ABSTRACT
Enteric pathogens employ sophisticated strategies to colonize and infect mammalian hosts. Gram-negative bacteria, such as Escherichia coli, Salmonella, and Campylobacter jejuni, are among the leading causes of gastrointestinal tract infections worldwide. The virulence strategies of many of these Gram-negative pathogens rely on type III secretion systems (T3SSs), which are macromolecular syringes that translocate bacterial effector proteins directly into the host cytosol. However, synthesis of T3SS proteins comes at a cost to the bacterium in terms of growth rate and fitness, both in the environment and within the host. Therefore, expression of the T3SS must be tightly regulated to occur at the appropriate time and place during infection. Enteric pathogens have thus evolved regulatory mechanisms to control expression of their T3SSs in response to specific environmental and host cues. These regulatory cascades integrate multiple physical and chemical signals through complex transcriptional networks. Although the power of bacterial genetics has allowed elucidation of many of these networks, the biochemical interactions between signal and sensor that initiate the signaling cascade are often poorly understood. Here, we review the physical and chemical signals that Gram-negative enteric pathogens use to regulate T3SS expression during infection. We highlight the recent structural and functional studies that have elucidated the biochemical properties governing both the interaction between sensor and signal and the mechanisms of signal transduction from sensor to downstream transcriptional networks.
KEYWORDS: T3SS, cell signaling, enteric pathogens, environmental cues, nutritional stress, pathogenesis, surface sensing
INTRODUCTION
Gastrointestinal (GI) tract infections caused by enteric pathogens affect over 1.7 billion individuals annually, with approximately 2.2 million cases ending in death (1). Severe diarrheal disease resulting from GI tract infections is a leading cause of death for children under 5 years (2). Many developed countries have significantly reduced their incidence of foodborne outbreaks, but factors including globalization, environmental change, and increasing antibiotic resistance are facilitating the rapid reemergence and spread of severe enteric pathogens (3). The causal agents of gastrointestinal tract infections include bacteria, viruses, and eukaryotic parasites (1, 2). Most GI tract infections, even those caused by bacteria, cannot be treated effectively by current antibiotic therapies because the antibiotics are either ineffective, cause severe dysbiosis of the intestinal microbiota, or trigger serious complications, such as septicemia from antibiotic-induced endotoxin release (3). Thus, new strategies must be employed to develop antimicrobial therapies that will be effective and safe in the treatment of infections caused by enteric bacterial pathogens (4, 5).
Enteric bacterial pathogens span several genera, including Escherichia, Salmonella, Shigella, Yersinia, Vibrio, and Campylobacter. These species occupy different environmental habitats and use diverse domestic and wild animals as reservoirs (6). The oral sources of infection by these pathogenic bacteria vary as widely as their environmental niches. Salmonella and Campylobacter infections are usually acquired from undercooked chicken and eggs, while GI tract infections caused by Yersinia species are usually acquired from undercooked pork and vegetables (3, 6–8). Many Vibrio parahaemolyticus infections are caused by contaminated raw or undercooked seafood, and Vibrio cholerae infections are typically caused by contaminated drinking water (9, 10). Enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC, respectively) infections, as well as Shigella infections, are linked to consumption of contaminated food and water (11). Interestingly, these pathogens colonize different parts of the GI tract. For example, V. cholerae and EPEC prefer the small intestine, whereas Campylobacter jejuni and EHEC infect the cecum and colon, respectively (12, 13).
Toxin delivery systems are key to the environmental survival and persistence of enteric pathogens, as well as to their mechanisms of pathogenesis within human hosts (14). An important toxin delivery system used by many Gram-negative pathogens is the type III secretion system (T3SS). The T3SS is a macromolecular injectisome that translocates bacterial toxins, termed effectors, directly from the bacterial cytoplasm into the host cytosol (15). The expression of the T3SS apparatus and effector proteins must be tightly regulated to coincide with both the appropriate host cell environment and correct stage and location of infection (16). To achieve this coordination, bacteria have evolved mechanisms to sense different physical and chemical host signals and integrate the sensing of these signals into the regulatory control of pathogens via the T3SS apparatus and effector expression. This review focuses on the signals and sensors mediating T3SS expression during gastrointestinal tract infections.
TEMPERATURE
For bacteria to survive and thrive in diverse environments, they must be able to sense and respond to changes in temperature (17). For intestinal pathogens, temperature change is a primary indicator for successful invasion of a mammalian host (18). Therefore, temperature-sensing mechanisms are almost ubiquitously integrated into the regulatory circuits governing virulence gene expression of human intestinal pathogens (18). Many temperature-sensing mechanisms exist in bacteria. These include temperature-induced structural changes in intrinsically bent or supercoiled DNA, thermosensing regulatory proteins, and RNA thermometers (RNATs) (18–20). RNATs are RNA sequences present in the 5′-untranslated region (UTR) of certain mRNAs that adopt ribosomal binding site-occluding secondary structures at restrictive temperatures but not at permissive temperatures (21). The intestinal pathogens Yersinia pseudotuberculosis and Yersinia enterocolitica utilize these temperature-sensing strategies to efficiently regulate the expression of virulence factors, including the T3SS, within mammalian hosts (22). In pathogenic Yersinia species, the T3SS genes are encoded on the pYV virulence plasmid. Like the majority of the pYV-encoded virulence genes, the T3SS genes are not expressed at temperatures below 30°C and are most highly expressed at 37°C (23). The expression of the T3SS-encoding ysc genes (yscA to yscL), as well as of the yop effector genes (yopE, yopH, yopK, and yopO), is activated by the AraC-type regulator LcrF, which responds to both temperature and host cell contact (24–26). The lcrF gene is in an operon with yscW, and their transcription is controlled by a promoter sequence upstream of yscW (Fig. 1A).
FIG 1 .
Thermosensing by intergenic RNAT and thermolabile protein YmoA in Y. pseudotuberculosis. (A) At moderate temperatures (25°C), the transcription of the yscW-lcrF operon is partially repressed by YmoA homodimers and/or YmoA–H-NS heterodimers. Translation is fully repressed by a two-stem-loop structure in the 5′-UTR of the lcrF mRNA, the RNAT. (B) At 37°C, the thermolabile protein YmoA is rapidly degraded by ClpP and Lon proteases, and this lifts the transcriptional repression of lcrF. High temperatures also melt the inhibitory two-stem-loop structure in the lcrF mRNA, allowing for enhanced translation of LcrF, the transcriptional activator of the pYV-encoded T3SS genes.
In Y. pseudotuberculosis, temperature regulation of the ysc/yop regulon is achieved by the combined action of temperature-dependent proteolysis of the transcriptional repressor YmoA and an RNAT within the lcrF 5′-UTR (20). At moderate temperatures, similar to what a bacterium might experience outside a mammalian host (25°C), expression of the yscW-lcrF operon is repressed by two distinct mechanisms (Fig. 1A). First, the transcriptional repressor YmoA, which can form a homodimer or a heterodimer with the nucleoid-associated protein H-NS, binds to sequences downstream of the transcription initiation site and partially represses transcription of the yscW-lcrF operon (Fig. 1A) (20, 27). Second, any transcribed lcrF mRNA is not translated because of a two-stem-loop structure that forms in the 5′-UTR at moderate temperatures. This RNA structure masks the Shine-Dalgarno sequence, thereby preventing the binding of the 30S ribosomal subunit, ribosome assembly, and translation of the lcrF mRNA (Fig. 1A) (20). Without the transcriptional activator LcrF, the expression of the genes encoding the T3SS apparatus and effectors is not induced (Fig. 1A) (23, 28). At higher temperatures, such as those that would be encountered within mammalian hosts (37°C), both types of repression of lcrF expression are relieved. Transcriptional repression of the yscW-lcrF operon is lifted by the rapid, temperature-dependent degradation of YmoA by both ClpXP and Lon proteases at 37°C (Fig. 1B). Several mechanisms for the temperature-dependent degradation of YmoA have been proposed. These include increased expression or activity of ClpXP or Lon proteases at 37°C, conformational changes in YmoA that make it a better substrate for ClpXP or Lon at higher temperatures, or direct modification and targeting of YmoA for degradation by an accessory protein at 37°C. Another possible mechanism is that Lon and ClpXP have differing affinities for YmoA homodimers and YmoA–H-NS heterodimers, and the higher-affinity species may dominate at elevated temperatures (29). Translational repression of the lcrF mRNA is also relieved at 37°C, because the inhibitory two-stem-loop structure melts at this temperature, thereby revealing the sequestered Shine-Dalgarno sequence and allowing translation of LcrF (Fig. 1B) (20). LcrF then induces production of the T3SS by binding to specific TTTaGYcTtTat (highly conserved nucleotides are in capital letters) DNA motifs in the promoter regions of many T3SS genes of the ysc/yop regulon and activating their expression (Fig. 1B) (30, 31).
HORMONES
The locus for enterocyte effacement (LEE)-encoded T3SS of EHEC, which is essential for formation of the characteristic attaching and effacing (AE) lesions, is regulated by two host hormones, epinephrine (Epi) and norepinephrine (NE), and by an autoinducer (AI-3) that is produced by the gut microbiota. Epi and NE are at the core of stress response signaling in humans and are recognized by G-protein-coupled receptors in mammalian cells (32). However, bacteria lack homologues of these mammalian adrenergic receptors and have instead evolved sensor histidine kinases (HKs) to recognize these host signals. These chemical signals are received and transmitted by two HKs, QseC and QseE (33–35). Why EHEC and other enteric pathogens have developed mechanisms to sense Epi and NE remains unclear, but given the important functions neurotransmitters play in gut homeostasis, it is thought that sensing these molecules may allow the pathogen to assess the fitness of the host and modulate its virulence program accordingly (36). In fact, it has been shown that a qseC qseE double mutant is attenuated in an EHEC infant rabbit infection model (36). The importance of qseC and qseE in virulence has also been demonstrated in Citrobacter rodentium, a murine pathogen that harbors a LEE-encoded T3SS (36). C. rodentium LEE expression and gut colonization are both diminished in Dbh−/− mice, because they do not produce Epi and NE (36).
For EHEC, AI-3, Epi, and NE are first sensed by the membrane-bound HK QseC, resulting in autophosphorylation and transfer of a phosphate to its cognate response regulator (RR), QseB. Phosphorylated QseB (P-QseB) then activates the transcription of the qseB-qseC operon as well as the flagellar motility operon flhD-flhC (Fig. 2A) (32, 34). QseC can also phosphorylate the noncognate RRs QseF and KdpE (32, 37). P-KdpE directly activates transcription of ler, which encodes the master transcriptional activator of all LEE genes (37). QseC autophosphorylation in response to AI-3, Epi, or NE also activates the expression of the sensor HK QseE, which can itself sense Epi, sulfate, and phosphate (Fig. 2A). Both QseC and QseE can phosphorylate the RR QseF, thereby regulating the expression of the non-LEE-encoded effector T3SS EspFu, which is directly involved in AE lesion formation. QseC also indirectly induces expression of the λ phage-encoded Shiga toxin (stxAB) through the QseF RR by activating the SOS response. During the SOS response, expression of recA is upregulated and, through its coprotease activity, RecA cleaves the λ cl repressor of the stxAB genes, allowing for their expression (Fig. 2A) (32, 38).
FIG 2 .
Regulation of the LEE-encoded T3SS by hormone and nutritional sensing in EHEC. (A) The histidine sensor kinase QseC binds host hormones Epi and NE as well as the microbiome-produced AI-3. Upon ligand binding, QseC autophosphorylates and transfers phosphate to the RRs QseB, QseF, and KdpE. P-KdpE activates expression of the LEE-encoded T3SS through Ler. P-QseB modulates flagellar operons, while P-QseF induces expression of the non-LEE T3SS effector EspFU and activates the SOS response, which in turn activates stx2 expression. QseF is also phosphorylated by the sensor HK QseE in response to Epi, SO42−, and PO43−. (B) Within the intestinal lumen, EHEC senses fucose produced by B. thetaiotaomicron through the HK/RR pair FusKR, which represses T3SS expression when fucose is present. To reach the intestinal epithelium, EHEC produces mucinases that obliterate the mucosal layer and create a gluconeogenic environment. B. thetaiotaomicron then switches to gluconeogenic metabolism and secretes large amounts of succinate, which EHEC senses through the transcriptional regulator Cra. Sensing of succinate by Cra induces T3SS expression and AE lesion formation. The gluconeogenic environment also triggers EHEC’s stringent response, during which the alarmone ppGpp is synthesized. ppGpp directly binds RNA polymerase and modulates its activity, such that LEE expression is upregulated.
The structural basis by which QseC or QseE senses AI-3, Epi, or NE remains undefined. Although the crystal structure of the cytoplasmic domain of QseC is available, it does not include the periplasmic sensor region. Recently, Parker et al. used software to generate a predicted structure of the periplasmic domain of QseC (39). The authors identified 8 conserved residues in the periplasmic domain that may mediate ligand binding. These residues reside within a potential binding pocket formed by a series of β-sheets and proximal α-helices. However, mutation of these residues did not abrogate the ability of QseC to sense Epi and NE (39). Future structural studies of QseC and its ligands, NE and Epi, are required to completely understand how these signals are sensed.
SUGARS AND NUTRITIONAL STRESS
While inside the host gastrointestinal tract, enteric pathogens must not only adapt to the nutritional conditions of the gut but also use these nutritional cues to direct the appropriate expression of virulence factors. Pathogenic bacteria that colonize the colon (e.g., EHEC) do not have access to simple dietary sugars because these sugars are absorbed in the small intestine (40). These pathogens must compete with a specialized microbiota that resides in the lumen and outer mucous layer of the intestine. Many members of the microbiota can use the abundant undigested plant polysaccharides and host glycans found in the colon (40). However, neither commensal nor pathogenic E. coli, such as EHEC, can use polysaccharides. EHEC avoids competition with commensal E. coli for monosaccharides and disaccharides by invading the mucosal layer and adhering to enterocytes of the intestinal epithelium (41). To achieve this, EHEC must sense and appropriately respond to a variety of environmental signals.
A prominent member of the luminal microbiota, Bacteroides thetaiotaomicron, produces multiple fucosidases that cleave fucose from the host glycans comprising the mucosal layer (42). Initially, EHEC senses the high concentration of fucose through the HK and RR pair FusKR (Fig. 2B). Activation of the FusKR signaling cascade by fucose represses transcription of several genes, including Ler, the master regulator of the LEE-encoded T3SS (43). The downregulation of the T3SS and other LEE-encoded virulence factors, which are unnecessary for EHEC’s survival in the lumen, is a crucial energy-saving measure that helps EHEC outcompete commensal E. coli. During the second stage of its virulence strategy, EHEC senses other B. thetaiotaomicron metabolites (e.g., succinate) and ramps up expression of mucinase, which clears the mucous layer of the intestine and opens a path to the intestinal epithelium (Fig. 2B). Without the mucous layer as a primary carbon source, B. thetaiotaomicron switches to a gluconeogenic metabolism and secretes large amounts of succinate. The high levels of succinate are sensed by EHEC via an unknown mechanism and induce the transcriptional regulator Cra, which in turn activates expression of the T3SS (44).
The gluconeogenic, nutrient-poor environment also stimulates EHEC’s stringent response (Fig. 2B). The stringent response is a universal bacterial transcriptional regulatory mechanism that involves synthesis of the alarmone ppGpp by the RelA and SpoT enzymes during times of nutrient deprivation (45). ppGpp, along with the starvation-induced small protein DksA, directly binds to and modulates the activity of RNA polymerase. This activates or represses transcription at specific promoters, resulting in upregulation of LEE expression in EHEC (Fig. 2B) (46). The double activation of LEE expression by both Cra and ppGpp in response to the nutrient-poor conditions at the intestinal epithelium ensures that the LEE-encoded T3SS is expressed at the appropriate stage of infection.
CATIONIC ANTIMICROBIAL PEPTIDES
Cationic antimicrobial peptides (CAMPs) are produced by the host as part of the immune response to bacterial, viral, protozoan, or fungal infections (47). Due to their net positive charge and amphipathicity, CAMPs can interact with and permeate phospholipid membranes. Many bacterial pathogens, such as Salmonella enterica serovar Typhimurium (48), several streptococcal species (49), Neisseria meningitidis (50), and Pseudomonas aeruginosa (51), have evolved mechanisms of CAMP resistance. S. Typhimurium achieves this by sensing the CAMPs and modifying the lipopolysaccharides (LPSs) that decorate its cell surface. In S. Typhimurium, the genes involved in LPS modification are regulated by the PhoPQ two-component system (TCS) (48, 52). This TCS is composed of PhoQ, a membrane-spanning HK, and PhoP, a DNA-binding RR. In addition, the PhoPQ TCS also controls the expression of Salmonella pathogenicity islands 1 (SPI-1) and SPI-2, each of which encodes a T3SS (Fig. 3A) (53). PhoQ can function as both an HK and a phosphatase and thereby modulate the phosphorylation status of PhoP in response to environmental cues, such as CAMPs, divalent cations, and acidic pH (54, 55). Phosphorylated PhoP induces expression of SPI-2 T3SS genes and represses the expression of most SPI-1 T3SS genes (Fig. 3A) (56, 57). The SPI-1 T3SS is used for the initial invasion of nonphagocytic cells, and the SPI-2 T3SS is essential for maintaining the Salmonella-containing vacuole (SCV) and survival within phagocytic cells (58–60).
FIG 3 .
SP1-1 and SPI-2 T3SS regulation by CAMPs in Salmonella. (A) CAMPs are sensed by the PhoQ HK. When CAMPs are not present, divalent cations (gray spheres) bridge the acidic patch (yellow) of the PhoQ PD and the negatively charged IM phosphate groups, tethering the PhoQ PD to the membrane and locking it in a repressed state. When CAMPs are present, they compete for binding to the PhoQ PD acidic patch and displace the bound divalent cations, which frees the PhoQ PD from its membrane-locked state and induces a conformational change that activates PhoQ HK activity. The activated HK domain of PhoQ then phosphorylates the RR PhoP, which in turn induces SPI-2 expression and represses SPI-1 expression. (B) The crystal structure of the S. Typhimurium PhoQ PD dimer in a divalent cation-bound (gray spheres) state (PDB ID 1YAX). The N- and C-terminal TM helices, HAMP domain, and HK domain are represented as rectangles. Acidic patch residues (yellow sticks) coordinate cations (gray spheres) at the IM outer leaflet interface.
Crystallographic studies have demonstrated that divalent cations bind to an acidic patch created by the PhoQ-DcuS-CitA (PDC) sensor fold within the periplasmic domain (PD) (Fig. 3B) (61). In vitro studies using recombinant truncations of PhoQ reconstituted in lipid vesicles have shown that PhoQ can directly sense CAMPs through its PD (55). Binding of divalent cations to this acidic patch represses PhoQ HK activity (61, 62). The acidic patch is also important for CAMP recognition, suggesting that CAMPs and divalent cations compete for binding to this region (55). In vivo, divalent cations are thought to be coordinated between the PhoQ PD and the phosphate groups of the outer leaflet of the inner membrane (IM) (Fig. 3A and B) (61). The divalent cations bridge the negative charges of the PD acidic patch and the IM phosphates, thus tethering the PhoQ PD to the membrane. Based on nuclear magnetic resonance studies, the metal-bound, and therefore membrane-tethered, PhoQ PD is believed to be limited in structural flexibility and locked in a state that inactivates the HK domain (Fig. 3A) (55). However, when Salmonella is exposed to host CAMPs, the positively charged peptides compete for binding to the PhoQ acidic patch and displace bound divalent cations. This displacement is thought to release the PhoQ sensor domain from its membrane-locked state and induce a conformational change that promotes PhoQ HK activity (Fig. 3A) (55, 61).
The exact nature of the conformational change induced by CAMPs, as well as the mechanism of PhoQ HK activation, remains unclear. In a recent study, however, multistate Bayesian modeling of disulfide cross-linking data and existing structures of PhoQ homologous domains predicted that PhoQ transitions between two major states. These states are defined by the insertion or displacement of the PD acidic patch in the IM (Fig. 3A) (63). The authors found that insertion or removal of the PD acidic patch within the membrane was coupled with scissoring transitions, or diagonally opposing displacements, in the PD sensor domain, transmembrane (TM) helical bundles, HAMP, and DHp domains. These changes in conformation are predicted to alter HK domain activity (Fig. 3A and B), but further studies are needed to confirm these changes and to elucidate how they mechanistically alter PhoQ’s HK activity.
FATTY ACIDS
Fatty acids (FAs) in the intestinal tract are generated through the combined action of the host metabolism and the gut flora. Long-chain fatty acids (LCFAs) enter the small intestine through the diet and via fat metabolism (64). The resident microbiota in the large intestine produces abundant quantities of short-chain fatty acids (SCFAs), including acetate, propionate, and butyrate (reviewed in reference 65). Thus, host and bacterial metabolism result in different LCFA and SCFA concentrations along the GI tract. In addition to their metabolic roles, LCFAs and SCFAs act as cues for bacteria to sense their location in the GI tract and modulate the expression of their virulence factors.
Individual FAs have differential effects on S. Typhimurium virulence. For example, acetate induces invasion genes, while propionate, butyrate, and LCFAs reduce invasiveness (66–68). Regulation of S. Typhimurium virulence by FAs is dependent on the multilayered regulatory network that controls the expression of the invasive SPI-1 T3SS (Fig. 4). The BarA/SirA TCS, which is composed of the hybrid HK BarA and the DNA-binding RR SirA (also known as UvrY), is at the top of this network. The BarA/SirA TCS is activated by various SCFAs through different mechanisms (Fig. 4). Acetate activates SirA via the metabolite acetyl-phosphate, which phosphorylates SirA directly (66). This sensing mechanism bypasses BarA, as it depends on the conversion of acetate to acetyl-phosphate by metabolic enzymes (66). While the role of BarA in SCFA sensing for S. Typhimurium is not clear, work on its homologue in E. coli demonstrated that BarA senses formate (69), a signal that induces SPI-1 in S. Typhimurium (68). In addition, deletion of acetyl-phosphate synthesis genes in E. coli still results in low-level activation of a SirA-dependent operon in a BarA-dependent manner when acetate is present, suggesting that BarA also senses acetate (69). Once phosphorylated, the SirA RR activates the expression of CsrB and CsrC, two regulatory RNAs that repress the RNA-binding protein CsrA (Fig. 4) (70, 71). CsrB and CsrC sequester CsrA, preventing it from binding to the hilD mRNA Shine-Dalgarno sequence. The free hilD mRNA can then be translated.
FIG 4 .
Long- and short-chain fatty acids regulate SPI-1 expression in Salmonella. The SCFAs acetate and formate activate BarA HK activity and phosphotransfer to the SirA RR. The acetate metabolite, acetyl-P, can also directly phosphorylate SirA independently of BarA HK activity. P-SirA induces expression of CsrB and CsrC RNAs, which sequester the CsrA protein, thereby alleviating CsrA-mediated repression of hilD translation. The HilD protein then induces expression of the regulators HilC and RtsA, which through a feed-forward loop further amplify HilD expression. HilD, HilC, and RtsA also activate expression of HilA, which in turn induces expression of SPI-1 T3SS genes. Conversely, both LCFAs and proprionate negatively repress HilD activity. LCFAs bind to HilD and reduce its affinity for target DNA, including the hilA promoter. The propionate metabolite propionyl-CoA destabilizes the HilD protein, thereby inhibiting its transcription factor activity and activation of SPI-1 expression.
LCFA and propionate negatively affect SPI-1 T3SS expression by modulating HilD activity via different mechanisms (Fig. 4) (67, 72). LCFAs bind to HilD and reduce its affinity for DNA, thus reducing transcription from HilD-dependent promoters (72). HilD controls its own expression, as well as that of the regulators hilC and rtsA, resulting in a feed-forward loop that further amplifies LCFA suppression of SPI-1 expression (Fig. 4) (3). The effect of propionate is less direct, as its metabolite, propionyl coenzyme A (CoA), destabilizes HilD and thereby limits its activity (Fig. 4) (67). Golubeva et al. (72) proposed that absorption along the small intestine results in a low concentration of LCFAs in the distal ileum, which would relieve HilD repression and allow the expression of the SPI-1 activator hilA at this site. Since the propionate concentration is high in the colon and cecum, this molecule has been proposed to serve as a signal for S. Typhimurium that it has left the optimal site, the ileum, for infection (67). The repressive effects of certain FAs in Salmonella virulence and their antimicrobial activities have resulted in their use as supplements in animal feed (73). In addition to their direct effect on bacterial pathogens, SCFAs also influence host physiology in diverse ways, including alteration of immune function (74). Thus, the effects of SCFAs on gut pathogens such as Salmonella are probably due to a combination of their inhibitory effect on virulence, their antimicrobial properties, and the regulation of the host immune response.
IRON
Along with their eukaryotic hosts, most bacterial species require access to free iron for survival and replication (75, 76). Due to the toxicity of free ferric Fe3+ and host iron-sequestering mechanisms, the concentration of free iron is low in most host tissues (76). However, in the lumen of the small intestine, which is the primary site of iron absorption, free iron concentrations are high (77). S. Typhimurium uses the iron concentration to sense when it has entered the intestinal lumen (78). Many of the genes that are repressed by high Fe2+ concentrations are also repressed when S. Typhimurium is confined to the intestinal lumen (79). Iron concentration has been found to directly affect the expression of the S. Typhimurium SPI-1 T3SS. The activation of SPI-1 expression in response to high iron concentrations depends on the ferric uptake regulator Fur (78). Both metal chelation by 2,2-dipryridyl and deletion of fur have been shown to greatly reduce the transcription of the SPI-1 master regulator hilA under otherwise-SPI-1-inducing conditions (78). One mechanism by which Fur modulates hilA is by repressing expression of the gene encoding the global regulator H-NS (80). Under iron-limiting conditions, such as outside the host or within host macrophages, H-NS directly binds and represses transcription from the hilA promoter (Fig. 5A). Under iron-replete conditions, such as in the intestinal lumen, iron-bound Fur (Fur ⋅ Fe2+) binds to a regulatory site, termed the Fur box, in the hns promoter region and represses hns expression. By this mechanism, Fur ⋅ Fe2+ indirectly activates HilA and downstream SPI-1 expression (Fig. 5B) (81). In addition to acting through H-NS, Fur ⋅ Fe2+ also directly activates expression of hilD, a gene encoding the major transcriptional activator for hilA (82). Fur ⋅ Fe2+ binds to a Fur box upstream of the hilD promoter (positions −191 to −163) and activates the expression of hilD, which in turn activates the expression of hilA (Fig. 5B) (82). In most bacterial species, Fur ⋅ Fe2+ binds in the −35 to +12 region of target promoters and represses transcription through steric hindrance (83). However, in Salmonella, Fur ⋅ Fe2+ binds farther upstream in the hilD promoter region, thereby possibly activating hilD expression by directly recruiting RNA polymerase (RNAP) or by altering the conformation of the promoter DNA to facilitate RNAP loading (82).
FIG 5 .
Iron sensing and SPI-1 regulation by Fur in Salmonella. (A) Under iron-limiting conditions, apo-Fur cannot bind the Fur boxes upstream of the hilD or hns promoters and does not activate or repress expression of HilD or H-NS, respectively. H-NS then blocks transcription of hilA, the primary transcriptional activator of the SPI-1 T3SS. (B) Under iron-rich conditions (e.g., within the intestinal lumen), Fur binds free iron (Fe2+), inducing a conformational change that enables binding to Fur boxes upstream of hilD and hns. DNA-bound Fur then represses H-NS expression and activates expression of HilD, a transcriptional activator of hilA, resulting in HilA expression and downstream SPI-1 T3SS expression. (C) Crystal structures of M. gryphiswaldense MSR-1 apo, holo, and DNA-bound Fur. Binding of divalent cations (Mn2+, Fe2+) induces hinge-like movement of the DNA-binding domains of each monomer, creating a two-fold rotational axis within the holo-dimer and promoting DNA binding (PDB IDs 4RAY, 4RAZ, and 4RB2).
Fur is a major regulator of gene expression that is conserved in several bacterial species. It controls the expression of iron homeostasis genes as well as diverse iron-sensitive cellular processes, including respiration, DNA synthesis, and redox stress resistance (84, 85). In E. coli as well as many other bacterial species, Fur binds the Fur box in an Fe2+-dependent manner (86). The structural basis for the activation of Fur as a transcription factor by metal ions has not been specifically studied in S. Typhimurium, but structural studies performed on Fur homologues have begun to elucidate this mechanism. The Fur domain structure consists of an N-terminal DNA-binding domain (DBD) and a C-terminal dimerization domain linked by a disordered hinge region (Fig. 5C). A recent study of Magnetospirillum gryphiswaldense MSR-1 Fur, which solved high-resolution structures of apo, holo, and operator-bound forms of MSR-1 Fur, demonstrated that metal ion binding induces a caliper-like rotation and movement of the DBDs, orienting them for efficient DNA binding and stabilizing the structure (Fig. 5C) (87). Since no completely apo structures of any additional Fur homologues have been published, it is not known if a similar conformational change is induced by metal ion binding in Fur proteins from other species. However, all published holo structures of Fur (i.e., for E. coli, P. aeruginosa, and V. cholerae) adopt the same V-shaped conformation with the dimerization domains at the nexus and DNA-binding domains at the periphery (88).
BILE SALTS
Bile salts are a component of biliary secretions that are released into the small intestine to solubilize fats. They also have antimicrobial properties against bacteria that inhabit or transit the gastrointestinal tract (89). Successful enteric pathogens have developed ways to tolerate bile salt-induced stress and to use these compounds as markers of intestinal location. The marine bacterium Vibrio parahaemolyticus infects humans through the consumption of contaminated raw or undercooked seafood, causing acute gastroenteritis. V. parahaemolyticus enterotoxicity is largely mediated by a pathogenicity island (Vp-PAI) which encodes T3SS2, one of two T3SSs in this bacterium. T3SS2 expression is induced by bile salts via signaling, which includes the IM proteins VtrA, VtrB, and VtrC (Fig. 6A) (90–92). VtrA and VtrB are transcription factors that are embedded in the IM by a single transmembrane α-helix (Fig. 6A). VtrA also contains a C-terminal periplasmic domain that interacts with the periplasmic domain of VtrC, which is also anchored to the inner membrane. VtrA and VtrC form a 1:1 complex in which eight β-strands from VtrC and a single β-strand from VtrA form a β-barrel (Fig. 6B). Binding of bile salts to the hydrophobic interior of the VtrA/C dimer displaces a loop that covers the β-barrel in the apo structure (Fig. 6B, inset). This heterodimeric receptor transmits a signal upon bile salt binding that activates the DNA-binding domain of VtrA to induce transcription from the vtrB promoter (Fig. 6A) (92). VtrB then promotes transcription of T3SS2-related genes (90, 91). Interestingly, the topologies of the VtrA and VtrB transcription factors tether them to the IM, which may serve to localize T3SS gene products to the membrane site, where they will be assembled and exported. The V. parahaemolyticus bile-sensing system is conserved in V. cholerae strains that contain a pathogenicity island similar to Vp-PAI. In V. cholerae AM-19226, which lacks toxin-coregulated pilus and cholera toxin genes but contains a Vp-PAI-like island, T3SS gene expression depends on the VtrA and VtrB homologues VttRA and VttRB (93). The encoded VtrC homologue is also likely to be part of this regulatory mechanism (93–95).
FIG 6 .
Bile salt sensing and regulation of the Vp-PAI by VtrABC. The V. parahaemolyticus IM proteins VtrA and VtrC form a complex through interactions between their PDs. As V. parahaemolyticus enters the small intestine, bile salts bind to the VtrA-VtrC complex and activate the DBD of VtrA to induce vtrB transcription. VtrB then directly activates the transcription of T3SS2 and other Vp-PAI genes. (B) The crystal structure of the VtrA (purple) and VtrC (teal) PD complex bound to the bile salt taurodeoxycholate (green sticks) (PDB ID 5KEW). TM helices of both VtrA and VtrC are represented as rectangles, as is the DBD of VtrA. An overlay of the bile salt-bound (teal) and apo (yellow) structures illustrates the conformational change that results from bile salt binding (inset).
The inner membrane proteins ToxR and ToxS also play roles in the regulation of T3SS2 expression in V. parahaemolyticus. ToxR and ToxS have been linked to bile salt sensing in V. cholerae; however, the sensing mechanism remains unclear (96, 97). ToxR has the same domain arrangement as VtrA, with a cytoplasmic DBD, a transmembrane helix, and a periplasmic domain (Fig. 6B). Like VtrC, ToxS is composed of a transmembrane helix and a periplasmic domain. Despite having similar architectures, the periplasmic domains of V. parahaemolyticus ToxR and ToxS have less than 25% sequence similarity to VtrA and VtrC, respectively. Recent work identified ToxR in a genetic screen for factors contributing to colonization of the mammalian intestine and showed that ToxR is necessary for vtrB expression in V. parahaemolyticus (98). This suggests that ToxR works with VtrA and VtrC to regulate T3SS2 expression via VtrB. Work with V. cholerae AM-19226 supported a similar role for ToxR in the regulation of this strain’s T3SS (99). ToxR and ToxS also interact through their periplasmic domains (100). Furthermore, Miggett et al. (100) demonstrated that bile salts destabilize the ToxR periplasmic domain, which promotes its interaction with ToxS (100). Additional studies are needed to determine if bile salts have any direct effect on ToxR’s partner, ToxS.
CONCLUSIONS
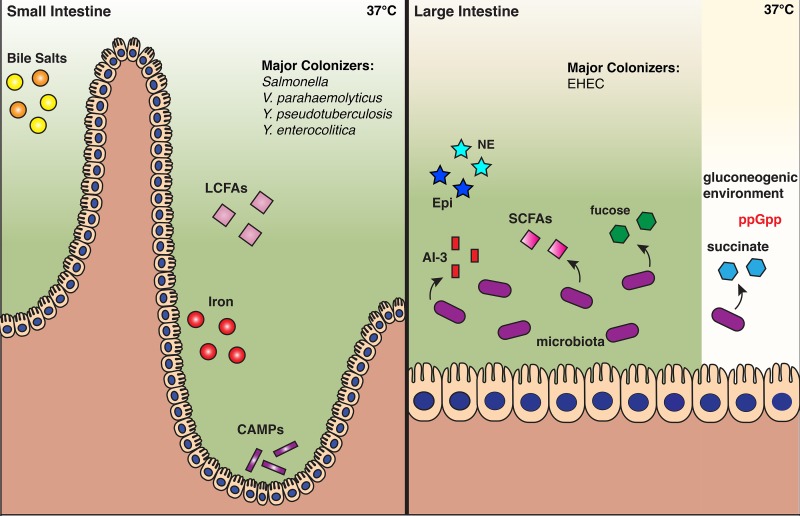
T3SSs are essential to many bacterial pathogens for successful host colonization and invasion. Because their production can reduce bacterial growth rates under certain conditions (101) and can trigger the host’s immune response (102), the timing of their expression must be tightly controlled. It is not surprising, then, that T3SS regulatory pathways are often multileveled and complex. Enteric pathogens have devised strategies to sense signals specifically related to the mammalian gut, and these strategies include molecules involved in host nutrition (iron, fatty acids), metabolism (bile salts), homeostasis (hormones), and microbiota by-products (fucose, succinate). These sensing mechanisms have been integrated into the regulatory networks controlling virulence and colonization factors, which often include T3SSs. As seen in the examples presented here, multiple signals often feed into a single regulatory cascade which has evolved complex feedback mechanisms that allow the bacterium to fine-tune T3SS expression in the signal-rich environment of the mammalian GI tract (Fig. 7).
FIG 7 .
Signals that regulate T3S in the mammalian GI tract. Spatial depiction of the signals that govern T3S in the small and large intestine. Certain signals, like bile salts and iron, are known to be constrained to a specific compartment, based on mammalian physiology, while others are depicted in the compartment where they are postulated to be at the greatest concentration and play the largest role in T3S regulation. Pathogens are listed based on their known major site of colonization within mammalian hosts. Although many host signals and enteric pathogens exist, this figure illustrates only the signals and specific pathogens discussed in this review.
Bacteria use T3SSs in different ways to promote their survival and proliferation within a host. Pathogens that use T3SSs to maintain their extracellular life cycle, like EHEC, inject effectors essential for establishing a strong attachment to intestinal epithelial cells (13). Other pathogens (e.g., S. flexneri and V. parahaemolyticus) use their T3SSs both extracellularly, to promote attachment and invasion, and intracellularly, to enhance their proliferation, facilitate vacuolar escape, and mediate cell-to-cell spread (103–106). Salmonella has two T3SSs that are used at different stages of infection and are often oppositely regulated by related signals to accommodate the complex wiring of downstream transcriptional regulatory networks.
There is still much to be learned about the signals and sensors that control T3SS expression in enteric bacterial pathogens. For example, it has been postulated that contact between a bacterium and its host cell plays a role in T3SS regulation. Recent work with EHEC suggested that the outer membrane lipoprotein NlpE may regulate T3SS expression through a surface-sensing mechanism involving the Cpx TCS (107, 108). However, NlpE is only required for activation of the LEE-encoded T3SS in response to attachment to abiotic surfaces and undifferentiated Caco-2 cells (107). It is possible that other lipoproteins, such as YafY, NlpA, Pal, or OsmB, sense adhesion to other cell types. However, further studies are needed to determine if these other lipoproteins are involved in the regulation of the T3SS in response to surface sensing (109). The mechanism by which the sensor domains NlpE and other outer membrane lipoproteins may sense hydrophobic surfaces is poorly understood and awaits further study. We anticipate that future work will further elucidate how biochemical signals from both the host and the environment are sensed by enteric pathogens to regulate T3S and other mechanisms of virulence. Elucidation of these sensing mechanisms could provide new potential therapeutic targets for the improved management of GI tract infections by enteric bacteria.
ACKNOWLEDGMENTS
We thank the members of the Orth lab for thoughtful discussion and revision of the manuscript.
This work was funded by Welch Foundation grant I-1561 and the Once Upon a Time… Foundation. N.J.D. was funded by NIH T32AI070116-10. K.O. is a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease, a Beckman Young Investigator, and a WW. Caruth, Jr., Biomedical Scholar and is an Earl A. Forsythe Chair in Biomedical Science.
The funders had no role in creating this work or in the decision to submit it for publication.
Footnotes
Citation De Nisco NJ, Rivera-Cancel G, Orth K. 2018. The biochemistry of sensing: enteric pathogens regulate type III secretion in response to environmental and host cues. mBio 9:e02122-17. https://doi.org/10.1128/mBio.02122-17.
REFERENCES
- 1.Humphries RM, Linscott AJ. 2015. Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev 28:3–31. doi: 10.1128/CMR.00073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dennehy PH. 2005. Acute diarrheal disease in children: epidemiology, prevention, and treatment. Infect Dis Clin North Am 19:585–602. doi: 10.1016/j.idc.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Erhardt M, Dersch P. 2015. Regulatory principles governing Salmonella and Yersinia virulence. Front Microbiol 6:949. doi: 10.3389/fmicb.2015.00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanj SS, Kanafani ZA. 2011. Current concepts in antimicrobial therapy against resistant gram-negative organisms: extended-spectrum beta-lactamase-producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae, and multidrug-resistant Pseudomonas aeruginosa. Mayo Clin Proc 86:250–259. doi: 10.4065/mcp.2010.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CR, Cho IH, Jeong BC, Lee SH. 2013. Strategies to minimize antibiotic resistance. Int J Environ Res Public Health 10:4274–4305. doi: 10.3390/ijerph10094274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fredriksson-Ahomaa M. 2012. Isolation of enteropathogenic Yersinia from non-human sources. Adv Exp Med Biol 954:97–105. doi: 10.1007/978-1-4614-3561-7_12. [DOI] [PubMed] [Google Scholar]
- 7.Hoelzer K, Moreno Switt AI, Wiedmann M. 2011. Animal contact as a source of human non-typhoidal salmonellosis. Vet Res 42:34. doi: 10.1186/1297-9716-42-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Sánchez L, Melero B, Jaime I, Hänninen ML, Rossi M, Rovira J. 2017. Campylobacter jejuni survival in a poultry processing plant environment. Food Microbiol 65:185–192. doi: 10.1016/j.fm.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, Thompson S, Wilson S, Bean NH, Griffin PM, Slutsker L. 2000. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis 181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 10.Ashbolt NJ. 2004. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 198:229–238. doi: 10.1016/j.tox.2004.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellies JL, Barron AMS, Carmona AM. 2007. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun 75:4199–4210. doi: 10.1128/IAI.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer JM, Frost JA, Bolton FJ, Wareing DR. 2000. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J Food Prot 63:1654–1659. doi: 10.4315/0362-028X-63.12.1654. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen Y, Sperandio V. 2012. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front Cell Infect Microbiol 2:90. doi: 10.3389/fcimb.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnenberg MS. 2000. Pathogenic strategies of enteric bacteria. Nature 406:768–774. doi: 10.1038/35021212. [DOI] [PubMed] [Google Scholar]
- 15.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB. 2017. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 17.Konkel ME, Tilly K. 2000. Temperature-regulated expression of bacterial virulence genes. Microbes Infect 2:157–166. doi: 10.1016/S1286-4579(00)00272-0. [DOI] [PubMed] [Google Scholar]
- 18.Steinmann R, Dersch P. 2013. Thermosensing to adjust bacterial virulence in a fluctuating environment. Future Microbiol 8:85–105. doi: 10.2217/fmb.12.129. [DOI] [PubMed] [Google Scholar]
- 19.O Cróinín T, Carroll RK, Kelly A, Dorman CJ. 2006. Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol Microbiol 62:869–882. doi: 10.1111/j.1365-2958.2006.05416.x. [DOI] [PubMed] [Google Scholar]
- 20.Böhme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, Dersch P. 2012. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog 8:e1002518. doi: 10.1371/journal.ppat.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kortmann J, Narberhaus F. 2012. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol 10:255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 22.Straley SC, Perry RD. 1995. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol 3:310–317. doi: 10.1016/S0966-842X(00)88960-X. [DOI] [PubMed] [Google Scholar]
- 23.Hoe NP, Minion FC, Goguen JD. 1992. Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J Bacteriol 174:4275–4286. doi: 10.1128/jb.174.13.4275-4286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bölin I, Forsberg A, Norlander L, Skurnik M, Wolf-Watz H. 1988. Identification and mapping of the temperature-inducible, plasmid-encoded proteins of Yersinia spp. Infect Immun 56:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson KE, Wolf-Watz H. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 26.Cornelis GR, Biot T, Lambert de Rouvroit C, Michiels T, Mulder B, Sluiters C, Sory MP, Van Bouchaute M, Vanooteghem JC. 1989. The Yersinia yop regulon. Mol Microbiol 3:1455–1459. doi: 10.1111/j.1365-2958.1989.tb00129.x. [DOI] [PubMed] [Google Scholar]
- 27.Nieto JM, Madrid C, Miquelay E, Parra JL, Rodríguez S, Juárez A. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J Bacteriol 184:629–635. doi: 10.1128/JB.184.3.629-635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoe NP, Goguen JD. 1993. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol 175:7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson MW, Silva-Herzog E, Plano GV. 2004. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol Microbiol 54:1364–1378. doi: 10.1111/j.1365-2958.2004.04353.x. [DOI] [PubMed] [Google Scholar]
- 30.Wattiau P, Cornelis GR. 1994. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J Bacteriol 176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skurnik M, Toivanen P. 1992. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J Bacteriol 174:2047–2051. doi: 10.1128/jb.174.6.2047-2051.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog 5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A 100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. 2006. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci U S A 103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reading NC, Rasko DA, Torres AG, Sperandio V. 2009. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc Natl Acad Sci U S A 106:5889–5894. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreira CG, Russell R, Mishra AA, Narayanan S, Ritchie JM, Waldor MK, Curtis MM, Winter SE, Weinshenker D, Sperandio V. 2016. Bacterial adrenergic sensors regulate virulence of enteric pathogens in the gut. mBio 7:e00826-16. doi: 10.1128/mBio.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. 2012. Virulence meets metabolism: cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. mBio 3:e00280-12. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pacheco AR, Sperandio V. 2012. Shiga toxin in enterohemorrhagic E. coli: regulation and novel anti-virulence strategies. Front Cell Infect Microbiol 2:81. doi: 10.3389/fcimb.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker CT, Russell R, Njoroge JW, Jimenez AG, Taussig R, Sperandio V. 2017. Genetic and mechanistic analyses of the periplasmic domain of the enterohemorrhagic Escherichia coli QseC histidine sensor kinase. J Bacteriol 199:e00861-16. doi: 10.1128/JB.00861-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreyra JA, Ng KM, Sonnenburg JL. 2014. The enteric two-step: nutritional strategies of bacterial pathogens within the gut. Cell Microbiol 16:993–1003. doi: 10.1111/cmi.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. 2014. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. 2010. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. 2006. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol 61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- 47.Hancock RE, Diamond G. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol 8:402–410. doi: 10.1016/S0966-842X(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 48.Matamouros S, Miller SI. 2015. S. Typhimurium strategies to resist killing by cationic antimicrobial peptides. Biochim Biophys Acta 1848:3021–3025. doi: 10.1016/j.bbamem.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaRock CN, Nizet V. 2015. Cationic antimicrobial peptide resistance mechanisms of streptococcal pathogens. Biochim Biophys Acta 1848:3047–3054. doi: 10.1016/j.bbamem.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol 187:5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidtchen A, Frick IM, Andersson E, Tapper H, Björck L. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol Microbiol 46:157–168. doi: 10.1046/j.1365-2958.2002.03146.x. [DOI] [PubMed] [Google Scholar]
- 52.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 53.Dalebroux ZD, Miller SI. 2014. Salmonelleae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castelli ME, García Véscovi E, Soncini FC. 2000. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J Biol Chem 275:22948–22954. doi: 10.1074/jbc.M909335199. [DOI] [PubMed] [Google Scholar]
- 55.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 56.Baxter MA, Jones BD. 2015. Two-component regulators control hilA expression by controlling fimZ and hilE expression within Salmonella enterica serovar Typhimurium. Infect Immun 83:978–985. doi: 10.1128/IAI.02506-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190:79–90. doi: 10.1534/genetics.111.132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galán JE, Curtiss R III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ochman H, Soncini FC, Solomon F, Groisman EA. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A 93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A 93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI, Xu W. 2006. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J Mol Biol 356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 62.García Véscovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 63.Molnar KS, Bonomi M, Pellarin R, Clinthorne GD, Gonzalez G, Goldberg SD, Goulian M, Sali A, DeGrado WF. 2014. Cys-scanning disulfide crosslinking and bayesian modeling probe the transmembrane signaling mechanism of the histidine kinase, PhoQ. Structure 22:1239–1251. doi: 10.1016/j.str.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi Y, Burn P. 2004. Lipid metabolic enzymes: emerging drug targets for the treatment of obesity. Nat Rev Drug Discov 3:695–710. doi: 10.1038/nrd1469. [DOI] [PubMed] [Google Scholar]
- 65.Louis P, Flint HJ. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 66.Lawhon SD, Maurer R, Suyemoto M, Altier C. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol 46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 67.Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, McClelland M, Ahmer BM, Altier C. 2013. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol Microbiol 87:1045–1060. doi: 10.1111/mmi.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. 2008. Formate acts as a diffusible signal to induce Salmonella invasion. J Bacteriol 190:4233–4241. doi: 10.1128/JB.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chavez RG, Alvarez AF, Romeo T, Georgellis D. 2010. The physiological stimulus for the BarA sensor kinase. J Bacteriol 192:2009–2012. doi: 10.1128/JB.01685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fortune DR, Suyemoto M, Altier C. 2006. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun 74:331–339. doi: 10.1128/IAI.74.1.331-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teplitski M, Goodier RI, Ahmer BM. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol 185:7257–7265. doi: 10.1128/JB.185.24.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Golubeva YA, Ellermeier JR, Cott Chubiz JE, Slauch JM. 2016. Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. mBio 7:e02170-15. doi: 10.1128/mBio.02170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Immerseel F, Russell JB, Flythe MD, Gantois I, Timbermont L, Pasmans F, Haesebrouck F, Ducatelle R. 2006. The use of organic acids to combat Salmonella in poultry: a mechanistic explanation of the efficacy. Avian Pathol 35:182–188. doi: 10.1080/03079450600711045. [DOI] [PubMed] [Google Scholar]
- 74.Morrison DJ, Preston T. 2016. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 76.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 77.Conrad ME, Umbreit JN. 2002. Pathways of iron absorption. Blood Cells Mol Dis 29:336–355. doi: 10.1006/bcmd.2002.0564. [DOI] [PubMed] [Google Scholar]
- 78.Ellermeier JR, Slauch JM. 2008. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J Bacteriol 190:476–486. doi: 10.1128/JB.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Janakiraman A, Slauch JM. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol Microbiol 35:1146–1155. doi: 10.1046/j.1365-2958.2000.01783.x. [DOI] [PubMed] [Google Scholar]
- 80.Troxell B, Sikes ML, Fink RC, Vazquez-Torres A, Jones-Carson J, Hassan HM. 2011. Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 193:497–505. doi: 10.1128/JB.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schechter LM, Jain S, Akbar S, Lee CA. 2003. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect Immun 71:5432–5435. doi: 10.1128/IAI.71.9.5432-5435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teixidó L, Carrasco B, Alonso JC, Barbé J, Campoy S. 2011. Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro. PLoS One 6:e19711. doi: 10.1371/journal.pone.0019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother’s repressor: the complex role of Fur in pathogenesis. Infect Immun 77:2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hantke K. 2001. Iron and metal regulation in bacteria. Curr Opin Microbiol 4:172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 85.Stojiljkovic I, Bäumler AJ, Hantke K. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol 236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 86.Escolar L, Pérez-Martín J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deng Z, Wang Q, Liu Z, Zhang M, Machado AC, Chiu TP, Feng C, Zhang Q, Yu L, Qi L, Zheng J, Wang X, Huo X, Qi X, Li X, Wu W, Rohs R, Li Y, Chen Z. 2015. Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat Commun 6:7642. doi: 10.1038/ncomms8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pérard J, Covès J, Castellan M, Solard C, Savard M, Miras R, Galop S, Signor L, Crouzy S, Michaud-Soret I, de Rosny E. 2016. Quaternary structure of fur proteins, a new subfamily of tetrameric proteins. Biochemistry 55:1503–1515. doi: 10.1021/acs.biochem.5b01061. [DOI] [PubMed] [Google Scholar]
- 89.Begley M, Gahan CG, Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 90.Gotoh K, Kodama T, Hiyoshi H, Izutsu K, Park KS, Dryselius R, Akeda Y, Honda T, Iida T. 2010. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS One 5:e13365. doi: 10.1371/journal.pone.0013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kodama T, Gotoh K, Hiyoshi H, Morita M, Izutsu K, Akeda Y, Park KS, Cantarelli VV, Dryselius R, Iida T, Honda T. 2010. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS One 5:e8678. doi: 10.1371/journal.pone.0008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li P, Rivera-Cancel G, Kinch LN, Salomon D, Tomchick DR, Grishin NV, Orth K. 2016. Bile salt receptor complex activates a pathogenic type III secretion system. eLife 5:e15718. doi: 10.7554/eLife.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shin OS, Tam VC, Suzuki M, Ritchie JM, Bronson RT, Waldor MK, Mekalanos JJ. 2011. Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. mBio 2:e00106-11. doi: 10.1128/mBio.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alam A, Tam V, Hamilton E, Dziejman M. 2010. vttRA and vttRB encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain. Infect Immun 78:2554–2570. doi: 10.1128/IAI.01073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rivera-Cancel G, Orth K. 2017. Biochemical basis for activation of virulence genes by bile salts in Vibrio parahaemolyticus. Gut Microbes 8:366–373. doi: 10.1080/19490976.2017.1287655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hung DT, Mekalanos JJ. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci U S A 102:3028–3033. doi: 10.1073/pnas.0409559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Provenzano D, Schuhmacher DA, Barker JL, Klose KE. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun 68:1491–1497. doi: 10.1128/IAI.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hubbard TP, Chao MC, Abel S, Blondel CJ, Abel Zur Wiesch P, Zhou X, Davis BM, Waldor MK. 2016. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc Natl Acad Sci U S A 113:6283–6288. doi: 10.1073/pnas.1601718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miller KA, Sofia MK, Weaver JW, Seward CH, Dziejman M. 2016. Regulation by ToxR-like proteins converges on vttRB expression to control type 3 secretion system-dependent Caco2-BBE cytotoxicity in Vibrio cholerae. J Bacteriol 198:1675–1682. doi: 10.1128/JB.00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Midgett CR, Almagro-Moreno S, Pellegrini M, Taylor RK, Skorupski K, Kull FJ. 2017. Bile salts and alkaline pH reciprocally modulate the interaction between the periplasmic domains of Vibrio cholerae ToxR and ToxS. Mol Microbiol 105:258–272. doi: 10.1111/mmi.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. 2011. The cost of virulence: retarded growth of Salmonella typhimurium cells expressing type III secretion system 1. PLoS Pathog 7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao Y, Shao F. 2015. The NAIP-NLRC4 inflammasome in innate immune detection of bacterial flagellin and type III secretion apparatus. Immunol Rev 265:85–102. doi: 10.1111/imr.12293. [DOI] [PubMed] [Google Scholar]
- 103.de Souza Santos M, Orth K. 2014. Intracellular Vibrio parahaemolyticus escapes the vacuole and establishes a replicative niche in the cytosol of epithelial cells. mBio 5:e01506-14. doi: 10.1128/mBio.01506-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Souza Santos M, Salomon D, Orth K. 2017. T3SS effector VopL inhibits the host ROS response, promoting the intracellular survival of Vibrio parahaemolyticus. PLoS Pathog 13:e1006438. doi: 10.1371/journal.ppat.1006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang L, Krachler AM, Broberg CA, Li Y, Mirzaei H, Gilpin CJ, Orth K. 2012. Type III effector VopC mediates invasion for Vibrio species. Cell Rep 1:453–460. doi: 10.1016/j.celrep.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coburn B, Sekirov I, Finlay BB. 2007. Type III secretion systems and disease. Clin Microbiol Rev 20:535–549. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shimizu T, Ichimura K, Noda M. 2015. The surface sensor NlpE of enterohemorrhagic Escherichia coli contributes to regulation of the type III secretion system and flagella by the Cpx response to adhesion. Infect Immun 84:537–549. doi: 10.1128/IAI.00881-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol 177:4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miyadai H, Tanaka-Masuda K, Matsuyama S, Tokuda H. 2004. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J Biol Chem 279:39807–39813. doi: 10.1074/jbc.M406390200. [DOI] [PubMed] [Google Scholar]