ABSTRACT
Mechanisms underlying mitochondrial genome maintenance have recently gained wide attention, as mutations in mitochondrial DNA (mtDNA) lead to inherited muscular and neurological diseases, which are linked to aging and cancer. It was previously reported that human RAD51, RAD51C, and XRCC3 localize to mitochondria upon oxidative stress and are required for the maintenance of mtDNA stability. Since RAD51 and RAD51 paralogs are spontaneously imported into mitochondria, their precise role in mtDNA maintenance under unperturbed conditions remains elusive. Here, we show that RAD51C/XRCC3 is an additional component of the mitochondrial nucleoid having nucleus-independent roles in mtDNA maintenance. RAD51C/XRCC3 localizes to the mtDNA regulatory regions in the D-loop along with the mitochondrial polymerase POLG, and this recruitment is dependent upon Twinkle helicase. Moreover, upon replication stress, RAD51C and XRCC3 are further enriched at the mtDNA mutation hot spot region D310. Notably, the absence of RAD51C/XRCC3 affects the stability of POLG on mtDNA. As a consequence, RAD51C/XRCC3-deficient cells exhibit reduced mtDNA synthesis and increased lesions in the mitochondrial genome, leading to overall unhealthy mitochondria. Together, these findings lead to the proposal of a mechanism for a direct role of RAD51C/XRCC3 in maintaining mtDNA integrity under replication stress conditions.
KEYWORDS: mitochondria, genome integrity, RAD51 paralogs, replication stress, POLG
INTRODUCTION
Mitochondria are essential for providing cellular energy by generating ATP via respiration (1). Each mitochondrion contains several copies of the 16.5-kb circular genome (mitochondrial DNA [mtDNA]), which encodes 13 essential subunits of electron transport chain (ETC) complexes, 2 rRNAs, and 22 tRNAs (2). The presence of mtDNA in close proximity to the ETC and a lack of histone protection make mtDNA more susceptible to oxidative damage (3). In addition to spontaneously induced damage by oxidative stress, mtDNA is also susceptible to other types of lesions induced by ionizing radiation or chemical carcinogens (3, 4). An unrepaired or misrepaired mitochondrial genome impairs crucial cellular functions of mitochondria and can lead to premature aging, tumorigenesis, and neurological disorders (5–8).
Nuclear DNA replication and repair have been extensively studied; however, the pathways and mechanisms of mitochondrial DNA repair and replication are not well understood. The base excision repair (BER) pathway of mtDNA repair has been well characterized and appears to be a predominant repair mechanism in mitochondria (4). In contrast, evidence for the existence of nucleotide excision repair in mitochondria is still lacking. There are a few reports that suggest that DNA double-strand breaks (DSBs) in mitochondria are predominantly repaired by microhomology-mediated end joining (MMEJ) (9–12). The homologous recombination (HR)-mediated repair of nuclear DNA breaks has been well characterized in all organisms, and this mechanism of DSB repair is also well established for mitochondrial genomes of yeast and plants (13, 14, 99). However, the conservation of the HR pathway of DSB repair in human mitochondria remains largely controversial.
Apart from DNA repair pathways, the mitochondrial genome is maintained by the mitochondrial replisome consisting of mitochondrion-specific replication factors, including the mtDNA polymerase (POLG), DNA helicase (Twinkle), single-stranded DNA (ssDNA)-binding protein (mtSSB), RNA polymerase (POLRMT), transcription factor A (TFAM), and exonuclease (MGME1) (15–17). Nuclear DNA replication occurs in the S phase, whereas mtDNA replication is known to occur in all phases of the cell cycle (18). In precancerous lesions, oncogene-induced replication stress occurs due to the depletion of deoxynucleoside triphosphates (dNTPs), which causes replication stalling, chromosome instability, and tumorigenesis (19, 20). It is conceivable that continued mtDNA replication in all stages of the cell cycle may cause replication stress in mitochondria due to the depletion of dNTPs in precancerous cells. Moreover, mtDNA is constantly exposed to reactive oxygen species (ROS) generated during oxidative phosphorylation, which majorly induce abasic sites and 8-oxo-7,8-dihydroguanine (8-oxo-G) in the mtDNA. It has been shown that such oxidative lesions stall the mitochondrial replisome, and decreasing concentration of dNTPs further reduces the progression of the mitochondrial replisome (21). In addition, the mitochondrial genome is GC rich and possesses ∼100 G-quadruplex (G4) DNA-forming motifs (22). These G4 motifs are potential road blocks to faithful DNA replication. Indeed, it has been shown that the mitochondrial DNA helicase Twinkle fails to efficiently unwind G4 DNA structures, and mtDNA deletion breakpoints have been mapped to G4 DNA-forming motifs (23). mtDNA deletions can be an outcome of replication slippage, aberrant DNA structures, and DSBs caused by stalled replication forks. Apart from secondary structures and dNTP imbalance, the transcription termination factor mTERF has been shown to pause mtDNA replication at certain sites in the mitochondrial genome (24). However, unlike those in the nucleus, the pathways and mechanisms by which mitochondria respond to replication stress and maintain the integrity of mtDNA are obscure.
RAD51 and its orthologs play a key role in the recombination-mediated repair of DSBs in all organisms (25, 26). The mammalian RAD51 paralogs RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3 have been shown to regulate HR-mediated repair of DSBs and DNA damage signaling (27–31). Biochemical and two-hybrid studies show that RAD51 paralogs form two distinct complexes: the RAD51B/RAD51C/RAD51D/XRCC2 (BCDX2) complex and the RAD51C/XRCC3 (CX3) complex (32, 33). RAD51 paralog deficiency severely affects the formation of RAD51 foci at the sites of DNA breaks (30, 34, 35). Mouse knockout of RAD51 paralogs causes early embryonic lethality (36–39), and germ line mutations in all RAD51 paralogs have been found to be linked to breast and ovarian cancers (40–45). Interestingly, deletions and alterations in mtDNA copy numbers have been reported for breast and ovarian cancers (5, 46). Previously, we reported that RAD51 paralogs in distinct complexes protect and restart stalled replication forks during nuclear DNA replication (47). Interestingly, RAD51, RAD51C, and XRCC3 have been shown to localize to mitochondria and participate in the maintenance of mtDNA during oxidative stress (48). However, it is unclear whether RAD51 paralogs are involved in the faithful duplication of the mitochondrial genome and in the maintenance of mtDNA integrity during replication stress. Here, we report nucleus-independent functions of RAD51C/XRCC3 in mtDNA maintenance. Using subfractionation and mtDNA immunoprecipitation (mtDNA-IP), we find that RAD51C/XRCC3 is an integral part of the mitochondrial nucleoid that binds to mtDNA control regions within the D-loop. Upon replication stress, RAD51, RAD51C, and XRCC3 are further enriched at the mutation hot spot region D310, which prevents spontaneous mtDNA damage during replication stress. Interestingly, RAD51C/XRCC3 also regulates the binding of POLG to mtDNA, and a defect in this recruitment of POLG leads to decreases in mtDNA synthesis/contents and aberrant mitochondrial functions.
RESULTS
RAD51, RAD51C, and XRCC3 are a part of the mitochondrial nucleoid.
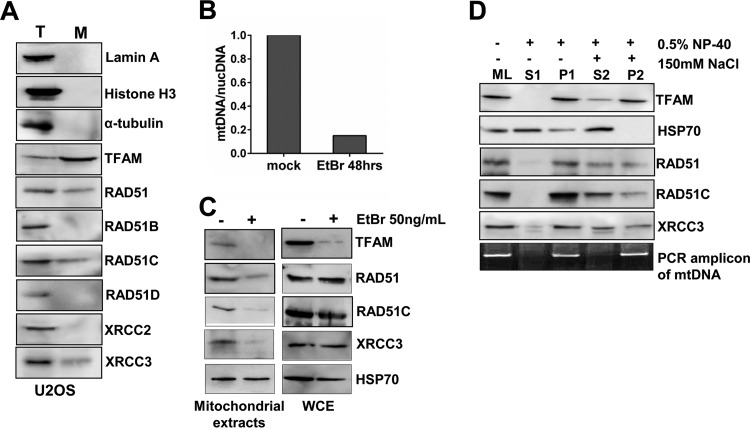
RAD51 and RAD51 paralogs are crucial for nuclear DNA repair by HR (31). A previous study (48) indicated that RAD51, RAD51C, and XRCC3 localize to mitochondria following oxidative stress and contribute to the protection of the mitochondrial genome. To understand additional roles of RAD51 paralogs in mitochondria under unperturbed conditions, we employed classical differential centrifugation and reexamined the localization of RAD51 paralogs in mitochondria. Interestingly, RAD51, RAD51C, and XRCC3 were present in the mitochondrial extracts of both U2OS (Fig. 1A) and HeLa (see Fig. S1A in the supplemental material) cells, whereas XRCC2, RAD51B, and RAD51D were absent (Fig. 1A and Fig. S1A).
FIG 1.
RAD51, RAD51C, and XRCC3 are a part of the mitochondrial nucleoid. (A) RAD51, RAD51C, and XRCC3 localize to mitochondria. Mitochondria were isolated from U2OS cells, and 20 μg of the mitochondrial fraction (M) was analyzed by immunoblotting along with 50 μg of the total cell lysate (T). Lamin A and histone H3 were taken as nuclear markers, α-tubulin was taken as a cytoplasmic marker, and TFAM was taken as a mitochondrial marker. (B) Assessment of mtDNA content in HeLa cells upon mtDNA depletion using EtBr for 48 h. (C) Western blots showing the levels of the indicated proteins in HeLa mitochondrial extracts and whole-cell extracts after EtBr treatment. (D) Isolated mitochondria from U2OS cells were divided into three parts. One part was lysed directly in 2× SDS buffer (mitochondrial lysate [ML]), and the second part was incubated in suspension buffer containing 0.5% NP-40 for 30 min on ice and centrifuged for 30 min to obtain the supernatant fraction (S1) and the pellet fraction (P1). The third part was incubated in suspension buffer with 0.5% NP-40 and 150 mM NaCl and separated similarly as supernatant (S2) and pellet (P2) fractions. Pellet fractions were lysed in 2× SDS buffer. Equal amounts of each fraction were loaded and analyzed by Western blotting.
We next asked whether the localization of RAD51, RAD51C, and XRCC3 is dependent upon the presence of mtDNA in mitochondria. Ethidium bromide (EtBr) treatment for longer periods leads to a loss of mtDNA (49, 50). Hence, we incubated HeLa cells with 50 ng/ml of EtBr for 48 h and confirmed mtDNA depletion using semiquantitative PCR (Fig. 1B). Strikingly, the levels of RAD51, RAD51C, and XRCC3 in mitochondria were diminished dramatically upon the depletion of mtDNA (Fig. 1C). However, their levels were unchanged in whole-cell extracts (Fig. 1C), except for TFAM, which has been reported to undergo Lon protease-mediated degradation upon mtDNA depletion (51). This observation is consistent with data from a previous study (52) where EtBr-mediated depletion of mtDNA in U2OS cells severely reduced the levels of RAD51 in mitochondria, indicating that the presence of mtDNA is a primary requirement for the mitochondrial localization of RAD51, RAD51C, and XRCC3. Next, we determined the localization of RAD51, RAD51C, and XRCC3 along with the mitochondrial genome by performing mitochondrial subfractionation as described previously (53), where isolated mitochondria were treated with a buffer containing a mild detergent (0.5% NP-40). RAD51, RAD51C, and XRCC3 were found in the pellet fractions containing the mtDNA, as monitored by PCR amplification. TFAM served as a nucleoid protein marker, whereas HSP70, a mitochondrial matrix protein, was released into the supernatant fraction (Fig. 1D). To examine the effect of ionic strength, we added 150 mM NaCl to the buffer. HSP70 was completely released into the supernatant, as opposed to TFAM, whose abundance in the pellet fraction was largely unaffected (Fig. 1D). Interestingly, substantial amounts of RAD51, RAD51C, and XRCC3 were still present in the nucleoid fraction even after salt extraction (Fig. 1D), indicating that RAD51, RAD51C, and XRCC3 are integral parts of the mitochondrial nucleoid.
Nucleus-independent function of RAD51C in mitochondrial genome maintenance.
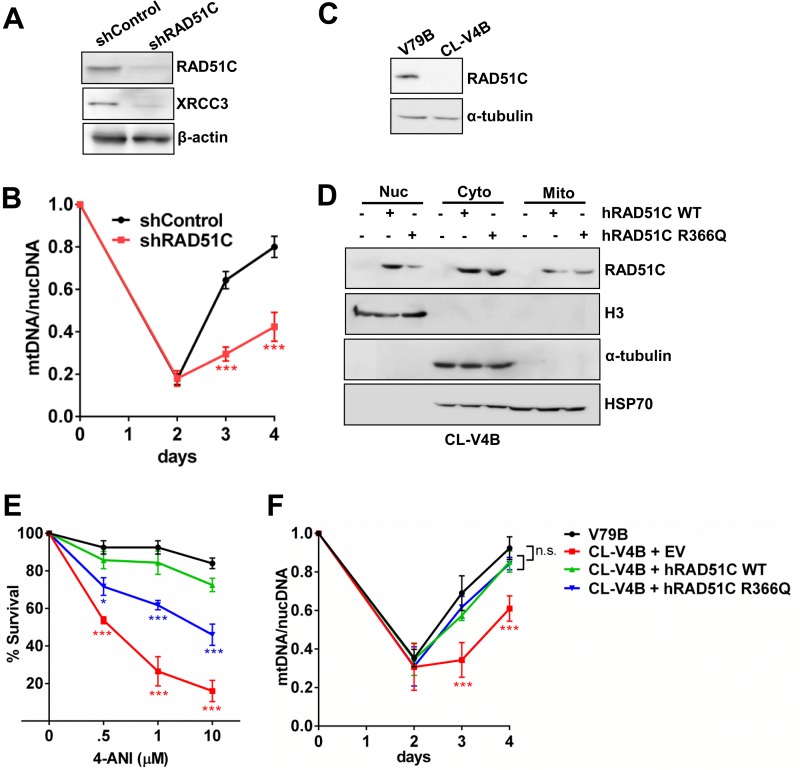
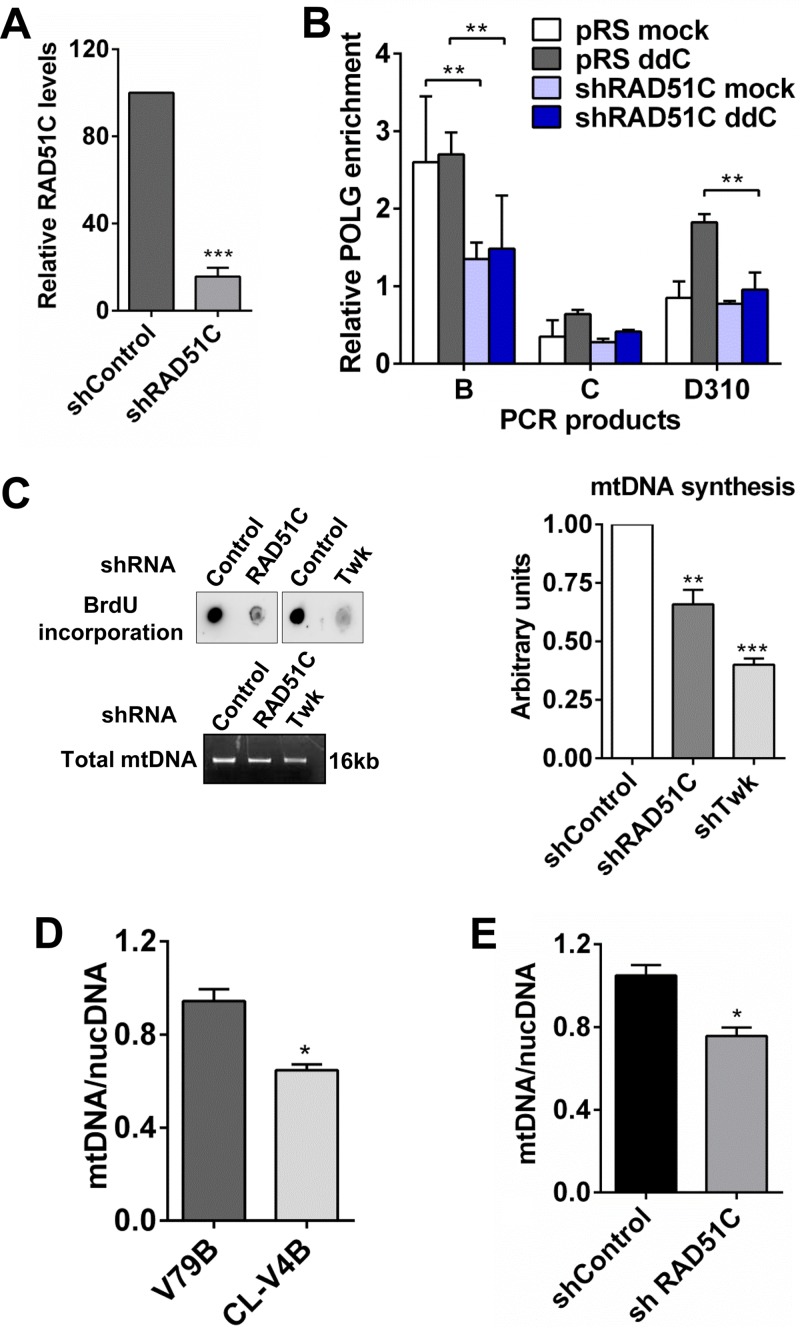
To understand whether RAD51C contributes to mtDNA replication, we performed depletion of mtDNA and studied its repopulation. RAD51C and XRCC3 form a stable complex (33), and consistent with data from previous reports (30, 54), the depletion of RAD51C reduced the levels of XRCC3 (Fig. 2A) and vice versa (see Fig. S1B in the supplemental material). After 48 h of treatment with EtBr, both RAD51C-proficient and -deficient HeLa cells underwent mtDNA depletion. However, upon EtBr withdrawal, mtDNA repopulation was severely impaired in both RAD51C-deficient (Fig. 2B) and XRCC3-deficient (Fig. S1C) cells, indicating perturbed mtDNA replication. Moreover, in agreement with data from a previous study (52), RAD51-deficient cells also exhibited a similar defect (Fig. S1B and S1C). In contrast, consistent with the lack of mitochondrial localization, the repopulation of mtDNA was unaffected in XRCC2-depleted cells (Fig. S1B and S1C).
FIG 2.
Nucleus-independent role of RAD51C in mtDNA maintenance. (A) Western blot showing levels of RAD51C and XRCC3 after shRNA-mediated depletion in HeLa cells. shRAD51C, short hairpin RNA targeting RAD51C. (B) mtDNA/nucDNA levels in control and RAD51C-depleted HeLa cells after recovery from mtDNA depletion using EtBr for 48 h. mtDNA/nucDNA levels were calculated by semiquantitative PCR using primers listed in Table S1 in the supplemental material. Data points represent means ± SD from three independent experiments. (C) Western blot for RAD51C in parental V79B and RAD51C-deficient CL-V4B hamster cells. (D) Western blot showing levels of RAD51C in the nucleus, cytoplasm, and mitochondria of CL-V4B cells transfected with WT human RAD51C (hRAD51C) and the RAD51C R366Q NLS mutant. Histone H3, α-tubulin, and HSP70 were taken as nuclear, cytoplasmic, and mitochondrial markers, respectively. (E) Sensitivities of V79B, CL-V4B, and CL-V4B cells expressing WT RAD51C and the RAD51C R366Q mutant at the indicated concentrations of the PARP inhibitor 4-amino-1,8-napthalimide (4-ANI). Data represent means ± SD from three independent experiments. (F) mtDNA/nucDNA levels in V79B, CL-V4B, and CL-V4B cells expressing WT RAD51C and the RAD51C R366Q mutant.
RAD51C knockout mice are embryonically lethal due to the essential functions of RAD51C in the DNA damage response and replication fork maintenance (27–29, 35, 39, 47). Thus, it is possible that defective nuclear functions of RAD51C contribute to its failure in recovery upon mtDNA depletion. To investigate this, we expressed a pathological RAD51C nuclear localization signal (NLS) mutant, R366Q (41), in RAD51C-deficient (CL-V4B) hamster cells (Fig. 2C). As expected, the RAD51C R366Q mutant showed a significantly reduced nuclear abundance compared to that of wild-type (WT) RAD51C (Fig. 2D). Interestingly, its localization in mitochondria remained unperturbed (Fig. 2D). Since RAD51C is critical for the HR-mediated repair of DSBs, we treated the cells with a poly(ADP-ribose) polymerase (PARP) inhibitor, which generates one-ended breaks, specifically requiring the HR pathway. In agreement with data from our previous study (35), the PARP inhibitor sensitized CL-V4B cells expressing the RAD51C R366Q mutant (Fig. 2E). Strikingly, cells expressing the RAD51C R366Q mutant were able to efficiently repopulate the depleted mtDNA after EtBr stress similarly to WT RAD51C-expressing cells (Fig. 2F), suggesting a nucleus-independent function of RAD51C in mitochondrial genome maintenance.
RAD51C/XRCC3 binds to mtDNA control regions along with POLG.
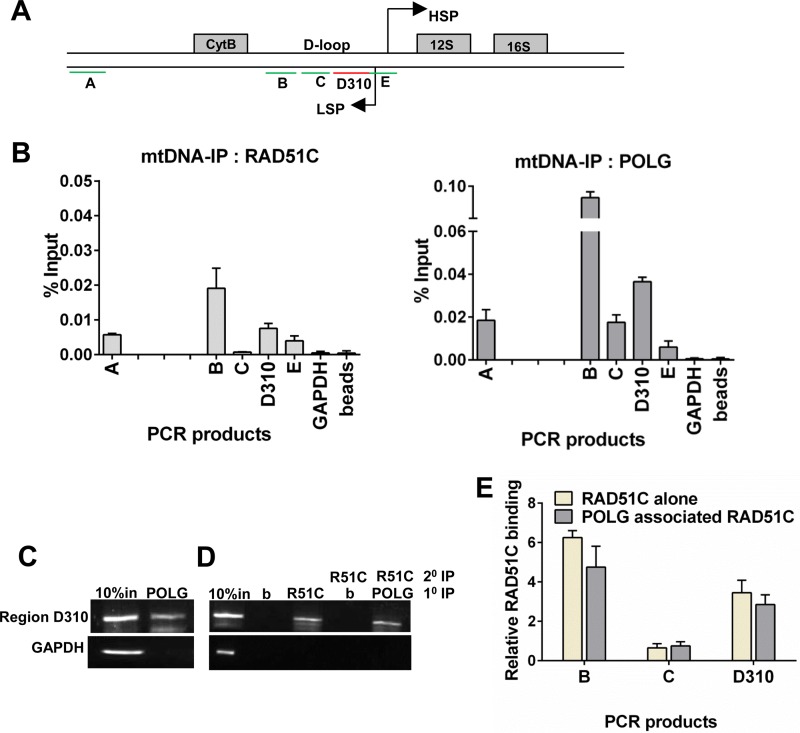
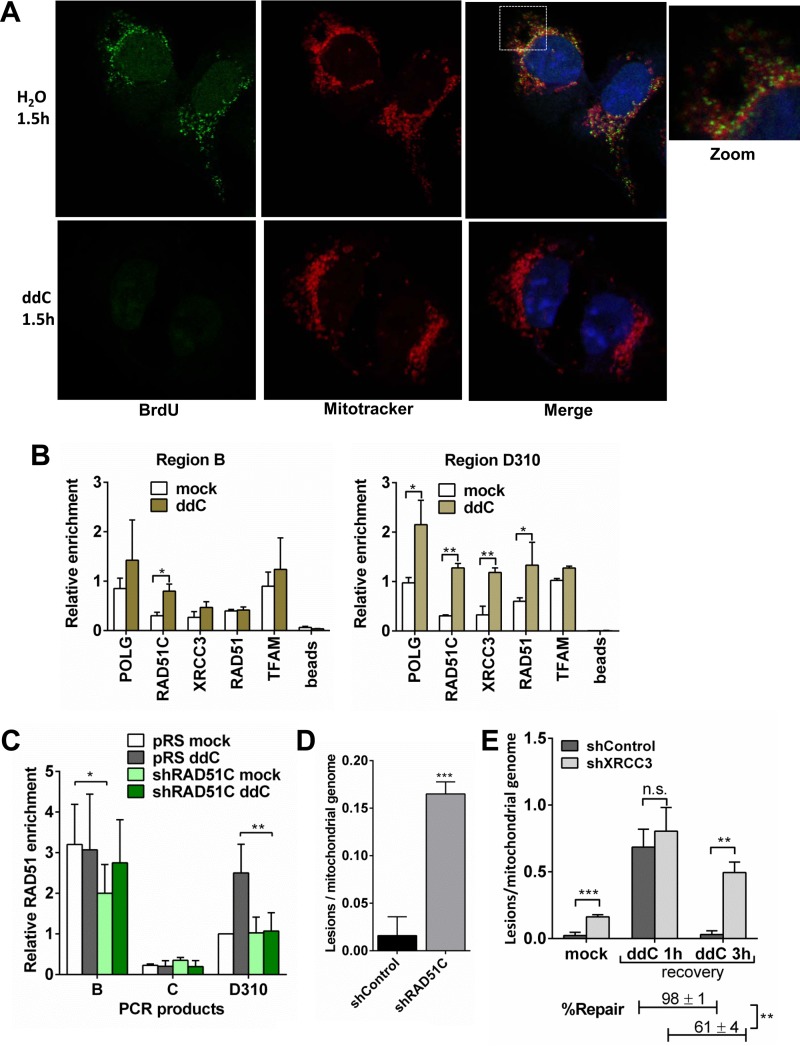
Given that RAD51C/XRCC3 was present in the mitochondrial nucleoid fraction and RAD51C/XRCC3 deficiency prevented the restoration of mtDNA upon its depletion, we asked whether RAD51C/XRCC3 binds to mtDNA control regions in the D-loop and participates in mtDNA replication. To test this, we used an mtDNA-IP protocol in combination with 5 primer pairs to screen a part of the human mtDNA genome (Fig. 3A) (24, 55). Region B (adjacent to the cytochrome b coding sequence), region C, region D310 (a cancer mutation hot spot), and region E are part of the regulatory D-loop region, whereas region A is outside the D-loop site (16, 56, 57). RAD51C exhibited maximum binding at region B and moderate binding at regions D310 and E within the D-loop (Fig. 3B). The binding at region C was minimal (Fig. 3B). Analysis of XRCC3 and RAD51 showed a binding profile similar to that of RAD51C (see Fig. S2A in the supplemental material). Interestingly, the pattern of binding of POLG to the regions of the D-loop was similar to those for RAD51, RAD51C, and XRCC3 (Fig. 3B), suggesting a possibility of the concomitant binding of POLG along with RAD51 and RAD51C/XRCC3 to these hot spots. To test this, the primary anti-POLG nucleoid immunocomplex (Fig. 3C) was reimmunoprecipitated with anti-RAD51C, anti-XRCC3, and anti-RAD51 antibodies. Strikingly, POLG-RAD51C (Fig. 3D and E), POLG-XRCC3, and POLG-RAD51 (Fig. S2B) were found to be present at the region B- and D310-bound DNA-binding complex. These interactions were not found at region C, which served as a negative control (Fig. 3E and Fig. S2B). Together, these data indicate that mitochondrial RAD51, RAD51C, and XRCC3 could contribute to mtDNA replication by occupying the regulatory D-loop site along with POLG.
FIG 3.
RAD51C/XRCC3 and POLG show sequence-specific recruitment on the mtDNA D-loop. (A) Schematic representation of primer pairs used for mtDNA-IP. D-loop regions and adjacent mtDNA genes are depicted (HSP, heavy strand promoter; LSP, light strand promoter). Isolated mitochondria from U2OS cells were subjected to mtDNA-IP using POLG- and RAD51C-specific antibodies. PCR products were resolved on an agarose gel and quantified by using ImageJ. (B) Quantification of data obtained from three independent experiments represented as the percent input for RAD51C (left) and POLG (right). (C) mtDNA-IP analysis with mitochondrial extracts prepared from U2OS cells, carried out by using a POLG antibody. (D) Double immunoprecipitation was carried out either using beads or against RAD51C antibody from the nucleoid collected from the POLG immunoprecipitate, as indicated. The input (IN) nucleoid is indicated; b, bead control. (E) Quantification of PCR amplicons obtained after single mtDNA-IP of RAD51C (RAD51C alone) and double mtDNA-IP of RAD51C from the POLG-immunoprecipitated nucleoid (POLG-associated RAD51C).
Twinkle associates with RAD51 to mediate the recruitment of RAD51 and RAD51C/XRCC3 to mtDNA control regions.
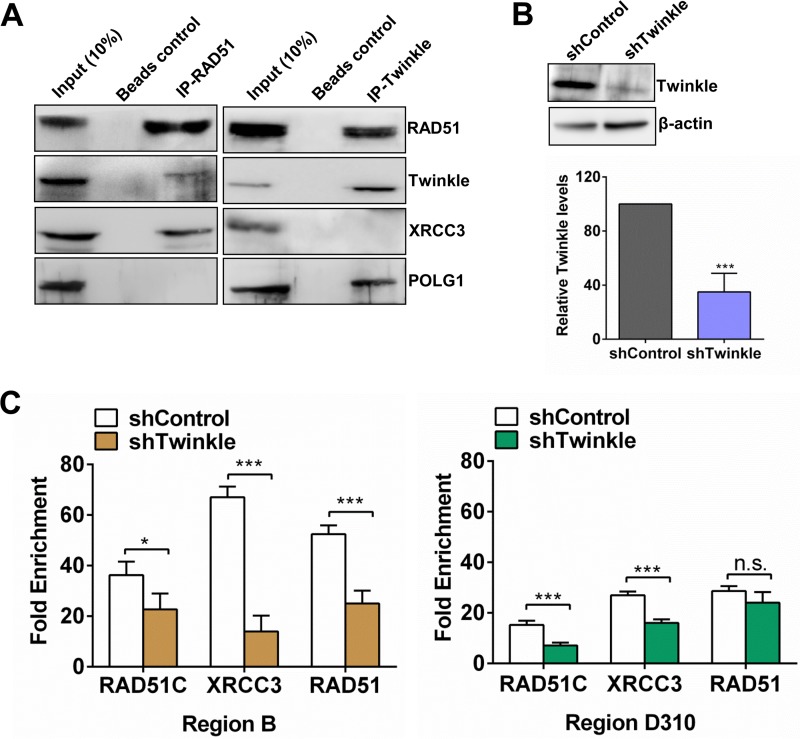
The colocalization data for POLG with RAD51, RAD51C, and XRCC3 on the mtDNA regulatory D-loop regions prompted us to hypothesize that RAD51, RAD51C, and XRCC3 could associate with mitochondrial replisome components. In order to test this, we performed immunoprecipitation of RAD51 from the mitochondrial extracts. Interestingly, we could not detect an association of POLG with RAD51. However, we found the mitochondrial helicase Twinkle along with XRCC3 in the RAD51 immunoprecipitate fraction (Fig. 4A), which was further validated by reciprocal IP, where Twinkle immunoprecipitates contained not only POLG but also RAD51 (Fig. 4A). Since Benzonase was included in the immunoprecipitation reaction mixture, we eliminated the possibility of any DNA-dependent interaction. The absence of a direct interaction between XRCC3 and Twinkle/POLG indicates an indirect association of RAD51 paralogs with the mtDNA replisome via RAD51.
FIG 4.
Twinkle associates with RAD51 to mediate RAD51 and RAD51C/XRCC3 recruitment on mtDNA control regions. (A) HeLa mitochondrial lysates containing Benzonase were immunoprecipitated with mouse anti-RAD51 and rabbit anti-Twinkle antibodies. (B, top) shRNA-mediated knockdown of Twinkle in U2OS cells. (Bottom) Quantification of Twinkle in U2OS cells transfected with shRNA targeting Twinkle (shTwinkle) (n = 3). (C) mtDNA-IP was performed with control and Twinkle-depleted U2OS cells against RAD51, RAD51C, and XRCC3. Data show fold enrichment of the indicated proteins in both the cells analyzed at region B (left) and those analyzed at region D310 (right). *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Because RAD51 associates with Twinkle, we speculated that Twinkle, being a helicase, might facilitate the loading of RAD51, RAD51C, and XRCC3 onto the mtDNA control regions. To test this, we carried out mtDNA-IP of RAD51C, XRCC3, and RAD51 in Twinkle-depleted U2OS cells (Fig. 4B). Indeed, the depletion of Twinkle helicase significantly decreased the enrichment of RAD51C, XRCC3, and RAD51 on mtDNA, suggesting Twinkle-mediated mtDNA recruitment of RAD51C/XRCC3 (Fig. 4C and Fig. S2C).
RAD51C/XRCC3 is enriched at region D310 to prevent and repair replication stress-induced mtDNA damage.
The mitochondrial genome is constantly exposed to ROS (58–61). By using catalytic POLG and Twinkle mutants, recent reports proposed that replication pausing or stalling can result in mtDNA lesions in the form of point mutations, rearrangements, and deletions (62–65). Dideoxycytidine (ddC) is known to cause the stalling of mtDNA replication specifically (64). To test this, U2OS cells were exposed to ddC, and mtDNA replication was measured by bromodeoxyuridine (BrdU) incorporation. Incubation with 100 μM ddC for 90 min almost completely abrogated the incorporation of BrdU in mtDNA (Fig. 5A). However, under these conditions, nuclear DNA replication was unperturbed (see Fig. S3A in the supplemental material). Also, ddC treatment did not result in nuclear DNA damage, as shown by γ-H2AX levels (Fig. S3B). To investigate whether RAD51, RAD51C, and XRCC3 respond to mtDNA replication stress, we performed mtDNA-IP upon treatment with ddC. Except for RAD51C, we did not find any change in the enrichment of RAD51 or XRCC3 at region B (Fig. 5B and Fig. S3C). However, at cancer mutation hot spot region D310, RAD51C and XRCC3 showed a nearly 4-fold enrichment, and RAD51 showed a nearly 3-fold enrichment, upon ddC-induced replication stress compared to mock-treated cells (Fig. 5B and Fig. S3C). In contrast, TFAM, which served as a control, did not show any change in its abundance in both regions (Fig. 5B and Fig. S3C). Overall, mitochondrial levels of RAD51C/XRCC3, RAD51, POLG, and TFAM were unaltered upon ddC treatment (Fig. S3D). Intriguingly, we also found a 2.5-fold enrichment of POLG at region D310 upon mtDNA replication stalling (Fig. 5B and Fig. S3C). One possible explanation for this would be a recently proposed replication-dependent repair pathway for the mtDNA lesions by POLG (66).
FIG 5.
RAD51C/XRCC3 shows enhanced enrichment at region D310 upon replication stalling to prevent and repair mtDNA lesions. (A) U2OS cells were treated with either water or 100 μM ddC for 1.5 h, followed by incubation with 10 μM BrdU for 2 h. Both cell types were previously treated with 2 μg/ml aphidicolin for 6 h to block nuclear DNA replication. Immunostaining was performed against BrdU; mitochondria were stained with MitoTracker, and nuclei were stained with DAPI. (B) Association of RAD51C/XRCC3 with mtDNA after ddC treatment. PCRs were carried out for region B and region D310 from immunoprecipitates of mtDNA with the indicated proteins in U2OS cells. Quantitative data from four independents are represented as means ± SD. **, P < 0.01; *, P < 0.05. (C) Relative enrichment of RAD51 on mtDNA control regions in control and RAD51C-depleted U2OS cells after ddC treatment. Data represent means ± SD. (D) RAD51C deficiency confers increased spontaneous mtDNA lesions. Genomic DNA was isolated from control and RAD51C-depleted U2OS cells, and mtDNA integrity was analyzed by using semiquantitative PCR. Data were normalized to the mtDNA content by using the amplification of 221-bp mtDNA. Data represent means ± SD for mtDNA lesions. (E) Replication stress-induced mtDNA lesions fail to repair in the absence of XRCC3. Control and XRCC3-depleted U2OS cells were treated with ddC and recovered in fresh medium at the indicated time intervals, and genomic DNA was isolated to calculate mtDNA lesions by using semiquantitative PCR as described above. Data represent means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Evidence from biochemical studies shows that RAD51C/XRCC3 promotes the formation of RAD51 nucleoprotein filament at the site of DSB lesions (34). Notably, RAD51C/XRCC3 is also critical for the recruitment and stabilization of RAD51 at the sites of stalled replication forks (47). To determine whether this function of RAD51C/XRCC3 is conserved in mitochondria, we performed mtDNA-IP assays with RAD51C-deficient U2OS cells. Spontaneously, RAD51 showed reduced binding at region B (Fig. 5C). Moreover, in response to replication stress, RAD51 failed to be enriched further at the D310 region, indicating that the localization of RAD51 on mtDNA is dependent upon RAD51C. The depletion of RAD51C, however, did not reduce the RAD51 levels in mitochondria (Fig. S3E).
Since RAD51, RAD51C, and XRCC3 showed enhanced enrichment at mtDNA region D310 upon replication stress, it was tempting to speculate that RAD51C/XRCC3 could prevent mtDNA damage during replication stress and participate in its repair. To examine this, genomic DNA was isolated from RAD51C-depleted U2OS cells and analyzed for mtDNA lesions by using a long-range PCR (LR-PCR) assay. The data presented in Fig. 5D indicate that there was a nearly 6-fold increase in mtDNA damage upon RAD51C deficiency. This increase in spontaneous mtDNA damage was also evident in XRCC3- and RAD51-deficient cells but not in XRCC2-depleted cells (see Fig. S4A in the supplemental material). This was further validated in hamster cells, where CL-V4B cells acquired a >7-fold increase in spontaneous mtDNA lesions compared to WT V79B cells (Fig. S4B). To test whether RAD51C/XRCC3 participates in the repair of replication-associated lesions, we treated control and XRCC3-depleted U2OS cells with 100 μM ddC, allowed the cells to recover for up to 3 h, and analyzed mtDNA integrity by using semiquantitative PCR. Strikingly, XRCC3-deficient cells were defective in repairing replication stress-induced mtDNA lesions (Fig. 5E and Fig. S4C).
RAD51C/XRCC3 stabilizes POLG on mtDNA.
Inspired by our observations that a deficiency of RAD51C/XRCC3 leads to increased mtDNA lesions and that RAD51C/XRCC3 shows a concomitant enrichment at mtDNA “along with POLG” during replication stress, we asked whether the stability of POLG on mtDNA is affected in the absence of RAD51C/XRCC3. To address this, we performed mtDNA-IP analysis for POLG upon the knockdown of RAD51C, XRCC3, and RAD51 in U2OS cells. Interestingly, the depletion of RAD51C and XRCC3 led to the reduced binding of POLG at region B (Fig. 6A and B and Fig. S4D and E) without affecting POLG protein levels in mitochondria (Fig. S3E). Upon replication stress, POLG failed to be enriched further at the D310 region in the absence of RAD51C/XRCC3 (Fig. 6A and B and Fig. S4D and E). Furthermore, the depletion of RAD51 dramatically decreased the recruitment of POLG both spontaneously and after ddC treatment (Fig. S4F and G). These data suggest that RAD51 and RAD51C/XRCC3 are required for the replication stress-induced recruitment of POLG to the D-loop sites.
FIG 6.
RAD51C stabilizes POLG on mtDNA to facilitate mtDNA replication. (A) Quantification of RAD51C levels in U2OS cells transfected with shRNA targeting RAD51C (n = 3). (B) Relative enrichment of POLG on mtDNA control regions in control and RAD51C-depleted U2OS cells after ddC treatment. Data represent means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.005. (C) Dot blot analysis showing the extent of BrdU incorporation in mtDNA. U2OS cells were transfected with the control vector and shRAD51C and shTwinkle plasmids. Twenty-four hours later, cells were incubated with 10 μM BrdU for 24 h, and mtDNA was isolated and normalized by visualization on the gel. Equal amounts of mtDNA were spotted onto a nitrocellulose membrane, cross-linked, and blotted with anti-BrdU antibody. (Left) Representative image of the dot blot; (right) quantitative data from three independent experiments represented as means ± SD. (D and E) mtDNA content measured as described above in RAD51C-deficient hamster (D) and U2OS (E) cells. Data represent means ± SD. **, P < 0.01; *, P < 0.05.
These results prompted us to consider that RAD51 and RAD51 paralogs might regulate overall mtDNA synthesis. To determine this, we carried out dot blot analysis with anti-BrdU antibody in U2OS cells following the depletion of RAD51, RAD51C, and XRCC3. There was a significant decrease in mtDNA synthesis in RAD51-, RAD51C-, and XRCC3-depleted U2OS cells compared to control empty vector cells (Fig. 6C and Fig. S5A). As expected, we did not find any alteration in mtDNA synthesis upon XRCC2 depletion (Fig. S5A). Knockdown of Twinkle helicase was taken as a positive control, which showed significantly low levels of mtDNA synthesis (Fig. 6C). For further validation, we measured the relative mtDNA levels in hamster cells lacking RAD51 paralogs (RAD51C, XRCC2, and XRCC3) with their WT counterparts using semiquantitative PCR for mtDNA and nuclear DNA (nucDNA). RAD51C-deficient CL-V4B cells and XRCC3-deficient irs1SF cells displayed modest reductions in the mtDNA/nucDNA ratio, whereas XRCC2 deficient irs1 cells did not show any reduction (Fig. 6D and Fig. S5B). Consistently, the knockdown of RAD51C and XRCC3 in U2OS cells showed modest reductions in the mtDNA/nucDNA ratio, whereas this ratio was unaffected in XRCC2-depleted cells (Fig. 6E and Fig. S5C). Moreover, consistent with data from a previous report (52), RAD51-deficient cells exhibited significantly low mtDNA levels (Fig. S5C).
RAD51C/XRCC3 deficiency leads to unhealthy mitochondria.
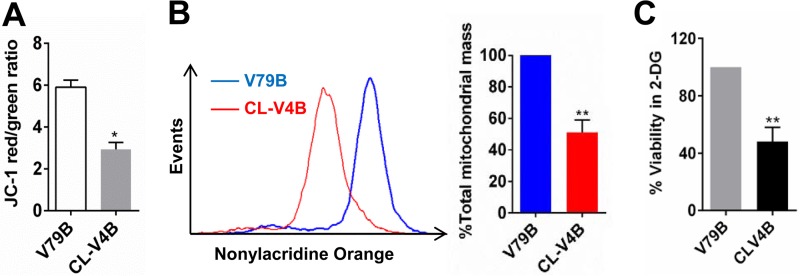
Finally, we measured the effects of RAD51C/XRCC3 deficiency on mitochondrial functions. As defects in mtDNA repair and replication can impair mitochondrial electron transport, we first used the JC-1 stain to study the mitochondrial membrane potential in RAD51 paralog-deficient hamster cells. RAD51C-deficient CL-V4B cells and XRCC3-deficient irs1SF cells had a significantly lower mitochondrial membrane potential than did WT V79B cells, whereas XRCC2-deficient irs1 cells did not show any such defect (Fig. 7A and Fig. S5D).
FIG 7.
RAD51C deficiency results in an unhealthy mitochondrial population. (A) Assessment of mitochondrial membrane potential by the JC-1 red-to-green ratio in RAD51C-deficient CL-V4B cells compared with WT V79B cells. (B) Determination of mitochondrial mass by nonyl-acridine orange staining. (C) CLV-4B cells are hypersensitive to cytotoxic effects of the glycolysis inhibitor 2-deoxyglucose (2-DG).
We next measured the overall mitochondrial mass in RAD51C-deficient CL-V4B cells using nonyl-acridine orange (NAO) stain. Interestingly, CL-V4B cells possessed a lower mitochondrial mass than did V79B cells (Fig. 7B). This could be explained by the induction of the PINK1/parkin-mediated pathway of mitophagy after the loss of membrane potential (67, 68).
Moreover, RAD51C-, XRCC3-, and RAD51-deficient cells were sensitive to 2-deoxyglucose, an inhibitor of glycolysis, suggesting an increased dependence on nonmitochondrial forms of ATP production (Fig. 7C and Fig. S5E). Taken together, our results demonstrate that the RAD51 paralogs RAD51C and XRCC3 facilitate mtDNA replication and are required for the prevention of mtDNA lesions during replication stress. Their deficiency leads to increased spontaneous mtDNA lesions, which affect the overall health of mitochondria and their critical functions.
DISCUSSION
In the present study, we show that RAD51C/XRCC3 and RAD51 are an integral part of the mitochondrial nucleoid and that their recruitment to mtDNA is dependent on Twinkle. Upon replication stress, RAD51C/XRCC3 and RAD51 are enriched at mutation hot spot region D310 along with POLG to prevent and possibly repair the mtDNA lesions induced by replication stress. The absence of these proteins destabilizes POLG on mtDNA, leading to reduced mtDNA synthesis and increased spontaneous and unrepaired replication stress-induced mtDNA lesions, thus resulting in unhealthy mitochondria.
The presence of the mitochondrial genome in close proximity to the electron transport chain makes it more susceptible to oxygen radicals generated during ATP production. Moreover, since mitochondria continue to replicate in all phases of the cell cycle, it is possible that changes in mitochondrial dNTPs lead to a slowing down or pausing of mtDNA replicating forks. Also, mtDNA contains numerous G-rich sequences, which can result in mtDNA replication stalling. Previously, it was reported that RAD51 facilitates mtDNA replication upon replication stress (52). However, the molecular mechanism by which RAD51 is recruited to mtDNA and promotes mtDNA replication is not known. Additionally, whether its function is independent of RAD51 paralogs also remains to be studied. In the present study, using mitochondrial subfractionation, we demonstrated the presence of RAD51C/XRCC3 in the NaCl-resistant pellet fraction of NP-40 lysis buffer along with TFAM and mtDNA. Moreover, similar to TFAM (69, 70), mitochondrial levels of RAD51C/XRCC3 and RAD51 were severely reduced upon mtDNA depletion, suggesting that RAD51C/XRCC3 is a new member of the mitochondrial nucleoid. Recently, many new proteins have been identified to constitute mitochondrial nucleoids that are majorly involved in controlling mtDNA copy numbers through direct or indirect interactions with mtDNA. These proteins include Lon protease, which regulates mitochondrial TFAM levels (51); ATAD3, which binds to the D-loop and is speculated to attach mtDNA to the mitochondrial inner membrane (71); and actin and myosin, which might promote mtDNA segregation or transmission (72). Our mtDNA-IP data show the concomitant binding of RAD51C/XRCC3 and the mitochondrial polymerase POLG at mtDNA control regions, thereby indicating the involvement of RAD51C/XRCC3 in the initiation of mtDNA replication.
Our data show that RAD51 associates with Twinkle, and the knockdown of Twinkle leads to the reduced enrichment of RAD51C/XRCC3 and RAD51, suggesting Twinkle-mediated recruitment of RAD51C/XRCC3 and RAD51 to mtDNA. This observation can be interpreted in two ways. First, since RAD51 paralogs show preferential binding to ssDNA (33), Twinkle helicase unwinds the mtDNA to provide favorable substrates for RAD51C/XRCC3 to load onto the mtDNA and thereby facilitates mtDNA replication. Second, as the knockdown of Twinkle severely reduced mtDNA synthesis (Fig. 6C), and Sage and Knight (52) demonstrated that the recruitment of RAD51 to mitochondria depends on ongoing DNA replication, it is conceivable that RAD51 and RAD51C/XRCC3 could be part of active replication forks. In addition, recently, a new member of the mitochondrial BER pathway, Polβ, was shown to interact with Twinkle and TFAM (73). These data imply that mtDNA repair factors cooperate with mitochondrial replication machinery to preserve the integrity of the mitochondrial genome.
RAD51C/XRCC3 deficiency leads to increased spontaneous lesions in the mitochondrial genome. Recently, using two-dimensional agarose gel electrophoresis, the presence of replication intermediates was detected in mtDNA of mice expressing an error-prone POLG mutant (62). Moreover, the expression of catalytic mutants of POLG and Twinkle in mammalian cells also showed the presence of replication-stalling phenotypes in addition to mtDNA rearrangements, point mutations, and deletions and low mtDNA copy numbers (63, 64). Since RAD51 associates with Twinkle, and RAD51 and RAD51C/XRCC3 are necessary to stabilize POLG on mtDNA, it is plausible that such spontaneous lesions upon RAD51C/XRCC3 and RAD51 deficiencies are an outcome of persistent endogenous replication stress. Recovery upon ddC treatment resulted in tremendous increases in mtDNA lesions. Such replication stress-induced lesions acquired in the mtDNA may be due to the activity of nucleases like MGME1 and DNA2, which create nicks to remove flap structures generated during mtDNA replication (74, 75).
RAD51 and RAD51C/XRCC3 showed nearly 3- to 4-fold enrichments at region D310 within CSBII (conserved sequence box II) upon ddC treatment. Mutation in CSBII affects premature transcription termination and initiation of mtDNA replication (76). In addition, the D310 mononucleotide repeat is a somatic insertion/deletion hot spot in many types of cancer (56, 77–80). Thus, RAD51C/XRCC3 not only promotes the initiation of mtDNA replication but also assists in the prevention of mtDNA damage. It is thought that oxidative stress is the major cause of mutations in region D310. However, this was challenged in an unbiased nucleotide sequence analysis of high-oxidative-stress-related chronic kidney disease (CKD) patients (81), which failed to show a significant correlation between oxidative stress and D310 mutations, further stressing our concept of replication stress as an inducer of mtDNA lesions. Interestingly, we also found POLG enrichment at region D310 upon replication stalling. Possible explanations for this are as follows: first, an imbalance in the overall turnover of POLG on mtDNA in the adjacent regions of the D-loop; second, a replication-mediated repair pathway of mtDNA lesions upon replication stress involving both POLG and RAD51C; and third, HR-mediated repair of DSBs generated during replication stress requiring the mitochondrial polymerase POLG.
The observation that RAD51C/XRCC3-deficient cells failed to replenish the lost mtDNA after mtDNA depletion is similar to previously reported observations of individuals with pathogenic POLG mutations (82), suggesting that a perturbation in mtDNA replication can lead to a defect in the restoration of mtDNA. This idea is supported by our mtDNA-IP data, which showed significantly reduced binding of POLG on mtDNA upon RAD51C/XRCC3 knockdown. RAD51C/XRCC3 has a high affinity for ssDNA, and Escherichia coli SSB was reported previously to participate in DNA replication by promoting the loading of DNA polymerase III (83). Recently, mitochondrial SSB was demonstrated to potentiate POLG activity by modifying the organization of template DNA (84). It will be interesting to study whether RAD51C/XRCC3 provides better substrates for POLG and thereby promotes its activity. It is generally believed that more relaxed template DNA provided by SSB augments the process of polymerization. However, studies by Ciesielski et al. (84) showed that, as opposed to insect POLG, the polymerization activity of human POLG was favored by compact template DNA. We speculate that the recombination function of RAD51 and RAD51C/XRCC3 may provide the desired compact mtDNA for its efficient replication. The reduced mitochondrial DNA content upon RAD51C/XRCC3 depletion may be due to a destabilization of POLG from mtDNA leading to decreased mtDNA synthesis. Alternatively, a trigger of mtDNA degradation that occurs upon an increased load of damage in the mitochondrial genome (85) is also a possible explanation for the reduced mtDNA content, since RAD51C/XRCC3-deficient cells acquire acute mtDNA lesions spontaneously.
RAD51C/XRCC3 participates in HR-mediated DSB repair, DNA damage signaling, and replication fork protection and restart (28–31, 47, 86), and monoallelic germ line mutations in RAD51C have been identified to cause breast and ovarian cancers (41, 42). Many studies have linked variation in mtDNA copy numbers with the risk of cancer subtypes (87, 88) where mutations in the D-loop are associated with reduced mtDNA copy numbers in breast cancer (89). Our study shows extended roles of RAD51C/XRCC3 in mitochondrial genome maintenance. A deficiency of this function along with the critical roles of RAD51C/XRCC3 in the nucleus may contribute to tumorigenesis in RAD51C/XRCC3-defective tissue. Although the breast cancer- and ovarian cancer-associated RAD51C NLS mutant failed to show any defect in recovery from mtDNA depletion, one cannot rule out the possibility that other breast cancer- and ovarian cancer-associated RAD51C mutants show defective functions in mitochondria. Also, since mitochondrial dysfunction is central to aging, our findings with RAD51 paralogs imply that RAD51C/XRCC3 dysfunction could lead to aging disorders.
In conclusion, we report here that RAD51C and XRCC3 are additional members of the mammalian mitochondrial nucleoid, which facilitates mtDNA replication. The enhanced enrichment of RAD51C/XRCC3 at mutation hot spot region D310 upon replication stress may be important for preventing toxic lesions generated during mtDNA replication stress. Since the absence of RAD51C/XRCC3 destabilized POLG on mtDNA, we hypothesize a direct role of RAD51C/XRCC3 in promoting POLG-mediated repair synthesis of stressed mtDNA forks. The discovery of the role of RAD51C/XRCC3 in augmenting mtDNA synthesis will help in understanding the regulation of mtDNA replication in physiological adaptations such as dNTP pool alterations, increased energy demand, mtDNA depletion, disease, and aging.
MATERIALS AND METHODS
Cell lines, cell culture, and transfections.
The human cell lines HeLa and U2OS and the Chinese hamster cell lines CL-V4B (RAD51C−/−), irs1 (XRCC2−/−), and irs1-SF (XRCC3−/−) and their respective parental cell lines V79B, V79, and CHO-AA8 were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in humidified air containing 5% CO2. WT and mutant RAD51C constructs were generated as described previously (35). All short hairpin RNA (shRNA) constructs were generated by using previously reported small interfering RNA (siRNA) sequences and cloned into the pRS shRNA vector. Various shRNA constructs were generated by using previously reported siRNA sequences: (i) 5′-CACCTTCTGTTCAGCACTAGA-3′ for RAD51C (86), (ii) 5′-GAATTATTGCTGCAATTAA-3′ for XRCC3 (86), (iii) 5′-TTGCAACGACACAAACTATAA-3′ for XRCC2 (90), (iv) 5′-GAAGAAAUUGGAAGAAGCUTT-3′ for RAD51 (91), and (v) 5′-GCAAAGCAUCAGGACUGUAAUAGAU-3′ for Twinkle (92). All plasmid transfections were performed by using a Bio-Rad Gene Pulsar X cell instrument (250 V and 950 μF).
Chemicals.
ddC, H2O2, and 2-deoxyglucose were purchased from Calbiochem. BrdU and Benzonase were purchased from Sigma. JC-1 stain and NAO were kind gifts from Patrick D'Silva.
Isolation of mitochondria and mitochondrial nucleoid.
Cells were washed twice in NKM buffer (0.13 M NaCl, 5 mM KCl, 7.5 mM MgCl2, and 10 mM Tris-HCl, pH 7.4) and allowed to swell in swelling buffer (10 mM Tris-HCl [pH 8], 10 mM NaCl, 0.15 mM MgCl2) for 15 min on ice, followed by disruption using a Dounce homogenizer and the immediate addition of sucrose to a final concentration of 0.25 M. Nuclei were pelleted upon centrifugation at 1,200 × g for 10 min. This step was repeated twice, and the supernatant was centrifuged at 16,000 × g for 10 min to pellet the mitochondria. The mitochondria were washed twice in suspension buffer (0.25 M sucrose, 10 mM Tris-HCl [pH 8], and 1.5 mM MgCl2) and lysed in Laemmli buffer for Western blotting. mtDNA nucleoids were isolated as described previously (53), by incubating the isolated mitochondria in suspension buffer with either 0.5% NP-40 or 0.5% NP-40 with 150 mM NaCl for 30 min on ice with gentle tapping. The lysates were later centrifuged at 16,000 × g for 30 min to collect the supernatant and mtDNA-containing pellet fractions. The purity of the fraction was checked by amplifying mtDNA-specific regions using 5-μl aliquots of each fraction.
mtDNA-IP analysis.
Cells were cultured overnight at a density of 1 × 107 cells per 150-mm petri dish and subjected to treatment with 100 μM ddC for 1.5 h. Isolated mitochondria from the cells were resuspended in suspension buffer and cross-linked with 1% formaldehyde for 10 min, and the reaction was stopped by adding 125 mM glycine to the mixture. The mitochondrial pellet was then washed three times with suspension buffer, resuspended in 400 μl of lysis buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.5% deoxycholate) freshly supplemented with a protease inhibitor cocktail (Roche), and sonicated on ice with 10-s pulses at 25% maximal power by using a sonicator. After centrifugation at 16,000 × g for 15 min, the supernatant was collected, and the protein concentration was determined by using Bradford reagent. For each immunoprecipitation, 50 μg of the mitochondrial lysate was incubated overnight at 4°C with 6 μg of antibodies specific for RAD51, RAD51C, XRCC3, TFAM, and POLG. A reaction mixture containing an equivalent amount of beads was included as the background control. Ten percent of the precleared chromatin was taken as the input control. Antibody-nucleoid complexes were pulled down by adding 50 μl of protein G beads, and the mixture was incubated for 4 h at 4°C. The beads were washed with chilled lysis buffer once, followed by chilled high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8], 500 mM NaCl), chilled LiCl wash buffer (250 mM LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0]), and chilled TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Finally, DNA was eluted with elution buffer (1% SDS, 100 mM NaHCO3). Eluates were incubated at 65°C overnight with the addition of 5 M NaCl to a final concentration of 200 mM to reverse the formaldehyde cross-linking and digested at 55°C for 3 h with proteinase K at a final concentration of 50 μg/ml. Following phenol-chloroform extraction and ethanol precipitation, sheared DNA fragments served as the templates for semiquantitative PCR analysis. As a negative control, the whole-cell lysate was used instead of the mitochondrial lysate, and a chromatin immunoprecipitation (ChIP) assay was performed by using anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody. For double mtDNA-IP, complexes from the primary immunoprecipitation step were eluted by incubation with 10 mM dithiothreitol (DTT) (37°C) and diluted 1:50 in radioimmunoprecipitation assay (RIPA) buffer, followed by reimmunoprecipitation with the second antibody as described above. After immunoprecipitation with the second antibody, complexes were washed, eluted, and purified as described above. Control mtDNA-IPs were treated in a similar fashion. Purified DNA samples were quantified, and PCR was performed with 15 ng of DNA. The primers that were used for mtDNA-IP analysis are listed in Table S2 in the supplemental material. Quantification of all PCR products was done by using ImageJ.
mtDNA synthesis by dot blot analysis.
DNA synthesis in mitochondria was measured by using a dot blot assay as described previously (93). Briefly, cells were incubated with 10 μM BrdU for 24 h, and isolated mitochondria were lysed in genomic DNA lysis buffer with proteinase K at 56°C, followed by precipitation with an equal volume of a chilled NaCl-ethanol solution. mtDNA was dissolved in water and quantified, equal amounts of mtDNA were denatured by adding 10 volumes of a 0.4 N NaOH solution to the mixture, and the mixture was incubated for 30 min at room temperature (RT). The denatured DNA was snap chilled on ice and renatured by using an equal volume of Tris-HCl (pH 7.5), followed by spotting 5 μl of each sample onto a nitrocellulose membrane. The membrane was dried by using a hair dryer, blocked in skimmed milk, and incubated with anti-BrdU antibody overnight. The blots were later washed in TBS-T (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 0.1% Tween 20) and developed.
Western blotting and antibodies.
Cells were harvested and lysed in RIPA buffer supplemented with complete protease inhibitor tablets (Roche). Protein concentrations were estimated by a standard Bradford assay (94). Proteins were resolved by 10% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were blocked by using 5% (wt/vol) dry milk in TBS-T. For analysis of γ-H2AX, 3% (wt/vol) bovine serum albumin (BSA) in TBS-T was used for blocking. The membranes were then incubated with the primary antibody overnight at 4°C. The primary antibodies against RAD51 (rabbit) (1:500 dilution; catalog number sc-8349), RAD51 (mouse) (1:200; catalog number sc-56212), RAD51B (1:250; catalog number sc-377192), RAD51C (1:250; catalog number sc-56214), RAD51D (1:100; catalog number sc-53432), XRCC2 (1:250; catalog number sc-365854), XRCC3 (1:250; catalog number sc-271714), MCM3 (1:500; catalog number sc-365616), TFAM (1:250; catalog number sc-28200), POLG (1:200; catalog number sc-390634), Twinkle (rabbit) (1:250; catalog number sc-293368), Twinkle (mouse) (1:100; catalog number sc-293368), KU70 (1:500; catalog number sc-12729), α-tubulin (1:500; catalog number sc-5286), lamin A/C (1:250; catalog number sc-6214), histone H3 (1:250; catalog number sc-10809), and HSP70 (1:1,000; catalog number sc-66048) that were used for Western blot analysis were purchased from Santa Cruz. γ-H2AX antibody (1:1,000; catalog number 560443) was purchased from BD Biosciences. The membranes were washed with TBS-T and incubated with the respective horseradish peroxidase (HRP)-conjugated secondary antibodies (1:8,000; Santa Cruz) for 1 h at 4°C. After washes with TBS-T, membranes were developed with a chemiluminescent HRP substrate (Millipore) and imaged by using a ChemiDoc imager (LAS 4000; GE Health Care).
Immunoprecipitation.
Isolated mitochondria were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8], 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 5% glycerol) supplemented with complete protease inhibitor tablets (Roche) along with Benzonase. After 1 h of incubation on ice, lysates were cleared by centrifugation. Where appropriate, antibodies were added to a lysate containing Benzonase and incubated for 12 to 16 h at 4°C. Lysates were then incubated with 25 μl of protein A/G beads (GE Healthcare) for 2 h at 4°C. Ig-antigen complexes were washed extensively and eluted in 2× Laemmli sample buffer at 90°C for 30 min before SDS-PAGE.
mtDNA integrity analysis.
DNA lesions in the mitochondrial genome were measured as described previously (95–97). Briefly, cells were harvested upon treatment, and genomic DNA was isolated by using a standard isolation protocol with proteinase K treatment. Fifteen nanograms of DNA was taken to amplify an 8.9-kb region from mtDNA by using primers listed in Table S1 in the supplemental material. The 8.9-kb amplicons were normalized by the mtDNA content. Primers that were used for assessing the mtDNA copy numbers are listed in Table S1 in the supplemental material. The numbers of lesions/mitochondrial genome were calculated by using the formula L = −log (AT/A0) × 1.85 (correction factor [1.85 = 16.5 kb/8.9 kb]), where A is LR-PCR/mtDNA content (AT is the A value for the treated sample, and A0 is the A value for the mock-treated sample). PCR products were visualized on an agarose gel and stained with EtBr, and band intensities were quantified by using ImageJ.
BrdU incorporation assay.
Cells were incubated with 50 μM BrdU and fixed in cold 70% ethanol at 4°C. DNA was denatured by using 2 N HCl and 0.5% Triton X-100 and then neutralized with phosphate-buffered saline (PBS). After blocking with 0.5% Tween 20 and 0.5% BSA in PBS, cells were incubated with anti-BrdU antibody (1:100) (catalog number 555627; BD Biosciences) for 2 h, followed by anti-mouse secondary antibody conjugated to fluorescein isothiocyanate (FITC) (1:100; Sigma-Aldrich) for 1 h. Later, after two washes with PBS, samples were incubated with RNase A-propidium iodide (PI) and analyzed on a FACS Verse flow cytometer (Becton Dickinson).
Immunofluorescence.
Exponentially growing U2OS cells were seeded onto coverslips and then treated with 2 μg/ml aphidicolin for 6 h to block nuclear DNA replication, followed by incubation with H2O or ddC as indicated. After treatment, the cells were washed with PBS and stained with 50 nM MitoTracker Red CMXROS (catalog number 7512; Invitrogen) for 15 min, followed by incubation in DMEM for 15 min at 37°C. Stained cells were fixed in 3.7% formaldehyde for 10 min at RT and permeabilized (0.2% Triton X-100 in PBS). DNA was denatured with 2 N HCl for 40 min, followed by neutralization with five PBS washes, and blocked (DMEM) for 30 min. The coverslips were incubated with the anti-BrdU antibody for 2 h at RT. After a wash with blocking buffer, the coverslips were incubated with the respective FITC-, tetramethylrhodamine isothiocyanate (TRITC)-, and Alexa Fluor-conjugated secondary antibodies for 1 h at RT and then stained with 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml; Sigma-Aldrich) for 10 min before mounting onto slides. Cells were acquired by using a confocal microscope (LSM510; Carl Zeiss), and images were processed by using Zeiss LSM image browser software. The primary antibody was same as the one that we used for fluorescence-activated cell sorter (FACS) analysis. FITC- and TRITC-conjugated (1:100) secondary antibodies were purchased from Sigma-Aldrich.
Miscellaneous.
Analysis of mitochondrial mass and mitochondrial membrane potential was performed as previously described (98). Briefly, cells were harvested, resuspended in PBS, stained with JC-1 (5 μM) for 5 min in the dark, and analyzed by FACS analysis. For NAO staining, resuspended cells were stained with NAO (5 nM) and analyzed immediately, without any incubation.
Statistical analysis.
Data are expressed as means ± standard deviations (SD). Two-tailed unpaired Student's t test was used to evaluate statistical significance, as indicated in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank Patrick D'Silva and Utpal Tatu for providing reagents for our studies. We thank Kumar Somyajit for stimulating discussions and critical reading of the manuscript. We are grateful to Jim Haber, Sagar Sengupta, and Wolf Heyer for their critical comments and suggestions on our data. We thank the divisional bioimaging and flow cytometry facility for assisting the work.
We greatly acknowledge funding support from the Department of Science and Technology (EMR/2015/001720), the Department of Biotechnology (DBT) (BT/PR5846/BRB/10/1099/2012), and the IISc-DBT partnership program (DBT/BF/PR/INS/IISc/2011–12). We also thank infrastructure support provided by funding from the DST-FIST and UGC. A.M. and S.S. are supported by fellowships from the DBT and IISc, respectively.
We have no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00489-17.
REFERENCES
- 1.Friedman JR, Nunnari J. 2014. Mitochondrial form and function. Nature 505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falkenberg M, Larsson N-G, Gustafsson CM. 2007. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem 76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 3.Kang D, Hamasaki N. 2002. Maintenance of mitochondrial DNA integrity: repair and degradation. Curr Genet 41:311–322. doi: 10.1007/s00294-002-0312-0. [DOI] [PubMed] [Google Scholar]
- 4.Kazak L, Reyes A, Holt IJ. 2012. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol 13:659–671. doi: 10.1038/nrm3439. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee A, Mambo E, Sidransky D. 2006. Mitochondrial DNA mutations in human cancer. Oncogene 25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 6.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, Törnell J, Jacobs HT, Larsson N-G. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 7.Park CB, Larsson N-G. 2011. Mitochondrial DNA mutations in disease and aging. J Cell Biol 193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J-I. 2008. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 9.Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A. 2011. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta 1813:186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Coffey G, Lakshmipathy U, Campbell C. 1999. Mammalian mitochondrial extracts possess DNA end-binding activity. Nucleic Acids Res 27:3348–3354. doi: 10.1093/nar/27.16.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakshmipathy U, Campbell C. 1999. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res 27:1198–1204. doi: 10.1093/nar/27.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tadi SK, Sebastian R, Dahal S, Babu RK, Choudhary B, Raghavan SC. 2016. Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions. Mol Biol Cell 27:223–235. doi: 10.1091/mbc.E15-05-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barr CM, Neiman M, Taylor DR. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol 168:39–50. doi: 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- 14.Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. 2007. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell Online 19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustafsson CM, Falkenberg M, Larsson N-G. 2016. Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem 85:133–160. doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- 16.Holt IJ, Reyes A. 2012. Human mitochondrial DNA replication. Cold Spring Harb Perspect Biol 4:a012971. doi: 10.1101/cshperspect.a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey LJ, Doherty AJ. 2017. Mitochondrial DNA replication: a PrimPol perspective. Biochem Soc Trans 45:513–529. doi: 10.1042/BST20160162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnusson J, Orth M, Lestienne P, Taanman JW. 2003. Replication of mitochondrial DNA occurs throughout the mitochondria of cultured human cells. Exp Cell Res 289:133–142. doi: 10.1016/S0014-4827(03)00249-0. [DOI] [PubMed] [Google Scholar]
- 19.Woo YH, Li W-H. 2012. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat Commun 3:1004. doi: 10.1038/ncomms1982. [DOI] [PubMed] [Google Scholar]
- 20.Poli J, Tsaponina O, Crabbé L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31:883–894. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stojkovič G, Makarova AV, Wanrooij PH, Forslund J, Burgers PM, Wanrooij S. 2016. Oxidative DNA damage stalls the human mitochondrial replisome. Sci Rep 6:28942. doi: 10.1038/srep28942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W-C, Tseng T-Y, Chen Y-T, Chang C-C, Wang Z-F, Wang C-L, Hsu T-N, Li P-T, Chen C-T, Lin J-J, Lou P-J, Chang T-C. 2015. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res 43:10102–10113. doi: 10.1093/nar/gkv1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bharti SK, Sommers JA, Zhou J, Kaplan DL, Spelbrink JN, Mergny JL, Brosh RM. 2014. DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative twinkle helicase. J Biol Chem 289:29975–29993. doi: 10.1074/jbc.M114.567073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hyvärinen AK, Pohjoismäki JLO, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. 2007. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res 35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 26.Sung P. 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 27.Nagaraju G, Hartlerode A, Kwok A, Chandramouly G, Scully R. 2009. XRCC2 and XRCC3 regulate the balance between short- and long-tract gene conversions between sister chromatids. Mol Cell Biol 29:4283–4294. doi: 10.1128/MCB.01406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagaraju G, Odate S, Xie A, Scully R. 2006. Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol Cell Biol 26:8075–8086. doi: 10.1128/MCB.01235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somyajit K, Subramanya S, Nagaraju G. 2012. Distinct roles of FANCO/RAD51C protein in DNA damage signaling and repair: implications for Fanconi anemia and breast cancer susceptibility. J Biol Chem 287:3366–3380. doi: 10.1074/jbc.M111.311241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somyajit K, Basavaraju S, Scully R, Nagaraju G. 2013. ATM- and ATR-mediated phosphorylation of XRCC3 regulates DNA double-strand break-induced checkpoint activation and repair. Mol Cell Biol 33:1830–1844. doi: 10.1128/MCB.01521-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somyajit K, Subramanya S, Nagaraju G. 2010. RAD51C: a novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer. Carcinogenesis 31:2031–2038. doi: 10.1093/carcin/bgq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lio YC, Schild D, Brenneman MA, Redpath JL, Chen DJ. 2004. Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phase cells. J Biol Chem 279:42313–42320. doi: 10.1074/jbc.M405212200. [DOI] [PubMed] [Google Scholar]
- 33.Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. 2001. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev 15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. 2001. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev 15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somyajit K, Mishra A, Jameei A, Nagaraju G. 2015. Enhanced non-homologous end joining contributes toward synthetic lethality of pathological RAD51C mutants with poly(ADP-ribose) polymerase. Carcinogenesis 36:13–24. doi: 10.1093/carcin/bgu211. [DOI] [PubMed] [Google Scholar]
- 36.Pittman DL, Schimenti JC. 2000. Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis 26:167–173. doi:. [DOI] [PubMed] [Google Scholar]
- 37.Shu Z, Smith S, Wang L, Rice MC, Kmiec EB. 1999. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53−/− background. Mol Cell Biol 19:8686–8693. doi: 10.1128/MCB.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deans B, Griffin CS, Maconochie M, Thacker J. 2000. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J 19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuznetsov SG, Haines DC, Martin BK, Sharan SK. 2009. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res 69:863–872. doi: 10.1158/0008-5472.CAN-08-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orr N, Lemnrau A, Cooke R, Fletcher O, Tomczyk K, Jones M, Johnson N, Lord CJ, Mitsopoulos C, Zvelebil M, McDade SS, Buck G, Blancher C, KConFab Consortium, Trainer AH, James PA, Bojesen SE, Bokmand S, Nevanlinna H, Mattson J, Friedman E, Laitman Y, Palli D, Masala G, Zanna I, Ottini L, Giannini G, Hollestelle A, van den Ouweland AMW, Novaković S, Krajc M, Gago-Dominguez M, Castelao JE, Olsson H, Hedenfalk I, Easton DF, Pharoah PDP, Dunning AM, Bishop DT, Neuhausen SL, Steele L, Houlston RS, Garcia-Closas M, Ashworth A, Swerdlow AJ. 2012. Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nat Genet 44:1182–1184. doi: 10.1038/ng.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, Freund M, Lichtner P, Hartmann L, Schaal H, Ramser J, Honisch E, Kubisch C, Wichmann HE, Kast K, Deissler H, Engel C, Muller-Myhsok B, Neveling K, Kiechle M, Mathew CG, Schindler D, Schmutzler RK, Hanenberg H. 2010. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 42.Osorio A, Endt D, Fernández F, Eirich K, De la Hoya M, Schmutzler R, Caldés T, Meindl A, Schindler D, Benitez J. 2012. Predominance of pathogenic missense variants in the RAD51C gene occurring in breast and ovarian cancer families. Hum Mol Genet 21:2889–2898. doi: 10.1093/hmg/dds115. [DOI] [PubMed] [Google Scholar]
- 43.Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, Bowden G, Kalmyrzaev B, Warren-Perry M, Snape K, Adlard JW, Barwell J, Berg J, Brady AF, Brewer C, Brice G, Chapman C, Cook J, Davidson R, Donaldson A, Douglas F, Greenhalgh L, Henderson A, Izatt L, Kumar A, Lalloo F, Miedzybrodzka Z, Morrison PJ, Paterson J, Porteous M, Rogers MT, Shanley S, Walker L, Eccles D, Evans DG, Renwick A, Seal S, Lord CJ, Ashworth A, Reis-Filho JS, Antoniou AC, Rahman N. 2011. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park DJ, Lesueur F, Nguyen-Dumont T, Pertesi M, Odefrey F, Hammet F, Neuhausen SL, John EM, Andrulis IL, Terry MB, Daly M, Buys S, Le Calvez-Kelm F, Lonie A, Pope BJ, Tsimiklis H, Voegele C, Hilbers FM, Hoogerbrugge N, Barroso A, Osorio A, Giles GG, Devilee P, Benitez J, Hopper JL, Tavtigian SV, Goldgar DE, Southey MC. 2012. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet 90:734–739. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golmard L, Caux-Moncoutier V, Davy G, Al Ageeli E, Poirot B, Tirapo C, Michaux D, Barbaroux C, d'Enghien CD, Nicolas A, Castéra L, Sastre-Garau X, Stern M-H, Houdayer C, Stoppa-Lyonnet D. 2013. Germline mutation in the RAD51B gene confers predisposition to breast cancer. BMC Cancer 13:484. doi: 10.1186/1471-2407-13-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yadav N, Chandra D. 2013. Mitochondrial DNA mutations and breast tumorigenesis. Biochim Biophys Acta 1836:336–344. doi: 10.1016/j.bbcan.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Somyajit K, Saxena S, Babu S, Mishra A, Nagaraju G. 2015. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res 43:9835–9855. doi: 10.1093/nar/gkv880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sage JM, Gildemeister OS, Knight KL. 2010. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J Biol Chem 285:18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leibowitz RD. 1971. The effect of ethidium bromide on mitochondrial DNA synthesis and mitochondrial DNA structure in HeLa cells. J Cell Biol 51:116–122. doi: 10.1083/jcb.51.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seidel-Rogol BL, Shadel GS. 2002. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res 30:1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsushima Y, Goto Y, Kaguni LS. 2010. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM). Proc Natl Acad Sci U S A 107:18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sage JM, Knight KL. 2013. Human Rad51 promotes mitochondrial DNA synthesis under conditions of increased replication stress. Mitochondrion 13:350–356. doi: 10.1016/j.mito.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. 2003. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res 31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chun J, Buechelmaier ES, Powell SN. 2013. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol 33:387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta S, De S, Srivastava V, Hussain M, Kumari J, Muniyappa K, Sengupta S. 2014. RECQL4 and p53 potentiate the activity of polymerase γ and maintain the integrity of the human mitochondrial genome. Carcinogenesis 35:34–45. doi: 10.1093/carcin/bgt315. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Jeronimo C, Jordan RCK, Nicol T, Koch WM, Schoenberg M, Mazzarelli P, Fazio VM, Sidransky D. 2001. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res 61:7015–7019. [PubMed] [Google Scholar]
- 57.Fish J, Raule N, Attardi G. 2004. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science 306:2098–2101. doi: 10.1126/science.1102077. [DOI] [PubMed] [Google Scholar]
- 58.Ballinger SW, Van Houten B, Jin GF, Conklin CA, Godley BF. 1999. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res 68:765–772. doi: 10.1006/exer.1998.0661. [DOI] [PubMed] [Google Scholar]
- 59.Ballinger SW, Patterson C, Yan CN, Doan R, Burow DL, Young CG, Yakes FM, Van Houten B, Ballinger CA, Freeman BA, Runge MS. 2000. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res 86:960–966. doi: 10.1161/01.RES.86.9.960. [DOI] [PubMed] [Google Scholar]
- 60.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. 2013. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harb Perspect Biol 5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. 2009. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res 37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey LJ, Cluett TJ, Reyes A, Prolla TA, Poulton J, Leeuwenburgh C, Holt IJ. 2009. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res 37:2327–2335. doi: 10.1093/nar/gkp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pohjoismäki JLO, Goffart S, Spelbrink JN. 2011. Replication stalling by catalytically impaired Twinkle induces mitochondrial DNA rearrangements in cultured cells. Mitochondrion 11:630–634. doi: 10.1016/j.mito.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 64.Wanrooij S, Goffart S, Pohjoismäki JLO, Yasukawa T, Spelbrink JN. 2007. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res 35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN. 2009. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet 18:328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Phillips AF, Millet AR, Tigano M, Dubois SM, Crimmins H, Babin L, Charpentier M, Piganeau M, Brunet E, Sfeir A. 2017. Single-molecule analysis of mtDNA replication uncovers the basis of the common deletion. Mol Cell 65:527.e6–538.e6. doi: 10.1016/j.molcel.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 67.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, Kimura M, Komatsu M, Hattori N, Tanaka K. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK. 2013. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell 49:121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garrido N. 2002. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell 14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, Reichelt S, Spelbrink JN, Walker JE, Holt IJ. 2007. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol 176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reyes A, He J, Mao CC, Bailey LJ, Di Re M, Sembongi H, Kazak L, Dzionek K, Holmes JB, Cluett TJ, Harbour ME, Fearnley IM, Crouch RJ, Conti MA, Adelstein RS, Walker JE, Holt IJ. 2011. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res 39:5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sykora P, Kanno S, Akbari M, Kulikowicz T, Baptiste BA, Leandro GS, Lu H, Tian J, May A, Becker KA, Croteau DL, Wilson DM, Sobol RW, Yasui A, Bohr VA. 30 May 2017. DNA polymerase beta participates in mitochondrial DNA repair. Mol Cell Biol doi: 10.1128/MCB.00237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uhler JP, Thörn C, Nicholls TJ, Matic S, Milenkovic D, Gustafsson CM, Falkenberg M. 2016. MGME1 processes flaps into ligatable nicks in concert with DNA polymerase γ during mtDNA replication. Nucleic Acids Res 44:5861–5871. doi: 10.1093/nar/gkw468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uhler JP, Falkenberg M. 2015. Primer removal during mammalian mitochondrial DNA replication. DNA Repair (Amst) 34:28–38. doi: 10.1016/j.dnarep.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 76.Xuan HP, Farge G, Shi Y, Gaspari M, Gustafsson CM, Falkenberg M. 2006. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J Biol Chem 281:24647–24652. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 77.Xu C, Tran-Thanh D, Ma C, May K, Jung J, Vecchiarelli J, Done SJ. 2012. Mitochondrial D310 mutations in the early development of breast cancer. Br J Cancer 106:1506–1511. doi: 10.1038/bjc.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parrella P, Seripa D, Matera MG, Rabitti C, Rinaldi M, Mazzarelli P, Gravina C, Gallucci M, Altomare V, Flammia G, Casalino B, Benedetti-Panici PL, Fazio VM. 2003. Mutations of the D310 mitochondrial mononucleotide repeat in primary tumors and cytological specimens. Cancer Lett 190:73–77. doi: 10.1016/S0304-3835(02)00578-5. [DOI] [PubMed] [Google Scholar]
- 79.Geurts-Giele WRR, Gathier GHGK, Atmodimedjo PN, Dubbink HJ, Dinjens WNM. 2015. Mitochondrial D310 mutation as clonal marker for solid tumors. Virchows Arch 467:595–602. doi: 10.1007/s00428-015-1817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang M, Baez S, Pruyas M, Diaz A, Calvo A, Riquelme E, Wistuba II. 2004. Mitochondrial DNA mutation at the D310 (displacement loop) mononucleotide sequence in the pathogenesis of gallbladder carcinoma. Clin Cancer Res 10:1041–1046. doi: 10.1158/1078-0432.CCR-0701-3. [DOI] [PubMed] [Google Scholar]
- 81.Chen JB, Lin TK, Liao SC, Lee WC, Lee LC, Liou CW, Wang PW, Tiao MM. 2009. Lack of association between mutations of gene-encoding mitochondrial D310 (displacement loop) mononucleotide repeat and oxidative stress in chronic dialysis patients in Taiwan. J Negat Results Biomed 8:10. doi: 10.1186/1477-5751-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stewart JD, Schoeler S, Sitarz KS, Horvath R, Hallmann K, Pyle A, Yu-Wai-Man P, Taylor RW, Samuels DC, Kunz WS, Chinnery PF. 2011. POLG mutations cause decreased mitochondrial DNA repopulation rates following induced depletion in human fibroblasts. Biochim Biophys Acta 1812:321–325. doi: 10.1016/j.bbadis.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 83.Glover BP, McHenry CS. 1998. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem 273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 84.Ciesielski GL, Bermek O, Rosado-Ruiz FA, Hovde SL, Neitzke OJ, Griffith JD, Kaguni LS. 2015. Mitochondrial single-stranded DNA-binding proteins stimulate the activity of DNA polymerase γ by organization of the template DNA. J Biol Chem 290:28697–28707. doi: 10.1074/jbc.M115.673707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shokolenko IN, Wilson GL, Alexeyev MF. 2013. Persistent damage induces mitochondrial DNA degradation. DNA Repair (Amst) 12:488–499. doi: 10.1016/j.dnarep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Badie S, Liao C, Thanasoula M, Barber P, Hill MA, Tarsounas M. 2009. RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. J Cell Biol 185:587–600. doi: 10.1083/jcb.200811079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bai R-K, Chang J, Yeh K-T, Lou MA, Lu J-F, Tan D-J, Liu H, Wong L-JC. 2011. Mitochondrial DNA content varies with pathological characteristics of breast cancer. J Oncol. 2011:496189. doi: 10.1155/2011/496189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kornblum C, Nicholls TJ, Haack TB, Schöler S, Peeva V, Danhauser K, Hallmann K, Zsurka G, Rorbach J, Iuso A, Wieland T, Sciacco M, Ronchi D, Comi GP, Moggio M, Quinzii CM, DiMauro S, Calvo SE, Mootha VK, Klopstock T, Strom TM, Meitinger T, Minczuk M, Kunz WS, Prokisch H. 2013. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet 45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R. 2007. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life 59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 90.Zheng Z, Ng WL, Zhang X, Olson JJ, Hao C, Curran WJ, Wang Y. 2012. RNAi-mediated targeting of noncoding and coding sequences in DNA repair gene messages efficiently radiosensitizes human tumor cells. Cancer Res 72:1221–1228. doi: 10.1158/0008-5472.CAN-11-2785. [DOI] [PubMed] [Google Scholar]
- 91.Quiros S, Roos WP, Kaina B. 2011. Rad51 and BRCA2—new molecular targets for sensitizing glioma cells to alkylating anticancer drugs. PLoS One 6:e27183. doi: 10.1371/journal.pone.0027183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. 2004. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum Mol Genet 13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 93.Ueda J, Saito H, Watanabe H, Evers BM. 2005. Novel and quantitative DNA dot-blotting method for assessment of in vivo proliferation. Am J Physiol Gastrointest Liver Physiol 288:G842–G847. doi: 10.1152/ajpgi.00463.2004. [DOI] [PubMed] [Google Scholar]
- 94.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 95.Sharma NK, Lebedeva M, Thomas T, Kovalenko OA, Stumpf JD, Shadel GS, Santos JH. 2014. Intrinsic mitochondrial DNA repair defects in ataxia telangiectasia. DNA Repair (Amst) 13:22–31. doi: 10.1016/j.dnarep.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. 2006. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol 314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 97.Hunter SE, Jung D, Di Giulio RT, Meyer JN. 2010. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods 51:444–451. doi: 10.1016/j.ymeth.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinha D, Srivastava S, Krishna L, D'Silva P. 2014. Unraveling the intricate organization of mammalian mitochondrial presequence translocases: existence of multiple translocases for maintenance of mitochondrial function. Mol Cell Biol 34:1757–1775. doi: 10.1128/MCB.01527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stein A, Kalifa L, Sia EA. 2015. Members of the RAD52 epistasis group contribute to mitochondrial homologous recombination and double-strand break repair in Saccharomyces cerevisiae. PLoS Genet 11(11):e1005664. doi: 10.1371/journal.pgen.1005664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.