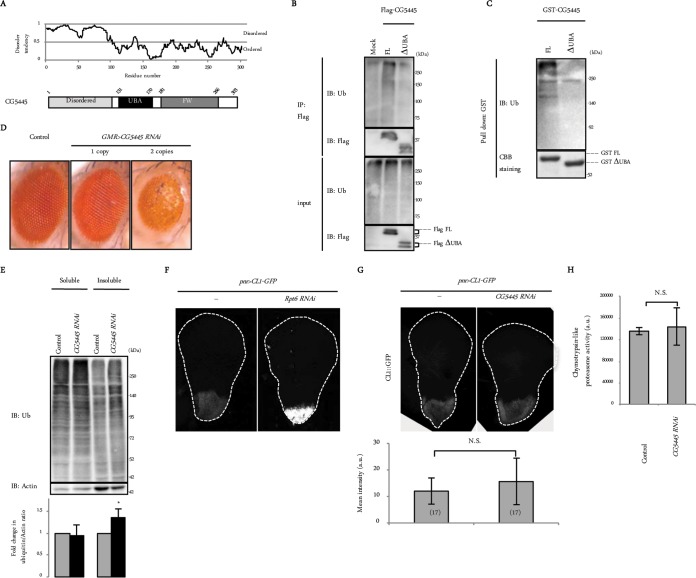
CG5445 is a protein of unknown function that contains a UBA domain, an FW (also known as NBR1) domain (16), and an N-terminal disordered region (Fig. 2A). We first examined whether CG5445 binds to ubiquitin. Full-length CG5445 (CG5445 FL), whether expressed in Schneider 2 (S2) cells or purified as a recombinant protein, pulled down ubiquitin conjugates, while CG5445 lacking the UBA domain (ΔUBA) did not (Fig. 2B and C), indicating that CG5445 interacts with ubiquitin conjugates via the UBA domain.
FIG 2.
CG5445 binds to ubiquitin conjugates. (A) Domain composition of CG5445. Disorder prediction was performed by using IUPred. (B) Flag-CG5445 (full length [FL] and ΔUBA) was expressed in S2 cells, immunoprecipitated (IP), and subjected to immunoblot analysis with the indicated antibodies. Ub, ubiquitin. (C) Recombinant GST-CG5445 (FL and ΔUBA) was incubated with whole-fly extracts, pulled down, and resolved by SDS-PAGE, followed by immunoblotting against ubiquitin and Coomassie brilliant blue (CBB) staining. (D) Effect of CG5445 knockdown on Drosophila eyes. Genotypes are GMR-Gal4/CyO (control), GMR-Gal4/CyO; UAS-CG5445 RNAi/TM6B (1 copy), and GMR-Gal4/CyO; UAS-CG5445 RNAi (2 copies). (E) Heads of control and CG5445 RNAi (2 copies) flies on day 10 posteclosion were separated into Triton-soluble and -insoluble fractions, followed by SDS-PAGE and immunoblotting with the indicated antibodies. Values represent means and standard deviations of the relative band intensities of ubiquitin (normalized to actin) obtained from four independent experiments. *, P < 0.05. (F and G) Detection of the UPS reporter in third-instar larva wing discs. Broken lines indicate edges of wing discs. Genotypes are UAS-lacZ/+; pnr-Gal4/UAS-CL1-GFP and UAS-Rpt6 RNAi/+; pnr-Gal4/UAS-CL1-GFP (F) and pnr-Gal4/UAS-CL1-GFP and pnr-Gal4 UAS-CG5445 RNAi/UAS-CL1-GFP (G). Values represent means ± standard deviations of the intensities of CL1-GFP, and values in parentheses indicate the number of flies examined. N.S., not significant; a.u., arbitrary units. (H) Head extracts were assayed for chymotrypsin-like peptidase activities. Genotypes are GMR-Gal4/+ and GMR-Gal4/+; UAS-CG5445 RNAi/+. Values represent means ± standard deviations of data from three independent experiments. N.S., not significant.
We next examined the effect of CG5445 knockdown in vivo. Knockdown of CG5445 under the control of the eye-specific driver GMR-Gal4 (GMR>CG5445 RNAi) caused a dosage-dependent disorganization of compound eyes (Fig. 2D). The heads of these flies were lysed with Triton X-100, fractionated into soluble and insoluble fractions, and then analyzed by immunoblotting with an antiubiquitin antibody. This revealed that the levels of Triton X-100-insoluble but not soluble ubiquitinated proteins were significantly increased in GMR>CG5445 RNAi flies (Fig. 2E), suggesting that CG5445 suppresses the accumulation of insoluble ubiquitinated proteins that are presumably deleterious in vivo.
One possibility for the accumulation of ubiquitinated proteins in CG5445 knockdown flies is that CG5445 alters proteasome function. To evaluate proteasome function, we measured the intensity of CL1-green fluorescent protein (GFP), a fluorescent reporter protein that is continually degraded by the proteasome (17, 18). CL1-GFP was expressed in the wing disc under the control of pnr-Gal4 (pnr>CL1-GFP). The fluorescent signal of CL1-GFP increased when Rpt6, one of the 26S proteasome subunits, was downregulated (Fig. 2F). Under the same conditions, the knockdown of CG5445 did not augment reporter levels of CL1-GFP (Fig. 2G). We also measured peptidase activity using head extracts of GMR>CG5445 RNAi flies. This analysis showed comparable peptidase activities between control and RNAi flies (Fig. 2H). These results suggest that CG5445 does not affect proteasome function.
CG5445 protects against ALS-linked TDP-43 toxicity in Drosophila.
Since CG5445 knockdown increased the levels of insoluble ubiquitinated proteins and compound eye disorganization, we explored the effect of CG5445 on the toxicity of aggregate-prone proteins in flies. Mutant TDP-43 is one of the major causative proteins in ALS (19). Mutant TDP-43 has been shown to be subjected to degradation via the ubiquitin-proteasome pathway and often forms ubiquitin-positive aggregates in patient tissues, which are hallmarks of neurodegenerative diseases (4, 8, 20). It has been shown that the expression of TDP-43 containing a familial ALS (fALS)-linked mutation (M337V) often generates high-molecular-weight (HMW) smearing of TDP-43 as well as a monomeric form of TDP-43 (∼43 kDa) (21), which is observed in patient tissues with TDP-43 proteinopathy (8, 22).
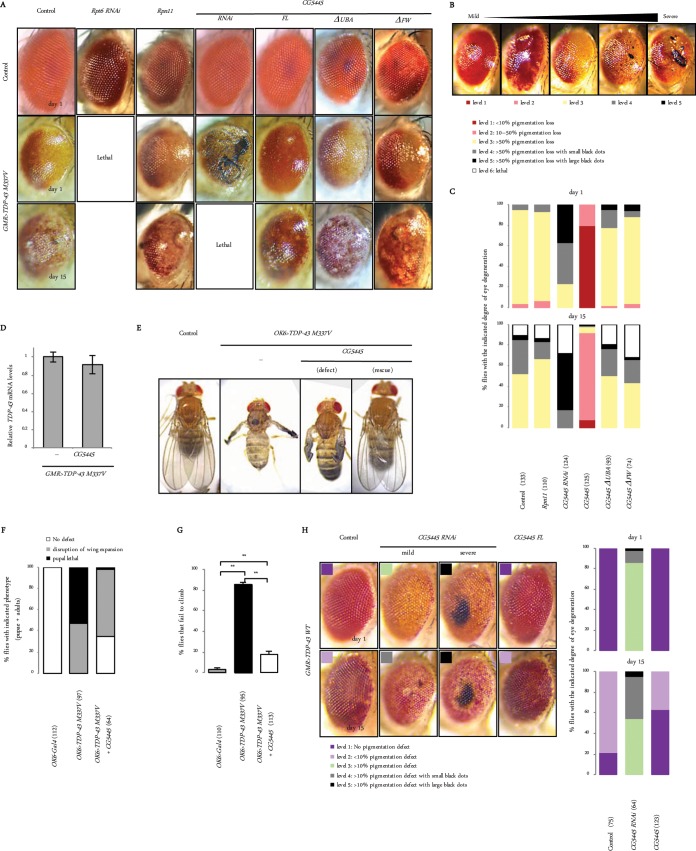
As characterized in a previous report, the overexpression of the TDP-43 M337V mutant under the control of GMR-Gal4 (GMR>TDP-43 M337V) in fly eyes resulted in age-dependent degeneration associated with pigmentation loss and necrotic patches (Fig. 3A) (23). To explore the genetic interactions with TDP-43 M337V, GMR>TDP-43 M337V flies were crossed with flies that overexpressed or downregulated the proteasome or CG5445 (Fig. 3A). We categorized the phenotypes according to the severity of eye degeneration, with level 1 being the mildest and level 5 being the most severe (Fig. 3B). Knockdown of Rpt6 exacerbated the phenotype, leading to lethality before eclosion (Fig. 3A and C). Overexpression of the proteasome subunit Rpn11 has been shown to increase proteasome activity and mitigate eye degeneration caused by Machado-Joseph disease protein with an expanded polyQ domain (MJDtr-Q78) (13). The ectopic upregulation of Rpn11 mildly decreased the number of flies with necrotic patches on day 15 posteclosion (Fig. 3A and C). These results suggest that the activity of the 26S proteasome is involved in alleviating the toxicity of TDP-43 M337V in Drosophila.
FIG 3.
CG5445 protects against TDP-43 toxicity. (A) Effect of knockdown and overexpression of proteasomes and CG5445 on TDP-43-mediated eye degeneration. Genotypes (top panels from the left column) are GMR-Gal4, UAS-Rpt6 RNAi/+; GMR-Gal4/+, UAS-Rpn11/+; GMR-Gal4/+, GMR-Gal4/+; UAS-CG5445 RNAi/+, UAS-CG5445/+; GMR-Gal4/+, UAS-CG5445ΔUBA/+; GMR-Gal4/+, and UAS-CG5445ΔFW/+; GMR-Gal4/+. Genotypes in middle and bottom panels from the left column are UAS-TDP-43 M337V-Myc/+; GMR-GAL4/+, UAS-Rpt6 RNAi/UAS-TDP-43 M337V-Myc; GMR-Gal4/+, UAS-Rpn11/UAS-TDP-43 M337V-Myc; GMR-Gal4/+, GMR-Gal4/UAS-TDP-43 M337V-Myc; UAS-CG5445 RNAi/+, UAS-CG5445/UAS-TDP-43 M337V-Myc; GMR-Gal4/+, UAS-CG5445ΔUBA/UAS-TDP-43 M337V-Myc; GMR-Gal4/+, and UAS-CG5445ΔFW/UAS-TDP-43 M337V-Myc; GMR-Gal4/+. Adult flies were aged at 29°C. (B) Flies expressing TDP-43 M337V in eyes were categorized according to the degree of eye degeneration as follows: level 1 for <10% pigmentation loss, level 2 for 10 to 50% pigmentation loss, level 3 for >50% pigmentation loss, level 4 for >50% pigmentation loss with small necrotic patches (black dots), level 5 for >50% pigmentation loss with large necrotic patches, and level 6 for lethal. Typical eyes representing each level (levels 1 to 5) are shown. (C) Percentage of each level of TDP-43 M337V expression in flies on day 1 (top) and day 15 (bottom) posteclosion. Values in parentheses indicate the number of flies examined. Genotypes are as described above for panel A. (D) Total RNAs from fly heads on day 2 posteclosion were analyzed by qRT-PCR. Genotypes (from the left) are UAS-TDP-43 M337V-Myc/+; GMR-GAL4/+ and UAS-TDP-43 M337V-Myc/UAS-CG5445; GMR-GAL4/+. Data are means ± standard deviations of data from three independent experiments. (E) Dorsal views of adult flies. Genotypes (from the left) are OK6-Gal4/+ (control), OK6-Gal4/UAS-TDP-43 M337V-Myc, and OK6-Gal4/UAS-TDP-43 M337V-Myc; UAS-CG5445/+. (F) Percentages of pupae and adults with the indicated phenotypes. Values in parentheses indicate the number of flies examined. Genotypes are as described above for panel E. (G) Percentage of flies that failed to climb. Data represent means and standard deviations of results from three independent experiments. Values in parentheses indicate the number of flies examined. Genotypes are as described above for panel E. **, P < 0.001. Flies were raised at 20°C (E to G). (H, left) Appearance of fly eyes on days 1 and 15 posteclosion. Genotypes (from the left column) are UAS-TDP-43 WT-Myc/+; GMR-GAL4/+, GMR-Gal4/UAS-TDP-43 WT-Myc; UAS-CG5445 RNAi/+, and UAS-CG5445/UAS-TDP-43 WT-Myc; GMR-Gal4/+. Flies expressing TDP-43 WT in eyes were categorized according to the degree of eye degeneration, similar to GMR>TDP-43 M337V flies, but with minor modifications, as follows: level 1 for no pigmentation loss, level 2 for <10% pigmentation loss, level 3 for >10% pigmentation loss, level 4 for >10% pigmentation loss with small necrotic patches (black dots), and level 5 for >10% pigmentation loss with large necrotic patches. (Right) Percentage of eyes of each level on days 1 and 15 posteclosion. Values in parentheses indicate the number of flies examined. Adult flies were aged at 29°C.
A more profound suppression of the phenotype was observed with CG5445 FL overexpression without affecting TDP-43 mRNA levels, while the phenotype was severely exacerbated by CG5445 knockdown; approximately 80% of CG5445 knockdown flies exhibited necrotic patches even on day 1 posteclosion, and the patches were enhanced with elevated lethality as the flies aged (Fig. 3A, C, and D). Overexpression of mutant CG5445ΔUBA failed to alleviate degeneration, demonstrating the importance of the ubiquitin-binding capacity of CG5445 for mitigating toxicity (Fig. 3A and C). Moreover, CG5445ΔFW did not suppress this phenotype, suggesting that the FW domain is also required for mitigation (Fig. 3A and C).
We further investigated the effect of full-length CG5445 overexpression on motor neurons. The ectopic expression of TDP-43 M337V under the control of OK6-Gal4 (OK6>TDP-43 M337V) in motor neurons resulted in defective eclosion or disruption of wing expansion in adults (Fig. 3E and F). In addition, OK6>TDP-43 M337V flies exhibited a progressive defect in climbing ability (Fig. 3G). The overexpression of CG5445 FL mitigated lethality, generated progenies with no obvious phenotype, and even ameliorated the adult climbing defect (Fig. 3E to G). These results demonstrate that CG5445 suppresses the toxicity of TDP-43 M337V in locomotor function.
To confirm the genetic interaction between CG5445 and TDP-43, wild-type TDP-43 (TDP-43 WT) was ectopically expressed in eyes (GMR>TDP-43 WT). Similarly to the TDP-43 M337V mutant, GMR>TDP-43 WT flies were categorized according to the severity of the eye phenotype (Fig. 3H). The expression of TDP-43 WT did not cause obvious eye defects on day 1 but induced weak pigmentation loss as the flies aged (Fig. 3H). CG5445 overexpression slightly mitigated the progression of pigmentation loss, while RNAi led to enhanced pigmentation loss and the appearance of necrotic patches (Fig. 3H). Taken together, these results demonstrate that CG5445 counteracts the toxicity of TDP-43.
CG5445 also alleviates toxicity induced by mutant FUS in Drosophila.
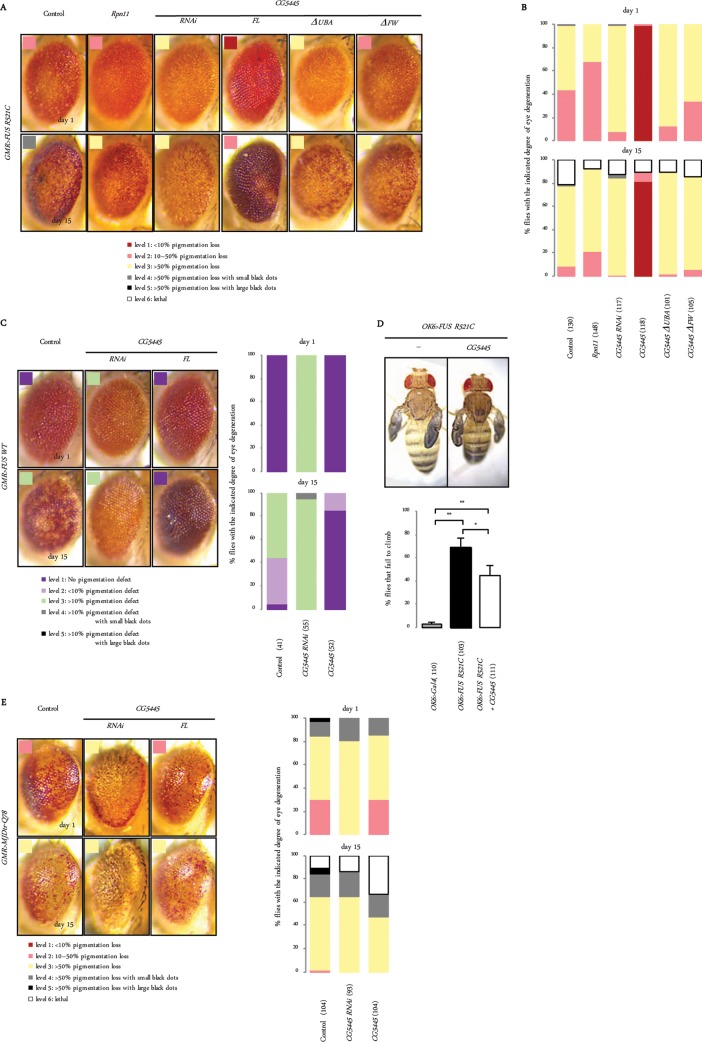
We examined whether CG5445 also modulates toxicity in other conformational disease models. Mutants of fused in sarcoma (FUS), another fALS-linked protein, have been shown to be ubiquitinated and form aggregates in cells, similarly to mutant TDP-43 (4, 24, 25). As shown in a previous report, the ectopic expression of FUS containing the fALS-linked R521C mutation (GMR>FUS R521C) in compound eyes caused age-dependent pigmentation loss (Fig. 4A) (24). GMR>FUS R521C flies were categorized according to the severity of eye degeneration (Fig. 4B). Rpn11 overexpression mildly suppressed the pigmentation loss. CG5445 RNAi exacerbated it, whereas CG5445 FL overexpression mitigated it. This improvement was not observed when CG5445 lacked the UBA or the FW domain (Fig. 4A and B). The ectopic expression of wild-type FUS in eyes also induced age-dependent pigmentation loss, which was enhanced and mitigated by CG5445 RNAi and overexpression, respectively (Fig. 4C).
FIG 4.
CG5445 mitigates toxicity of FUS but not of the polyQ protein. (A) Representative images of eyes associated with mutant FUS-mediated degeneration. Genotypes (from the left column) are GMR-GAL4/+; UAS-HA-FUS R521C/+, GMR-GAL4/UAS-Rpn11; UAS-HA-FUS R521C/+, GMR-GAL4/+; UAS-HA-FUS R521C/UAS-CG5445 RNAi, GMR-GAL4/UAS-CG5445; UAS-HA-FUS R521C/+, GMR-GAL4/+; UAS-HA-FUS R521C/UAS-CG5445ΔUBA, and GMR-GAL4/+; UAS-HA-FUS R521C/UAS-CG5445ΔFW. Flies expressing FUS R521C in eyes were categorized according to the degree of eye degeneration, as described in the legend to Fig. 3B. Adult flies were aged at 29°C. (B) The percentage of FUS R521C-expressing flies with each level of degeneration on days 1 and 15 posteclosion. Values in parentheses indicate the number of flies examined. Genotypes are as described above for panel A. (C, left) Representative images of eyes expressing FUS WT. (Right) The phenotypes were categorized as described in the legend to Fig. 3H, and the percentage of flies with each level of degeneration is shown. Parentheses indicate the number of flies examined. Genotypes (from the left column) are GMR-GAL4/+; UAS-HA-FUS WT/+, GMR-GAL4/+; UAS-HA-FUS WT/UAS-CG5445 RNAi, and GMR-GAL4/UAS-CG5445; UAS-HA-FUS WT/+. Adult flies were aged at 29°C. (D, top) Dorsal views of adult flies. Genotypes (from the left) are OK6-Gal4/+; UAS-HA-FUS R521C/+ and OK6-Gal4/+; UAS-HA-FUS R521C/UAS-CG5445. All adults showed wing expansion defects for both genotypes. (Bottom) Percentage of flies that failed to climb. Data represent means and standard deviations of results from three independent experiments. The control data are the same as those described in the legend to Fig. 3G. Values in parentheses indicate the number of flies examined. *, P < 0.05; **, P < 0.001. Flies were raised at 20°C. (E, left) Representative images of eyes expressing MJDtr-Q78. (Right) The phenotypes were categorized as described in the legend to Fig. 3H, and the percentage of eyes with each level of degeneration is shown (right). Values in parentheses indicate the number of flies examined. Genotypes (from the left column) are GMR-Gal4 UAS-MJDtr-Q78/+, GMR-Gal4 UAS-MJDtr-Q78/UAS-CG5445 RNAi, and UAS-CG5445/+; GMR-Gal4 UAS-MJDtr-Q78/+. Adult flies were aged at 29°C.
The ectopic expression of FUS R521C in motor neurons (OK6>FUS R521C) did not lead to any eclosion defect but caused a wing expansion defect in all adults, which was associated with increased climbing defects (Fig. 4D). The overexpression of CG5445 FL did not rescue the FUS R521C-induced wing defect but ameliorated the climbing defect (Fig. 4D). These results suggest that CG5445 also protects against FUS-mediated neurodegeneration.
We next examined the effect of CG5445 on polyQ-mediated neurodegeneration. Although Rpn11 overexpression has been shown to mitigate polyQ-mediated eye degeneration (13), CG5445 FL overexpression did not ameliorate polyQ-mediated pigmentation loss (Fig. 4E). CG5445 RNAi enhanced pigmentation loss on day 1 but did not increase the formation of necrotic patches on day 15 (Fig. 4E). These results suggest that CG5445 does not protect against polyQ-mediated toxicity.
Taken together, our results show that the effect of CG5445 on reducing the cytotoxicity of aberrant proteins is not limited to TDP-43 but is also valid for a number of other aggregate-prone proteins such as FUS, although this effect is not total.
CG5445 counteracts accumulation of insoluble forms of ALS-linked TDP-43.
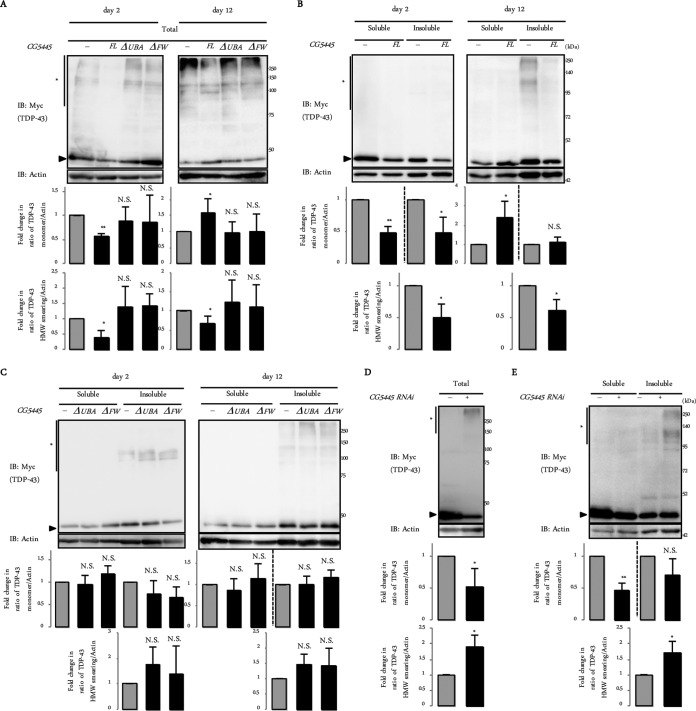
To clarify the effect of CG5445 on the TDP-43 M337V mutant biochemically, we first examined whether CG5445 affects the solubility and clearance of mutant TDP-43. Heads of GMR>TDP-43 M337V flies were fractionated into Triton X-100-soluble and -insoluble fractions. We also prepared total lysates without fractionation. In flies that do not overexpress CG5445, TDP-43 M337V was observed in monomeric and HMW smearing forms, and on day 12 posteclosion, the HMW smearing of TDP-43 M337V became obvious (Fig. 5A). Monomeric TDP-43 was observed in both Triton X-100-soluble and -insoluble fractions, whereas HMW smearing of TDP-43 was especially observed in the insoluble fraction on day 12 (Fig. 5B).
FIG 5.
CG5445 inhibits accumulation of detergent-insoluble TDP-43 M337V. (Top panels) Fly heads expressing TDP-43 M337V on day 2 and day 12 (A to C) and day 1 (D and E) posteclosion were subjected to immunoblotting with the indicated antibodies, with (B, C, and E) or without (A and D) separation into Triton-soluble and -insoluble fractions. Arrowheads and asterisks indicate monomeric TDP-43 M337V and HMW smearing, respectively. (Bottom panels) Values represent means and standard deviations of the relative band intensities of TDP-43 (normalized to actin) obtained from four independent experiments. *, P < 0.05; **, P < 0.005; N.S., not significant. Genotypes are UAS-TDP-43 M337V-Myc/+; GMR-Gal4/+, UAS-TDP-43 M337V-Myc/UAS-CG5445; GMR-Gal4/+, UAS-TDP-43 M337V-Myc/+; GMR-Gal4/UAS-CG5445ΔUBA, and UAS-TDP-43 M337V-Myc/+; GMR-Gal4/UAS-CG5445ΔFW (A to C) and UAS-TDP-43 M337V-Myc/GMR-Gal4 and UAS-TDP-43 M337V-Myc/GMR-Gal4; UAS-CG5445 RNAi/+ (D and E). Adult flies were aged at 29°C.
The overexpression of CG5445 FL decreased both the monomeric and HMW smearing forms of TDP-43 on day 2 (Fig. 5A, left). The overexpression of CG5445ΔUBA or CG5445ΔFW failed to reduce TDP-43 levels (Fig. 5A, left, and C, left). These results suggest that CG5445 promotes the clearance of mutant TDP-43. On day 12, the overexpression of CG5445 FL increased the level of the monomeric form, while it decreased the level of the HMW smearing form (Fig. 5A, right). These changes were attributed to an increase in the level of the soluble monomeric form and a decrease in the level of the insoluble HMW smearing form (Fig. 5B, right). Again, the overexpression of CG5445ΔUBA or CG5445ΔFW did not improve the amount and solubility of each form of TDP-43 (Fig. 5A, right, and C, right).
In contrast, CG5445 knockdown increased the HMW smearing of TDP-43 M337V by as early as day 1, while it decreased monomeric TDP-43 levels (Fig. 5D). The increase in HMW smearing was a result of the accumulation of the insoluble fraction, while CG5445 knockdown decreased the levels of soluble monomeric TDP-43 (Fig. 5E). These results, together with the results shown in Fig. 2E, suggest that CG5445 increases the solubility of ubiquitinated mutant TDP-43 and thus may facilitate its degradation.
CG5445 physically associates with ALS-linked TDP-43.
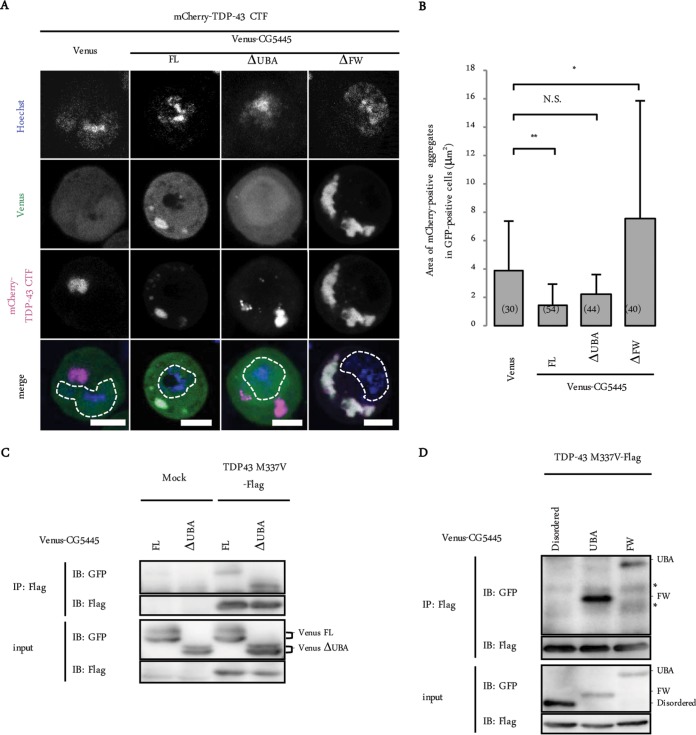
We next investigated whether CG5445 colocalizes with TDP-43. The C-terminal fragment of TDP-43 (TDP-43 CTF), which has been linked to ALS (8), exists predominantly in the cytosol and often forms aggregates (4). Venus-tagged CG5445 and mCherry-tagged TDP-43 CTF were coexpressed in S2 cells, and the cells were treated with bortezomib, a reversible proteasome inhibitor, to induce cytosolic aggregates of TDP-43 CTF (Fig. 6A). CG5445 FL was distributed in both the cytosol and the nuclei and colocalized with cytosolic TDP-43 CTF aggregates (Fig. 6A). CG5445ΔUBA failed to colocalize with the aggregates, suggesting that CG5445 associates with TDP-43 aggregates via the UBA domain (Fig. 6A). CG5445ΔFW colocalized with TDP-43 CTF, forming large cytosolic aggregates (Fig. 6A).
FIG 6.
CG5445 colocalizes and interacts with mutant TDP-43. (A) Representative images of S2 cells coexpressing TDP-43 CTF and CG5445 (FL, ΔUBA, and ΔFW). The cells were treated with 50 nM bortezomib for 12 h. Bar, 5 μm. Broken lines indicate edges of nuclei. (B) The area of mCherry-positive aggregates in Venus-positive cells was quantified. Values represent means and standard deviations. Values in parentheses indicate the number of aggregates examined. *, P < 0.05; **, P < 0.0001; N.S., not significant. (C and D) Flag–TDP-43 M337V was coexpressed with Venus-CG5445 (FL and ΔUBA [C] and the disordered region, the UBA domain, and the FW domain [D]) in S2 cells, immunoprecipitated, and subjected to immunoblot analysis with the indicated antibodies. Asterisks indicate nonspecific bands.
We then quantified the size of the TDP-43 CTF aggregates in order to evaluate the effect of each domain (Fig. 6B). The coexpression of CG5445 FL moderately but significantly reduced the aggregate size, whereas CG5445ΔUBA did not show a significant effect on size (Fig. 6B). Conversely, the TDP-43 CTF aggregates became larger in CG5445ΔFW-expressing cells, suggesting that the FW domain is involved in the inhibition of TDP-43 aggregation (Fig. 6B).
To further examine the interaction, Flag-tagged TDP-43 M337V was coexpressed with Venus-tagged CG5445 in S2 cells and then immunoprecipitated with anti-Flag antibody. CG5445 FL and even CG5445ΔUBA associated with TDP-43, implying that CG5445 possesses additional domains that recognize TDP-43 M337V, other than the UBA domain (Fig. 6C). To test this possibility, TDP-43 M337V coexpressed with each CG5445 domain was immunoprecipitated. Both the UBA and FW domains coimmunoprecipitated with TDP-43, suggesting that CG5445 also interacts with mutant TDP-43 via the FW domain (Fig. 6D). It is possible that the UBA domain interacts with ubiquitin chains tagged onto TDP-43, whereas the FW domain might recognize an aberrant region of TDP-43.
CG5445 does not interact with ALS-linked FUS.
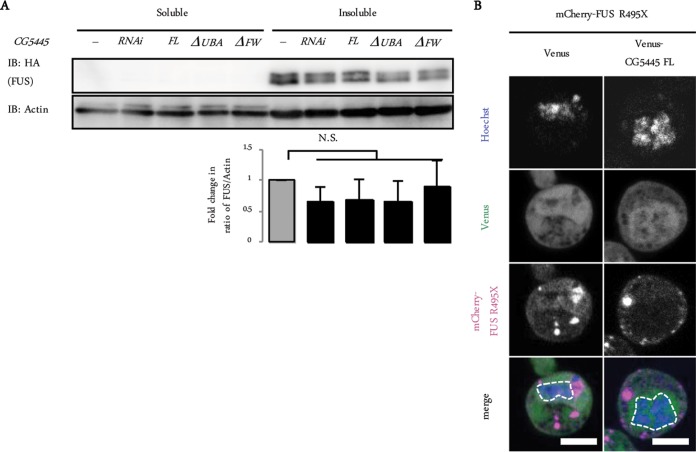
We next tested the effect of CG5445 on another ALS-linked aberrant protein, FUS. Immunoblot analysis of head extracts of GMR>FUS R521C flies showed that FUS R521C was detected as Triton-insoluble doublet bands, consistent with data from previous reports (Fig. 7A) (26, 27). However, although CG5445 FL overexpression and knockdown mitigated and augmented toxicity in FUS-expressing flies, respectively (Fig. 4), neither the overexpression nor the knockdown of CG5445 affected the amount and solubility of FUS (Fig. 7A). We also examined the colocalization of CG5445 with mutant FUS. FUS R495X is one of the ALS-linked mutants lacking C-terminal residues containing a putative nuclear localization signal (28). Consistent with data from previous reports, Venus-tagged FUS R495X was detected as cytosolic aggregates (Fig. 7B) (4, 28); however, CG5445 did not colocalize with the aggregates (Fig. 7B). Therefore, mutant FUS does not seem to be a direct target of CG5445. Rather, it might indirectly ameliorate neurodegeneration phenotypes caused by mutant FUS by as-yet-unclear mechanisms, unlike TDP-43, which are regulated by CG5445 with respect to protein levels and solubility status (Fig. 4A to E).
FIG 7.
CG5445 does not affect the solubility of mutant FUS. (A, top) Fly heads expressing FUS R521C on day 2 posteclosion were separated into Triton-soluble and -insoluble fractions and subjected to immunoblotting with the indicated antibodies. (Bottom) Values represent means and standard deviations of the relative band intensities of insoluble FUS (normalized to actin) obtained from four independent experiments. N.S., not significant. Genotypes are GMR-GAL4/+; UAS-HA-FUS R521C/+, GMR-GAL4/+; UAS-HA-FUS R521C/UAS-CG5445 RNAi, GMR-GAL4/UAS-CG5445; UAS-HA-FUS R521C/+, GMR-GAL4/+; UAS-HA-FUS R521C/UAS-CG5445ΔUBA, and GMR-GAL4/+; UAS-HA-FUS R521C/UAS-CG5445ΔFW. Adult flies were aged at 29°C. (B) Representative images of S2 cells coexpressing FUS R495X and CG5445. Bar, 5 μm. Broken lines indicate edges of nuclei.
DISCUSSION
The UPS plays a pivotal role in intracellular protein quality control to prevent neurodegeneration. Soluble misfolded proteins that are unable to be refolded are ubiquitinated and subsequently degraded by the proteasome (25). However, the proteasome cannot degrade insoluble and nondissociable aggregates, which are usually substrates for autophagy (25). It was recently shown that ubiquitination renders substrates prone to aggregation (29). Therefore, the ubiquitination of aggregate-prone misfolded proteins should further increase the possibility of forming cytotoxic ubiquitin-positive aggregates before proteasomal degradation. This probably holds true not only for cases with genetic mutations but also for sporadic cases of neurodegenerative diseases, since CG5445 knockdown increased the accumulation of ubiquitinated proteins in an insoluble state in tissues not expressing specific aberrant proteins. It would be reasonable to suppose that some mechanisms that prevent the aggregation of ubiquitinated proteins exist. We propose that CG5445 prevents the aggregation of ubiquitinated proteins, which consequently promotes their proteasomal degradation and decreases cytotoxic aggregate formation. The physical interaction of CG5445 with ubiquitinated aberrant proteins via the UBA domain may be a key step for increasing the solubility of aberrant proteins, since TDP-43 M337V-mediated eye degeneration was not rescued by mutant CG5445 lacking the UBA domain. The FW domain also seems to play an important role in increasing the solubility of aberrant proteins, as the expression of CG5445 lacking the FW domain augmented TDP-43 aggregate size and failed to rescue eye degeneration, even though CG5445ΔFW showed an ability to interact with TDP-43. Importantly, proteins with the same domain composition as that of CG5445 are conserved in metazoans, including the human orthologue C6orf106, although the position of its disordered region is located at the C terminus (16). Recent studies suggest the involvement of C6orf106 in cancer progression (30, 31), but its role in human cells remains unknown. Further studies of CG5445 and C6orf106 will shed new light on the pathogenic mechanisms underlying neurodegenerative diseases and will provide insights into understanding the pathogenesis of ALS.
MATERIALS AND METHODS
Expression constructs.
A cDNA encoding CG5445 was synthesized from total RNA isolated from the w1118 strain. cDNAs of TDP-43 and FUS were synthesized from total RNA isolated from HEK293T cells. The M337V mutation in TDP-43 was introduced by site-directed mutagenesis. The amplified fragments were cloned into the pGEX6p-1 vector (Amersham) or pUAST (32).
Fly stocks and genetic screening.
All Drosophila stocks were maintained on standard medium at 25°C unless otherwise stated. The following fly strains were used in this study: GMR-Gal4(II), GMR-Gal4(III), Da-Gal4, tub-Gal80ts, OK6-Gal4, and UAS-lacZ (Bloomington Drosophila Stock Center); pnr-Gal4 (33); UAS-Rpn11 (13); UAS-TDP-43 WT-Myc and UAS-TDP-43 M337V-Myc (21); UAS-HA-FUS WT and UAS-HA-FUS R521C (24); UAS-MJDtr-Q78 (34); UAS-CL1-GFP (17); UAS-Txl RNAi (transformant 5495R-1) and UAS-CG5445 RNAi (transformant 5445R-4) (National Institute of Genetics); UAS-Rpt6 RNAi (transformant 100620), UAS-DTH RNAi (transformant 108879), and UAS-Ddc RNAi (transformant 109881) (Vienna Drosophila Resource Center); and GS11377 (used in the genetic screen) and GS1003 (Drosophila Genetic Resource Center strains). UAS-CG5445 flies (full length, ΔUBA, and ΔFW) were generated and balanced by using standard techniques (BestGene Inc.). The gain-of-function screen was performed at 20°C by using GS lines (14) and the “local hop” technique (15), as described previously (13). In brief, pnr>Txl RNAi flies were crossed with GS lines in which a GS vector containing a UAS is randomly inserted into the fly genome, and progenies in which melanization is suppressed were isolated. The insertion sites for the GS vector in the suppressor strains were determined by inverse PCR.
Reverse transcription-quantitative PCR (qRT-PCR).
Total RNAs were isolated by using a High Pure RNA isolation kit (Roche) and reverse transcribed by using ReverTra Ace (Toyobo). Quantitative PCR was performed by using Universal ProbeLibrary probes (Roche). Each mRNA level was normalized by Rpl32 mRNA levels. Primers used were as follows: 5′-GAACAGTGTGCAGGATCTAAGC-3′ and 5′-TTGACGTAGCGCAGATTCAC-3′ for Txl, 5′-GCAATAGACAGTTAGAAAGAAGTGGA-3′ and 5′-CTGATTCCCAAAGCCACCT-3′ for human TDP-43, and 5′-CGGATCGATATGCTAAGCTGT-3′ and 5′-CGACGCACTCTGTTGTCG-3′ for Rpl32.
Cell culture and transfection.
S2 cells were cultured in Schneider's Drosophila medium supplemented with 10% fetal bovine serum (MBL), 100 U/ml penicillin, and 100 μg/ml streptomycin at 25°C. Transfection with the pUAST and pWAGAL4 vectors was performed by using Effectene transfection reagent (Qiagen), and cells were analyzed 48 h after transfection. Bortezomib (LC Laboratories) was added at 50 nM for 12 h. Cells were treated with Hoechst 33342 (Life Technologies) before microscopic analysis.
Antibodies.
Polyclonal antibodies against Txl were raised by immunizing rabbits with the peptide C+MNDFKRVVGKKGESH (residues 275 to 289 of TXNL1, a human orthologue of Txl) (Eurofins Scientific). Antibodies for actin (C4; Chemicon), the Flag epitope (F1804; Sigma), the Myc epitope (A-14; Santa Cruz), the hemagglutinin (HA) epitope (5D8; MBL), GFP (1E4; MBL), and ubiquitin (FK2; LifeSensors) were purchased.
Immunoblotting and immunoprecipitation.
For fractionation into detergent-soluble and -insoluble fractions, cells and fly heads were homogenized in phosphate-buffered saline (PBS) with 0.5% Triton X-100 and separated into supernatant (soluble) and pellet (insoluble) fractions by centrifugation at 20,000 × g for 15 min. For immunoprecipitation, cells were lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 0.2% NP-40, and 150 mM NaCl, and lysates were incubated with anti-DYKDDDDK tag antibody beads (Wako). Samples were separated by SDS-PAGE, transferred onto a polyvinylidene difluoride (PVDF) membrane, and subjected to immunoblot analysis. Band intensities were quantified by using ImageJ software.
GST pulldown assay.
Glutathione S-transferase (GST)-fused CG5445 (full length and ΔUBA) was purified from Escherichia coli by using glutathione-Sepharose 4B beads (GE Healthcare) and incubated with lysates of the w1118 fly strain.
Measurement of proteasome activity.
Fly heads were homogenized in ice-cold buffer containing 25 mM Tris-HCl (pH 7.5), 0.2% NP-40, 1 mM dithiothreitol, 2 mM ATP, and 5 mM MgCl2. The proteasome chymotrypsin-like peptidase activities in clarified lysates were measured by using the synthetic peptide substrate succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-MCA) (Peptide Institute), as described previously (35).
Histology.
Fluorescence microscopy images were captured on a TCS SP5 or SP8 confocal laser scanning microscope (Leica). Light microscopy images were captured on an M165 FC fluorescence stereomicroscope (Leica). Dissected wing discs were fixed in 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature (RT). The area of the cytosolic aggregates was quantified by using ImageJ software.
Climbing assay.
Climbing ability was analyzed by negative geotaxis as described previously, with minor modifications (36). In brief, 30 to 40 flies were gently tapped to the bottom of a vertical plastic vial, and the number of flies remaining at the bottom (less than 1 cm) was counted after 20 s. Ten trials were performed in each experiment.
Statistical analysis.
Statistical significance was calculated by using an unpaired two-tailed t test, a one-sample t test, Tukey's test, or a Mann-Whitney U test.
ACKNOWLEDGMENTS
We thank J. Paul Taylor, Udai Bhan Pandey, and Nancy M. Bonini for providing fly strains and Y. Sakurai for critical comments on the manuscript. We also thank Enago for the English language review.
This work was supported by JSPS KAKENHI grants JP25221102 and JP26111704 and the AMED-CREST from Japan Agency for Medical Research and Development.
We declare no competing financial interests.
REFERENCES
- 1.Glickman MH, Ciechanover A. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Murata S, Yashiroda H, Tanaka K. 2009. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol 10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 3.Rubinsztein DC. 2006. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 4.Deng H-X, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. 2011. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravid T, Hochstrasser M. 2008. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz AL, Ciechanover A. 2009. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol 49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 7.Forman MS, Trojanowski JQ, Lee VM-Y. 2004. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med 10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 8.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM-Y. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 9.Bruijn LI, Miller TM, Cleveland DW. 2004. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci 27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 10.Wiseman RL, Chin KT, Haynes CM, Stanhill A, Xu CF, Roguev A, Krogan NJ, Neubert TA, Ron D. 2009. Thioredoxin-related protein 32 is an arsenite-regulated thiol reductase of the proteasome 19 S particle. J Biol Chem 284:15233–15245. doi: 10.1074/jbc.M109.002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen KM, Madsen L, Prag S, Johnsen AH, Semple CA, Hendil KB, Hartmann-Petersen R. 2009. Thioredoxin Txnl1/TRP32 is a redox-active cofactor of the 26 S proteasome. J Biol Chem 284:15246–15254. doi: 10.1074/jbc.M900016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H. 2009. Regulation and function of the melanization reaction in Drosophila. Fly (Austin) 3:105–111. doi: 10.4161/fly.3.1.7747. [DOI] [PubMed] [Google Scholar]
- 13.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. 2009. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol 29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toba G, Ohsako T, Miyata N, Ohtsuka T, Seong KH, Aigaki T. 1999. The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tower J, Karpen GH, Craig N, Spradling AC. 1993. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics 133:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchbank K, Waters S, Roberts RG, Solomon E, Whitehouse CA. 2012. MAP1B interaction with the FW domain of the autophagic receptor Nbr1 facilitates its association to the microtubule network. Int J Cell Biol 2012:208014. doi: 10.1155/2012/208014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao T-P, Baehrecke EH, Taylor JP. 2007. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 18.Bence NF, Sampat RM, Kopito RR. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 19.Robberecht W, Philips T. 2013. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci 14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 20.Scotter EL, Vance C, Nishimura AL, Lee Y-B, Chen H-J, Urwin H, Sardone V, Mitchell JC, Rogelj B, Rubinsztein DC, Shaw CE. 2014. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J Cell Sci 127:1263–1278. doi: 10.1242/jcs.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, Lee VM-Y, Forman MS, Taylor JP. 2010. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci 30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann M, Mackenzie IR, Cairns NJ, Boyer PJ, Markesbery WR, Smith CD, Taylor JP, Kretzschmar HA, Kimonis VE, Forman MS. 2007. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J Neuropathol Exp Neurol 66:152–157. doi: 10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, Fushimi K, Wu JY. 2010. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci U S A 107:3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanson NA, Maltare A, King H, Smith R, Kim JH, Taylor JP, Lloyd TE, Pandey UB. 2011. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet 20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dantuma NP, Bott LC. 2014. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front Mol Neurosci 7:70. doi: 10.3389/fnmol.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, MacKenzie IRA. 2009. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miguel L, Avequin T, Delarue M, Feuillette S, Frébourg T, Campion D, Lecourtois M. 2012. Accumulation of insoluble forms of FUS protein correlates with toxicity in Drosophila. Neurobiol Aging 33:1008.e1–1008.e15. doi: 10.1016/j.neurobiolaging.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Sapp P, Mckenna-Yasek D, Brown RH, Hayward LJ. 2010. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morimoto D, Walinda E, Fukada H, Sou Y-S, Kageyama S, Hoshino M, Fujii T, Tsuchiya H, Saeki Y, Arita K, Ariyoshi M, Tochio H, Iwai K, Namba K, Komatsu M, Tanaka K, Shirakawa M. 2015. The unexpected role of polyubiquitin chains in the formation of fibrillar aggregates. Nat Commun 6:6116. doi: 10.1038/ncomms7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Miao Y, Yu X, Zhang Y, Jiang G, Liu Y, Yu J, Han Q, Zhao H, Wang E. 2015. C6orf106 enhances NSCLC cell invasion by upregulating vimentin, and downregulating E-cadherin and P120ctn. Tumor Biol 36:5979–5985. doi: 10.1007/s13277-015-3274-9. [DOI] [PubMed] [Google Scholar]
- 31.Jiang G, Zhang X, Zhang Y, Wang L, Fan C, Xu H, Miao Y, Wang E. 2015. A novel biomarker C6orf106 promotes the malignant progression of breast cancer. Tumor Biol 36:7881–7889. doi: 10.1007/s13277-015-3500-5. [DOI] [PubMed] [Google Scholar]
- 32.Brand AH, Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415. [DOI] [PubMed] [Google Scholar]
- 33.Calleja M, Herranz H, Estella C, Casal J, Lawrence P, Simpson P, Morata G. 2000. Generation of medial and lateral dorsal body domains by the pannier gene of Drosophila. Development 127:3971–3980. [DOI] [PubMed] [Google Scholar]
- 34.Warrick JM, Paulson HL, Gray-Board GL, Bui QT, Fischbeck KH, Pittman RN, Bonini NM. 1998. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell 93:939–949. doi: 10.1016/S0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 35.Koizumi S, Irie T, Hirayama S, Sakurai Y, Yashiroda H, Naguro I, Ichijo H, Hamazaki J, Murata S. 2016. The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. eLife 5:e18357. doi: 10.7554/eLife.18357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L-B, Yu Z, Teng X, Bonini NM. 2008. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]