ABSTRACT
Human antigen R (ELAVL1; HuR) is perhaps the best-characterized RNA-binding protein. Through its overexpression in various tumor types, HuR promotes posttranscriptional regulation of target genes in multiple core signaling pathways associated with tumor progression. The role of HuR overexpression in pancreatic tumorigenesis is unknown and led us to explore the consequences of HuR overexpression using a novel transgenic mouse model that has a >2-fold elevation of pancreatic HuR expression. Histologically, HuR-overexpressing pancreas displays a fibroinflammatory response and other pathological features characteristic of chronic pancreatitis. This pathology is reflected in changes in the pancreatic gene expression profile due, in part, to genes whose expression changes as a consequence of direct binding of their respective mRNAs to HuR. Older mice develop pancreatic steatosis and severe glucose intolerance. Elevated HuR cooperated with mutant K-rasG12D to result in a 3.4-fold increase in pancreatic ductal adenocarcinoma (PDAC) incidence compared to PDAC presence in K-rasG12D alone. These findings implicate HuR as a facilitator of pancreatic tumorigenesis, especially in the setting of inflammation, and a novel therapeutic target for pancreatitis treatment.
KEYWORDS: HuR, cancer, inflammation, pancreas
INTRODUCTION
Human antigen R (ELAVL1), also known as HuR, is an RNA-binding protein that posttranscriptionally regulates the expression of many genes, including various proinflammatory genes and tumor-promoting factors associated with the survival of cancer cells. It is located primarily in the nucleus, but in response to cellular stressors (e.g., hypoxia, DNA damage, nutrient depletion, and exposure to chemotherapeutic drugs), HuR selectively binds to AU-rich elements (AREs) primarily in the 3′ untranslated region (UTR) of specific target transcripts (1–7). Under these conditions, HuR and its bound transcripts are observed in the cytoplasm, where stabilization of HuR-bound mRNAs can occur, allowing for their enhanced translation. In this way, HuR regulates the expression of specific target genes known to enhance cell growth and survival (e.g., COX-2, VEGF, c-Myc, WEE1, and PIM1) (1, 3, 8–10). While this ability of HuR to influence gene expression is needed when normal cells are stressed, most cancer cells, including pancreatic ductal adenocarcinoma (PDAC) cells, take advantage of this aspect of HuR's function by increasing HuR expression, thus allowing for enhanced cytoplasmic localization of key transcripts related to prosurvival and oncogenic stimuli.
Nearly all PDACs that have been analyzed have been shown to harbor multiple mutations affecting core signaling pathways (11). We have determined that HuR-associated transcripts in PDACs are involved in multiple pro-oncogenic pathways, including cellular proliferation, invasion, apoptosis, and DNA damage control, thus implicating HuR as an important player in PDAC tumorigenesis and a potential therapeutic target (1, 3, 7, 12, 13). Aiming to assess the consequences of HuR overexpression in normal pancreatic cells and its potential role in the progression of pancreatic tumorigenesis, we generated transgenic mice that overexpress HuR specifically in the developing pancreas and in the pancreas of adult mice. Here, we characterize the histopathology, changes in gene expression, metabolic impact, and impact on tumorigenesis associated with elevated HuR levels in the pancreas of these mice.
RESULTS
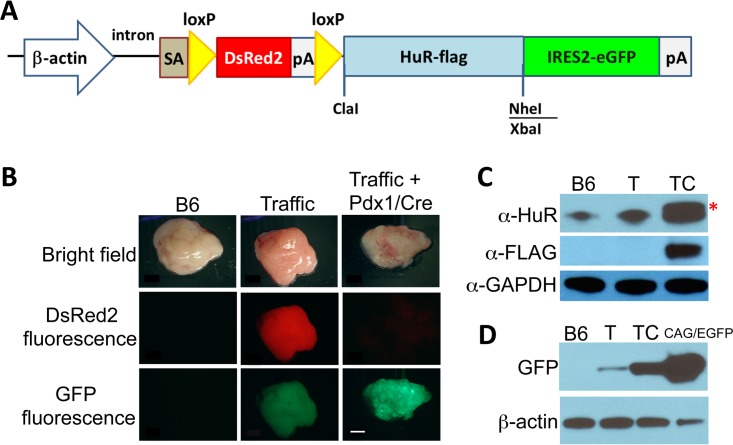
Overexpression of pancreatic HuR in Traffic/HuR transgenic mice.
We generated transgenic mice bearing a transgene called Traffic/HuR that allows for targeted HuR overexpression in the pancreas (Fig. 1A). The transgenic HuR sequence encodes human HuR protein containing a Flag epitope tag. In mice bearing this transgene, the strong CAG promoter/enhancer drives ubiquitous expression of floxed DsRed2. The transgene can be activated in a tissue-specific manner upon Cre recombinase-mediated recombination between two loxP sites that flank the DsRed2-STOP cassette, resulting in excision of the cassette, expression of HuR-Flag, and the gain of enhanced green fluorescent protein (eGFP) fluorescence, which is expressed from an internal ribosomal entry sequence (IRES). We bred Traffic/HuR (T) mice to Pdx1/Cre (C) mice to generate mice with HuR overexpression in the pancreas. The promoter of the PDX1 (pancreatic and duodenal homeobox 1) gene is active specifically in pancreatic precursor cells in mouse embryos at embryonic day 8.5 (14), resulting in Cre-mediated recombination and HuR overexpression in all three cell types in the pancreas: exocrine, endocrine, and ductal. Here, we report our studies of the pancreas of adult double-transgenic Traffic/HuR Pdx1/Cre mice (here called TC mice).
FIG 1.
Traffic/HuR transgene structure and expression. (A) Diagram of Traffic/HuR transgene. (B) Pancreas of B6, Traffic/HuR only (T), and Traffic/HuR + Pdx1/Cre (TC) mice observed under bright light, red fluorescent protein filter, and GFP filter. Note low GFP expression in T mice. (C) Immunoblots of protein extracts of pancreas of B6, T, and TC pancreas. Asterisk indicates HuR-Flag protein. (D) Immunoblot showing GFP expression in TC pancreas and low GFP expression in T pancreas. CAG/EGFP plasmid DNA serves as a positive control for GFP. GAPDH and β-actin serve as gel loading controls. Scale bar, 2 mm.
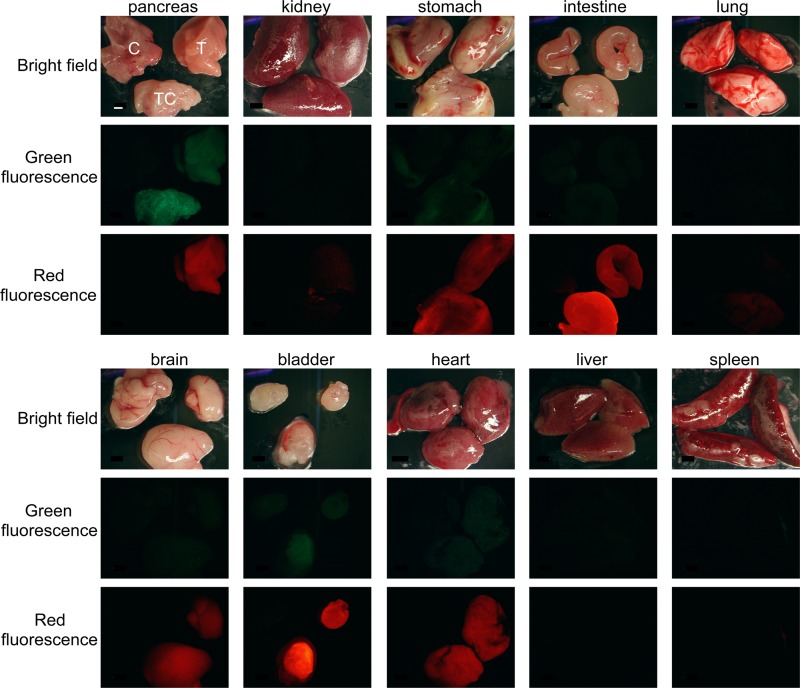
We observed HuR-Flag expression and strong GFP fluorescence in the pancreas of TC mice (Fig. 1B to D and 2). Strong GFP fluorescence in the pancreas typically was observed in 50 to 90% of pancreatic cells, reflecting the incomplete penetrance of Cre recombinase-mediated recombination. Very low fluorescent GFP expression was observed in the pancreas of T mice and confirmed by a weak GFP band on Western blots (Fig. 1B and D). We attribute this low GFP expression to transcripts generated from the IRES2-GFP cistron regulated by the distal CAG promoter. HuR-Flag protein was not detected in pancreas of T mice. Extremely low fluorescent GFP expression was also observed in some other organs but was not detectable on Western blots (data not shown). As expected, DsRed2, driven by the CAG promoter/enhancer, was expressed in multiple organs in T mice (Fig. 2).
FIG 2.
Traffic/HuR transgene expression in pancreas and other organs. DsRed2 protein is expressed in multiple organs in T and TC mice (red fluorescence) but not in C mice. Elevated GFP expression is observed in large amounts only in the pancreas of TC mice (green fluorescence). Low background GFP fluorescence is also observed in T pancreas and TC bladder, while no GFP expression is detected in other organs. In each image, C is in the upper left, T is in the upper right, and TC is on the bottom. The scale bar, indicating 2 mm, is shown in the pancreas bright-field image and applies to all panels.
No significant difference in body weight of either gender between C and TC mice was observed (measured weekly between 2 and 8 months of age) (data not shown).
HuR overexpression in pancreas elevates cytoplasmic HuR expression.
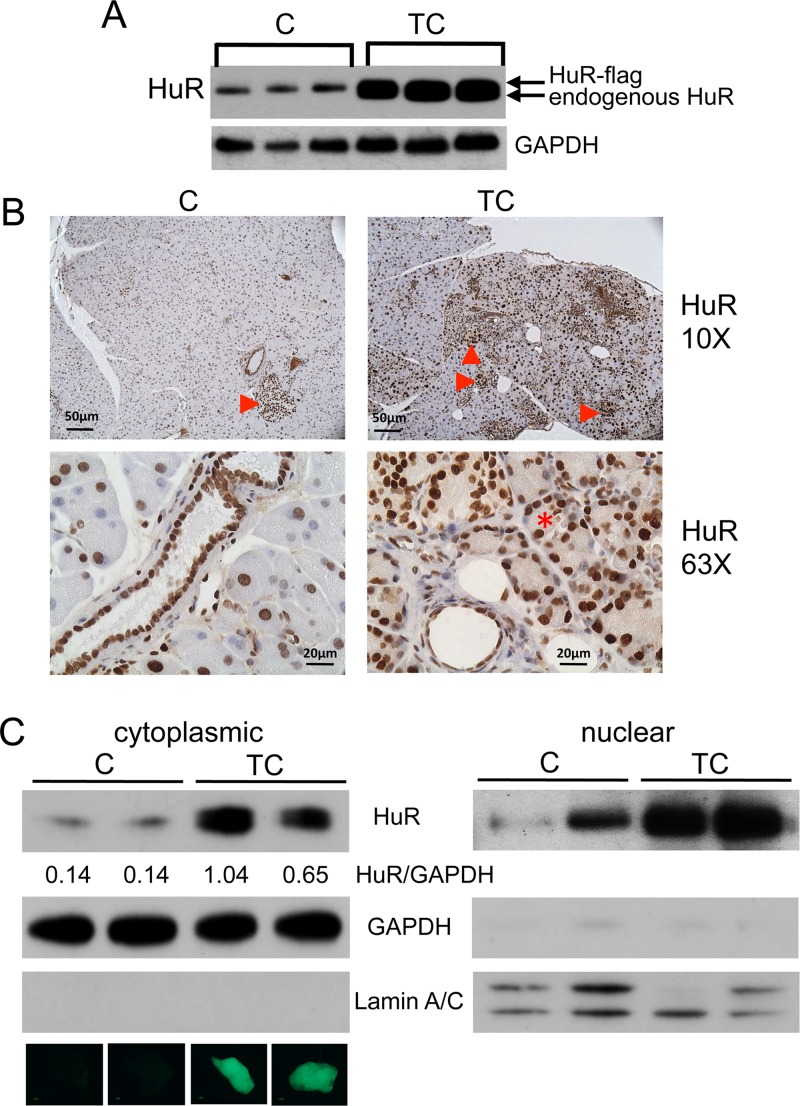
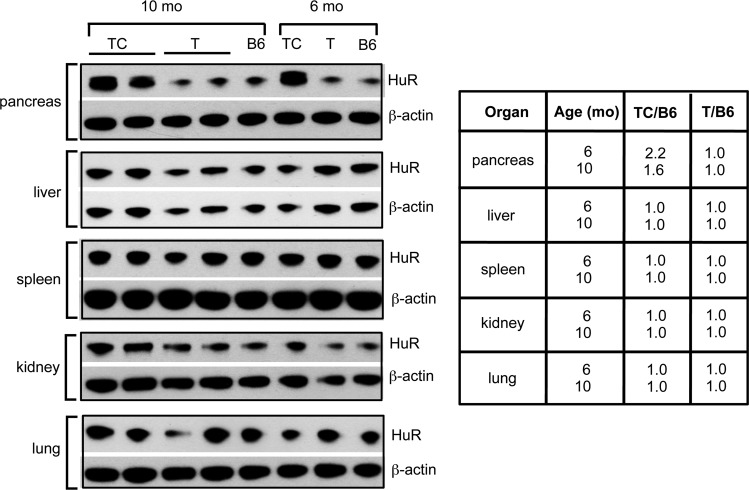
The transgene product, human HuR-Flag protein, was easily identified and distinguishable from endogenous mouse HuR in pancreas protein extracts of TC mice on immunoblots and showed a significant increase in total HuR expression compared to that in C, T, and C57BL/6 control mice (Fig. 1C, 3A, and 4). Quantitative analysis of total HuR (endogenous mouse HuR plus transgenic human HuR-Flag) showed a 2.2-fold and 1.6-fold increase in HuR in the pancreas of 6-month-old and 10-month-old TC mice, respectively, compared to pancreas of C57BL/6 control mice (Fig. 4). No HuR-Flag expression was observed by immunoblot analysis of other organ protein extracts we tested (Fig. 4).
FIG 3.
HuR expression in C and TC pancreas. (A) Immunoblot of protein extracts pancreas from C and TC mice. GAPDH serves as gel loading control. (B) Pancreas sections from C and TC pancreas immunostained for HuR. Arrowheads indicate large amounts of HuR in islet cells. Asterisk indicates cytoplasmic HuR in acinus cells in TC pancreas. (C) Immunoblots of cytoplasmic and nuclear protein lysates prepared from C and TC pancreas. Loading control GAPDH was used for quantitative comparison of HuR amounts in C and TC cytoplasmic lysates (values indicated beneath HuR panel). GAPDH and lamin A/C were used as markers for cytoplasmic and nuclear extract preparations, respectively. Whole-mount GFP fluorescent images of the pancreas from which each preparation was made are shown below the cytoplasmic lamin A/C panel.
FIG 4.
Pancreas-specific overexpression of HuR. HuR immunoblots showing pancreas-specific expression of Traffic-HuR transgene in 6-mo- and 10-mo-old TC mice. Scanned blots show total HuR amount (endogenous plus HuR-Flag) is ∼2-fold larger in TC mice than in pancreas of B6 and T control mice; in other organs, the amount of HuR in TC pancreas is the same as that in control pancreata.
In agreement with the Western blot analyses, elevated HuR expression was also observed in immunostained sections of TC pancreas compared to C pancreas (Fig. 3B). HuR expression was high in nuclei of TC acinar cells and ductal epithelial cells and especially high in islet cells in both TC and C pancreas. Importantly, cytoplasmic HuR (i.e., activated HuR) was observed in some, but not all, acinar cells in TC pancreas, suggesting incomplete Cre recombinase-mediated recombination of the Traffic transgene. A stochastic expression pattern of Pdx1-regulated Cre recombinase in the pancreas of Pdx1/Cre transgenic mice was reported previously (15). A 5- to 10-fold increase in cytoplasmic HuR in TC pancreas compared to that of C pancreas was observed on Western blots of cytoplasmic protein extracts; the fold increase appeared to correlate with the amount of GFP fluorescence observed in whole-mount pancreata (Fig. 3C).
HuR overexpression in pancreas results in a fibroinflammatory reaction and metabolic disease.
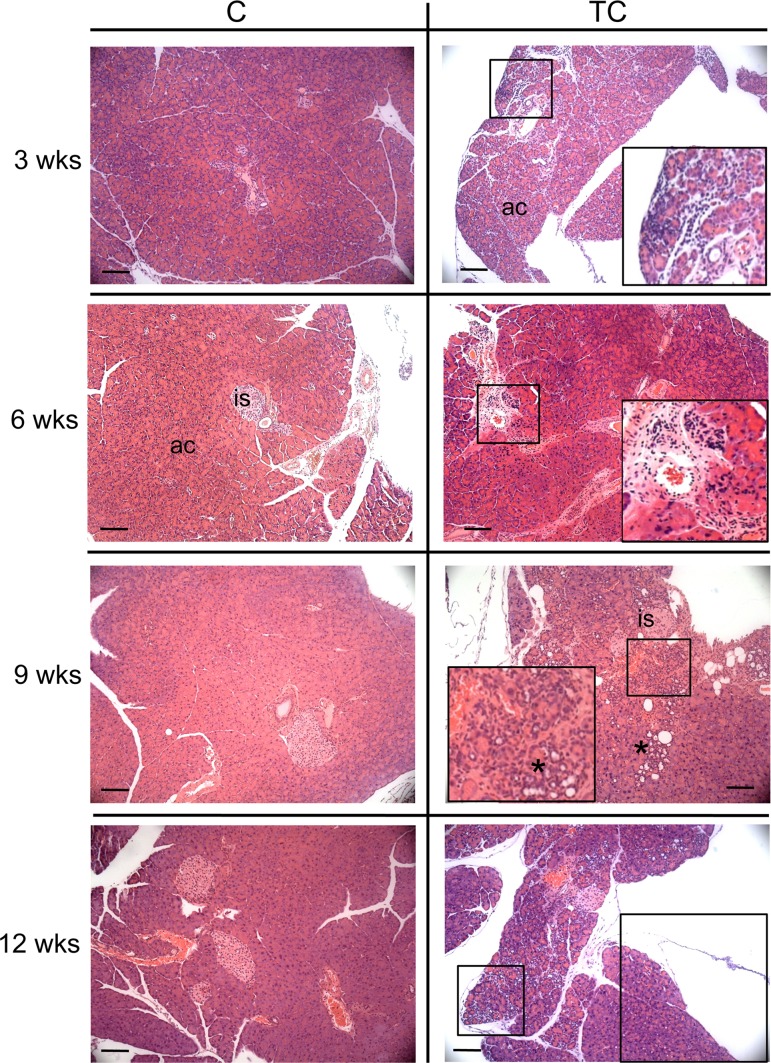
We examined hematoxylin and eosin (H&E)-stained pancreas sections from 3-, 6-, 9-, and 12-week-old C and TC mice for evidence of pathology associated with elevated HuR expression. We observed chronic inflammation in TC pancreata as early as 3 weeks of age, and the amount of inflammatory infiltrates increased with age (Fig. 5). No inflammation was observed in C mice at the ages examined. Prominent pancreatic ductal proliferation was first observed in 9-week-old TC mice. There was no evidence of ductal proliferation in C mice or precancerous pancreatic intraepithelial neoplasia (PanIN) lesions in TC or C mice at the ages examined.
FIG 5.
H&E-stained pancreas of C and TC mice at 3, 6, 9, and 12 weeks of age. Boxes show immune infiltrates in TC pancreas. An asterisk indicates ductal proliferation. is, islet of Langerhans; ac, acinar cells. Scale bars, 50 μm.
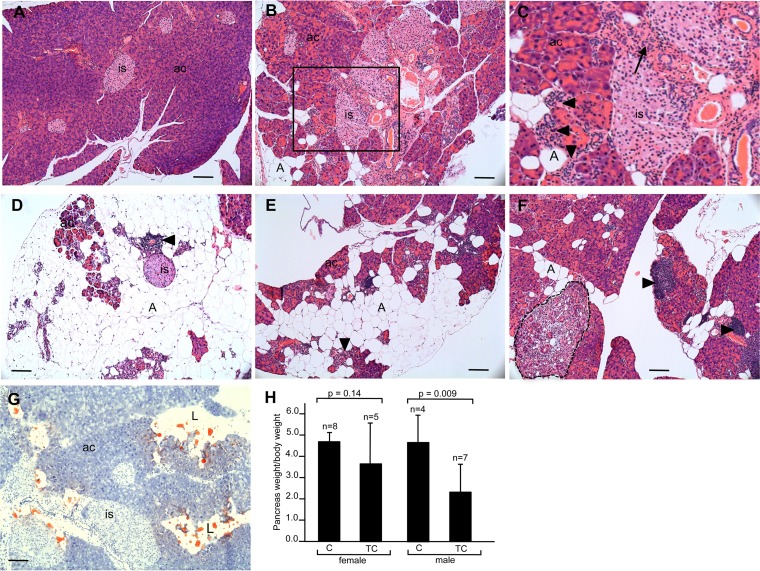
A marked increase in inflammatory infiltrates, fibrosis, and ductal complexes in H&E-stained pancreas sections is observed in older TC mice (8 months) compared to levels for C mice (Fig. 6A to C). Sections stained with Oil Red O, a lipid stain, show acinar cell loss and their replacement by accumulating adipocytes to various degrees in different mice, in some cases leaving normal-appearing islet cells in a field of fat cells (Fig. 6D to G). The smaller size, as well as a reduction in weight, of pancreata in older TC mice compared to age-matched control C mice is attributed to acinar atrophy and adipocyte accumulation resulting from HuR overexpression (Fig. 6H).
FIG 6.
Histopathology and weight of pancreas of older C and TC mice. (A to F) H&E-stained pancreas sections of 8-month-old C mouse (A) and TC mice (B to F). Panel C is a higher magnification of the boxed area in panel B. Extreme acinus cell atrophy is seen in panel D. (G) Oil Red O-stained pancreas section from 8-month-old TC mouse. (H) Pancreas weight/body weight of C and TC male and female mice. is, islet of Langerhans; ac, acinar cells; A, adipocytes; L, lipid. Arrowheads indicate immune infiltrates. The arrow indicates area of fibrosis. The area circumscribed by the dotted line in panel F is a ductal proliferative complex. Scale bars, 50 μm.
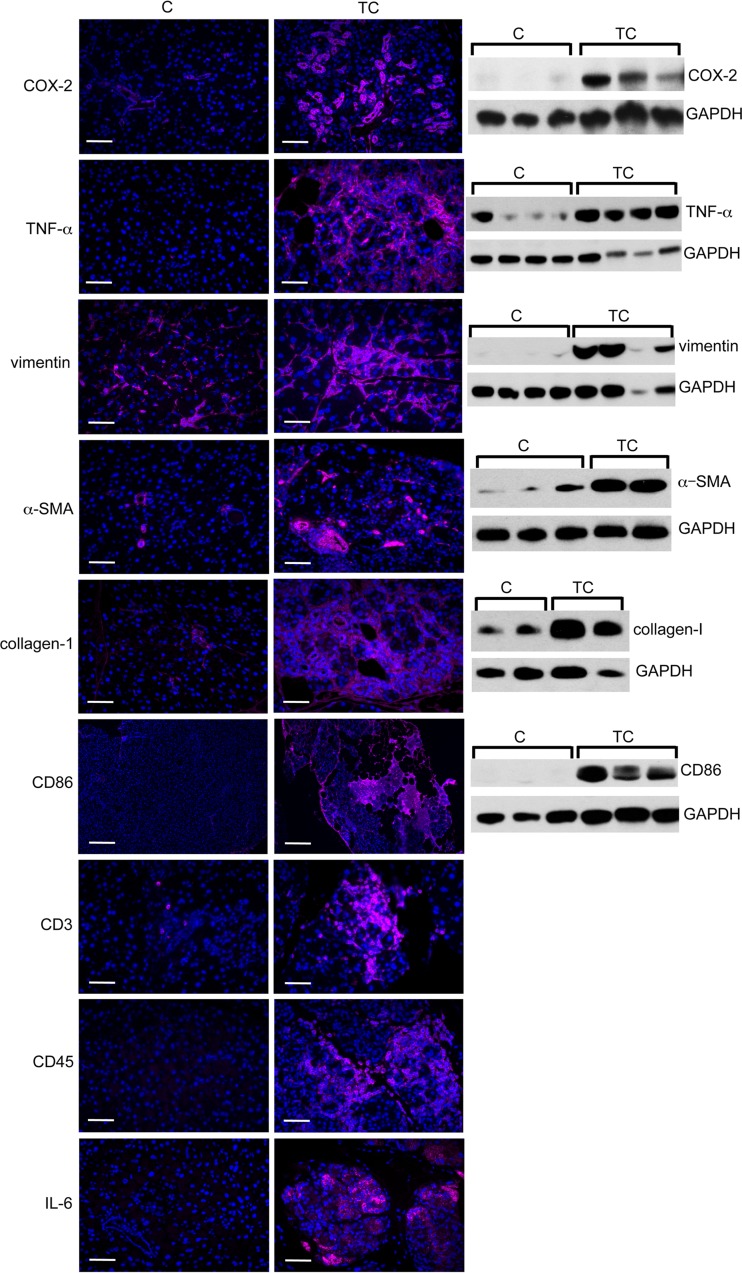
Immunostaining of pancreas sections from TC and C mice shows increased expression of COX-2 (inflammatory enzyme), tumor necrosis factor alpha (TNF-α; a proinflammatory cytokine), vimentin (fibroblasts), α-smooth muscle actin (α-SMA; stellate cells), and collagen 1 (fibroblasts) in TC compared to C pancreata (Fig. 7). CD86 (antigen-presenting cells), CD3 (T cells), CD45 (B cells), and interleukin-6 (IL-6) (proinflammatory cytokine) expression also is elevated in TC compared to C pancreata (Fig. 7). Western blot analyses of these proteins confirm immunofluorescent staining observations (Fig. 7). These results provide evidence for activation of pancreatic stellate cells contributing to a fibroinflammatory reaction as a consequence of high HuR expression in the pancreas. The histopathology we have observed in TC pancreata, namely, inflammatory infiltrates, ductal proliferation, increased fibrosis, and acinar atrophy, are features characteristic of chronic pancreatitis (16).
FIG 7.
Fibroinflammatory gene expression in HuR-overexpressing pancreas. Immunostaining and immunoblots of markers of inflammation (COX-2 and TNF-α), fibrosis (vimentin, α-SMA, and collagen 1), immune cells (CD86, CD3, and CD45), and IL-6 in pancreas of T and TC mice. GAPDH serves as a gel loading control. Scale bars, 50 μm.
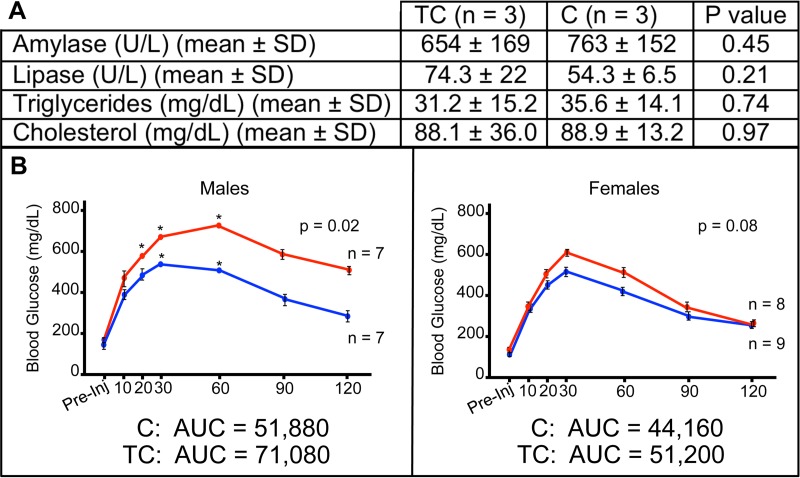
While serum markers that are often elevated in chronic pancreatitis (amylase, lipase, triglycerides, and cholesterol) showed no significant differences between 8-month-old TC and C mice (Fig. 8A), we tested 8-month-old TC and C mice of both genders for diabetes using a glucose tolerance test to further examine metabolic consequences associated with HuR overexpression in the pancreas. The peak glucose concentration in whole blood of TC compared to C males was significantly higher and its clearance slower, indicative of severe glucose intolerance (Fig. 8B). A nonsignificant increase in peak glucose concentration was observed in TC females compared to C females. An additional test of a single 11-month-old female indicated glucose intolerance, suggesting a delay in females compared to males, perhaps due to delayed accumulation of overall body fat mass.
FIG 8.
Metabolic measurements in C and TC pancreas. (A) Levels of serum amylase, lipase, triglycerides, and cholesterol in TC and C mice. (B) Concentrations of blood glucose at various times following administration of glucose bolus to 8-month-old TC (red) and C (blue) male and female mice. Asterisks indicate time points at which the glucose concentration exceeded the range of the GTT assay.
HuR overexpression in pancreas results in global changes in gene expression.
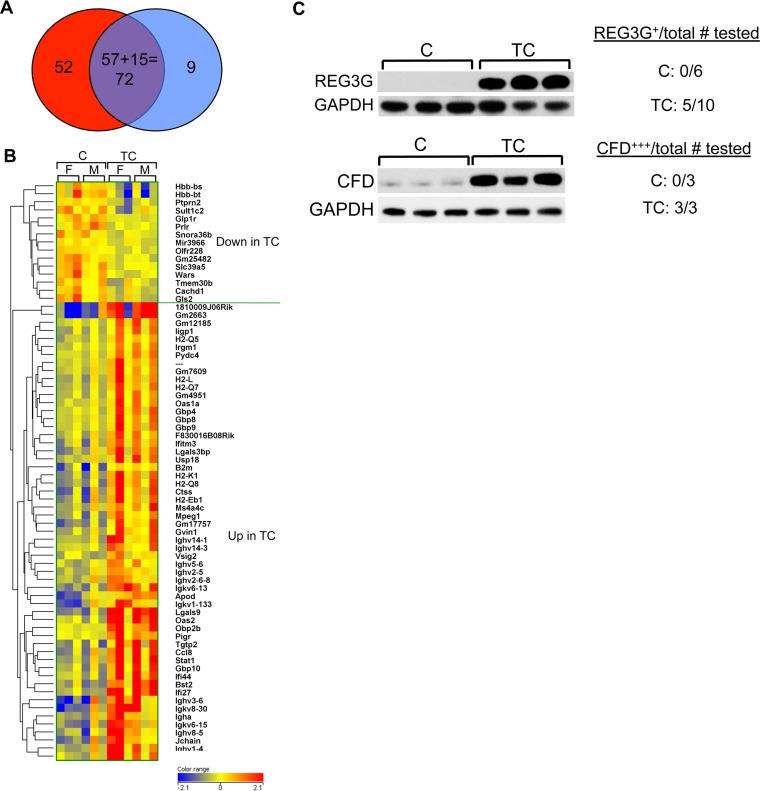
Total RNA isolated from pancreata of 6-month-old TC and C male and female mice was processed and subjected to microarray analysis to identify changes in gene expression associated with pancreatic HuR overexpression. In a comparison of TC versus C mice (pooled data from both genders), 210 known genes showed differential expression (≥1.5-fold; P ≤ 0.05) (178 up and 32 down). A similar comparison of female TC and C mice yielded 273 genes (181 up and 92 down), while a comparison of male TC and C yielded 152 genes (72 up and 80 down). Using pooled data from both genders, we identified 72 genes that had the largest fold change in expression (either up- or downregulated) as well as the most significance associated with that change (Table 1 and Fig. 9A and B). The resulting data on this set of genes was analyzed using Ingenuity Pathway Analysis software. In keeping with HuR's known role in proliferation, cell growth, and survival, 26 genes associated with cell death and survival and 17 genes associated with cellular growth and proliferation were identified as upregulated in TC pancreas (Table 2). In agreement with our observations of pancreas histopathology, observed changes in gene expression were most strongly associated with five disease classes: endocrine system disorders (23 genes), gastrointestinal disease (27 genes), immunological disease (28 genes), metabolic disease (26), and organismal injury and abnormalities (33 genes) (Table 3). Several genes were also associated with antimicrobial response and inflammatory response. Genes having highly significant changes in expression were detected in several canonical pathways, including allograft rejection signaling, OX40 signaling, antigen presentation, interferon signaling, and phagosome maturation. To partially validate microarray analysis results, we chose to assay expression of two genes, Reg3g and CFD (also known as adipsin or adiponectin), in cell lysates prepared from TC and C pancreata on Western blots. On microarrays, both genes displayed significantly increased expression in TC pancreas compared to C pancreas (Reg3g, 3.58-fold, P = 0.03; CFD, 2.93-fold, P = 0.05). Both REG3G and CFD showed elevated expression in HuR-overexpressing TC pancreata compared to C pancreata on Western blots (Fig. 9C), thus validating the microarray analyses. REG3G is an antimicrobial C-type lectin secreted primarily by the pancreas and testis (17, 18). IL-6 induction of Reg3g suggests that elevated REG3G is associated with tissue inflammation and acute pancreatitis (17). CFD is a chymotrypsin that functions as an adipokine, a cell signaling protein secreted by adipocytes. It regulates insulin in mice and improves β cell function in diabetes, most likely reflecting its function as a potent anti-inflammatory molecule (19, 20).
TABLE 1.
Differences in gene expression in pancreas of TC and C micea
| Gene designation and regulation status | Gene name | Fold change | Disease(s) or function |
|---|---|---|---|
| Upregulated | |||
| 1810009J06Rik | RIKEN cDNA 1810009J06 | 21.6 | |
| Gm2663 | Predicted gene 2663 | 20.1 | |
| Lgals9 | Lectin, galactose binding, soluble 9 | 12.2 | Metabolic |
| Igkv8-30 | Immunoglobulin kappa chain variable 8-30 | 7.5 | |
| Ighv3-6 | Immunoglobulin heavy variable 3-6 | 6.0 | |
| Oas2 | 2-5 Oligoadenylate synthetase 2 | 5.5 | Organismal injury |
| Igkv6-13 | Immunoglobulin kappa variable 6-13 | 5.0 | |
| Ifi27 | Interferon, alpha-inducible protein 27 | 4.9 | |
| Bst2 | Bone marrow stromal cell antigen 2 | 4.5 | |
| Stat1 | Signal transducer and activator of transcription 1 | 4.1 | |
| Tgtp2 | T cell specific GTPase2 | 3.7 | |
| Obp2b | Odorant binding protein 2B | 3.6 | |
| Ighv1-4 | Immunoglobulin heavy variable 1-4 | 3.6 | |
| Ccl8 | Chemokine (C-C motif) ligand 8 | 3.6 | Endocrine |
| Ifi44 | Interferon-induced protein 44 | 3.5 | Gastrointestinal |
| Ighv1-7 | Immunoglobulin heavy variable V1-7 | 3.4 | |
| H2-Q8 | Histocompatibility 2, Q region locus 8 | 3.4 | |
| Igkv6-15 | Immunoglobulin kappa variable 6-15 | 3.4 | |
| Ighv8-5 | Immunoglobulin heavy variable V8-5 | 3.4 | |
| Gbp10 | Guanylate-binding protein 10 | 3.4 | |
| Jchain | Immunoglobulin joining chain | 3.3 | Gastrointestinal, immunological, organismal injury |
| Ctss | Cathepsin S | 3.3 | Endocrine, gastrointestinal, immunological, metabolic, organismal injury, inflammatory response |
| Gm17757 | Predicted gene, 17757, noncoding RNA | 3.2 | |
| Ighv14-1 | Immunoglobulin heavy variable 14-1 | 3.1 | |
| Igkv1-133 | Immunoglobulin kappa variable 1-133 | 3.0 | |
| H2-K1 | Histocompatibility 2, K1, K region | 2.9 | |
| Apod | Apolipoprotein D | 2.7 | |
| H2-Eb1 | Histocompatibility 2, class II antigen E beta | 2.7 | |
| Lgals3bp | Lectin, galactose-binding, soluble, 3 binding protein | 2.7 | |
| Downregulated | |||
| Mir3966 | MicroRNA 3966 | −1.4 | |
| Olfr228 | Olfactory receptor 228 | −1.4 | |
| Cachd1 | Cache domain containing 1 | −1.6 | |
| Snora36b | Small nucleolar RNA, H/ACA box 36B | −1.7 | |
| Slc39a5 | Solute carrier family 39 (metal ion transporter), member 5 | −1.7 | Metabolic |
| Tmem30b | Transmembrane protein 30B | −1.7 | |
| Glp1r | Glucagon-like peptide 1 receptor | −1.7 | Organismal injury |
| Ptprn2 | Protein tyrosine phosphatase, receptor type, N polypeptide | −1.7 | |
| Prlr | Prolactin receptor | −1.7 | |
| Gm25482 | Predicted gene, 25482 | −1.8 | |
| Wars | Tryptophanyl-tRNA synthetase | −1.8 | |
| Gls2 | Glutaminase 2 (liver, mitochondrial) | −1.9 | Organismal injury |
| Sult1c2 | Sulfotransferase family, cytosolic, 1C, member 2 | −2.0 | |
| Hbb-bs | Hemoglobin, beta adult s chain | −2.2 | |
| Hbb-bt | Hemoglobin, beta adult t chain | −3.1 |
The top 30 genes that have the largest fold increase and all genes with the largest fold decrease in expression, as well as the lowest P value, are listed. Genes having function in the 7 diseases or functions most affected by HuR overexpression are in boldface.
FIG 9.
Microarray analysis comparing gene expression in pancreata of TC and C mice. (A) Venn diagram. Of 124 genes having the largest fold difference in expression (100 increased in TC [red], 24 decreased [blue]), 72 genes also had the lowest P values (57 increased, 15 decreased). (B) Heat map indicating 57 genes with increased expression in TC and 15 genes with decreased expression. Pancreata of 3 females (F) and 3 males (M) for each genotype were analyzed. (C) Immunoblots of TC and C pancreas protein extracts probed for REG3g and CFD. GAPDH serves as gel loading control.
TABLE 2.
Genes having altered expression (TC vs C pancreas) associated with cell death, survival, growth, and proliferationa
| Functions | Genes |
|---|---|
| Cell death and survival | JUN, Cxcl9, IRF9, PLA2G7, TP53INP1, GLP1R*, MAT2A, VCAM1, H2-K2/H2-Q9, STAT1, CDKN1A, DCDC2, PTPRN2*, ATP1B3, B2 M, Ms4a4b, Oas1b, GLIPR1, HLA-A, PRLR*, Irgm1, IFIH1, IFI1b, CFD, FCGR3A/FCGR3B |
| Cell growth and proliferation | JUN, B2 M, TP53INP1, IFITM3, NFKB1A, VCAM1, Ms4a4b, HLA-A, PRLR, Igm1, STAT1, IFIH1, CDKN1A, Slfn2, LGALS9B, IFI1b, FCGR3A/FCGR3B |
Genes among the top 30 genes having the largest fold increase in expression are in boldface. Genes with the largest fold decrease in expression are designated with an asterisk.
TABLE 3.
Genes having altered expression (TC versus C pancreas) associated with disease classesa
| Disease class | Genes |
|---|---|
| Endocrine disorder | CTSS, SLC39A5*, OAS1, Ccl8, APOD, Tgtp1/Tgtp2, B2 M, IFITM3, GLP1R*, Ms4a4b, PRLR*, HLA-A, Irgm1, Gbp8, STAT1, GBP6, HLA-DRB5, IFI44, LGALS9B, IFI16, Gm4951, ligp1, MPEG1 |
| Gastrointestinal disease | JCHAIN, Ccl8, PIGR, Tgtp1/Tgtp2, IFITM3, GLP1R*, Gbp8, STAT1, HLA-DRB5, MPEG1, CTSS, SLC39A5*, OAS1, Hbb-b2, APOD, Hbb-b1, B2 M, Ms4a4b, HLA-A, Irgm1, PRLR*, GBP6, IFI44, LGALS9B, IFI16, Gm4951, ligp1 |
| Immunological disease | JCHAIN, Ccl8, PIGR, Tgtp1/Tgtp2, IFITM3, USP18, GLP1R*, Gbp8, STAT1, HLA-DRB5, LGALS3BP, PTPRN2*, MPEG1, CTSS, OAS1, Hbb-b2, B2 M, Ms4a4b, HLA-A, Irgm1, Bst2, GBP6, IFI44, IFI16, Gm4951, ligp1 |
| Metabolic disease | Ccl8, PIGR, Tgtp1/Tgtp2, IFITM3, GLP1R*, Gbp8, STAT1, HLA-DRB5, MPEG1, CTSS, SLC39A5*, OAS1, Hbb-b2, B2 M, Ms4a4b, HLA-A, Irgm1, PRLR*, GBP6, IFI44, LGALS9B, IFI16, Gm4951, ligp1 |
| Organismal injury and/or abnormality | JCHAIN, Ccl8, PIGR, Tgtp1/Tgtp2, USP18, IFITM3, GLP1R*, Gbp8, STAT1, HLA-DRB5, LGALS3BP, PTPRN2*, MPEG1, CTSS, SLC39A5*, OAS1, Hbb-b2, APOD, OAS2, Hbb-b1, B2 M, SULT1C2*, Ms4a4b, HLA-A, PRLR*, Irgm1, Bst2, GBP6, IFI44, LGAL69B, IFI16, Gm4951, ligp1 |
Genes among the top 30 genes having the largest fold increase in expression are in boldface. Genes with the largest fold decrease in expression are designated with an asterisk.
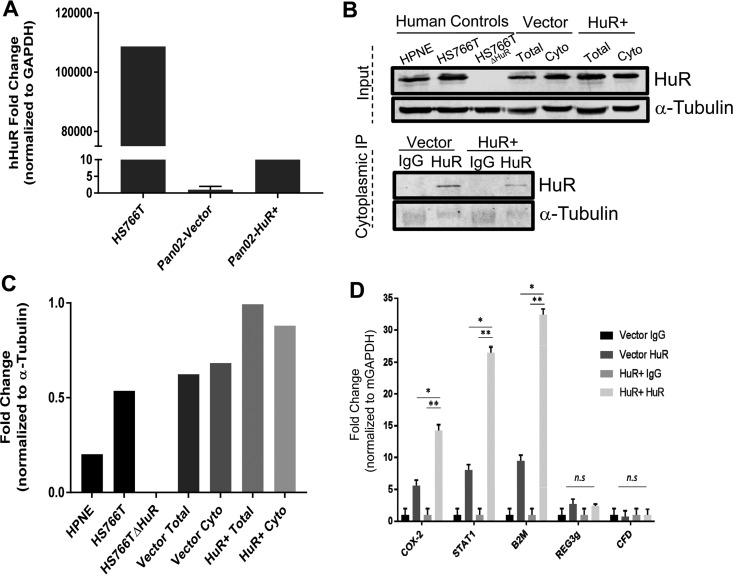
To further validate microarray results (Fig. 9) and determine whether mRNAs are direct or indirect HuR targets, we transfected Pan02 mouse pancreatic tumor cells with human HuR (hHuR) cDNA and vector control plasmids. Overexpression of HuR mRNA was confirmed using DNase-treated total mRNA extracts and probes specific against hHuR (Fig. 10A). Western blot analyses of input protein and immunoprecipitation (IP) lysates confirm overexpression of HuR, and specific IP with the HuR antibody was observed in hHuR-transfected cells compared to vector control-transfected cells (Fig. 10B and C). A messenger ribonucleoprotein IP (mRNP-IP) assay was performed on fractionated, cytoplasmic lysates of transfected cells to identify HuR-bound target mRNAs (Fig. 10D). Assessment of previously predicted HuR targets, Stat1 and B2m (21), along with top microarray hits, Reg3g and CFD, were analyzed for their relative expression in RNA extracted from cytoplasmic IP samples (Fig. 10D). In addition to an established HuR target, Cox-2 (8), only the predicted HuR targets, Stat1 and B2m, showed significant binding to HuR compared to the IgG control, whereas REG3g and CFD did not. Taken together with microarray data (Fig. 9), these data support a role for HuR in the direct regulation of some genes associated with the proinflammatory response. We conclude that elevated expression of other genes (e.g., Reg3g and CFD) is an indirect consequence of HuR overexpression.
FIG 10.
HuR RNP-IP assays of selected proinflammatory mRNAs. (A) qRT-PCR analysis of murine Pan02 cells transfected with a human HuR expression plasmid or empty vector using probes that detect human HuR. RNA extracted from human PDAC cell line Hs766T serves as positive control. (B) Validation of mRNP-IP performed with total and cytoplasmic fractions of Pan02 cells transfected with either HuR overexpression plasmid or vector control; α-tubulin was used as a loading control for the input and a negative control for the immunoprecipitation samples. Hs766T, Hs766TΔHuR (60), and a human normal pancreatic line, HPNE, serve as controls. (C) Quantification of total HuR protein relative to α-tubulin levels. (D) Relative binding of mRNA targets to HuR, normalized to respective IgG controls, determined by qRT-PCR using GAPDH mRNA as a loading control and Cox-2 as a positive control. *, P < 0.01; **, P < 0.001; n.s., not significant.
HuR overexpression in pancreas does not initiate tumorigenesis.
It is well documented that HuR is highly expressed in many tumor types, including oral, gastric, colorectal, lung, breast, renal, skin carcinoma, mesothelioma, ovary, and pancreas (22–24), supporting a role for HuR overexpression in tumor progression. To determine whether HuR overexpression can facilitate tumor initiation, we examined H&E-stained sections of pancreata from TC mice ≥10 months old (n = 17 mice aged 10 to 15 months). In all TC pancreata examined, no precancerous PanIN lesions or adenocarcinomas were observed, consistent with our observation in 8-month-old TC mice (Fig. 6). The absence of Alcian blue staining in TC pancreata from mice of all ages examined confirmed the absence of PanINs (data not shown). In the absence of driver mutations (i.e., KRAS), HuR overexpression alone does not appear to initiate tumor development in the pancreas.
HuR overexpression, in combination with KrasG12D/+−, accelerates pancreatic dysplasia and enhances tumorigenesis.
KRAS mutations are commonly observed in pancreatic tumors and accepted as an early tumor-initiating event (25, 26). Our data showing pancreatic HuR overexpression elicits an inflammatory phenotype without evidence of PanIN involvement suggested a role for HuR in PDAC progression downstream of a mutant KRAS initiating event. To test this, we generated [LSL-KrasG12D/+ + Traffic/HuR + Pdx1/Cre] triple transgenic mice (KTC) with the hypothesis that KrasG12D in combination with HuR overexpression would enhance the development of pancreatic tumors.
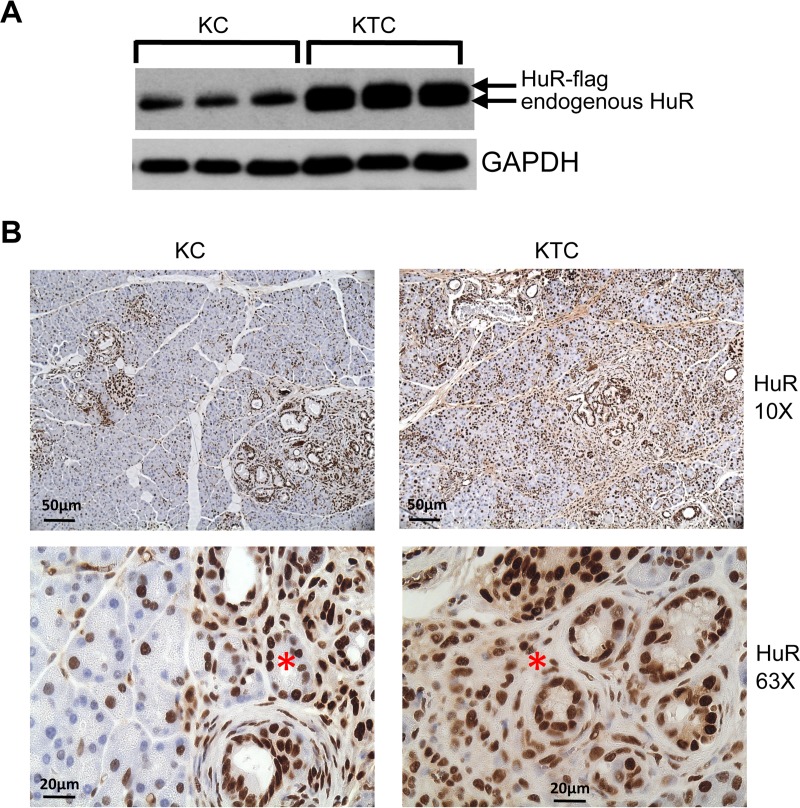
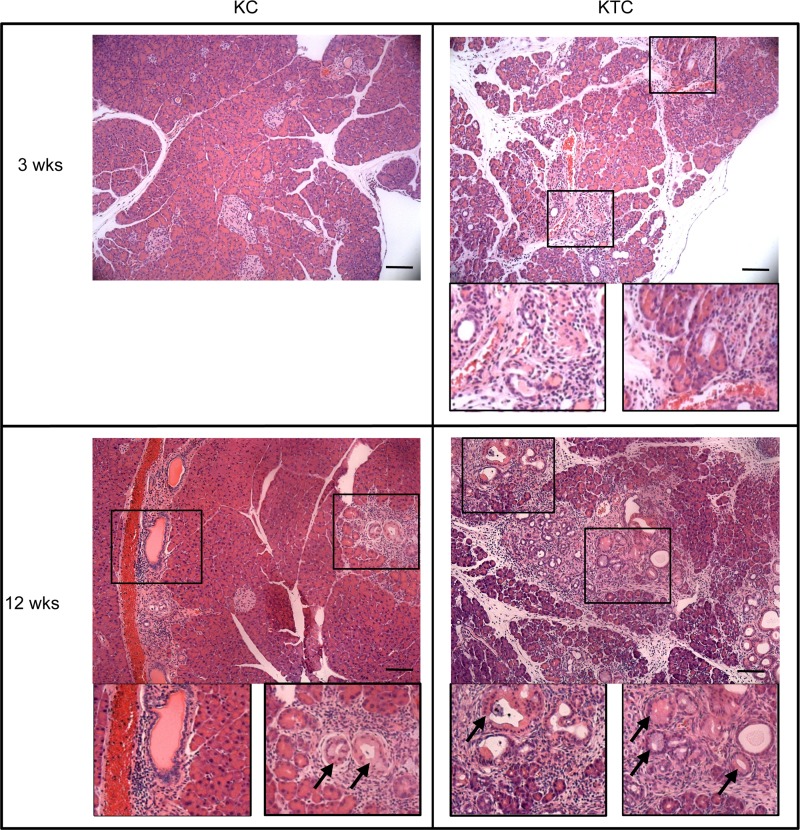
As expected, HuR protein expression is increased in pancreata of KTC mice compared to KC mice (Fig. 11A). Cytoplasmic HuR was observed in both KC and KTC pancreata of 9-week-old mice in areas of abnormal pathology resulting from expression of KrasG12D (Fig. 11B). Significant chronic inflammation was observed in H&E-stained sections of KTC pancreata as early as 3 weeks of age and increased in both KC and KTC pancreas with age (Fig. 12). Ductal proliferation was also prominent in pancreata of KC and KTC beginning at 6 weeks of age. Ducts displaying precancerous PanIN-1A lesions were present in <50% of ducts in KC and KTC pancreata beginning at 6 weeks of age. By 12 weeks, >50% of ducts in KTC mice displayed PanIN-1A, whereas the percent was still <50% in KC pancreas. In young mice (3 to 12 weeks old), protein markers of inflammation and fibrosis were enhanced in KTC mice compared to KC mice (Fig. 13). As both KTC and KC mice age, pancreatic histology becomes increasingly disorganized and dysmorphic; in addition to an increased number and grade of PanIN lesions, there is extensive chronic inflammation, fibrosis, ductal proliferation, and neutrophilic inflammation (Fig. 14A and B). We monitored 10- to 18-month-old KTC and KC mice for the presence of pancreas tumors. We identified PDACs in 5/29 (17%) KC mice and in 8/14 (57%) KTC mice (P < 0.001) (Fig. 14C and D). These data support the idea that the inflammatory microenvironment in KTC pancreas enhances KrasG12D-promoted tumorigenesis.
FIG 11.
HuR expression in KC and KTC mice. (A) Immunoblot of protein extracts from pancreas from KC and KTC mice. GAPDH serves as gel loading control. (B) Pancreas sections from 9-week-old KC and KTC pancreas immunostained for HuR. Asterisks indicate cytoplasmic HuR in regions of ductal proliferation in KC and KTC pancreas.
FIG 12.
H&E-stained sections of KC and KTC pancreas from 3-week- and 12-week-old mice. Examples of areas of immune infiltrates and ductal proliferation are enlarged in boxed images. Arrows indicate PanIN-1A lesions. Scale bars, 20 μm.
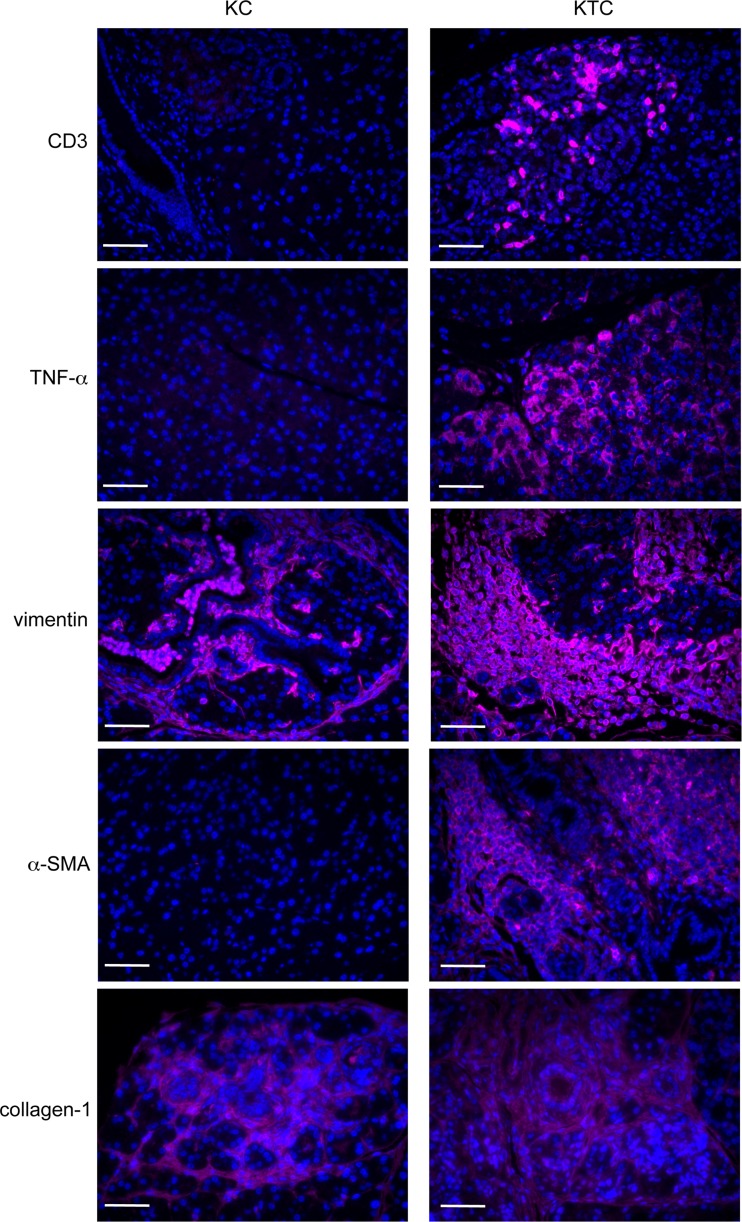
FIG 13.
Fibroinflammatory gene expression in KC and KTC pancreata. Sections of pancreas of KC and KTC mice immunostained for markers of immune cells (CD3), inflammation (TNF-α), and fibrosis (vimentin, α-SMA, and collagen 1). Scale bars, 20 μm.
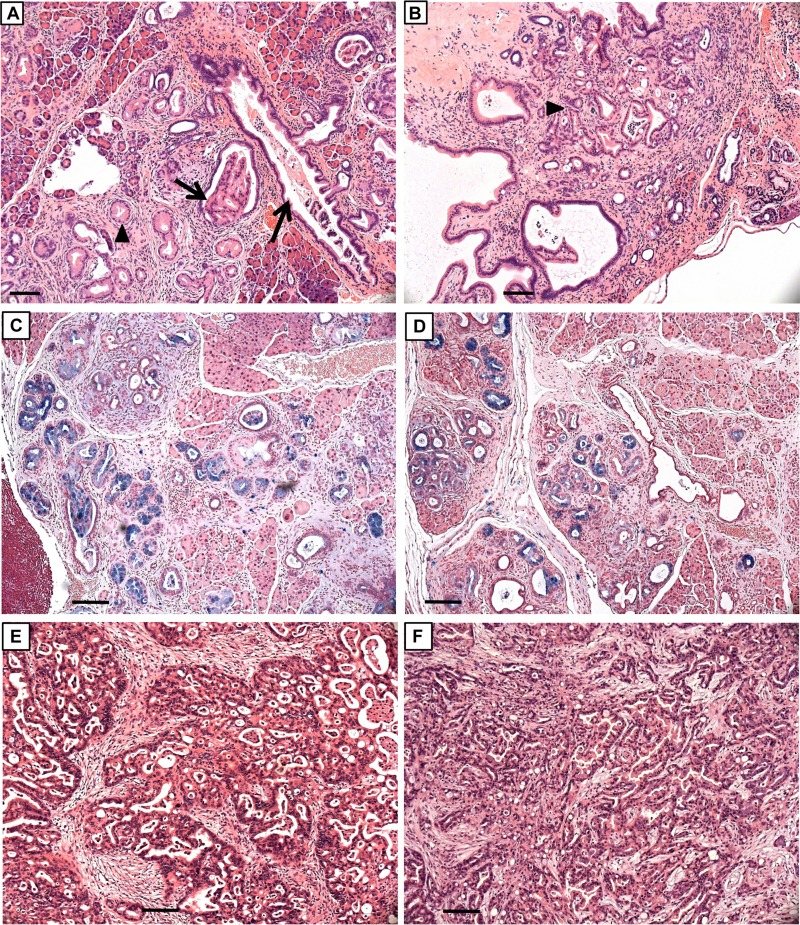
FIG 14.
Histopathology and PDACs in KC and KTC pancreas. H&E-stained sections of 32-week-old (A and C) KC and (B and D) KTC pancreas. PanIN-1A (arrowheads) and PanIN-1B (arrows) lesions are present. Alcian blue staining in panels C and D highlights mucus-containing PanIN lesions. H&E-stained sections of well-differentiated PDACs in 44-week-old KC mouse (E) and 49-week-old KTC mouse (F). Note extensive fibrosis in tumors. Scale bars, 50 μm.
Extrapancreatic Traffic/HuR transgene expression reduces the incidence of papillomas.
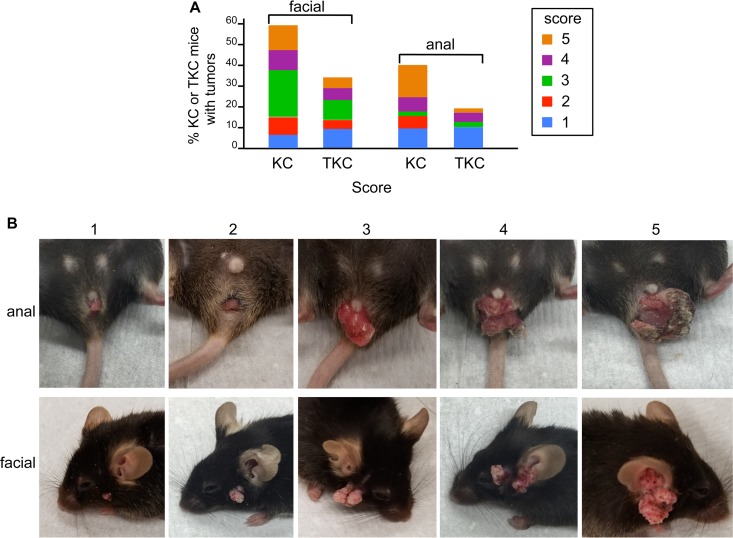
The presence of mucocutaneous papillomas in KC mice has previously been reported and attributed to activity of the Pdx1 promoter that regulates Cre recombinase expression (27). We also observed facial and anal papillomas in KC and KTC mice. Interestingly, the percentage of KC mice with facial and anal tumors was significantly higher (P < 0.0001) than the percentage of KTC mice (for facial tumors, KC, 41/73 [56%], KTC, 17/48 [35%]; for anal tumors, KC, 29/73 [40%], KTC, 9/48 [19%]). We assigned a score from 1 to 5 to each tumor based upon its size. KC tumors were larger than KTC tumors, but this increase in size was not significant (Fig. 15A and B).
FIG 15.
Anal and facial papillomas in KC and KTC mice. (A) Incidence of facial and anal tumors of various sizes in KC and TKC mice. (B) Images of anal and facial papillomas representative of the scoring system that was adopted to rate tumor size (1, smallest; 5, largest).
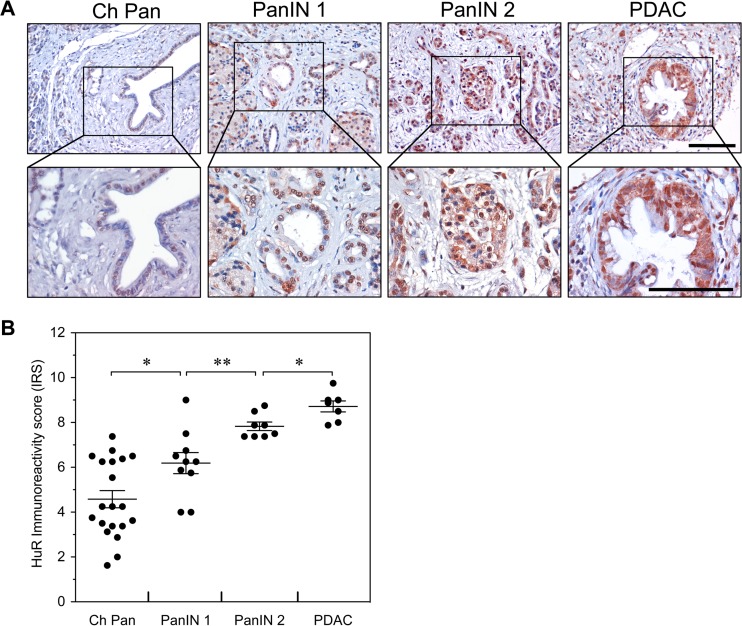
HuR expression is elevated in human pancreatitis and PDAC tumorigenesis.
We have previously shown that weak to moderate nuclear HuR expression occurs in normal pancreatic ductal and acinar cells, whereas elevated nuclear expression of HuR was observed in PDACs and was associated with cytoplasmic HuR accumulation (2). To determine the state of HuR expression in human pancreatitis and PanIN lesions, HuR expression was evaluated by immunohistochemistry (IHC) using human tissue arrays containing pancreatitis, PanIN, and PDAC tissues (Fig. 16A). HuR immunoreactivity was elevated in chronic pancreatitis and progressively increased in PanIN-1, PanIN-2, and PDACs, with elevated ductal cell staining. The cytoplasmic abundance of HuR was increased in PanIN lesions and PDACs. To evaluate expression patterns of HuR, tissue sections were assigned immunoreactivity scores (IRS) and grouped as low IRS of 0 to 6 or high IRS of 7 to 12 (Fig. 16B). In chronic pancreatitis, HuR immunoreactivity ranged from low (65%) to high (35%) IRS, whereas HuR expression displayed a shift toward high immunoreactivity in 60% of the PanIN-1 samples and in all of the PanIN-2 and PDAC samples.
FIG 16.
Expression of HuR in human PDAC progression. (A) Immunohistochemical detection of HuR expression in chronic pancreatitis (Ch Pan), PanIN-1, PanIN-2, and PDAC. Representative tissue sections were examined for HuR expression and counterstained with hematoxylin. Scale bars, 100 μm. (B) Immunoreactivity scores (IRS) for HuR expression in tissue sections of Ch Pan, PanIN-1, PanIN-2, and PDAC. *, P < 0.05; **, P < 0.01.
DISCUSSION
We present histologic, metabolic, and gene expression evidence that targeted overexpression of the RNA-binding protein HuR to the pancreas in a novel transgenic mouse model, called Traffic-HuR/Cre (TC), elicits a fibroinflammatory reaction characteristic of the pathological hallmarks of chronic pancreatitis. We note that human and mouse HuR protein sequences are 98.5% homologous. While several genetic models of chronic pancreatitis exist that alter individual gene expression (28, 29), the TC model uniquely alters expression of the RNA-binding protein HuR that can impact expression of hundreds of genes through posttranscriptional regulation. It is interesting that several of the existing mouse models for chronic pancreatitis are based on altered expression of HuR target genes (e.g., Cox-2 [30], K-ras [31], Eif2ak-2 [32], IL-1β [33], and transforming growth factor β1 [34] genes).
HuR is located primarily in the nucleus of healthy cells, and it functions in posttranscriptional RNA trafficking and maintaining cellular homeostasis. When cells are stressed due to environmental insult of normal cells (e.g., smoking and chronic alcohol abuse) or to cancer-related stress (e.g., hypoxia, chemotherapeutic exposure, and nutrient deprivation), HuR is activated and translocates to the cytoplasm. Prior to transit, it binds to selected mRNA transcripts, most of which are known to be proinflammatory, thereby stabilizing them and transporting them to the cytoplasm. In TC mice, HuR overexpression is genetically induced in normal pancreas cells, and we observed elevated cytoplasmic HuR. Given our understanding of HuR's role in posttranscriptional regulation of many genes under stressful conditions, it is reasonable to surmise that constitutively activated HuR in the pancreas leads to the observed fibroinflammatory reaction in an attempt to overcome a stressful imbalance in gene expression. Thus, HuR overexpression in the pancreas of TC mice is relevant to physiological stresses imposed on the pancreas in humans, thereby providing an alternative to the widely used chemical induction of acute and chronic pancreatitis by cerulein induction (35–37).
HuR has a recognized role in inflammation (38). Through its regulation of mRNAs encoding both proinflammatory proteins (e.g., COX-2, TNF-α, and IL-6) as well as proteins that inhibit production of anti-inflammatory factors (e.g., thrombomodulin), HuR has been implicated in promoting inflammation associated with several disease conditions, including rheumatoid arthritis, inflammatory bowel disease, asthma, vascular inflammation, and atherosclerosis, in HIV patients undergoing protease inhibitor therapy. This study is the first report to demonstrate directly an inflammatory response to HuR overexpression resulting in chronic pancreatitis. Double transgenic mouse models combining the Traffic-HuR transgene with transgenes in which Cre recombinase is driven by tissue-specific promoters other than Pdx1 may provide useful models for studying HuR's role in other inflammatory diseases.
The level of HuR protein in the pancreas of TC mice is >2-fold higher than that in pancreas of nontransgenic mice. This increase in HuR expression is particularly evident in acinar and ductal epithelial cells upon immunostaining of pancreas of TC mice of all ages. Progressive pancreatic histopathology ensues as mice age, resulting in extensive dysplasia in the exocrine parenchyma, including ductal proliferation, cystic duct formation, and a massive diminution and replacement of acinus cells by adipocytes. We confirmed histological observations of chronic inflammation and fibrosis by immunostaining and Western blot analyses of specific protein markers of fibrosis (α-smooth muscle actin, vimentin, and collagen 1), immune infiltrates (CD3+ T cells, CD45+ B cells, and CD86+ antigen-presenting cells), and markers of inflammation (COX-2, TNF-α, and IL-6 expression).
Together with known environmental and physiological risk factors (e.g., cigarette smoking, alcoholic abuse, Helicobacter pylori infection, and obesity) (16, 39), chronic pancreatitis in its most advanced state in humans can lead to endocrine dysfunction and diabetes (40). We observed severe glucose intolerance in 8-month-old male TC mice, but insulin serum levels were normal, suggesting that overt diabetes had not yet evolved. Glucose intolerance was first detected in female TC mice at 11 months of age. Pancreas weight relative to body weight in 8-month-old male TC mice is less than that in females of the same age (Fig. 6) and may account for the observed difference in the onset of glucose intolerance between genders. Similarly, lipase, amylase, triglycerides, and cholesterol serum levels were also within the normal range in 8-month-old mice despite significant exocrine dysplasia. This may reflect the great functional reserve of the exocrine pancreas that has been noted by others (41, 42). Production of lipase by other organs in the intestinal tract, including esophagus, duodenum, stomach, and colon (43), also may mask reduction of lipase produced by the pancreas.
Chronic pancreatitis is also a known risk factor for pancreatic cancer (39, 44–48). We did not observe preneoplastic PanIN lesions and PDACs in HuR-overexpressing pancreas, even in TC mice over a year old. We conclude that overexpression of HuR in itself is not oncogenic. However, in the context of an oncogenic initiating event (i.e., K-rasG12D mutation), HuR overexpression promotes development of numerous high-grade PanINs in triple transgenic KTC mice (activated KrasG12D plus HuR overexpression). Similarly, 8/14 (57%) KTC mice developed PDACs, whereas only 5/29 (17%) KC mice had PDACs. HuR overexpression promoted a more severe fibroinflammatory environment in KTC pancreas than in KC mice (Fig. 12 and 14).
Thus, in addition to showing that HuR overexpression in the pancreas results in clinical features of pancreatitis, this study also establishes that HuR drives enhancement of PDAC tumorigenesis when combined with the K-rasG12D oncogene. These results are consistent with the observation that PDACs develop from a combination of genetic mutations (e.g., activated K-rasG12D) and nongenetic events (e.g., chronic fibroinflammation) (49). These results are also consistent with our observations during colorectal tumor progression using intestine-specific HuR overexpression. When overexpressed in intestinal epithelial cells, HuR did not initiate tumor formation but served to promote tumor progression downstream with loss of the APC tumor suppressor or during colitis-associated carcinogenesis (unpublished data). The fact that HuR overexpression in combination with K-rasG12D in KTC pancreas increases the frequency of PDACs compared to that in KC mice appears at first to be at odds with recent evidence suggesting that HuR posttranscriptional regulation of KRAS itself results in suppression of its expression (50). This observation may serve to reinforce the strong influence of an inflammatory microenvironment on tumorigenesis.
HuR overexpression in the pancreas by itself did not result in the development of PDAC. Nevertheless, the intense fibroinflammatory response elicited by HuR overexpression may shed light on the role that HuR plays in the extensive desmoplastic deposition characteristic of PDAC. It further suggests that therapies that target HuR to pancreatic tumor cells, especially those that have a K-rasG12D activating mutation, may be effective in suppressing proliferation of both tumor cells and desmoplastic stromal cells. Finally, it also validates that mutant KRAS is a strong driving force in pancreatic tumorigenesis and that additional nongenetic insults (like HuR overexpression) can cooperate to form tumors.
An interesting aspect of this study is evidence suggesting that while HuR overexpression in the pancreas results in cellular atypia and loss of normal tissue organization, HuR may also attempt to maintain homeostasis and reduce cellular dysfunction resulting from its overexpression in noncancerous tissues. It may do so by posttranscriptionally regulating expression of selective genes. Thus, in contrast to a decrease in serum CFD observed in obese mice (51), we observed increased expression of CFD, an anti-inflammatory adipokine, in the inflammatory microenvironment of HuR-overexpressing pancreas. Similarly, the reduced frequency and mass of benign facial and anal papillomas in KTC mice compared to KC mice (Fig. 15) may be attributed to differences in the tumor microenvironment that support HuR suppression of tumor growth of some tumors compared to others. Expression of Cox-2, an enzyme that mediates prostaglandin synthesis, is posttranscriptionally regulated by HuR (8, 52). Cox-2 overexpression in the murine exocrine pancreas results in chronic inflammation and pancreatitis-like dysplastic features similar to those we have observed in HuR-overexpressing pancreas (30). Lesions in older mice in the Cox-2 model, however, showed ductal metaplasia and PanINs more typical of overt pancreatic cancer, again suggesting that HuR overexpression in TC pancreas results in a fibroinflammatory stromal response and at the same time aims to suppress neoplastic progression. Our data suggest that in the presence of both activated KRAS and HuR overexpression in the pancreas, the ability of a chronic inflammatory microenvironment to initiate and support tumorigenesis often wins out over tumor suppression. Therapeutics that target HuR expression in the pancreas may effectively reduce multiple factors contributing to inflammation, thereby stemming PDAC growth.
Comparison of microarray analyses of RNAs prepared from pancreas of 6-month-old TC and C mice revealed multiple genes whose expression was different, either elevated or suppressed, as a consequence of HuR overexpression. A majority of these genes fell into functional groups that were consistent with the observed TC pancreas histopathology. mRNP-IP analysis of selected genes showed that some (Cox-2, Stat1, and B2m) are direct targets of HuR, while others (Reg3g and CFD) appear to be indirect targets. Of note, poly(ADP-ribosyl)ation (PARylation) of HuR by PARP1 has been shown to stabilize mRNAs of proinflammatory cytokines/chemokines (53). We observed elevated expression in HuR-overexpressing TC pancreata of two other poly(ADP-ribose) polymerase family members, PARP12 and PARP14.
Interestingly, we also observed a 2.7-fold increase in apolipoprotein D (ApoD) expression in TC pancreas. This is noteworthy because transgenic mice that overexpress ApoD in liver develop hepatic steatosis (54), suggesting that ApoD plays a role in the observed pancreatic steotosis in TC mice as well. It also should be noted that due to the chimeric nature of Cre-recombinase-mediated recombination, the effect of HuR overexpression on gene expression in acinar, ductal epithelial, and islet cells may be underestimated. Laser capture analyses of RNAs isolated from exo- and endocrine cells and from stromal cells in TC and C pancreas will be informative about the cross talk between cell populations that affects gene expression and cell function. Ultimately, informed analysis of microarray data may provide leads to identify new therapeutic targets, in addition to HuR itself, for the treatment of pancreatitis, diabetes, and PDAC.
Finally, the TC mouse model provides an excellent preclinical tool with which to evaluate novel pancreas-targeted therapeutics for the treatment of pancreatitis and for increasing our understanding of the role of inflammation in the progressive changes leading to endocrine dysfunction, diabetes, and cancer.
MATERIALS AND METHODS
Mice.
Fourteen founder Traffic-HuR transgenic mice were generated by oocyte DNA microinjection (Cyagen) of Flag epitope-tagged HuR (human) cDNA (8) cloned into the inducible transgenic vector pTraffic (55). Based upon high DsRed2 expression, two founders were selected for expansion to generate transgenic lines maintained on a C57BL/6J genetic background (C57BL/6J mice from Jackson Laboratory). Initial characterization of transgene expression and associated pathology was done on TC mice generated from both founder lines. No observable differences between lines were detected. Pdx1/Cre and B6.129-Krastm4Tyj mice were from Jackson Laboratory and the NCI-Frederick Mouse Repository, respectively. Animals were maintained under the guidelines of the Guide for the Care and Use of Laboratory Animals (61) and under the evaluation and approval of the Institutional Animal Care and Use Committee (Lankenau Institute for Medical Research).
Cell culture and cell transfection.
Hs766T and HPNE cells were purchased from the American Type Culture Collection (ATCC), and Pan02 cells were obtained from the NCI DTP Tumor Repository (Frederick, MD). All cells were grown in media as recommended by the supplier. Transient transfection of Pan02 cells with HuR cDNA plasmid was performed as previously described (1).
RT-qPCR and mRNA expression analysis.
Total RNA was extracted from transfected cells and then analyzed by reverse transcription-quantitative PCR (RT-qPCR) to assess transfection efficiency. RNA was isolated 48 h posttransfection. A Turbo DNase kit was used to eliminate residual plasmid from extracted total RNA (Ambion) in an effort to avoid detection of exogenous cDNA plasmid remaining after posttransfection washes. Total RNA extraction was reverse transcribed, and RT-qPCR was performed as previously described (56). Relative quantification was performed using the 2−ΔΔCT method (1, 56).
Immunoblot analysis.
Whole-cell pancreas extracts were prepared in radioimmunoprecipitation assay (RIPA) buffer containing proteinase inhibitors. Extract protein concentrations were determined using the bicinchoninic acid (BCA) protein assay kit (Pierce). Three pancreata each from C and TC mice were assayed. Cytoplasmic extracts of pancreas and cell lines were isolated using the NE-PER nuclear and cytoplasmic extraction kit (Thermo-Scientific, Waltham, MA) per the manufacturer's instructions. Soluble proteins were separated on 10% SDS-PAGE gels and analyzed by Western blotting using HuR monoclonal antibody (MAb) clone 3A2 (Santa Cruz Biotechnology) and others as previously described (56) (Table 4 provides a complete list of antibodies used in this study). Membranes were developed and quantified with either ECL (Pierce) and ImageJ or with the Odyssey infrared imaging system (LI-COR Biosciences).
TABLE 4.
Antibodies used for Western blot analyses and immunostaining
| Antibody | Source | Product no. | Primary antibody dilution |
|---|---|---|---|
| Anti-HuR (3A2) (Western blots) | Santa Cruz | sc-5261 | 1:500–1:1,000 |
| Anti-HuR (19F12) (mouse pancreas immunostaining) | Clonegene | CGF12 | 1:5,000 |
| Anti-HuR (19F12) (human pancreas microarray IHC) | Molecular Probes | A21277 | 1:250 |
| Anti-GAPDH (6C5) | Ambion | AM4300 | 1:8,000 |
| Anti-lamin A/C | Santa Cruz | Sc-6215 | 1:500 |
| Anti-beta actin, clone AC-15 | Sigma-Aldrich | A5441 | 1:10,000 |
| Antiactin, alpha-smooth muscle | Sigma-Aldrich | A5228 | 1:1,000 |
| Anti-REG3G | Abcam | ab198216 | 1:250–1:500 |
| Antiadipsin (C3) | Santa Cruz | sc-373860 | 1:500 |
| Anti-Cox2 (EPR12012) | Abcam | ab179800 | 1:1,000 |
| Anti-PDCD4 (D29C6) XP | Cell Signaling | 9535 | 1:1,000 |
| Anti-collagen type I | Millipore | ABT123 | 1:2,000 |
| Anti-CD45 | BioLegend | 103121 | 1:200 |
| Anti-CD80 | Abcam | ab64116 | 1:1,000 |
| Anti-CD86 | Abcam | ab213044 | 1:1,000 |
| Anti-CD3 (SP7) | Abcam | ab16669 | 1:200 |
| Anti-TNF-α (52B83) | Abcam | ab1793 | 1:500 |
| Anti-HIF-1 alpha Ab-4 (Clone H1alpha67) | Thermo Scientific | MS-1164-P | 1:500 |
| Antivimentin (EPR3776) | Abcam | ab92547 | 1:2,000 |
| Anti-IL-6 (D5W4V) XP | Cell Signaling | 12912 | 1:500 |
Ribonucleoprotein immunoprecipitation assay (mRNP-IP).
HuR-transfected and control cells were fractionated, and immunoprecipitates of cytoplasmic lysates were prepared. HuR-bound mRNAs versus IgG-bound control were detected and quantified by qRT-PCR, as previously described (1, 3, 57, 58), using the following specific probes from Thermo Fisher Scientific: STAT1 (Mm01257286_m1), B2M (Mm00437762_m1), REG3g (Mm00441127_m1), COX-2 (Mm03294838_g1), CFD (Mm01143935_g1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Mm99999915_g1 and Hs02786624_g1), and HuR (ELAVL1; Mm00516011_m1 and Hs00171309_m1).
Histology and immunostaining.
Pancreata were fixed in 10% formalin for 2 h and processed for paraffin embedding. Four-micrometer sections were deparaffinized, and hematoxylin and eosin staining and immunostaining were performed. Prior to immunostaining, antigen retrieval was performed by steam heating for 30 min in citrate buffer, followed by endogenous peroxidase quenching with 3% H2O2–methanol for 20 min. After blocking, sections were incubated with primary antibody overnight at 4°C and then biotinylated secondary antibody was added for 30 min at room temperature. Signals were amplified and visualized either using the TSA-plus fluorescence system (PerkinElmer), according to the manufacturer's instructions, or using the VECTASTAIN elite ABC avidin/biotin complex kit (Vector Laboratories) followed by diaminobenzidine visualization and hematoxylin counterstaining. Slides were imaged with a Zeiss Axiovert 200M microscope or Zeiss Axioplan microscope. Primary antibodies included HuR MAb 19F12 (1:5,000) (Clonegene). For each genotype and age, pancreas samples from 3 to 4 mice were examined. All histological data were evaluated in a blind manner by a pathologist (W. Jian); immunostaining data were evaluated in an independent and blind manner by two individuals (W. Peng and J. A. Sawicki).
Pancreas sections also were stained at room temperature for 30 min with Alcian blue 8GX (Sigma-Aldrich), a stain for acetic mucins, followed by counterstaining with nuclear-fast red for 5 min.
Immunohistochemical analysis of HuR expression was performed using pancreas intraepithelial neoplasia, pancreatitis, and cancer tissue array BIC14011a (US Biomax Inc., Rockville, MD) that contained 48 tissue cores from chronic pancreatitis (Ch Pan), pancreatic intraepithelial neoplasia 1/2 (PanIN-1 and PanIN-2), and PDAC graded by histology. HuR immunostaining was performed using HuR 19F12 monoclonal antibody (A21277; Molecular Probes, Eugene, OR) at 0.5 μg/ml overnight at 4°C. A standard staining protocol was performed, and stained tissue sections were evaluated for intensity of staining as described previously (8, 59) using two investigators blinded to the staining procedure (V. Vishwakarma and E. Peters). For each tissue core, five different areas were evaluated for HuR staining intensity and percentage of area stained. The percentage of HuR-positive cells was scored on a scale of 0 to 4 (0 [0% positive cells], 1 [<25%], 2 [25 to 50%], 3 [50 to 75%], and 4 [>75%]). Staining intensity was scored on a scale of 0 to 3 (0 [negative], 1 [weak], 2 [moderate], and 3 [strong]). The two scores were multiplied to give an immunoreactivity score (IRS) ranging from 0 to 12.
Gene expression microarray analysis.
A protocol was adopted that avoided RNA digestion resulting from very large RNase amounts in the pancreas. Immediately following euthanasia, pancreas was injected with RNAlater (Qiagen). Following dissection, the pancreas was kept in RNAlater on ice until ready for preparation. The tissue was homogenized in 8 ml of QIAzol (Qiagen), and then 1.5 ml lysate was centrifuged (at 12,000 × g) for 5 min at 4°C. RNA was purified from 700 μl supernatant using the Qiagen total RNA extraction protocol (Qiagen). RINe values for RNA preparations were determined, and only those of sufficient quality were prepared and used for microarray analysis. cRNAs were prepared from total RNAs and then used to generate cDNAs. cDNAs were labeled using the Affymetric WT plus kit (250 ng/sample). Gene level differential expression analysis was performed on an Affymetrix mouse transcriptome array 1.0. Software used to analyze microarray data included Gene Expression Console, Transcriptome Analysis Console, and Ingenuity Pathway Analysis. Microarray data were normalized using the SST-RMA algorithm. RNAs from pancreas of TC and C mice were analyzed (3 females and 3 males for each genotype; 1 microarray/specimen).
Glucose tolerance test (GTT).
Eight-month-old male and female TC and C mice were fasted overnight before assay (n = 7 to 9 mice for each genotype/gender). Glucose (1 g/kg of body weight) was injected intraperitoneally, and blood samples from mouse tail were taken before and at 10, 20, 30, 60, 90, and 120 min after glucose injection for glucose measurement with a glucometer (Alpha TRAK2).
Insulin, amylase, lipase, triglyceride, and cholesterol assays.
Serum insulin level was measured using the mouse insulin enzyme-linked immunosorbent assay kit (Thermo Scientific). Serum triglyceride and cholesterol concentrations were assayed using L-type triglyceride M and cholesterol E kits (Wako Diagnostics). Each sample was measured in triplicate. Serum amylase and lipase assays were performed by IDEXX BioResearch (North Grafton, MA). All assays were done in triplicate.
Lipid staining.
Oil Red O staining was performed with the Pico Sirius red stain kit (Abcam).
Statistical analysis.
Differences in final blood glucose concentrations in GTTs were determined using a two-tailed Student t test. A Microsoft Excel analysis based on Reimann sums was used to approximate the area under the curve (AUC) of GTT data. Pearson's chi-square test was used to evaluate differences in facial and anal papilloma prevalence between populations of genetically different mice. HuR IRS values were plotted as groups based on tissue core pathology and compared using two-tailed Student t test.
Accession number(s).
The microarray data have been deposited in the GEO database under accession number GSE102508.
ACKNOWLEDGMENTS
We thank Lindsay Courtney for technical assistance and Lisa Laury-Kleintop and Summer Reilly for triglyceride and cholesterol analyses.
This work was supported by the W. W. Smith Charitable Foundation (J.A.S.), the Lankenau Medical Center Foundation (J.A.S.), NIH R01 CA134609 (D.A.D.), NIH/NCI Cancer Center support grant P30 CA168524 (D.A.D.), NIH 1R01CA212600-01 (J.R.B., J.A.S., and D.A.D.), and the Mary Halinski Pancreatic Cancer Research Fund (J.R.B.). Microarray analyses were supported in part by the Thomas Jefferson Cancer Center support grant 5P30CA056036-17.
J.A.S., D.A.D., and J.R.B. conceived and designed the experiments; W.P., N.F., L.A., Y.-H.H., S.Z.B., V.V., E.P., and S.A. performed the experiments; D.A.D., W.P., W.J., S.A., and J.A.S. analyzed the data; and J.A.S., D.A.D., and J.R.B. wrote the manuscript.
We have no competing financial interests to declare.
REFERENCES
- 1.Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, Meisner-Kober N, Londin ER, Rigoutsos I, Sawicki JA, Risud MV, Witkiewicz AK, McCue PA, Jiang W, Rui H, Yeo CJ, Petricoin EF, Winter JM, Brody JR. 2016. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene 35:2529–2541. doi: 10.1038/onc.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, Keen JC, Yeo CJ, Gorospe M, Brody JR. 2009. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res 69:4567–4572. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lal S, Burkhart RA, Bhattacharjee V, Beeharry N, Londin ER, Cozzitorto JA, Romeo C, Jimbo M, Norris ZA, Yeo CJ, Sawicki JA, Winter JM, Rigoutsos I, Yen TJ, Brody JR. 2014. HuR post-transcriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res 74:1128–1140. doi: 10.1158/0008-5472.CAN-13-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lal S, Zarei M, Chand SN, Dylgjeri E, Mambelli-Lisboa NC, Pishvaian MJ, Yeo CJ, Winter JM, Brody JR. 2016. Wee1 inhibition in pancreatic cancer cells is dependent on DNA repair status in a context dependent manner. Sci Rep 6:33323. doi: 10.1038/srep33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McAllister F, Pineda DM, Jimbo M, Lal S, Burkhart RA, Moughan J, Winter KA, Abdelmohsen K, Gorospe M, Acosta AdJ, Lankapalli RH, Winter JM, Yeo CJ, Witkiewicz AK, Iacobuzio-Donahue CA, Laheru D, Brody JR. 2014. dCK expression correlates with 5-fluorouracil efficacy and HuR cytoplasmic expression in pancreatic cancer. Cancer Biol Ther 15:688–698. doi: 10.4161/cbt.28413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pineda DM, Valley CC, Cozzitorto JA, Burkhart R, Leiby B, Winter JM, Weber MC, Londin ER, Rigoutsos I, Yeo CJ, Gorospe M, Brody JR. 2012. HuR's post-transcriptional regulation of death receptor 5 in pancreatic cancer cells. Cancer Biol Ther 13:946–955. doi: 10.4161/cbt.20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romeo C, Weber MC, Zarei M, DeCicco D, Chand SN, Lobo AD, Winter JM, Sawicki JA, Sachs JN, Meisner-Kober N, Yeo CJ, Vadigepalli R, Tykocinski ML, Brody JR. 2016. HuR contributes to TRAIL resistance by restricting death receptor 4 expression in pancreatic cancer cells. Mol Cancer Res 14:599–611. doi: 10.1158/1541-7786.MCR-15-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. 2009. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osera C, Martindale JL, Amadio M, Kim J, Yang X, Moad CA, Indig FE, Govoni S, Abdelmohsen K, Gorospe M, Pascale A. 2015. Induction of VEGFA mRNA translation by CoCl2 mediated by HuR. RNA Biol 12:1121–1130. doi: 10.1080/15476286.2015.1085276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Ouyang M, Rao JN, Zou T, Xiao L, Chung HK, Wu J, Donahue JM, Gorospe M, Wang JY. 2015. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell 15:1797–1810. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyana H, Jimeno A, Hong S-M, Fu B, Lin M-T, Calhoun ES, Kamiyama M, Walter K, Nikolskaua T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarei M, Lal S, Parker SJ, Nevier A, Vziri-Gohar A, Dukleska K, Mambelli-Lisboa NC, Moffat C, Bianco FF, Chand SN, Jimbo M, Cozzitorto JA, Jiang W, Yeo CJ, Londin ER, Seifert EL, Metallo CM, Brody JR, Winter JM. 2017. Posttranscriptional upregulation of IDH1 by HuR establishes a powerful survival phenotype in pancreatic cancer cells. Cancer Res 77:4460–4471. doi: 10.1158/0008-5472.CAN-17-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H-L, Telonis AG, Jing Y, Xia NL, Biederman L, Jimbo M, Blanco F, Londin ER, Brody JR, Rigoutsos I. 2016. GPRC5A is a potential oncogene in pancreatic ductal adenocarcinoma cells that is upregulated by gemcitabine with help from HuR. Cell Death Dis 7:e2294. doi: 10.1038/cddis.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu G, Dubauskaite J, Melton DA. 2002. Direct evidence for the pancreatic lineage: Ngn3+ cells are islet progenitors and are distinct from duct progenitors. Development 129:2447–2457. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. 2002. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 16.Kloppel G. 2007. Chronic pancreatitis, pseudotumors and other tumor-like lesions. Mod Pathol 20:S113–S131. doi: 10.1038/modpathol.3800690. [DOI] [PubMed] [Google Scholar]
- 17.Nata K, Liu Y, Xu L, Ikeda T, Akiyama T, Noguchi N, Kawaguchi S, Yamauchi A, Takahashi I, Shervani NJ, Onogawa T, Takawawa S, Okamoto H. 2004. Molecular cloning, expression and chromosomal localization of a novel human REG family gene, REG III. Gene 340:161–170. doi: 10.1016/j.gene.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIII gamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fantuzzi G. 2008. Adiponectin and inflammation: consensus and controversy. J Allergy Clin Immunol 121:326–330. doi: 10.1016/j.jaci.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Lo JC, Ljubicic S, Leibiger B, Kern M, Leibiger IB, Moede T, Kelly ME, Bhowmick DC, Murano I, Cohen P, Banks AS, Khandekar MJ, Dietrich A, Flier JS, Cinti S, Bluher M, Danial NN, Berggren P-O, Spiegelman BM. 2014. Adipsin is an adipokine that improves b cell function in diabetes. Cell 158:41–53. doi: 10.1016/j.cell.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A 101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinman MN, Lou H. 2008. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. 2003. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 24.Lopez de Silanes I, Lai A, Gorospe M. 2005. HuR: post-transcriptional paths to malignancy. RNA Biol 2:11–13. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- 25.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, Hruban RH, Maitra A, Kinzler K, Vogelstein B, Goggins M. 2012. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 142:730–733. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris JPT, Wang SC, Hebrok M. 2010. KRAS, Hedgehog, Wnt, and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer 10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowry AM, Tuveson DA. 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4:437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 28.Aghdassi AA, Mayerle J, Christochowitz S, Weiss FU, Sendler M, Lerch MM. 2011. Animal models for investigating chronic pancreatitis. Fibrogen Tiss Repair 4:26. doi: 10.1186/1755-1536-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lerch MM, Gorelick FS. 2013. Models of acute and chronic pancreatitis. Gastroenterology 144:1180–1193. doi: 10.1053/j.gastro.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 30.Colby JKL, Klein RD, McArthur MJ, Conti CJ, Kiguchi K, Kawamoto T, Riggs PK, Pavone AI, Sawicki J, Fischer SM. 2008. Progressive metaplastic and dysplastic changes in mouse pancreas induced by cyclooxygenase-2 overexpression. Neoplasia 10:782–796. doi: 10.1593/neo.08330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji B, Tsou T, Wang H, Gaiser S, Chang DZ, Daniluk J, Bi Y, Grote T, Longnecker DS, Logsdon CD. 2009. Ras activity levels control the development of pancreatic diseases. Gastroenterology 137:e1071–e1076. doi: 10.1053/j.gastro.2009.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iglesias M, Murga M, Laresgoiti U, Skoudy A, Bernales I, Fullaondo A, Moreno B, Lloreta J, Field SJ, Real FX, Zubiaga AM. 2004. Diabetes and exocrine pancreatic insufficiency in E2F1/E2F2 double-mutant mice. J Clin Investig 113:1398–1407. doi: 10.1172/JCI200418879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marrache F, Tu SP, Bhagat G, Pendyala S, Österreicher CH, Gordon S, Ramanathan V, Penz-Österreicher M, Benz KS, Sing Z, Wang TC. 2008. Overexpression of interleukin-1beta in the murine pancreas results in chronic pancreatitis. Gastroenterology 135:1277–1287. doi: 10.1053/j.gastro.2008.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee MS, Gu D, Feng L, Curriden S, Arnush M, Krahl T, Gurushanthaiah D, Wilson C, Loskutoff DL, Fox H, Savrvetnick N. 1995. Accumulation of extracellular matrix and deelopmental dysregulation in the pancreas by transgenic production of transforming growth factor-beta 1. Am J Pathol 147:42–52. [PMC free article] [PubMed] [Google Scholar]
- 35.Lampel M, Kern H. 1977. Acute interstitial pancreatitis in the rat induced by excessive doses of pancreatic secretagogue. Virchows Arch A Pathol Anat Histol 373:97–117. doi: 10.1007/BF00432156. [DOI] [PubMed] [Google Scholar]
- 36.Niederau C, Ferrell CD, Grendell JH. 1985. Caerulein-induced acute necrotizing pancreatitis in mice: protective effects of proglumide, benzotript, and secretin. Gastroenterology 88:1192–1204. doi: 10.1016/S0016-5085(85)80079-2. [DOI] [PubMed] [Google Scholar]
- 37.Neuschwander-Tetri BA, Bridele KR, Wells LD, Marcu M, Ramm GA. 2000. Repetitive acute pancreatic injury in the mouse induces procollagen alpha1. (l) expression colocalized to pancreatic stellate cells. Lab Investig 80:143–150. doi: 10.1038/labinvest.3780018. [DOI] [PubMed] [Google Scholar]
- 38.Srikantan S, Gorospe M. 2012. HuR function in disease. Front Biosci 17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duell EJ, Lucenteforte E, Olson SH, Bracci PM, Lim D, Risch HA, Silverman DT, Ji BT, Gallinger S, Holly EA, Fontham EH, Maisonneuve P, Bueno-de-Mesquita HB, Ghadiarian P, Kurtz RC, Ludwig E, Yu H, Lowenfels AB, Seminara D, Petersen GM, La Vecchia C, Boffetta P. 2012. Pancreatitis and pancreatic cancer risk: a pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4). Ann Oncol 23:2964–2970. doi: 10.1093/annonc/mds140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamm BH. 1984. Incidence and diagnostic significance of minor pathologic changes in the adult pancreas at autopsy: a systematic study of 112 autopsies in patients without known pancreatic disease. Hum Pathol 15:677–683. doi: 10.1016/S0046-8177(84)80294-4. [DOI] [PubMed] [Google Scholar]
- 41.Ferrone M, Raimondo M, Scolapio JS. 2007. Pancreatic enzyme pharmacotherapy. Pharmacotherapy 27:910–920. doi: 10.1592/phco.27.6.910. [DOI] [PubMed] [Google Scholar]
- 42.DiMagno EP, Go VL, Summerskill WH. 1973. Relations between pancreatic enzyme outputs and malabsorption in sever pancreatic insufficiency. N Engl J Med 288:813–815. doi: 10.1056/NEJM197304192881603. [DOI] [PubMed] [Google Scholar]
- 43.Tietz NW, Shuey DF. 1993. Lipase in serum–the elusive enzyme: an overview. Clin Chem 39:746–756. [PubMed] [Google Scholar]
- 44.Kolodecik T, Shugrue C, Ashat M, Thrower EC. 2014. Risk factors for pancreatic cancer: underlying mechanisms and potential targets. Front Physiol 4:415. doi: 10.3389/fphys.2013.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andren-Sandberg A, Domellof L. 1993. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med 328:1433–1437. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 46.Malka D, Hammel P, Maire F, Rufat P, Madeira I, Pessione F, Levy P, Ruszniewski P. 2002. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut 51:849–852. doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raimondi S, Lowenfels AB, Morselli-Labate AM, Maisonneuve P, Pezzilli R. 2010. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Prac Res Clin Gastroenterol 24:349–358. doi: 10.1016/j.bpg.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Whitcomb DC, Pogue-Geile K. 2002. Pancreatitis as a risk for pancreatic cancer. Gastroenterol Clin N Am 31:663–678. doi: 10.1016/S0889-8553(02)00004-3. [DOI] [PubMed] [Google Scholar]
- 49.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubas P, Sandgren EP, Barbacid M. 2007. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Kim M, Kogan N, Slack FJ. 2016. Cis-acting elements in its 3′UTR mediate post-transcriptional regulation of KRAS. Oncotarget 7:11770–11784. doi: 10.18632/oncotarget.7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zyromski NJ, Mathur A, Pitt HA, Wade TE, Wang S, Nakshatri P, Swartz-Basile DA, Nakshatri H. 2009. Obesity potentiates growth and dissemination of pancreatic cancer. Surgery 146:258–263. doi: 10.1016/j.surg.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 52.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. 2001. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Investig 108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, Tian YX, Ba X, Boldogh I, Zeng X. 2017. PARP1 promotes gene expression at the post-transcriptional level by modulating the RNA-binding protein HuR. Nat Commun 8:14632. doi: 10.1038/ncomms14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Labrie M, Lalonde S, Najyb O, Thiery M, Daneault C, Des Rosiers C, Rassart E, Mounier C. 2014. Apolipoprotein D transgenic mice develop hepatic steatosis through activation of PPARg and fatty acid uptake. PLoS One 10:e0130230. doi: 10.1371/journal.pone.0130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basheer WA, Harris BS, Mentrup HL, Abreha M, Thames EL, Lea JB, Swing DA, Copeland NG, Jenkins NA, Price RL, Matesic LE. 2015. Cardiomyocyte-specific overexpression of the ubiquitin ligase Wwp1 contributes to reduction in connexin 43 and arrhythmogenesis. J Mol Cell Cardiol 88:1–13. doi: 10.1016/j.yjmcc.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jimbo M, Blanco FF, Huang Y-H, Telonis AG, Screnci BA, Cosma GL, Alexeev V, Gonye GE, Yeo CJ, Sawicki JA, Winter JM, Brody JR. 2015. Targeting the mRNA-binding protien HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget 6:27312–27331. doi: 10.18632/oncotarget.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cozzitorto JA, Jimbo M, Chand SN, Blanco F, Lal S, Gilbert M, Winter JM, Gorospe M, Brody JR. 2015. Studying RNA-binding protein interactions with target mRNAs in eukaryotic cells: native ribonucleoprotein immunopreciptation (RIP) assays). Methods Mol Biol 1262:239–246. doi: 10.1007/978-1-4939-2253-6_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang Y-H, Peng W, Furuuchi N, DuHadaway JB, Jimbo M, Pirritano A, Dunton CJ, Daum GS, Leiby BE, Brody JR, Sawicki JA. 2016. Insights from HuR biology point to potential improvement for second-line ovarian cancer therapy. Oncotarget 7:21812–21824. doi: 10.18632/oncotarget.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denkert C, Koch I, von Keyserlingk N, Noske A, Niesporek S, Dietel M, Weichert W. 2006. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol 9:1261–1269. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- 60.Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC, Romeo C, Stout MC, Londin ER, Goetz A, Lowder CY, Nevier A, Yeo CJ, Campbell PM, Winter JM, Dixon DA, Brody JR. 2017. CRISPR knockout of the HuR gene causes a xenograft lethal phenotype. Mol Cancer Res 15:696–707. doi: 10.1158/1541-7786.MCR-16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]