ABSTRACT
Secondary metabolites are synthesized by many microorganisms and provide a fitness benefit in the presence of competitors and predators. Secondary metabolism also can be costly, as it shunts energy and intermediates from primary metabolism. In Pseudomonas spp., secondary metabolism is controlled by the GacS-GacA global regulatory system. Intriguingly, spontaneous mutations in gacS or gacA (Gac− mutants) are commonly observed in laboratory cultures. Here we investigated the role of secondary metabolism in the accumulation of Gac− mutants in Pseudomonas protegens strain Pf-5. Our results showed that secondary metabolism, specifically biosynthesis of the antimicrobial compound pyoluteorin, contributes significantly to the accumulation of Gac− mutants. Pyoluteorin biosynthesis, which poses a metabolic burden on the producer cells, but not pyoluteorin itself, leads to the accumulation of the spontaneous mutants. Interspecific competition also influenced the accumulation of the Gac− mutants: a reduced proportion of Gac− mutants accumulated when P. protegens Pf-5 was cocultured with Bacillus subtilis than in pure cultures of strain Pf-5. Overall, our study associated a fitness trade-off with secondary metabolism, with metabolic costs versus competitive benefits of production influencing the evolution of P. protegens, assessed by the accumulation of Gac− mutants.
KEYWORDS: interspecific competition, Pseudomonas, secondary metabolism, spontaneous mutations, GacS-GacA
IMPORTANCE
Many microorganisms produce antibiotics, which contribute to ecologic fitness in natural environments where microbes constantly compete for resources with other organisms. However, biosynthesis of antibiotics is costly due to the metabolic burdens of the antibiotic-producing microorganism. Our results provide an example of the fitness trade-off associated with antibiotic production. Under noncompetitive conditions, antibiotic biosynthesis led to accumulation of spontaneous mutants lacking a master regulator of antibiotic production. However, relatively few of these spontaneous mutants accumulated when a competitor was present. Results from this work provide information on the evolution of antibiotic biosynthesis and provide a framework for their discovery and regulation.
INTRODUCTION
Secondary metabolites play important roles in physiological adaptation and ecologic fitness of the organisms producing secondary metabolites (1, 2). These metabolites vary widely in structure and biological activity, with some having valuable pharmaceutical and agricultural applications (3, 4). Diverse classes of secondary metabolites are produced by plants, animals, and microorganisms, but knowledge of the natural functions of secondary metabolites is still limited. Secondary metabolites are thought to represent important adaptive characters of the organisms producing secondary metabolites in their native environments (2, 5). For example, a secondary metabolite with antimicrobial activity may allow organisms to inhibit competitors that occupy the same niches (6). On the other hand, secondary metabolism poses metabolic burdens to the producer by diverting precursors, cofactors, and energy from primary metabolism (7). Consequently, unnecessary expression of secondary metabolites can be costly to organisms producing secondary metabolites. To balance the benefits and costs, secondary metabolism is controlled by complex regulatory networks that regulate production in response to environmental factors and physiological status of the organism producing secondary metabolites (8, 9). These networks involve both global and pathway-specific regulators operating to coordinate secondary metabolite biosynthesis pathways at transcriptional and posttranscriptional levels (10–12).
The production of a large spectrum of secondary metabolites is a characteristic of bacteria in the genus Pseudomonas (13). Secondary metabolism in Pseudomonas spp. requires the GacS-GacA two-component regulatory system, composed of the sensor kinase GacS and the cognate cellular response regulator GacA (14). In the presence of an unknown signal(s), the sensor kinase GacS autophosphorylates and transfers a phosphate group to the response regulator GacA, which governs a complex signal transduction pathway involving regulatory RNAs and translational repression (14). The GacS-GacA system controls the expression of hundreds of genes, including those required for the production of secondary metabolites and extracellular enzymes (15–19). Accordingly, mutation of gacA and/or gacS has pleiotropic effects on the bacterial cell, influencing motility, iron acquisition, biofilm formation, aspects of primary metabolism, and many other phenotypes (15, 16, 20). Given the broad influence of the GacS-GacA system on bacterial physiology, it is striking that spontaneous gacS and gacA mutants (i.e., Gac− mutants) emerge and accumulate in cultures of Pseudomonas spp. (17, 21–27). Colonies of these mutants can be distinguished from those of the wild type visually by their expanded colony size, flattened appearance, increased fluorescence under UV light, and lack of extracellular protease production (21, 24).
In this study, we compared several species of Pseudomonas spp. for accumulation of spontaneous Gac− mutants in a nutrient-rich broth medium. One strain, Pseudomonas protegens Pf-5, was selected for further studies. Strain Pf-5 is a soil bacterium known for its capacity to inhibit the growth of phytopathogenic bacteria and fungi, suppress plant diseases, and kill certain insects through production of a large spectrum of secondary metabolites and toxins (10, 28–32). Here, we evaluated the role of secondary metabolism in the relative fitness of Gac− mutants versus wild-type Pf-5 in culture. We found that secondary metabolism contributes to the evolution of P. protegens Pf-5 in culture, assessed as the proportion of spontaneous mutants in this important regulatory system. More importantly, the accumulation of Gac− mutants was significantly affected by both biotic and abiotic environmental factors, including medium composition and interspecific competition, suggesting that spontaneous mutation of gacS or gacA can be an approach by which bacteria adapt to different environmental conditions.
RESULTS
P. protegens Pf-5 accumulates a high proportion of spontaneous Gac− mutants in culture.
Certain strains of Pseudomonas spp. are known to accumulate spontaneous Gac− mutants in culture. We compared the frequency with which spontaneous Gac− mutants accumulate in four species of Pseudomonas, including Pseudomonas protegens Pf-5, Pseudomonas fluorescens SS101, Pseudomonas aeruginosa PAO1, and Pseudomonas chlororaphis 30-84. These strains were cultured in NYB (nutrient broth supplemented with 0.5% yeast extract), a medium in which P. protegens CHA0 was previously shown to accumulate Gac− mutants (21). Bacterial cultures were diluted and spread on LMA (litmus milk agar) plates at 6 days postinoculation (dpi) and assessed for exoprotease activity. Production of exoprotease requires the GacS-GacA system (33) and is a readily visualized phenotype (27) that has been used reliably to estimate the proportion of Gac−mutants in cultures of P. protegens Pf-5 (17). As such, colonies deficient in exoprotease activity are referred to as Gac− mutants throughout this report.
All four tested strains grew well in NYB medium and accumulated spontaneous Gac− mutants by 6 dpi (Fig. 1). However, large variations in the proportion of spontaneous Gac− mutants that accumulated in cultures were observed among the four evaluated strains. Strains SS101 and PAO1 accumulated spontaneous Gac− mutants to a level of <2% of the total CFU in the culture. Strain 30-84 was more variable between experimental replicates, but on average, cultures were composed of <5% Gac− mutants. P. protegens Pf-5 was strikingly different, with 99% of total CFU on average being spontaneous Gac− mutants. Due to the high proportion of Gac− mutants in cultures of strain Pf-5, we focused on this strain to identify possible causes for the accumulation of mutants.
FIG 1 .
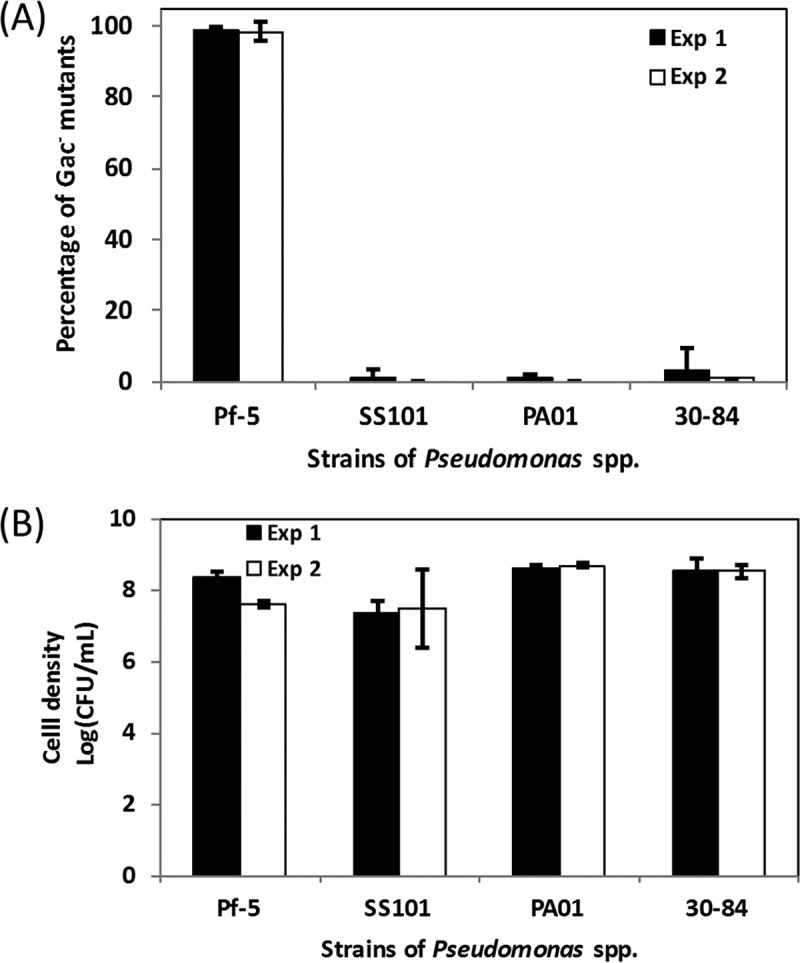
Accumulation of spontaneous Gac− mutants of Pseudomonas spp. in NYB cultures. (A) Four strains, P. protegens Pf-5, P. fluorescens SS101, P. aeruginosa PAO1, and P. chlororaphis 30-84, were grown independently in NYB for 6 days, and dilutions of cultures were spread on LMA to assess exoprotease production. The percentage of the Gac− mutants in the bacterial population was estimated from counts of exoprotease-deficient colonies relative to total colonies on LMA. At least 400 colonies were screened for each strain. The results of two experiments are shown. (B) Total cell density of the four strains in NYB cultures at 6 dpi. For both panels A and B, results are from two independent experiments (experiment 1 [Exp 1] and Exp 2). Each experiment had at least three replicates. Means ± standard deviations (error bars) are shown.
To understand the rate at which Gac−mutants accumulate in cultures of strain Pf-5, we estimated the proportion of mutants daily over 6 days. No Gac− mutant was observed at 1 dpi, whereas 2% of the population were Gac− mutants at 2 dpi (Fig. 2A). The population size of the wild type decreased after 1 dpi, while the population size of Gac− mutants increased gradually between 1 and 4 dpi (Fig. 2B). At 4 dpi, the Gac− mutant population size also began to decline, but the population size of the wild type declined at a higher rate than the Gac− mutants. By 5 dpi, the Gac− mutants dominated the cultures (94% on average).
FIG 2 .
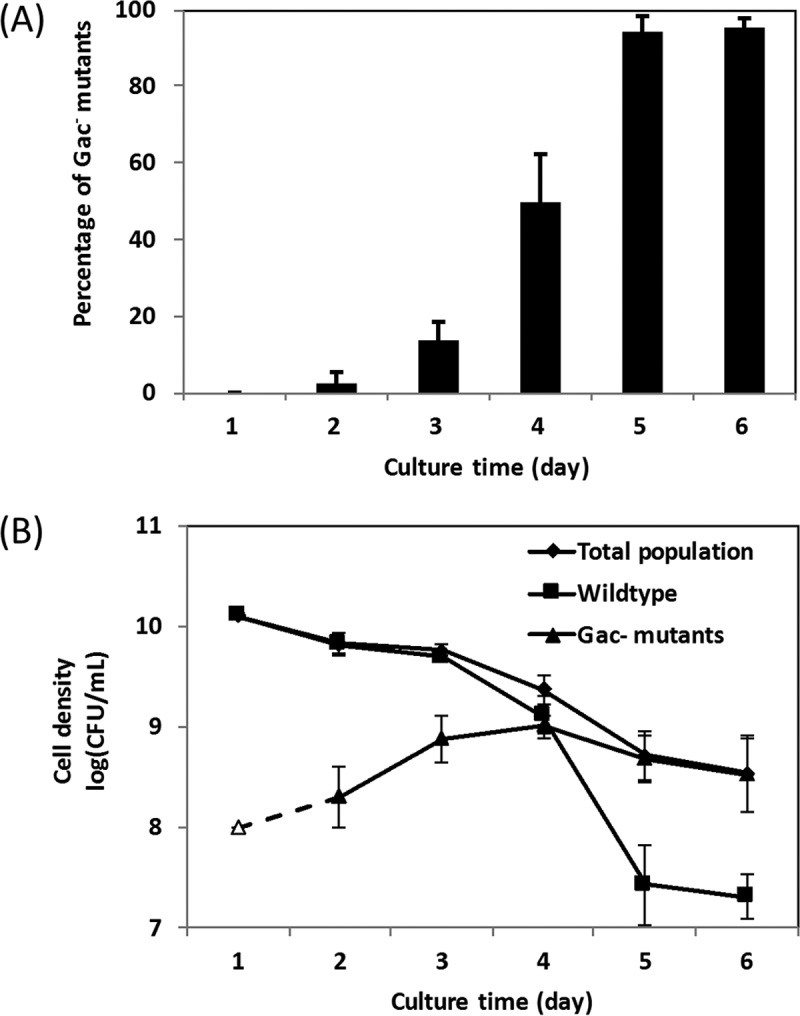
Accumulation of spontaneous Gac− mutants in NYB cultures of P. protegens Pf-5 over time. Wild-type P. protegens Pf-5 was cultured in NYB for 6 dpi. (A and B) The percentage of spontaneous Gac− mutants in the bacterial population (A) and the bacterial cell density (B) were recorded daily. In panel B, the cell density of Gac− mutants at 1 dpi is shown as an open triangle, which represents the detection limit, as no Gac− mutant was detected by screening more than 400 colonies. Each experiment had at least three replicates. Means ± standard errors (error bars) are shown.
Secondary metabolism affects the accumulation of spontaneous Gac− mutants.
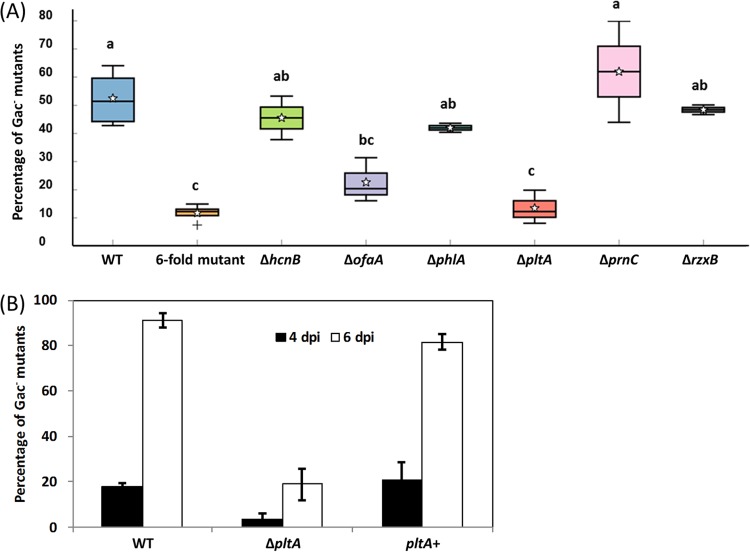
A prominent feature of P. protegens Pf-5 is the production of a large number of secondary metabolites (32, 34). Importantly, the production of these secondary metabolites is known to be positively regulated by the GacS-GacA system (15). We hypothesized that secondary metabolism contributes to the accumulation of spontaneous Gac− mutants. To test this hypothesis, we evaluated the accumulation of Gac− mutants of a Pf-5 derivative strain that contains mutations in hcnB, ofaA, phlA, pltA, prnC, and rzxB (referred to as the 6-fold mutant hereafter) (35). This 6-fold mutant does not produce any of the six secondary metabolites: hydrogen cyanide, orfamide A, 2,4-diacetylphloroglucinol, pyoluteorin, pyrrolnitrin, or rhizoxin (35). After 6 days of growth, the proportion of Gac− mutants was significantly lower in cultures of this 6-fold mutant relative to wild-type Pf-5 (Fig. 3A), which supports our hypothesis that secondary metabolism plays a role in the accumulation of spontaneous Gac− mutants in strain Pf-5.
FIG 3 .
Accumulation of spontaneous Gac− mutants of P. protegens Pf-5 and its secondary metabolism knockout mutants. Wild-type P. protegens Pf-5 and derivatives that contain mutations in the biosynthetic genes for six secondary metabolites (A) and the complemented ΔpltA mutant (B) were cultured in NYB. The percentage of Gac− mutants in the bacterial population was calculated at 4 dpi (A and B) and 6 dpi (B). WT, wild-type Pf-5; 6-fold mutant, Pf-5 mutant containing mutations in prnC, rzxB, pltA, hcnB, ofaA, and phlA; pltA+, ΔpltA complemented strain. Each experiment had at least three replicates. In panel A, different letters above the boxes indicate that the mean values differ significantly (P < 0.05) according to Tukey’s pairwise comparison test. In panel B, means ± standard errors are shown.
To pinpoint the biosynthetic pathway(s) that influences the accumulation of spontaneous Gac− mutants of P. protegens Pf-5, we evaluated six independent mutants, each containing a single mutation in a biosynthetic gene for one of the six secondary metabolites. Of the six mutants, only the ΔpltA mutants and ΔofaA mutants, which are unable to produce pyoluteorin and orfamide A, respectively, significantly differed from wild-type Pf-5 in the proportion of Gac− mutants that accumulated in culture (Fig. 3A). The proportions of Gac− mutants in cultures of wild-type Pf-5 and ΔpltA and ΔofaA mutants were also assessed in another independent experiment, in which the production of secondary metabolites was quantified over time (see Fig. S1 in the supplemental material). Again, cultures of the ΔpltA and ΔofaA mutants accumulated a smaller proportion of Gac− mutants than a culture of the wild type in NYB medium. Results also confirmed that strain Pf-5 produced pyoluteorin and orfamide A in the NYB cultures, whereas the ΔpltA and ΔofaA mutants did not produce pyoluteorin and orfamide A, respectively. Additionally, 2,4-diacetylphloroglucinol and rhizoxin were not detected in the cultures of wild-type Pf-5, the ΔpltA mutant, or the ΔofaA mutant, which is consistent with results that the ΔphlA and ΔrzxB mutants accumulated proportions of Gac− mutants similar to those accumulated by the wild type when cultured in NYB medium (Fig. 3A). We also noticed that pyrrolnitrin was produced by Pf-5 in NYB cultures (Fig. S1), but the ΔprnC mutant did not differ significantly from the wild type in the accumulation of Gac− mutants (Fig. 3A). Therefore, it appears that the biosynthesis of some, but not all, secondary metabolites influences the accumulation of Gac− mutants of Pf-5.
(A to C) Production of pyoluteorin (A), orfamide A (B), or pyrrolnitrin (C) by P. protegens Pf-5 and derivative strains (ΔpltA and ΔofaA mutants) in NYB cultures over time. (D) Accumulation of Gac− mutants in NYB cultures at 4 dpi. Values are the means of three replicate cultures, and error bars show the standard errors. Download FIG S1, TIF file, 0.3 MB (301.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Most spontaneous Gac− mutants have point mutations in gacS or gacA.
We sequenced the gacS and gacA alleles from Gac− mutants that accumulated in cultures of the following four strains: wild-type Pf-5, ΔpltA mutant, ΔofaA mutant, and 6-fold mutant. A total of 24 exoprotease-deficient colonies were evaluated: two colonies were randomly selected from LMA plated with three independent populations of each strain, for a total of six colonies per strain. Of the 24 exoprotease-deficient colonies, 23 have a single missense or nonsense mutation, an insertion, or a short deletion in the open reading frame (ORF) or ribosome binding site (RBS) of gacA or gacS (Table 1). This result substantiates that counting colonies lacking exoprotease production provided an accurate estimate of the number of gacS and gacA mutants in a culture of P. protegens Pf-5, as shown earlier (17). The mutations of 23 colonies mapped to 17 different sites of gacS or gacA. Ten colonies were affected in gacA, with nine having a point mutation in the ORF, and one having a point mutation in the predicted RBS. Thirteen colonies have mutations in gacS, of which two have small deletions that cause a frameshift and one has a single-nucleotide insertion that introduces a stop codon (Table 1). Two colonies were sampled from each of the 12 cultures of this experiment; 3 of the 12 pairs of colonies had identical mutant alleles and are probably siblings (Table 1). The observation that most pairs from the same population have different mutations, and the identification of 17 different mutation sites show that different mutants were selected within populations over the course of the experiment.
TABLE 1 .
Sites of mutations in spontaneous Gac− mutants sampled from cultures of wild-type P. protegens Pf-5 and derivativesa
| Strain | Replicate | Colony |
gacSb |
gacAb |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1A | T254S | T307P | A447V | A463D | L476F | Q727* | 2419^2420 insG* | Δ500-504 | Δ2422-2426 | RBS | V4E | V4G | I29N | T103I | Q155R | K190R | |||
| Pf-5 | 1 | 1 | X | ||||||||||||||||
| 1 | 2 | X | |||||||||||||||||
| 2 | 1 | X | |||||||||||||||||
| 2 | 2 | X | |||||||||||||||||
| 3 | 1 | X | |||||||||||||||||
| 3 | 2 | X | |||||||||||||||||
| ΔofaA | 1 | 1 | X | ||||||||||||||||
| 1 | 2 | X | |||||||||||||||||
| 2 | 1c | ||||||||||||||||||
| 2 | 2 | X | |||||||||||||||||
| 3 | 1 | X | |||||||||||||||||
| 3 | 2 | X | |||||||||||||||||
| ΔpltA | 1 | 1 | X | ||||||||||||||||
| 1 | 2 | X | |||||||||||||||||
| 2 | 1 | X | |||||||||||||||||
| 2 | 2 | X | |||||||||||||||||
| 3 | 1 | X | |||||||||||||||||
| 3 | 2 | X | |||||||||||||||||
| LK147 | 1 | 1 | X | ||||||||||||||||
| 1 | 2 | X | |||||||||||||||||
| 2 | 1 | X | |||||||||||||||||
| 2 | 2 | X | |||||||||||||||||
| 3 | 1 | X | |||||||||||||||||
| 3 | 2 | X | |||||||||||||||||
P. protegens Pf-5 and derivatives (ΔofaA, ΔpltA, and the 6-fold mutant LK147) were each grown in three replicate flasks containing NYB for 4 days, dilutions of the cultures were spread on LMA, and two exoprotease-deficient colonies were selected from each replicate. gacS and gacA were PCR amplified and sequenced to map mutations.
Abbreviations of amino acids show the results of point mutations in the gacS or gacA gene. RBS indicates a mutation in the predicted ribosome binding site. An asterisk indicates a stop codon introduced by a point mutation or by an insertion.
No mutation in gacS or gacA was found in this isolate.
Biosynthesis of pyoluteorin contributes to the accumulation of spontaneous Gac− mutants.
To further investigate the relationship of secondary metabolism to the accumulation of spontaneous Gac− mutants, we focused on the pyoluteorin biosynthetic pathway because it had the greatest effect on the accumulation of Gac− mutants in our experiments (Fig. 3A). To confirm the role of pyoluteorin biosynthesis in the accumulation of spontaneous Gac− mutants, we complemented the ΔpltA mutant with a wild-type pltA gene. Data showed that the complemented mutant accumulated spontaneous Gac− mutants at a level similar to that of the wild type (Fig. 3B). We also evaluated cultures of P. protegens Pf-5 growing in NBG (nutrient broth supplemented with 2% glucose). This medium supports bacterial growth but not pyoluteorin biosynthesis by strain Pf-5 (36). No spontaneous Gac− mutant was detected when strain Pf-5 was cultured in NBG (193, 155, and 148 colonies were screened in three independent cultures), which further supports the conclusion that pyoluteorin biosynthesis contributes to accumulation of spontaneous Gac− mutants of Pf-5.
Accumulation of spontaneous Gac− mutants is associated with the expression of pyoluteorin biosynthetic genes, but not the product pyoluteorin.
We then hypothesized that the presence of the compound pyoluteorin increased the accumulation of Gac− mutants in cultures of P. protegens Pf-5. To test this hypothesis, we cultured the ΔpltA mutant in NYB broth spiked with different concentrations of purified pyoluteorin. Our data showed that pyoluteorin did not increase the proportion of Gac− mutants in cultures of the ΔpltA mutant (Fig. 4). To the contrary, the proportion of Gac− mutants decreased with the increased concentration of pyoluteorin added to the ΔpltA mutant cultures. The addition of pyoluteorin had no significant effect on the growth of the ΔpltA mutant (Fig. 4B). Because pyoluteorin is present in cultures of wild-type Pf-5 grown in NYB (Fig. S2), reduced sensitivity of Gac− mutants to pyoluteorin could enhance their fitness relative to the wild-type strain in this medium. To test this possibility, we compared the ΔgacA mutant to the wild-type Pf-5 in their sensitivity to pyoluteorin. No significant difference was observed between the ΔgacA mutant and the wild-type Pf-5 in sensitivity to pyoluteorin either in NYB broth or on NYA (NYB with 1.5% agar) plates (Fig. S3). Furthermore, the addition of pyoluteorin to the wild-type Pf-5 cultures also resulted in a decreased proportion of Gac− mutants (Fig. 4C). These data indicate that pyoluteorin, the product of the biosynthesis pathway, is not responsible for the accumulation of Gac− mutants in cultures of Pf-5.
FIG 4 .
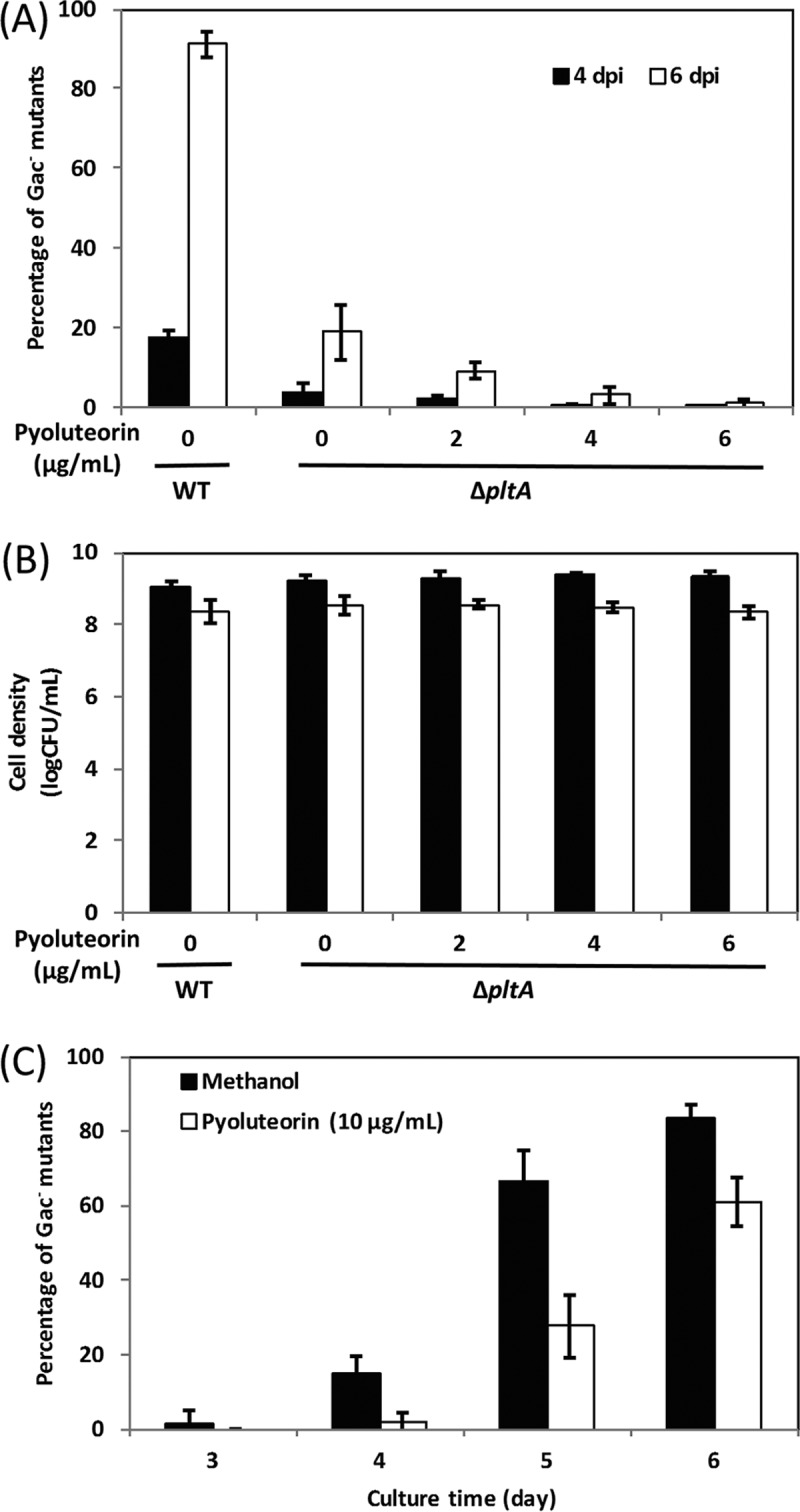
Influence of purified pyoluteorin on accumulation of Gac− mutants of P. protegens Pf-5. Wild-type Pf-5 (WT) and the ΔpltA mutant were cultured in NYB with or without the addition of purified pyoluteorin at different concentrations. (A and B) The percentage of Gac− mutants in the bacterial population (A) and the density of the total bacterial population (B) were recorded at 4 dpi and 6 dpi. (C) Wild-type Pf-5 was cultured in NYB with the addition of purified pyoluteorin at 10 μg/ml compared to the equivalent methanol control. The percentage of Gac− mutants was estimated at different time points. Each experiment had least three replicates. Means ± standard deviations are shown.
Concentrations of pyoluteorin produced by P. protegens Pf-5 in NYB over time. Values are the means of three replicate cultures, and error bars show the standard deviations. Download FIG S2, TIF file, 0.1 MB (66.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Influence of pyoluteorin on the growth of wild-type P. protegens Pf-5 and the ΔgacA mutant. (A) Sterilized filter papers containing 3 µl of different concentrations of pyoluteorin were placed on the top of NYA seeded with wild-type Pf-5 or the ΔgacA mutant. The results were imaged at 24 h after inoculation. (B) Wild-type Pf-5 and the ΔgacA mutant were cultured in a 96-well plate with wells containing NYG and pyoluteorin at the indicated concentrations. The plate was incubated at 27°C with shaking at 200 rpm. The cell density (OD600) was measured at 20 h after inoculation. The experiment had three replicates. Means ± standard deviations are shown. Download FIG S3, TIF file, 0.2 MB (262KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
These results (showing that deletion of pltA led to a decreased accumulation of Gac− mutants, whereas addition of pyoluteorin failed to restore the proportion of mutants to the wild-type Pf-5 level) imply that the process and the product of pyoluteorin biosynthesis contribute differentially to the accumulation of Gac− mutants. Therefore, we hypothesized that the differential expression of pyoluteorin biosynthetic genes in the wild-type Pf-5 and the ΔpltA mutant drives the selection favoring Gac− mutants. To test this hypothesis, a double mutant, the pltR* ΔpltA mutant, was constructed. The pltR gene encodes a positive transcriptional regulator of pyoluteorin biosynthesis genes, and the pltR* allele is an engineered variant in which all rare codons are substituted with optimized codons that increase its expression as well as that of its regulon (11). As expected, the promoter activity of pltL, the first gene in the plt biosynthetic gene cluster, increased significantly in the pltR* ΔpltA mutant versus the ΔpltA mutant (Fig. 5A). More importantly, the proportion of Gac− mutants accumulated in cultures of the pltR* ΔpltA mutant was significantly higher than that of the ΔpltA mutant (Fig. 5B), indicating that overexpression of pyoluteorin biosynthetic genes increased the accumulation of spontaneous Gac− mutations.
FIG 5 .
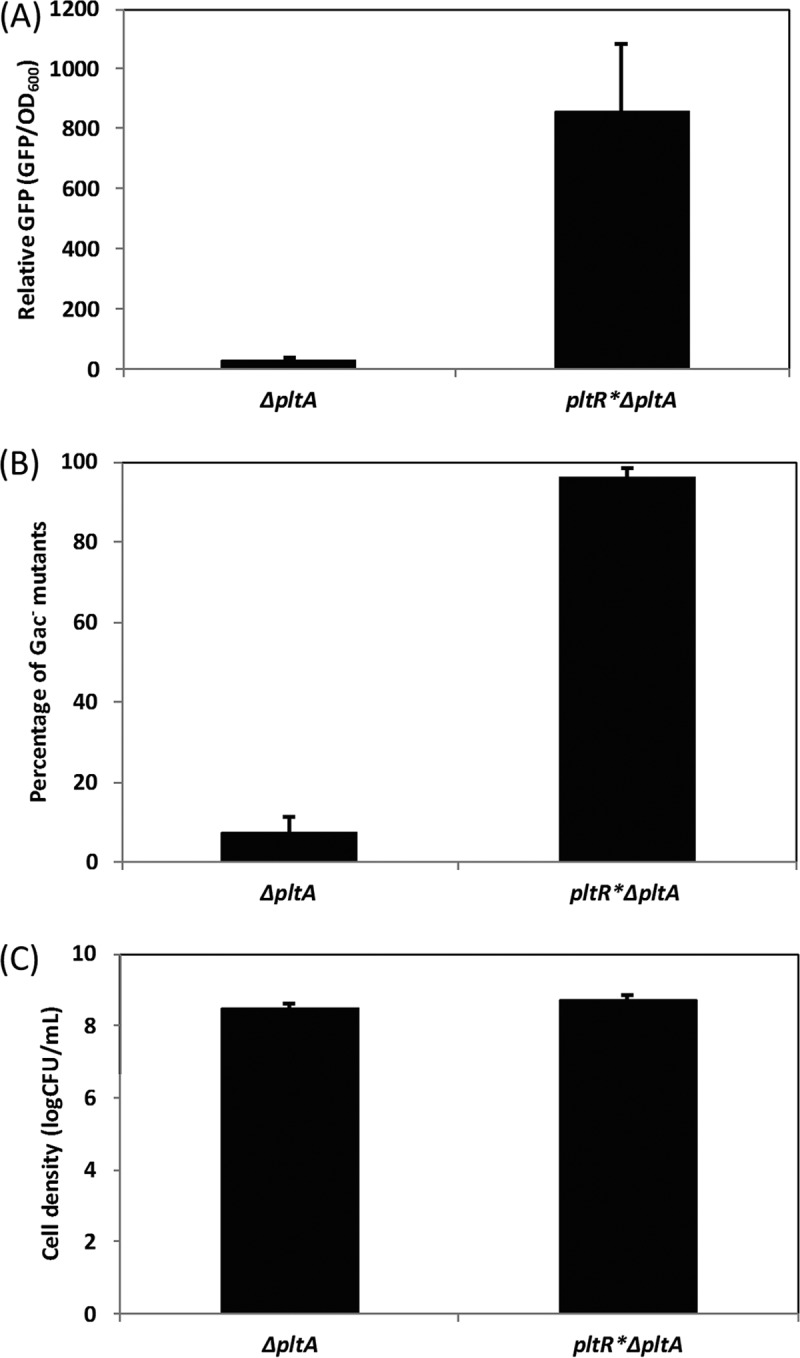
Effect of overexpression of the pyoluteorin biosynthetic genes on the accumulation of spontaneous Gac− mutants of P. protegens Pf-5. P. protegens Pf-5 derivative ΔpltA and pltR* ΔpltA strains were cultured in NYB. To test the expression level of pyoluteorin biosynthetic genes, plasmid ppltL-gfp containing a transcriptional fusion pltL::gfp, in which the promoter of pltL was fused with a promoterless gfp (11), was transformed into both strains, and the GFP level was assayed at 1 dpi (A). The accumulation of Gac− mutants (B) and the total bacterial cell density (C) were recorded at 4 dpi. Each experiment had at least three replicates. Means ± standard deviations are shown.
Collectively, our results suggest that the expression of pyoluteorin biosynthetic genes, but not the pyoluteorin product, contributes to accumulation of spontaneous Gac− mutants in strain Pf-5.
Interspecific competition reduced the accumulation of spontaneous Gac− mutants.
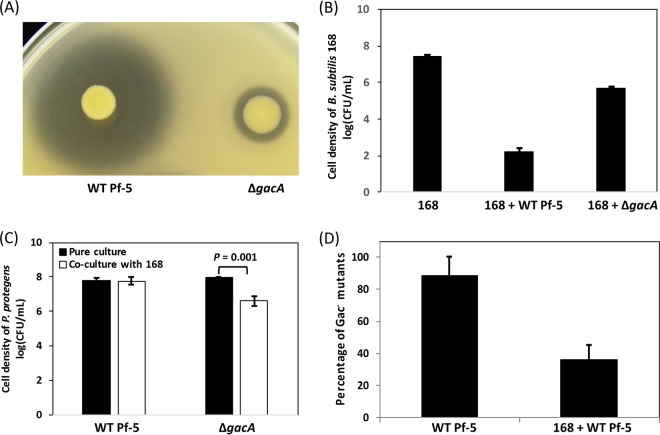
The high numbers of spontaneous Gac− mutants in P. protegens Pf-5 cultures prompted us to ask the question: does competitive pressure affect the accumulation of spontaneous Gac− mutants? Bacillus spp. and Pseudomonas spp. are commonly identified from the same environments, and secondary metabolites are known to mediate interactions between these bacteria (37). To test whether interspecific competition occurs between P. protegens Pf-5 and Bacillus subtilis strain 168, these two strains were cocultured on NYA and NYB. Our results showed that wild-type P. protegens Pf-5 strongly inhibited the growth of B. subtilis 168 in both solid and broth cultures (Fig. 6A and B). Importantly, a functional GacS-GacA system is essential for P. protegens Pf-5 to inhibit B. subtilis strain 168, as indicated by a strong inhibitory effect of the wild-type Pf-5, but a much lower effect of the ΔgacA mutant, on the growth of B. subtilis 168 (Fig. 6A and B). The role of the GacS-GacA system in the interspecific competition is further validated by the result that, relative to the single culture, the ΔgacA mutant had reduced survival in cocultures with B. subtilis 168 (Fig. 6C). However, coculturing with B. subtilis 168 had no significant effect on the growth of wild-type Pf-5 (Fig. 6C).
FIG 6 .
Influence of interspecific competition on the accumulation of spontaneous Gac− mutants of P. protegens Pf-5. Different inhibitory effects of the wild-type Pf-5 strain and the ΔgacA mutant of strain Pf-5 on the growth of B. subtilis strain 168 on NYA (A) and NYB (B). In panel B, strain 168 was inoculated in NYB or coinoculated with Pf-5 strains with an initial ratio 1:1 in NYB, and the cell density of strain 168 was tested at 6 dpi. (C) Cell density of P. protegens Pf-5 and the ΔgacA mutant grown in pure culture or in coculture with B. subtilis strain 168 in NYB at 6 dpi. P value was calculated by a Student’s t test. (D) The percentage of Gac− mutants that accumulated in the pure culture of wild-type Pf-5 or the coculture of wild-type Pf-5 and strain 168 in NYB at 6 dpi is shown. Each experiment had at least three replicates. Means ± standard deviations are shown.
We then compared the proportion of spontaneous Gac− mutants that accumulated in pure cultures started with P. protegens Pf-5 to those in the cocultures started with equal densities of P. protegens Pf-5 and B. subtilis 168 to test whether the presence of a competitor influenced the accumulation of the mutants. Our data showed that coculturing with B. subtilis 168 reduced the proportion of Gac− mutants of strain Pf-5 (36% on average) relative to pure cultures of strain Pf-5 (88% on average) (Fig. 6D).
Overall, these data show that a functional GacS-GacA system is important for strain Pf-5 to compete with B. subtilis and that interspecies competition led to a decreased accumulation of spontaneous Gac− mutants of P. protegens Pf-5.
DISCUSSION
It has been known for decades that spontaneous Gac− mutants accumulate in nutrient-rich cultures of some Pseudomonas spp. (17, 21, 22, 25, 26, 38–41), but the reasons for this phenomenon have remained obscure. In this study, we focused on P. protegens Pf-5, which accumulated a very high proportion of Gac− mutants in NYB, outnumbering wild-type cells by at least 9:1 in 6-day cultures (Fig. 2A). P. protegens Pf-5 is also known for the production of a large number of secondary metabolites (32, 34, 42). The results of this study showed that secondary metabolism influences the accumulation of spontaneous Gac− mutants in cultures of P. protegens Pf-5. The study also showed that secondary metabolism does not have a uniform role in the evolution of P. protegens Pf-5. Of six secondary metabolic pathways possessed by strain Pf-5, only two (pyoluteorin and orfamide A pathways) had a significant influence on the accumulation of Gac− mutants, determined by evaluating a series of mutants deficient in production of one or six of the secondary metabolites. Rhizoxin and 2,4-diacetylphloroglucinol were not produced to detectable levels in NYB medium, so it is possible that the production of these secondary metabolites could influence the proportion of Gac− mutants of Pf-5 in a different medium. However, no Gac− mutants were detected when strain Pf-5 was cultured in NBG, which is conductive to 2,4-diacetylphloroglucinol production (36), indicating that production of 2,4-diacetylphloroglucinol may not influence the accumulation of Gac− mutants. Pyrrolnitrin was produced by strain Pf-5 on NYB (see Fig. S1C in the supplemental material), but the ΔprnC mutant did not differ from the wild type in the accumulation of Gac− mutants (Fig. 3A). These results suggest that secondary metabolism does not have a uniform role in the accumulation of Gac− mutants. Of the two secondary metabolites that influenced the accumulation of Gac− mutants of Pf-5 in NYB medium, our study focused on pyoluteorin.
A mutant deficient in pyoluteorin biosynthesis accumulated spontaneous Gac− mutations to a level that is significantly lower than that of the wild type (Fig. 3), so we initially hypothesized that the compound pyoluteorin enhances the accumulation of Gac− mutants. Contrary to this expectation, the addition of purified pyoluteorin to cultures of a ΔpltA mutant or wild-type Pf-5 decreased the proportion of Gac− mutants in the culture (Fig. 4). These results suggest that the process of pyoluteorin production, instead of the pyoluteorin compound, contributes to the accumulation of spontaneous Gac− mutants. One possible explanation for these results is that the biosynthesis of pyoluteorin imposes a metabolic load on the bacterial cell such that Gac− mutants, relieved of that metabolic load, have a fitness advantage over the wild type in culture. In support of this explanation, a derivative of P. protegens Pf-5 (pltR* ΔpltA mutant) that overexpressed the pyoluteorin biosynthesis genes but did not synthesize pyoluteorin, accumulated a high proportion of Gac− mutants (Fig. 5B). Therefore, increasing the expression of pyoluteorin biosynthesis genes, which presumably increased the metabolic load on the cell, resulted in enhanced accumulation of the Gac− mutants. These results support the idea that the fitness disadvantage of the wild type to Gac− mutants in culture (Fig. 2) is due to the metabolic load imposed by pyoluteorin synthesis in the wild type, which is absent in the mutants.
We recognize that several alternative explanations for our results exist. For example, we evaluated the possibility that the Gac− mutants and wild-type Pf-5 differ in sensitivity to pyoluteorin, which could impose a selective pressure on the relative abundance of the two genotypes; however, no detectable difference was observed in pyoluteorin sensitivities of Gac− mutants versus wild-type Pf-5 (Fig. S3). It is also possible that intermediates accumulated during pyoluteorin biosynthesis (Fig. S4) could influence the relative fitness of Gac− mutants versus wild-type cells in culture. Pyoluteorin is composed of a pyrrole moiety, which is synthesized via a nonribosomal peptide synthetase (NRPS), and a resorcinol ring synthesized by a polyketide synthase (PKS). Of the eight enzymes (PltLABCDEFG [Fig. S4A]) required for the biosynthesis of pyoluteorin (30), two (PltE and PltF [Fig. S4B]) synthesize the pyrrolyl-2-thioester on the peptidyl carrier protein PltL (43). Two chlorines are added to the pyrrolyl substrate on the carrier protein scaffold by the FADH2-dependent halogenase PltA (44). The dichloropyrrolyl-S-PltL is the likely substrate for the PKS steps catalyzed by PltB and PltC, in which malonyl-coenzyme A (CoA) monomers are added to an elongating carbon chain, released from the PKS by the thioesterase PltG, and cyclized to form the resorcinol ring (30). The mutation of pltA used in this study is not expected to interfere with the expression of downstream genes because a promoter is predicted upstream of pltB (45). The PltA substrate is pyrrolyl-S-PltL (Fig. S4B), which is tethered to the peptidyl carrier protein so is unlikely to accumulate in the cytosol of the ΔpltA mutant (44). Therefore, it is unlikely that the pltA mutation causes an increased concentration of an intermediate that influences the accumulation of Gac− mutants in cultures of Pf-5. In wild-type Pf-5, the dichlorinated pyrrole is proposed to be transferred from PltL to the acyl carrier protein domains in the polyketide synthases PltB and PltC. The dichlorinated pyrrole, which is present in wild-type Pf-5 and absent in the ΔpltA mutant, is not thought to be released into the cytosol (44), so it is unlikely to influence the proportion of Gac− mutants in cultures of P. protegens Pf-5. On the basis of the current model for pyoluteorin biosynthesis, it seems unlikely that a pathway intermediate is responsible for the accumulation of Gac− mutants in cultures of strain Pf-5, but the model has not yet been fully evaluated in vivo. Therefore, we cannot exclude the possibility that an intermediate(s) in pyoluteorin biosynthesis, which is not present in the ΔpltA mutant, contributes to the differential fitness of Gac− mutants and wild-type Pf-5 in NYB cultures. Our recent work also showed that the intermediates of secondary metabolic pathways can function as signals regulating gene expression of P. protegens (10, 36, 46). Future experiments are needed to investigate the possible contribution of intermediates in pyoluteorin biosynthesis to the accumulation of spontaneous Gac− mutants.
(A and B) Diagrams of the pyoluteorin biosynthetic gene cluster (A) and the proposed biosynthetic pathway (B) of P. protegens Pf-5. Arrows represent gene locations and orientations in the biosynthetic gene clusters and are colored according to their functions. These diagrams were modified from previous work (10, 43, 44). Download FIG S4, TIF file, 0.1 MB (156.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Bacteria living in natural environments must compete with other organisms for nutrients and space, and competition is often a strong selective pressure influencing bacterial evolution. In this study, the proportion of spontaneous Gac− mutants in cultures of P. protegens Pf-5 was higher in pure culture than in coculture with B. subtilis (Fig. 6B). From these results, we speculate that phenotype(s) positively regulated by GacS-GacA were required for P. protegens Pf-5 to compete successfully with B. subtilis. Indeed, strain Pf-5 inhibited growth of B. subtilis on NYA, whereas a ΔgacA mutant showed much less inhibition (Fig. 6A). Production of pyoluteorin, which is positively controlled by GacS-GacA (15), is required for full inhibition of B. subtilis (Fig. S5). Also, the population size of strain Pf-5 was similar in pure culture or coculture with B. subtilis, whereas the population size of a ΔgacA mutant was significantly higher in pure culture than in coculture with B. subtilis (Fig. 6C). Consistent with our results, it has been reported that a ΔgacA mutant of P. aureofaciens 30-84 has a competitive advantage against the wild type in sterile rhizosphere soil, but the wild type has a competitive advantage in natural rhizosphere soil, where the bacteria must compete with other organisms (22). Taken together, these results indicate that competitive pressure influences the evolution of these Pseudomonas species, favoring wild-type cells and reducing the proportion of spontaneous Gac− mutants, which show reduced fitness in the presence of competing organisms.
Inhibitory effect of wild-type P. protegens Pf-5 and its derivatives on the growth of B. subtilis strain 168 on NYA at 4 dpi. The pltR* mutant is an overproducer of pyoluteorin (Table 2). The small zone of inhibition surrounding the ΔgacA mutant could be due to siderophore or bacterocin production, which do not require GacS-GacA (15, 16). Determining the nature of this inhibition was beyond the scope of this study. The experiment was repeated three times with similar results. One representative replicate of four replicates is shown. Download FIG S5, TIF file, 3.3 MB (3.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We commonly observe colony variants that exhibit a Gac− phenotype (expanded colony size, flat, increased fluorescence under UV light, orange color following many days of incubation) (21, 24) in 3- to 5-day agar cultures of P. protegens Pf-5. These observations suggest that Pf-5 cultures typically consist of subpopulations of the wild type and Gac− mutants that vary in proportion under different conditions. Populations of Pseudomonas spp. in the rhizosphere also contain variants with gacS or gacA mutations (24, 47, 48). The Gac− mutant subpopulation may dominate in nutrient-rich conditions, such as NYB evaluated in this study, to achieve a maximum proliferation of the whole population. The wild-type subpopulation may dominate in other conditions, such as in the presence of competitors when GacS-GacA is needed to optimize fitness. Given the known role of GacS-GacA in controlling diverse cellular traits, including motility, biofilm formation, and secondary metabolism (15, 16, 20), spontaneous mutation of gacS or gacA provides a mechanism to regulate bacterial adaption at both cellular and population levels.
In this study, medium composition (NYB versus NBG) and competition with B. subtilis influenced the predominance of Gac− mutants in culture in a manner that supports the role of secondary metabolism as a primary phenotype influencing bacterial fitness. We recognize, however, that the influence of the GacS-GacA system reaches beyond secondary metabolism, controlling the expression of approximately 10% of the genes in the genome of P. protegens Pf-5 (15). Accordingly, different GacS-GacA-regulated phenotypes are likely to influence the relative fitness of Gac− mutants versus wild type in different conditions. For example, the motility of strain Pf-5 is influenced by the GacS-GacA system (16), and the predominance of Gac− mutants at the advancing edge of swarming colonies of P. protegens Pf-5 has been attributed to their hyperflagellation (17). Biofilm formation is also influenced by the GacS-GacA system (16), and Driscoll et al. (23) reported evidence for a mutualism between Gac− mutant and wild-type cells of P. chlororaphis 30-84 in laboratory biofilms. Duffy and Défago (24) proposed that the osmotic potential of the medium influenced the proportion of Gac− mutants in cultures of P. protegens CHA0, recognizing that GacS-GacA influences the response of bacterial cells to environmental stress (20, 27). Secondary metabolism influenced the proportion of Gac− mutants in cultures of P. protegens Pf-5 in this study, but other GacS-GacA-controlled phenotypes are likely to mediate the proportion of Gac− mutant versus wild-type cells in other Pseudomonas species and in other environments.
P. protegens, P. chlororaphis, and other species in the Pseudomonas fluorescens group have characteristics, including their capacities to colonize plant surfaces and produce antifungal secondary metabolites, that make them well suited as biological control agents for the management of plant diseases (49–52). Bacteria in the P. fluorescens group are known to contribute to the disease suppressive characteristics of certain agricultural soils, and some strains have been developed as commercial products that can be applied to plant surfaces to suppress plant disease (53). The genetic instability of these bacteria is a concern in the development of biological control for use in agriculture: if Gac− mutants predominate in cultures used to produce inoculum, biocontrol efficacy could be lost because secondary metabolites essential for their biological control activity are not produced (20, 21, 24). The results of this study substantiate previous conclusions that the emergence and accumulation of Gac− mutants are context dependent (17, 24). Accordingly, media for inoculum development can be selected to minimize the possibility that Gac− mutants will develop.
Secondary metabolite production is an adaptive characteristic subject to natural selection. It contributes to ecologic fitness of the producers (1, 2) but can be metabolically costly (7). Secondary metabolism in Pseudomonas spp. is controlled by intricate regulatory circuits that include pathway-specific and global regulators that respond to environmental cues and physiological status. For example, in addition to the positive regulation imposed by GacS-GacA (54) and the pathway-specific regulator PltR (30), synthesis of pyoluteorin is also kept in check by numerous negative regulators that operate at the transcriptional and posttranscriptional levels. Specifically, pyoluteorin production by P. protegens Pf-5 and other strains of Pseudomonas is moderated by the presence of nonpreferred codons in PltR (11), transcriptional repressor PltZ (55), RNA-binding protein RsmE (45), stationary-phase sigma factor RpoS (29), Lon protease (56), and phloroglucinol, an intermediate in the biosynthesis of 2,4-diacetylphloroglucinol (36, 46). The presence of multiple layers of negative regulation suggests that controlling pyoluteorin production to moderate levels is important for the fitness of the bacterial cell. When conditions allow relatively high levels of pyoluteorin production, such as the NYB cultures evaluated in this study, Gac− mutants accumulate, thereby stopping secondary metabolism.
Overall, our data showed that P. protegens Pf-5 accumulates spontaneous Gac− mutants in a nutrient-rich medium conducive to the synthesis of pyoluteorin and other secondary metabolites. We showed that secondary metabolism, specifically pyoluteorin synthesis, has an important role in the accumulation of Gac− mutants. Furthermore, interspecific competition with B. subtilis, which requires a functional GacS-GacA system and its controlled secondary metabolism (pyoluteorin synthesis), influences the accumulation of Gac− mutants. We propose that both the metabolic load and the fitness advantage associated with secondary metabolism are important factors contributing to the accumulation of Gac− mutants. The results of this study support the contention that the accumulation of Gac− mutants is an approach by which bacteria shut off secondary metabolism under conditions in which cells pay more in metabolic cost than gained in ecologic fitness to the population.
MATERIALS AND METHODS
Strains of Pseudomonas spp. and cultural conditions.
Bacterial strains used in this study are listed in Table 2. Four species of Pseudomonas were evaluated for accumulation of spontaneous Gac− mutants: P. fluorescens SS101 (57), P. aeruginosa PAO1 (58), P. chlororaphis 30-84 (50), and P. protegens Pf-5 (49). Bacillus subtilis strain 168 was evaluated for its impact on the growth and accumulation of Gac− mutants of P. protegens Pf-5. All strains were stored at −80°C and grown in NYB (nutrient broth supplemented with 0.5% yeast extract), NYA (NYB with 1.5% agar), NBG (nutrient broth supplemented with 2% glucose), and/or KB (King’s medium) (59). The bacteria were cultured at 27°C unless specifically indicated.
TABLE 2 .
Bacterial strains, plasmids, and oligonucleotide primers used in this study
| Bacterial strain, plasmid, or oligonucleotide primer |
Description (genotype and/or relevant characteristicsa) or sequence of oligonucleotide primerb |
Reference or source |
|---|---|---|
| Bacterial strains | ||
| P. protegens | ||
| LK099 | Wild-type Pf-5 | 49 |
| LK147 | 6-fold mutant; contains mutations in hcnB, ofaA, phlA, pltA, prnC, and rzxB of strain Pf-5 described below; HCN− Ofa− DAPG− MAPG− Plt− Prn− Rzx− |
35 |
| JL4909 | ΔhcnB mutant; contains a 239-bp deletion in hcnB of Pf-5; HCN− | 52 |
| JL4807 | ΔofaA mutant; contains 1,143-bp deletion in ofaA (PFL_2145) of Pf-5; contains FRT scar in deleted ofaA frame; Ofa− |
15 |
| LK023 | ΔphlA mutant; contains a 639-bp deletion in phlA (PFL_5954) of Pf-5; MAPG− DAPG− | 36 |
| JL4805 | ΔpltA mutant; contains a 275-bp deletion in pltA (PFL_2787) of Pf-5; Plt− | 60 |
| LK415 |
pltA+ complemented mutant; contains a wild-type pltA gene replacing the mutated pltA in the chromosome of strain JL4805; Plt+ |
This study |
| JL4793 | ΔprnC mutant; contains an 86-bp insertion of FRT site in prnC (PFL_3606) of Pf-5; Prn− | 60 |
| JL4808 | ΔrzxB mutant; contains a 1,342-bp deletion in rzxB (PFL_2989) of Pf-5; Rzx− | 60 |
| LK298 |
pltR* mutant; contains codon-modified pltR (PFL_2785) in the chromosome of Pf-5; overexpresses plt biosynthesis genes and overproduces pyoluteorin |
11 |
| LK417 |
pltR* ΔpltA double mutant; contains a 275-bp deletion in pltA and the codon-modified pltR* in the chromosome of Pf-5; overexpresses plt biosynthesis genes; Plt− |
This study |
| JL4975 | ΔgacA mutant; contains a 612-bp deletion in gacA (PFL_3563) of Pf-5; altered in the secondary metabolism and many other phenotypes regulated by GacA |
60 |
| P. fluorescens SS101 | Wild-type strain | 57 |
| P. aeruginosa PAO1 | Wild-type strain | 58 |
| P. chlororaphis 30-84 | Wild-type strain | 50 |
| B. subtilis 168 | Wild-type strain | 61 |
| Plasmids | ||
| pEX18Tc 168 | Gene replacement vector with MCS from pUC18; sacB+ Tcr | 62 |
| P18Tc-pltA | pEX18Tc containing wild-type pltA in a 1,359-bp BamHI fragment | This study |
| ppltL-gfp | pPROBE′-gfp (tagless) contains the intergenic region between pltR and pltL, including the promoter of pltL fused with a promoterless gfp |
11 |
| pEX18km-pltR-MCod3 | pEX18Km with a 1,160-bp synthesized DNA fragment, containing pltR of Pf-5 with modifications in 35 types of rare codons |
11 |
| Oligonucleotide primers | ||
| plt UpF-Bam | GTGTGGTAGTGGATCCTCCAGGACTGTCGAGCAAC | |
| plt DnR-Bam | GCAGAAGAGAGGATCCTACTTGTGCCAGAGGTGTTC | |
| gacA-seqF | CGGTCTTGCGGAAATAGCTG | |
| gacA-seqR | TAGGACCGTTATTGCGCCC | |
| gacS-5’F | CCAAGATCAGCCCCCGGCAA | |
| gacS-Reverse (1) | ATCCAGCTCCTGGCTGCCCA | |
| gacS-Middle Forward (2) | GCCGCACAATCAACAACCCGC | |
| gacS-Middle Reverse (2) | GCGCAGTTGCACGCTGTCTT | |
| gacS-Middle Forward (3) | CGCTGCGGCTCAAGCAGATTC | |
| gacS-Middle Reverse (3) | TCGACACACAGCACTCGCGG | |
| gacS-Forward (1) | CAGCCAGTTGCAGGCCAAGC | |
| gacS-3’R | AGCGCCGAGGAAACTCTCGC | |
Ofa, orfamide A; DAPG, 2,4-diacetylphloroglucinol; MAPG, monoacetylphloroglucinol; Plt, pyoluteorin; Prn, pyrrolnitrin; Rzx, rhizoxin analogs; HCN, hydrogen cyanide; Tox, toxoflavin; Smr, streptomycin resistance; Tpr, trimethoprim resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; MCS, multiple-cloning site.
Restriction sites used for cloning are underlined in oligonucleotide primer sequences.
Colonies of Pseudomonas spp. were transferred to LMA (litmus milk agar) to ensure that bacteria produced extracellular protease, indicating the integrity of the GacS-GacA regulatory systems (17, 27). The presence of a halo surrounding the colonies on the LMA plates means that the extracellular protease is produced and confirms the lack of mutations in the gacS or gacA gene. Strains were used in experiments only after their exoprotease production was confirmed.
To evaluate the accumulation of Gac− mutants in cultures over time, bacteria were grown in culture tubes in NYB overnight with shaking (200 rpm). After that, optical densities (optical density at 600 nm [OD600]) were measured, and cultures were adjusted to have equivalent starting OD600 values. Then, 20 µl of the culture was transferred to 20 ml NYB in a 125-ml flask, which was incubated with shaking (200 rpm). For all strains tested, at least three replicate cultures were evaluated. At 1 day postinoculation (dpi) to 6 dpi, 10 µl was sampled from each flask and serially diluted, and dilutions were spread on KB and LMA plates. The numbers of CFU were counted on KB plates after 24-h incubation to estimate the population size in each flask. After 48-h incubation, the number of colonies with or without a halo on LMA was counted. Data were exported to the PAST software (University of Oslo, Norway), where an analysis of variance (ANOVA) and Tukey pairwise comparison tests were used to analyze the existence of significant differences between the treatments.
Construction of Pf-5 derivative strains.
To complement the ΔpltA mutant, the deleted pltA in the chromosome was repaired by the wild-type copy as follows. pltA was amplified from wild-type Pf-5 by using primers plt UpF-Bam/plt DnR-Bam (F stands for forward, R stands for reverse, and Dn stands for down) and then cloned into plasmid pEX18Tc, which is a suicide vector in Pf-5 (Table 2). The resultant plasmid, p18Tc-pltA, was then transformed into the ΔpltA mutant, and the repaired derivative strain was selected after two rounds of homologous recombination as described previously (11). The resultant pltA-repaired strain was identified by PCR and confirmed by sequence analysis.
To make the pltR* ΔpltA mutant, we used plasmid pEX18km-pltR-MCod3 (11), which contains pltR*, a derivative of pltR with all rare codons substituted with preferred synonymous codons. The approach described above was used to replace pltR with pltR* in the chromosome of the ΔpltA mutant.
Identification of the spontaneous mutation sites by PCR and DNA sequencing.
P. protegens Pf-5 and three derivative strains (ΔofaA, ΔpltA, and 6-fold mutants) were grown in NYB as described above. At 4 dpi, 10 µl was sampled from each flask and serially diluted, and dilutions were spread on LMA agar. The plates were incubated for 2 days, and exoprotease-deficient colonies, identified by the lack of a halo on LMA, were selected randomly. Genomic DNA was isolated from six exoprotease-deficient colonies of each strain (two colonies from each of the three replicate flasks). The gacA and gacS ORFs and flanking sequences were amplified by using primers gacA-seqF/gacA-seqR, and gacS-5’F/gacS-3’R (Table 2), respectively. The amplified products were cleaned up using the ExoSAP-IT enzyme and submitted for Sanger sequencing. Primers gacA-seqF and gacA-seqR (seq stands for sequencing) were used to sequence the PCR products of the gacA gene. Multiple sequencing reactions and seven primers, including gacS-5’F, gacS-3’R, gacS-Reverse (1), gacS-Middle Forward (2), gacS-Middle Forward (3), gacS-Middle Reverse (3), gacS-Forward (1) (Table 2) were used to sequence the full length (2,754 bp) of gacS ORF. The DNA sequences were analyzed in the Geneious software (Newark, NJ) to detect single-nucleotide polymorphisms (SNPs), deletions or insertions in gacS or gacA.
Quantification of secondary metabolites.
P. protegens Pf-5 and derivative strains were cultured in 20 ml NYB with shaking at 200 rpm as described above. Five milliliters of the culture were extracted twice with 2.5-ml ethyl acetate. The ethyl acetate extracts were dried under vacuum and solubilized in 100 μl methanol. A portion (10 μl) of the methanol solution was analyzed by high-performance liquid chromatography (HPLC) using an Agilent 1100 HPLC instrument consisting of a quaternary pump, autosampler, column heater (set at 30°C), and diode array detector. Separation was achieved using a Luna C18 column (4.6 mm by 150 mm; 5 μm; Phenomenex, Torrance, CA) with a flow rate of 1 ml/min with the following steps where solvent A was water plus 0.1% (vol/vol) formic acid, and solvent B was acetonitrile plus 0.1% (vol/vol) formic acid. The column was preequilibrated in 90% solvent A–10% solvent B, and upon injection, this composition was held for 2 min. The composition of the mobile phase was then changed to 0% solvent A–100% solvent B for 28 min utilizing a linear gradient. This composition was held for 6 min and then changed to 90% solvent A–10% solvent B for 2 min. The column was equilibrated in 90% solvent A–10% solvent B for 6 min prior to the next injection. Under these chromatographic conditions, pyoluteorin eluted at 15.1 min. The HPLC was operated with and data were viewed using ChemStation (version B.04.03; Agilent, Santa Clara, CA). Quantification was performed by integrating the area under the curve at 300 nm and comparing to a standard curve prepared by injection of purified pyoluteorin, 2,4-diacetylphloroglucinol, monoacetylphloroglucinol, orfamide A, rhizoxin WF-1360 F (the predominant rhizoxin analogue produced by P. protegens Pf-5), and pyrrolnitrin. Data were processed with GraphPad Prism (GraphPad Software, Inc., San Diego, CA).
Transcriptional expression assay using a green fluorescent protein (GFP) reporter.
To test the expression level of pyoluteorin biosynthetic genes, a gfp-based reporter plasmid ppltL-gfp (11) was transformed into P. protegens Pf-5 and its derivatives. The plasmid ppltL-gfp contains a promoterless gfp fused with the promoter of pltL, which is the first gene in the pyoluteorin biosynthetic gene cluster.
P. protegens Pf-5 strains containing ppltL-gfp were cultured overnight in NYB with kanamycin (50 μg/ml) with shaking at 200 rpm. The cells were washed once with NYB and inoculated in triplicate into 200 μl NYB in wells of a 96-well plate to a final optical density (OD600) of 0.01. The plate was incubated in a 96-well plate reader (Tecan Infinite 200 Pro; Männedorf, Switzerland) with shaking at approximately 200 rpm. Growth of the bacteria was monitored by measuring the OD600. The green fluorescence of bacteria was monitored by measuring the emission wavelength at 535 nm with an excitation wavelength at 485 nm and corrected for background by subtracting fluorescence emitted by the growth medium.
Assay for sensitivity to pyoluteorin.
Wild-type Pf-5 and the ΔgacA mutant were cultured in NYB for 24 h with shaking (200 rpm). Cells were washed one time and suspended in fresh NYB broth to an OD600 of 1.0. The bacterial suspensions were diluted 1:100 vol/vol into melted, warm (45°C) NYA medium before pouring the medium into petri plates. The solidified agar was air dried for 1 h before use in the sensitivity assay. Purified pyoluteorin was dissolved in methanol to concentrations ranging from 20 μg/ml to 20 mg/ml. Three microliters of the pyoluteorin solution was placed on a sterilized filter paper disk (10-mm diameter). Dried filter paper disks were placed on the top of NYA seeded with bacterial cells of P. protegens Pf-5 and derivatives. The plates were incubated at 27°C. Results were recorded at 24 h after incubation. At least four replicate plates were evaluated in these experiments.
Assay for inhibition of P. protegens Pf-5 against B. subtilis strain 168.
P. protegens Pf-5 (wild type and mutants) and B. subtilis 168 were cultured individually in NYB for 24 h. Cells were washed one time and suspended in fresh NYB broth to an OD600 at 1.0.
To test the inhibitory effect of P. protegens Pf-5 and its derivatives on the growth of B. subtilis 168 on agar plates, the washed cell suspensions of B. subtilis 168 were diluted 1:100 (vol/vol) into melted, warm (45°C) NYA before the medium was poured into petri plates. The solidified agar was air dried for 1 h before use. Three-microliter portions of the washed wild-type Pf-5 or ΔgacA, ΔpltA, or pltR* mutant cell suspensions were spotted onto the solidified plates. The plates were incubated at 27°C and imaged at 4 dpi. To test the inhibitory effect of P. protegens Pf-5 and its derivatives on the growth of B. subtilis 168 in liquid cultures, washed cells of B. subtilis 168 were inoculated alone or coinoculated (1:1) with strain Pf-5 (wild type or ΔgacA mutant) in NYB to an OD600 at 0.01. The bacterial cells were sampled at 6 dpi and diluted on NYA plates. The plates were incubated at 40°C which allows only the growth of B. subtilis 168 but not P. protegens Pf-5. The colonies were counted after 2-day incubation. At least three biological replicates were used in these experiments.
ACKNOWLEDGMENTS
We are grateful to Ashley Hamilton, Marcella Henkels, Richelle Castro, Brenda Villanueva, and Max Kohen for their assistance; Jeff Anderson and Tal Pupko for helpful discussions; and the Center for Genomics Research and Biocomputing at Oregon State University for sequencing services. P. aeruginosa strain PAO1 was kindly provided by Pierre Cornelis, P. chlororaphis strain 30-84 was kindly provided by Leland S. Pierson III, and B. subtilis strain 168 was kindly provided by Ping Ma.
This work was supported by Agriculture and Food Research Initiative Competitive grant 2011-67019-30192 from the U.S. Department of Agriculture National Institute of Food and Agriculture (to J.E.L.), College of Pharmacy start-up funds (to B.P.), and by a fellowship from the Organization for Economic Cooperation and Development (to J.M.R.).
Footnotes
Citation Yan Q, Lopes LD, Shaffer BT, Kidarsa TA, Vining O, Philmus B, Song C, Stockwell VO, Raaijmakers JM, McPhail KL, Andreote FD, Chang JH, Loper JE. 2018. Secondary metabolism and interspecific competition affect accumulation of spontaneous mutants in the GacS-GacA regulatory system in Pseudomonas protegens. mBio 9:e01845-17. https://doi.org/10.1128/mBio.01845-17.
REFERENCES
- 1.Price-Whelan A, Dietrich LE, Newman DK. 2006. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol 2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien J, Wright GD. 2011. An ecological perspective of microbial secondary metabolism. Curr Opin Biotechnol 22:552–558. doi: 10.1016/j.copbio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Demain AL, Sanchez S. 2009. Microbial drug discovery: 80 years of progress. J Antibiot 62:5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain MS, Fareed S, Ansari S, Rahman MA, Ahmad IZ, Saeed M. 2012. Current approaches toward production of secondary plant metabolites. J Pharm Bioallied Sci 4:10–20. doi: 10.4103/0975-7406.92725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wink M. 2003. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64:3–19. doi: 10.1016/S0031-9422(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 6.Jousset A, Rochat L, Péchy-Tarr M, Keel C, Scheu S, Bonkowski M. 2009. Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J 3:666–674. doi: 10.1038/ismej.2009.26. [DOI] [PubMed] [Google Scholar]
- 7.Kleijn RJ, Liu F, van Winden WA, van Gulik WM, Ras C, Heijnen JJ. 2007. Cytosolic NADPH metabolism in penicillin-G producing and non-producing chemostat cultures of Penicillium chrysogenum. Metab Eng 9:112–123. doi: 10.1016/j.ymben.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Haas D, Keel C. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41:117–153. doi: 10.1146/annurev.phyto.41.052002.095656. [DOI] [PubMed] [Google Scholar]
- 9.Brakhage AA. 2013. Regulation of fungal secondary metabolism. Nat Rev Microbiol 11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 10.Yan Q, Philmus B, Chang JH, Loper JE. 2017. Novel mechanism of metabolic co-regulation coordinates the biosynthesis of secondary metabolites in Pseudomonas protegens. eLife 6:e22835. doi: 10.7554/eLife.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Q, Philmus B, Hesse C, Kohen M, Chang JH, Loper JE. 2016. The rare codon AGA is involved in regulation of pyoluteorin biosynthesis in Pseudomonas protegens Pf-5. Front Microbiol 7:497. doi: 10.3389/fmicb.2016.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Hüttenhofer A, Haas D, Bläsi U. 2011. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol 80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- 13.Gross H, Loper JE. 2009. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26:1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 14.Lapouge K, Schubert M, Allain FH, Haas D. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 15.Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LDH, Hartney S, Duboy R, Goebel NC, Zabriskie TM, Paulsen IT, Loper JE. 2010. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ Microbiol 12:899–915. doi: 10.1111/j.1462-2920.2009.02134.x. [DOI] [PubMed] [Google Scholar]
- 16.Kidarsa TA, Shaffer BT, Goebel NC, Roberts DP, Buyer JS, Johnson A, Kobayashi DY, Zabriskie TM, Paulsen I, Loper JE. 2013. Genes expressed by the biological control bacterium Pseudomonas protegens Pf-5 on seed surfaces under the control of the global regulators GacA and RpoS. Environ Microbiol 15:716–735. doi: 10.1111/1462-2920.12066. [DOI] [PubMed] [Google Scholar]
- 17.Song C, Kidarsa TA, van de Mortel JE, Loper JE, Raaijmakers JM. 2016. Living on the edge: emergence of spontaneous gac mutations in Pseudomonas protegens during swarming motility. Environ Microbiol 18:3453–3465. doi: 10.1111/1462-2920.13288. [DOI] [PubMed] [Google Scholar]
- 18.Wang D, Lee S-H, Seeve C, Yu JM, Pierson LS, Pierson EA. 2013. Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30-84. MicrobiologyOpen 2:505–524. doi: 10.1002/mbo3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei X, Huang X, Tang L, Wu D, Xu Y. 2013. Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18. J Bacteriol 195:3387–3400. doi: 10.1128/JB.00214-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heeb S, Haas D. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol Plant Microbe Interact 14:1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- 21.Bull CT, Duffy B, Voisard C, Défago G, Keel C, Haas D. 2001. Characterization of spontaneous gacS and gacA regulatory mutants of Pseudomonas fluorescens biocontrol strain CHAO. Antonie Van Leeuwenhoek 79:327–336. doi: 10.1023/A:1012061014717. [DOI] [PubMed] [Google Scholar]
- 22.Chancey ST, Wood DW, Pierson EA, Pierson LS. 2002. Survival of GacS/GacA mutants of the biological control bacterium Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. Appl Environ Microbiol 68:3308–3314. doi: 10.1128/AEM.68.7.3308-3314.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Driscoll WW, Pepper JW, Pierson LS, Pierson EA. 2011. Spontaneous Gac mutants of Pseudomonas biological control strains: cheaters or mutualists? Appl Environ Microbiol 77:7227–7235. doi: 10.1128/AEM.00679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duffy BK, Défago G. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol 66:3142–3150. doi: 10.1128/AEM.66.8.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Broek D, Chin-A-Woeng TF, Eijkemans K, Mulders IH, Bloemberg GV, Lugtenberg BJ. 2003. Biocontrol traits of Pseudomonas spp. are regulated by phase variation. Mol Plant Microbe Interact 16:1003–1012. doi: 10.1094/MPMI.2003.16.11.1003. [DOI] [PubMed] [Google Scholar]
- 26.van den Broek D, Chin-A-Woeng TF, Bloemberg GV, Lugtenberg BJ. 2005. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. J Bacteriol 187:593–600. doi: 10.1128/JB.187.2.593-600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whistler CA, Corbell NA, Sarniguet A, Ream W, Loper JE. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor sigmaS and the stress response in Pseudomonas fluorescens Pf-5. J Bacteriol 180:6635–6641. http://jb.asm.org/content/180/24/6635.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowak-Thompson B, Gould SJ, Kraus J, Loper JE. 1994. Production of 2, 4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can J Microbiol 40:1064–1066. doi: 10.1139/m94-168. [DOI] [Google Scholar]
- 29.Sarniguet A, Kraus J, Henkels MD, Muehlchen AM, Loper JE. 1995. The sigma factor σS affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc Natl Acad Sci U S A 92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowak-Thompson B, Chaney N, Wing JS, Gould SJ, Loper JE. 1999. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J Bacteriol 181:2166–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loper JE, Henkels MD, Shaffer BT, Valeriote FA, Gross H. 2008. Isolation and identification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. Appl Environ Microbiol 74:3085–3093. doi: 10.1128/AEM.02848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philmus BJ, Shaffer BT, Kidarsa TA, Yan Q, Raaijmakers JM, Begley TP, Loper JE. 2015. Investigations into the biosynthesis, regulation, and self-resistance of toxoflavin in Pseudomonas protegens Pf-5. ChemBioChem 16:1782–1790. doi: 10.1002/cbic.201500247. [DOI] [PubMed] [Google Scholar]
- 33.Sacherer P, Défago G, Haas D. 1994. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol Lett 116:155–160. doi: 10.1111/j.1574-6968.1994.tb06694.x. [DOI] [PubMed] [Google Scholar]
- 34.Loper JE, Gross H. 2007. Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf-5. Eur J Plant Pathol 119:265–278. doi: 10.1007/s10658-007-9179-8. [DOI] [Google Scholar]
- 35.Loper JE, Henkels MD, Rangel LI, Olcott MH, Walker FL, Bond KL, Kidarsa TA, Hesse CN, Sneh B, Stockwell VO, Taylor BJ. 2016. Rhizoxin analogs, orfamide A and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ Microbiol 18:3509–3521. doi: 10.1111/1462-2920.13369. [DOI] [PubMed] [Google Scholar]
- 36.Kidarsa TA, Goebel NC, Zabriskie TM, Loper JE. 2011. Phloroglucinol mediates cross-talk between the pyoluteorin and 2, 4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol Microbiol 81:395–414. doi: 10.1111/j.1365-2958.2011.07697.x. [DOI] [PubMed] [Google Scholar]
- 37.Powers MJ, Sanabria-Valentín E, Bowers AA, Shank EA. 2015. Inhibition of cell differentiation in Bacillus subtilis by Pseudomonas protegens. J Bacteriol 197:2129–2138. doi: 10.1128/JB.02535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han B, Pain A, Johnstone K. 1997. Spontaneous duplication of a 661 bp element within a two-component sensor regulator gene causes phenotypic switching in colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol Microbiol 25:211–218. doi: 10.1046/j.1365-2958.1997.4511815.x. [DOI] [PubMed] [Google Scholar]
- 39.Sánchez-Contreras M, Martín M, Villacieros M, O’Gara F, Bonilla I, Rivilla R. 2002. Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J Bacteriol 184:1587–1596. doi: 10.1128/JB.184.6.1587-1596.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossignol G, Sperandio D, Guerillon J, Poc CD, Soum-Soutera E, Orange N, Feuilloley M, Merieau A. 2009. Phenotypic variation in the Pseudomonas fluorescens clinical strain MFN1032. Res Microbiol 160:337–344. doi: 10.1016/j.resmic.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Marvig RL, Sommer LM, Molin S, Johansen HK. 2015. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 42.Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS, Thomashow LS, Loper JE. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas MG, Burkart MD, Walsh CT. 2002. Conversion of l-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem Biol 9:171–184. doi: 10.1016/S1074-5521(02)00100-X. [DOI] [PubMed] [Google Scholar]
- 44.Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT. 2005. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc Natl Acad Sci U S A 102:13843–13848. doi: 10.1073/pnas.0506964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Huang X, Liu Y, Yang G, Liu Y, Zhang X. 2017. GacS/GacA activates pyoluteorin biosynthesis through Gac/Rsm-RsmE cascade and RsmA/RsmE-driven feedback loop in Pseudomonas protegens H78. Mol Microbiol 105:968–985. doi: 10.1111/mmi.13749. [DOI] [PubMed] [Google Scholar]
- 46.Clifford JC, Buchanan A, Vining O, Kidarsa TA, Chang JH, McPhail KL, Loper JE. 2016. Phloroglucinol functions as an intracellular and intercellular chemical messenger influencing gene expression in Pseudomonas protegens. Environ Microbiol 18:3296–3308. doi: 10.1111/1462-2920.13043. [DOI] [PubMed] [Google Scholar]
- 47.Chancey ST, Wood DW, Pierson LS. 1999. Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl Environ Microbiol 65:2294–2299. http://aem.asm.org/content/65/6/2294.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt-Eisenlohr H, Gast A, Baron C. 2003. Inactivation of gacS does not affect the competitiveness of Pseudomonas chlororaphis in the Arabidopsis thaliana rhizosphere. Appl Environ Microbiol 69:1817–1826. doi: 10.1128/AEM.69.3.1817-1826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howell CR, Stipanovic R. 1979. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69:480–482. doi: 10.1094/Phyto-69-480. [DOI] [Google Scholar]
- 50.Thomashow LS, Weller DM, Bonsall RF, Pierson LS. 1990. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol 56:908–912. http://aem.asm.org/content/56/4/908.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen X, Hu H, Peng H, Wang W, Zhang X. 2013. Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics 14:271. doi: 10.1186/1471-2164-14-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, Elbourne LD, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Kidarsa TA, Wilson NL, van de Mortel JE, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS, Pierson EA, Lindow SE, Kobayashi DY, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen IT. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8:e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stockwell VO, Stack JP. 2007. Using Pseudomonas spp. for integrated biological control. Phytopathology 97:244–249. doi: 10.1094/PHYTO-97-2-0244. [DOI] [PubMed] [Google Scholar]
- 54.Corbell N, Loper JE. 1995. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol 177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang X, Zhu D, Ge Y, Hu H, Zhang X, Xu Y. 2004. Identification and characterization of pltZ, a gene involved in the repression of pyoluteorin biosynthesis in Pseudomonas sp. M18. FEMS Microbiol Lett 232:197–202. doi: 10.1016/S0378-1097(04)00074-6. [DOI] [PubMed] [Google Scholar]
- 56.Whistler CA, Stockwell VO, Loper JE. 2000. Lon protease influences antibiotic production and UV tolerance of Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 66:2718–2725. doi: 10.1128/AEM.66.7.2718-2725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Souza JT, de Boer M, de Waard P, van Beek TA, Raaijmakers JM. 2003. Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl Environ Microbiol 69:7161–7172. doi: 10.1128/AEM.69.12.7161-7172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrook-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 59.King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307. [PubMed] [Google Scholar]
- 60.Henkels MD, Kidarsa TA, Shaffer BT, Goebel NC, Burlinson P, Mavrodi DV, Bentley MA, Rangel LI, Davis EW, Thomashow LS, Zabriskie TM, Preston GM, Loper JE. 2014. Pseudomonas protegens Pf-5 causes discoloration and pitting of mushroom caps due to the production of antifungal metabolites. Mol Plant Microbe Interact 27:733–746. doi: 10.1094/MPMI-10-13-0311-R. [DOI] [PubMed] [Google Scholar]
- 61.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, Haiech J, Harwood CR, Henaut A, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 62.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A to C) Production of pyoluteorin (A), orfamide A (B), or pyrrolnitrin (C) by P. protegens Pf-5 and derivative strains (ΔpltA and ΔofaA mutants) in NYB cultures over time. (D) Accumulation of Gac− mutants in NYB cultures at 4 dpi. Values are the means of three replicate cultures, and error bars show the standard errors. Download FIG S1, TIF file, 0.3 MB (301.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Concentrations of pyoluteorin produced by P. protegens Pf-5 in NYB over time. Values are the means of three replicate cultures, and error bars show the standard deviations. Download FIG S2, TIF file, 0.1 MB (66.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Influence of pyoluteorin on the growth of wild-type P. protegens Pf-5 and the ΔgacA mutant. (A) Sterilized filter papers containing 3 µl of different concentrations of pyoluteorin were placed on the top of NYA seeded with wild-type Pf-5 or the ΔgacA mutant. The results were imaged at 24 h after inoculation. (B) Wild-type Pf-5 and the ΔgacA mutant were cultured in a 96-well plate with wells containing NYG and pyoluteorin at the indicated concentrations. The plate was incubated at 27°C with shaking at 200 rpm. The cell density (OD600) was measured at 20 h after inoculation. The experiment had three replicates. Means ± standard deviations are shown. Download FIG S3, TIF file, 0.2 MB (262KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(A and B) Diagrams of the pyoluteorin biosynthetic gene cluster (A) and the proposed biosynthetic pathway (B) of P. protegens Pf-5. Arrows represent gene locations and orientations in the biosynthetic gene clusters and are colored according to their functions. These diagrams were modified from previous work (10, 43, 44). Download FIG S4, TIF file, 0.1 MB (156.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Inhibitory effect of wild-type P. protegens Pf-5 and its derivatives on the growth of B. subtilis strain 168 on NYA at 4 dpi. The pltR* mutant is an overproducer of pyoluteorin (Table 2). The small zone of inhibition surrounding the ΔgacA mutant could be due to siderophore or bacterocin production, which do not require GacS-GacA (15, 16). Determining the nature of this inhibition was beyond the scope of this study. The experiment was repeated three times with similar results. One representative replicate of four replicates is shown. Download FIG S5, TIF file, 3.3 MB (3.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.