Abstract
Among various isoforms of Apolipoprotein E (ApoE), the E4 isoform (ApoE4) is considered to be the strongest risk factor for Alzheimer’s disease, whereas the E3 isoform (ApoE3) is neutral to the disease. Interestingly, the sequence of ApoE4 differs from its wild-type ApoE3 by a single amino acid C112R in the 299-amino-acid-long sequence. Hence, the puzzle remains: how a single-amino-acid difference between the ApoE3 and ApoE4 sequences can give rise to structural dissimilarities between the two isoforms, which can potentially lead to functional differences with significant pathological consequences. The major obstacle in addressing this question has been the lack of a 3D atomistic structure of ApoE4 to date. In this work, we resolve the issue by computationally modeling a plausible atomistic 3D structure of ApoE4. Our microsecond-long atomistic simulations elucidate key structural differences between monomeric ApoE3 and ApoE4, which renders ApoE4 thermodynamically less stable, less structured, and topologically less rigid compared to ApoE3. Consistent with an experimental report of the molten globule state of ApoE4, simulations identify multiple partially folded intermediates for ApoE4, which are implicated in the stronger aggregation propensity of ApoE4.
Introduction
One of the most common forms of dementia is Alzheimer’s disease (AD) (1), characterized by the formation of neurofibrillary tangles and plaques in the brain, leading to neuronal dysfunction, neuronal loss, and finally, death. The increased risk of developing AD is associated with the Apolipoprotein E (ApoE) gene (2, 3, 4), which is located on the long arm of chromosome 19 (5). The concentration of ApoE in the brain and cerebrospinal fluid is 10–20% of that in the plasma (6). The E4 isoform of ApoE (abbreviated as ApoE4) is considered to be the principal genetic factor in developing late-onset AD (7). It is one of the three common isoforms of ApoE, namely ApoE2, ApoE3, and ApoE4, which are coded by same gene locus. All three isoforms consist of 299 amino acid residues with the only mutual difference being single amino acid changes. Specifically, these isoforms differ only by two residues at positions 112 and 158 of the protein sequence, where C112/C158, C112/R158, and R112/R158 are present in ApoE2, ApoE3, and ApoE4, respectively (7). It is, however, intriguing that such a small sequence difference among the ApoE isoforms can lead to significant functional and biophysical differences.
ApoE is known for its lipid transport function and is associated with multiple lipoprotein classes, including very low-density lipoprotein and high-density lipoprotein (HDL) (8). There exists a reversibility between lipid-free and lipid-bound states of ApoE isoforms (9). More significantly, ApoE isoforms have different binding preferences toward different lipoprotein classes: ApoE4 prefers very low-density lipoprotein, whereas ApoE2 and ApoE3 prefers HDL (10). The lipid-bound structure of ApoE is likely to be different from its lipid-free structure (9). The different preferential binding of ApoE3 and ApoE4 isoforms with lipid raises the fundamental question of whether the structure of these isoforms mutually differs in their respective lipid-free forms, even before one asks the question of their structure in their respective lipid-bound forms. Moreover, many biophysical experiments to date have been performed with the recombinant lipid-free form of these isoforms (11, 12, 13). Hence, resolving the structure of these isoforms in the lipid-free form is of paramount interest even before we explore their lipid-bound state and structural changes occurring during the lipid binding process.
In a lipid-free state, ApoE3 and ApoE4 exist as a slow equilibrium mixture of monomer, tetramer, octamer, and a small proportion of higher oligomers (8). This work, using computer simulations, provides unprecedented insights into the structural differences between lipid-free ApoE3 and ApoE4 isoforms in their monomeric form, which can potentially contribute to their functional differences. This article also lays the groundwork for future investigation of the multimeric lipid-free as well as the lipid-bound forms.
Although these isoforms mutually differ by only one amino acid in their respective sequence, ApoE4, among them, is known to be the strongest causative factor for late-onset AD, whereas, ApoE2 and ApoE3 have been found to be protective and neutral, respectively, in terms of propensity to develop the disease (2, 3, 4, 14). The population of allele APOE ε4 increases from 14% in control to 40% in an AD population (15). Hence, the question arises how a single-residue change in an amino acid causes this huge difference in physiological behavior. Experimental studies indicate that ApoE4 is less stable than ApoE3 and binds more strongly to lipid receptors (16), which is often implicated in the increased risk of cardiovascular disease (17, 18). ApoE4 also leads to reduced clearance of extracellular amyloid plaque deposits in the brain (14), increases neuronal degeneration (19), and reduces longevity (20). It is known that ApoE can bind to amyloid-β peptide (11), but further mechanistic detail is lacking.
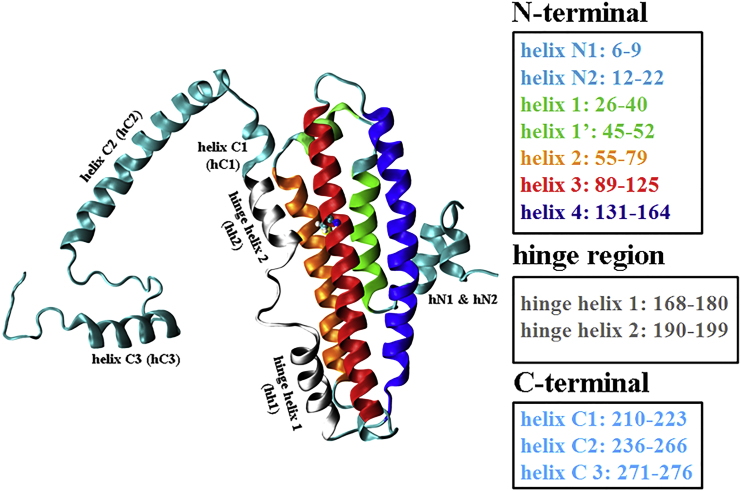
The most critical question concerns how the single-amino-acid difference (C112R) on ApoE3 → ApoE4 isoform affects protein structure or dynamics, which can potentially cause this profound physiological and pathological variation. Because the functional differences between ApoE3 and ApoE4 are generally hypothesized (21, 22) to be dictated by their respective structure, deciphering the atomistic details of their structure is of paramount interest on many aspects. Although it is already established by multiple studies that ApoE has two domains—one 22-kDa N-terminal domain (residues 1–191) that contains the low density lipoprotein receptor binding site, and the other a 10-kDa C-terminal domain (216–299) that binds to lipids (21), the progress in the area of structure determination has been thwarted by the lack of reported structure of any full-length wild-type isoform of ApoE3 or ApoE4. The major cause behind lack of monomeric structure has been that both ApoE3 and ApoE4 aggregate at the concentration needed for crystallographic determination and also form oligomers at micromolar concentration, which makes NMR studies highly improbable. A crystal structure of only N-terminal domain (residues 1–191) of ApoE was reported in 1991, and it exists as an elongated four-helix bundle (23). The structure of C-terminal domain (residues 216–299) is not yet known, but from circular dichroism spectroscopy it has been predicted to be highly α-helical (24). It has also been proposed that the N-terminal and C-terminal domains are connected by a flexible hinge region (21). In 2011, the structure of a full-length monomeric mutated ApoE3 was proposed by Chen et al. (25) (Fig. 1). There are several indications, based on hydrogen-deuterium exchange experiments and other biophysical characterizations, that this monomeric mutated form of ApoE3 is very nearly identical to the wild-type ApoE3, and thus is considered to be a true representation of wild-type ApoE3 in its lipid-free monomeric form (25, 26).
Figure 1.
Structure of ApoE3 based on the NMR report (25). Model of the four-helix N-terminal domain, connected to the C-terminal domain by a hinge region. (Inset) Detailed description of each helix is given. To see this figure in color, go online.
To date, no NMR or crystal structure of ApoE4 could be obtained, mainly because at 37°C, ApoE4 displays the greatest tendency to form high-molecular-weight aggregates as compared to ApoE3 or ApoE2 (27). However, two key properties of ApoE4 have been suggested to underlie its association with the disease: 1) domain interactions and 2) reduced stability relative to ApoE3 (9). Since structural details underlie the function, we hypothesize that the origin of functional differences between ApoE3 and ApoE4 might be rooted in their differences in molecular structures. Toward this end, our current work employs extensive computer simulations at an atomistic scale to explore the potential structural differences between monomeric mutants of ApoE3 and ApoE4.
Starting with the NMR structure of monomeric mutant ApoE3 (25) and via in silico C112R mutation, we perform multi-microsecond-long unbiased MD simulation in the presence of explicit water molecules, carry out free energy simulation, and construct a Markov state model to atomistically delineate the structural differences between ApoE3 and ApoE4. Although our free energy simulations in general confirm that ApoE4 is thermodynamically less stable as compared to ApoE3, our results delve further into the atomistic differences of both the secondary structure and the tertiary folds between the two isoforms. Specifically, we find that the hinge region corresponding to residues 190–199 is completely unstructured in ApoE4. Furthermore, our simulation result also reveals that helix 3 (residues 89–125) of the N-terminal four-helix bundle, containing the mutation site 112, is bent in ApoE4, whereas it remains straight in ApoE3. The Markov state model constructed based on simulation trajectories predicted a single major topologically rigid conformation for ApoE3 while multiple metastable macrostates for ApoE4, showing inherent structural heterogeneity. Our work also sheds light on the differential existence of N- and C-terminal domain interaction between ApoE3 and ApoE4. This work represents the most extensive structural elucidation of ApoE3 and ApoE4 in their monomeric lipid-free forms to date.
Methods
Simulation model
The starting structure for ApoE3 isoform was directly taken from the RCSB Protein Data Bank (PDB: 2L7B), which is the NMR structure of monomeric mutant ApoE3 published by Chen et al. (25). The starting structure for ApoE4 isoform was obtained by mutating Cysteine to Arginine at position 112 in silico using CHARMM-GUI (28, 29, 30). The two systems, i.e., ApoE3 and ApoE4 isoforms, were simulated independently using the GROMACS 5.1.4 package (31, 32) with the CHARMM36 force field (33).
Simulation method
In each of the simulation, the protein was explicitly solvated with TIP3P water molecules in a rectangular periodic box whose dimensions were at least 1.5 nm larger than the size of the corresponding solute molecules and were subsequently charge-neutralized by adding NaCl at 0.15 M concentration. The entire setup was generated using CHARMM-GUI. Each system, with a box size of 9.5 nm, consisted of ∼25,120 water molecules and a total of ∼80,300 particles including protein atoms and ions.
The initial round of simulation with explicit solvent and ions involved 50,000 steps of steepest descent energy minimization. After energy minimization, the respective systems were gently heated from 0 to 300 K in 25 ps with a time step of 0.001 ps followed by a final production run with the NPT ensemble using a 0.002-ps time step. The particle-mesh Ewald summation method (34) was used to treat long-range electrostatic interactions (with Fourier spacing of 0.12 nm and interpolation order four) and force-switch method was applied for nonbonded interactions (van der Waals) with a cutoff of 1.2 nm. The real-space cutoff was set to 1.2 nm. The Verlet cutoff scheme was implemented. We used the Nosé-Hoover thermostat (35) for maintaining the average temperature of 300 K and the Parrinello-Rahman (36, 37) barostat for maintaining the average pressure of 1 bar. All the hydrogen atoms bonded to the protein heavy atoms are constrained using LINCS algorithm (38) to their respective equilibrium bond-length. All the water molecules were simulated as rigid molecules using SETTLE (39). Both the isoforms were first individually simulated multiple times by varying initial velocity distributions of the systems. Further, to ensure more efficient sampling, we divided all the conformations of these simulated trajectories into 20 clusters using the k-center clustering algorithm (40) and then selected one conformation randomly from each cluster as an initial conformation to run subsequent unbiased short MD runs (each 50 ns). Overall, an aggregated total of 1.725- and 2.175-μs simulations have been performed for ApoE3 and ApoE4 isoforms, respectively. All probability distributions plotted in this article are normalized such that the area under the individual curve is unity.
We also performed two independent simulations for ApoE3 and two for ApoE4 considering only the first 200 residues, i.e., considering only the N-terminal domain. Each MD run was 300 ns long. All simulations were performed using GROMACS 5.1.4 and the simulation protocols applied for truncated proteins were same as those applied for simulating the entire ApoE isoforms, described earlier in this article. In these simulations, the system box size was 9.0 nm with ∼23,500 water molecules.
Markov state model construction and validation
The complex nature of aggregated trajectories was statistically analyzed for key structural insights by constructing Markov state models (MSMs). MSMs (41, 42, 43) were constructed using all the aggregated trajectories. In all the trajectories, each frame was converted to a single vector of 299 elements where elements were defined as the distances of Cα atoms from a reference structure after removing rotational and translational motions. The time-structure-based independent component analysis method (44) was used for dimensionality reduction. The high-dimensional simulated data was then projected onto the slowest varying four independent component analyses considering a 1-ns lag time. The k-means clustering algorithm (45) was used to discretize this 4D data into 70 microstates. A kinetic network, i.e., MSMs in this discretized space, was then constructed by computing the transition probability matrix at variable lag-times to determine the implied timescales that correspond to the timescale required for transition between different microstates. To obtain the transition probability matrix, T(τ), i.e., probability of transitioning between microstates, the number of transitions between the microstates at an interval of a certain lag time τ was counted and the transition count matrix was then symmetrized and normalized by column. We have used a lag time of 1 ns to construct the MSM as the implied timescale leveled off at ∼1 ns, which ensures the Markovianity of the model (Fig. S1). These MSMs were then used to calculate the equilibrium state properties. To better understand the structure and dynamics of ApoE3 and ApoE4, we further lumped the microstates model into three macrostate model (metastable states) using the Perron cluster-cluster-analysis-plus (46) lumping method. The MSM construction was performed using the software PyEMMA (47).
Umbrella sampling simulation
An umbrella sampling simulation was performed to quantitatively compare the free energy profile of the helical propensity of the hinge helix 2 region (residue 188–200) between ApoE3 and ApoE4. We used a popular free-energy plugin PLUMED 2.1 (48) patched with GROMACS 5.0.6 (31, 32) to perform the necessary umbrella sampling simulation. Within PLUMED, a reaction-coordinate or collective variable (CV) termed “alpharmsd” was chosen to define the total helical content. The details of the alpharmsd CV can be found in its original implementation by Pietrucci and Laio (49) and in the PLUMED user manual (50). Briefly, this CV probing the total helical content of protein structure is built on the premise that any chain of six contiguous residues in a protein chain can form an α-helix. This CV thus generates the set of all possible six-residue sections and calculates the root mean-squared deviation (RMSD) distance between the configuration in which the residues find themselves and an idealized α-helical structure. In this simulation, the helical segment of hinge helix 2, as present in the NMR-derived structure of ApoE3 and the in silico mutated structure of ApoE4, is considered to be the reference structure. Within the PLUMED implementation of alpharmsd, this is done by calculating the following sum of functions of the RMSD distances:
where the summation runs over all possible segments of α-helix; and parameters of the switching function n and m are set to the default values 8 and 12, as d0 = 0 nm and r0 = 0.08 nm, respectively.
A total number of 15 equally spaced umbrella windows ranging between 0 and 100% helical content were used for umbrella-sampling the helical content of the hinge helix 2 regions of ApoE3 and ApoE4. The initial configuration corresponding to each window was extracted from prior equilibrium simulations for each of the respective isoforms. A harmonic force constant of 60 kJ/mol was found to be optimum for having Gaussian distribution around each window and for maintaining sufficient overlap in the distribution between two adjacent windows. Each of the umbrella-sampling windows were subjected to 500 ps of equilibration in the NPT ensemble followed by 2 ns of production run in the NVT ensemble, using protocols similar to those previously discussed in this article. Finally, the weighted histogram analysis method (51, 52) was used over the last 10 ns of each of the umbrella-sampled trajectories to generate unbiased histograms and the corresponding potentials of mean force or free energies.
Free energy perturbation
The relative thermodynamic stability between ApoE3 and ApoE4 isoforms, which differ by single residue mutation C112R, were assessed by utilizing the techniques of free energy perturbation (FEP) as implemented within the software GROMACS 5.0.6. We calculated the relative binding free energy for mutation of Cysteine to Arginine at 112th residue of ApoE isoform by performing two sets of FEP simulations: free energy change due to C112R mutation within ApoE protein (ΔGcomplex) and free energy change due to C112R mutation in pure water (ΔGsolution). The relative thermodynamic stability is thereby assessed by ΔΔG = ΔGcomplex − ΔGsolution.
The FEP simulation was carried out in discrete steps for a series of coupling parameters λ with values ranging from λ = 0 (corresponding to the protein with Cys112) to λ = 1 (corresponding to the protein with Arg112). The topology files for FEP calculation were prepared using an automated Python tool, FESETUP (53). A total of 21 λ-values were used in both FEP calculations. At each λ-value, both Coulomb and LJ interactions are decoupled simultaneously. The nonbonding interactions, bond length, and angle were linearly interpolated. To avoid divergence near λ = 0 or λ = 1, a soft-core potential is used to modify the LJ interactions. Each λ-window is subjected to a cycle of energy minimization, followed by equilibration for 2 ns in the NVT ensemble and finally 5 ns of production runs in the NPT ensemble at 300 K and 1 bar pressure using a Langevin dynamics integrator at a time step of 0.002 ps. All simulations were performed using GROMACS 5.0.6, and simulation protocols similar to those in previously described equilibrium simulations were implemented. Finally, ΔG values for each of the respective FEP calculations were obtained using the implementation of the multiple Bennett acceptance ratio provided with the Python package PYMBAR (54). The statistical uncertainty associated with the computed free energy change was obtained by block-averaging of the data.
Results
Overall, an aggregated total of 1.725- and 2.175-μs MD simulations have been performed for ApoE3 and ApoE4 isoforms, respectively. As mentioned in the Methods, initially we ran three independent MD runs for each isoform starting from different initial velocity distributions. An assessment regarding the dynamics and overall behavior of the simulation trajectories was obtained by calculating the RMSD of each snapshot with respect to the corresponding NMR structure considering only Cα atoms. The RMSD plots for all six MD runs are shown in Fig. S2. The RMSD versus time plots indicated that the systems reach a plateau for different time periods in different trajectories. Therefore, we have carried out all the analysis considering only the time periods where the systems maintain equilibrium population; this time period is also shown in Fig. S2. As for the 20 independent short 50-ns MD runs, done for ApoE3 and ApoE4 each, we have considered the first 25 ns as equilibration time and performed all analysis for the last 25 ns of each 50-ns-long trajectory. Before moving further with investigating the conformations of both the isoforms from the respective simulation trajectories, the quality of ApoE3 simulations was validated by comparing the available experimental chemical shift of the corresponding Cα atoms with our calculated values. Average chemical shifts of Cα atoms of each residue were calculated from simulated ensemble (considering all the frames) using the program SPARTA+ (55). A very good correlation between the calculated and experimental chemical shift with R = 0.94 and MSD 2.4 ppm (Fig. S3) suggested that an adequate sampling of the conformational space has been achieved for ApoE3, and that the choice of force field is appropriate.
The structures elucidated by x-ray crystallography (23) and NMR spectroscopy (26) clearly represent N- and C-terminal domains connected by a hinge region (approximately residues 168–200) (19, 56, 57) formed by two short helices, i.e., hinge helix 1 (residues 168–180) and hinge helix 2 (residues 190–199). In 3D-representation of ApoE3, one might notice that these two helices are positioned close to helix 3 (residues 87–122), which contains the mutation site 112, with hinge helix 2 lying very close to the mutation point (Fig. 1). We reiterate that our starting structure of ApoE3 is obtained from the NMR-derived structure reported by Chen et al. (25) (PDB: 2L7B), which is the monomeric mutant of a wild-type ApoE3 sequence that is currently touted as the most complete structure of ApoE3 in a monomeric lipid-free state. Specifically, this monomeric mutant of ApoE3 contains the mutations V269A, L279Q, and V287E in the loop after helix C3 and mutations F257A and W264R at the end of helix C2. These mutations were introduced by Chen et al. (25) to avoid self-aggregation and shows nearly identical biophysical features to the wild-type ApoE3, and thus is considered to be a true representation of the wild-type (58, 59).
An initial comparison of secondary structure, obtained using the DSSP algorithm (60) and a contact map (based on Cα-Cα distances) of the ApoE3 isoform as obtained from an ensemble-average of 20 members of the NMR-reported structure (25) and our time-averaged structure, indicated substantial differences in a few locations. As illustrated in Fig. S4, certain regions, namely, hinge helix 2 (residues 190–199), helix C1 (residues 210–223), and helix C3 (residues 271–276) have far less per-residue helical fraction in the time-averaged structure obtained from our MD simulations compared to the reported NMR structure (Fig. S4 a). This is in-line with the observation from contact map analysis, which shows a loss in overall compactness of the structure from its initial configuration (Fig. S4 b). This observation is also in line with that reported by Williams and Convertino (61) using implicit-solvent-based computer simulation. All these structural features, such as secondary structure, contact map, and compactness, have been well compared for ApoE3 and ApoE4 isoforms on the basis of our simulation trajectories, and are extensively discussed in the following sections.
Secondary structures of ApoE3 and ApoE4 have considerable differences
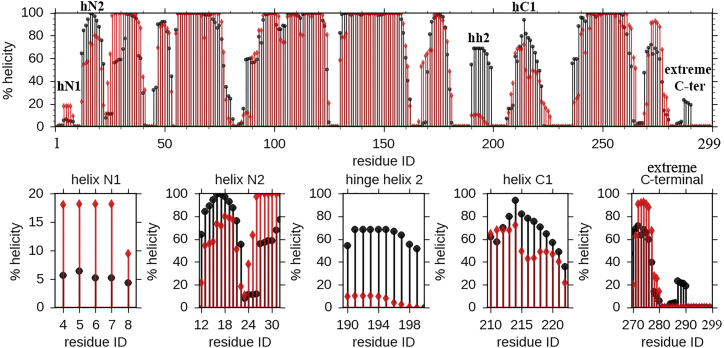
Along with the already available crystal structure of the N-terminal domain of ApoE3 (23) and NMR-reported structure of full-length monomeric mutated ApoE3 (25), a very recent report based on hydrogen exchange and mass spectrometry also suggests that the N-terminal domains of both ApoE3 and ApoE4 contains four helices (62) and our observations are in-line with this observation. In addition, we have found some strong differences between ApoE3 and ApoE4 in terms of per residue α-helicity. Fig. 2 compares percentage (%) helicity of each residue of ApoE3 and ApoE4 isoforms averaged over all trajectories; nearly 0% helicity indicates completely unstructured residue, whereas helicity of 100% implies a fully helix-forming residue. As evident from Fig. 2, the majority of the regions of the two isoforms are structurally very similar. The N-terminal domain formed by a four-helix bundle of helix 1 (residues 26–40), helix 2 (residues 55–79), helix 3 (residues 89–125), and helix 4 (residues 131–164) has α-helix conformations for all the four helices in both ApoE3 and ApoE4. This observation is consistent with the earlier mentioned report based on hydrogen exchange and mass spectrometry (62). However, certain key regions of ApoE4 are found by our computer simulation to be significantly structurally different from those of ApoE3. Our simulation results specifically point toward hinge helix 2 (residues 190–199), which shows major structural differences between ApoE3 and ApoE4. The hinge helix-2 region, which topologically lies close to the mutation site 112, is found to be completely unstructured in ApoE4 (nearly 0% helicity for all the residues), whereas in ApoE3, it is nearly 80% helical (Fig. 2).
Figure 2.
Differences in secondary structures between ApoE3 and ApoE4. Averaged secondary structures of ApoE3 (black) and ApoE4 (red) are as obtained from MD runs. The hinge helix 2 (residues 190–199) shows difference in helicity, with ApoE3 having much higher helicity compared to ApoE4. To see this figure in color, go online.
Apart from the hinge helix-2 region (residues 190–199), we also find that part of the C-terminal domain, namely, the helix-C3 region (residues 271–279), is structurally different for ApoE3 and ApoE4. As shown in Fig. 2, the helix-C3 region is completely unstructured in ApoE4, whereas it retains 25% helicity in ApoE3. This result is consistent with the experimental observation of Frieden and Garai (22), where it was proposed that due to propagation of the arginine-to-cysteine change at 112 the region 271–279, i.e., helix C3, is structurally different for ApoE3 and ApoE4 (22). Moreover, Fig. 2 also depicts nonnegligible differences in helicity in helix N2 region (residues 12–24) between the two isoforms, in congruence with the experimental observation of Frieden and Garai. In summary, our simulation result predicts significant differences between ApoE3 and ApoE4 with respect to the region, which is structurally or topologically very close to the mutation site. We have also observed differences with respect to regions that are both sequentially and topologically distant (residues 12–24 and 271–279) from the mutation site 112, and these observations are consistent with the earlier experimental work (21).
Markov state models quantify the structural heterogeneity in ApoE4 isoform
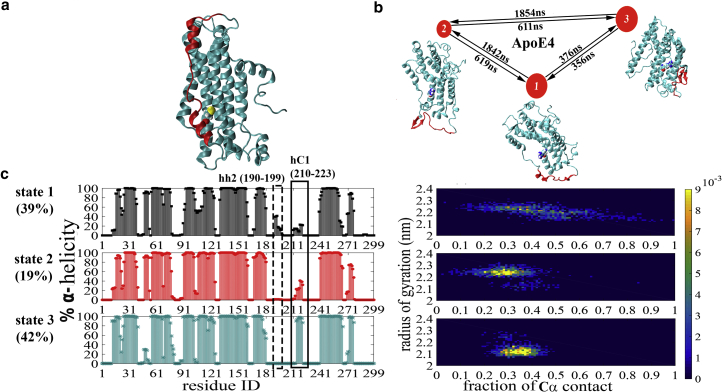
MSM-based analysis of the cumulative simulated trajectories of both ApoE isoforms quantified their inherent structural heterogeneities. The MSM-based analysis showed that equilibrium population of ApoE3 is highly dominated by only one state (Fig. 3 a) with a highly stable hinge helix-2 region, which is ∼80% helical.
Figure 3.
Macrostates identified by Markov state models. (a) Given here is a representative snapshot of the highly populated metastable state of ApoE3 (∼98%) obtained from MSM. (b) Shown here is the MSM of three metastable states found in ApoE4: State 1, State 2, and State 3, with respective populations of 39, 19, and 42%. Here, the graph represents the transitions between metastable states (i.e., nodes) where the node size represents the equilibrium population of a particular state and the thickness of the edges is proportional to the probability of transition. A representative snapshot for each state is given in cartoon representation. The region 190–200 is colored red and the 112 residue is drawn in CPK representation. (c) Given here are the time-averaged secondary structures and joint probability distribution of radius of gyration versus fraction of contacts of three states of ApoE4 obtained from MSM. To see this figure in color, go online.
In contrast, for ApoE4, the Markov state model identified an ensemble of three macrostates with equilibrium populations being ∼39, ∼19, and ∼42%, respectively (Fig. 3 b). We also computed the mean first passage time (63). The extracted rate of exchange shows that the transition between metastable states 1 → 2 and 3 → 2 is slower, with the values ∼1.84 and ∼1.85 μs, whereas transitions between states 1 → 3, 3 → 1, 2 → 3, and 2 → 1 are comparatively faster with the values ∼376, ∼356, ∼611, and ∼619 ns. All three metastable states of the ApoE4 isoform show significant unfolding of hinge helix 2 (residues 190–199) and can be characterized by respective secondary structures (Fig. 3 c). Previous experimental evidence has pointed toward greater propensity of formation of the so-called “molten globule conformation” by ApoE4, and often implicated in the formation of these molten globule states to the pathological relevance of ApoE4 (7, 64). The significant population of these partially unfolded conformations of ApoE4 as predicted by our simulation provides a structural view of those molten globulelike states for ApoE4. Hinge helix 2 (residues 190–199) is seen to have nearly 50% helicity in state 1 but is completely unstructured (0%) in the other macrostates. It is also evident from Fig. 3 c that macrostate 2 and macrostate 3 show formation of β-sheet structure near the hinge helix-2 region (Fig. S5). These observations are consistent with experimental characterization of the molten globule state by Morrow et al. (64), which predicted the molten globule of ApoE4 to have a partially unfolded four-helix bundle, with a loss of helical content and increase in β-structure. There are other structural differences among the three macrostates, such as helix C1 (residues 210–223) is in an α-helix conformation in state 3 but it does not maintain such a helicity in state 1 and state 2, and helix h1′ (residues 45–52) has nearly 100% helicity in state 1 and state 2, but <40% in state 3.
Furthermore, Arg112 is highly solvent-exposed in state 1, indicated by its time-averaged solvent-accessible surface area of 0.84 nm2 (0.4). In state 2 and state 3, the average solvent-accessible surface area values for the same residue are 0.36 nm2 (0.2) and 0.35 nm2 (0.2), respectively. This feature is also clear from the simulation snapshots corresponding to individual macrostates obtained from MSM of ApoE4 (Fig. 3 c). The joint probability distribution of radius of gyration and fraction of contacts also shows differences between the three states (discussed in detail later) (Fig. 3 c). It is to be mentioned here that we have considered all possible pairs of Cα atoms present within 0.65-nm distance in the initial ApoE3 structure and in the in silico mutated ApoE4 structure as the key contacts, and the total number of these so-called “Cα contacts” present in the initial structures have been counted during the progress of the simulation. This definition of “Cα contacts” is maintained throughout the article.
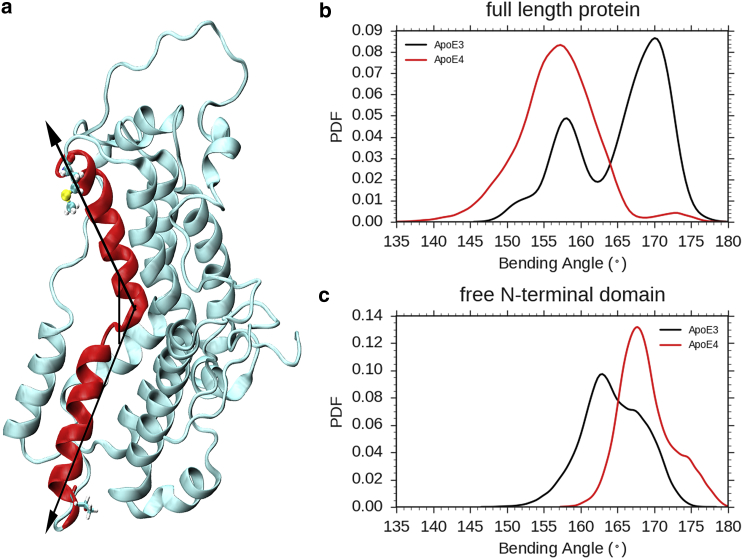
Helix 3, containing the mutation C112R, is bent in ApoE4
The mutation point 112 is accommodated in helix 3 of the ApoE isoforms. Initial visualization of our simulation trajectories indicate that helix 3 of ApoE4 frequently attains a bent conformation (Fig. 4 a), whereas the same helix in ApoE3 maintains a straightened structure during the period of simulation. As depicted in Fig. 4 b, the normalized distribution of bending angle (defined as the angle formed at the center of mass of helix 3 by two vectors passing through the two extreme residues, 89 and 125, of the same helix) is significantly left-shifted in ApoE4 with a lower average bending angle of 156.78° (5.4) corresponding to a bent helix, compared to ApoE3, which has an angular distribution centered around a higher bending angle of 165.30° (6.3) corresponding to a more straightened conformation. We have also analyzed our trajectory for kink or bending in helix 3 using the available standard program HELANAL (65). HELANAL characterizes the overall geometry of each helix, on the basis of their Cα atoms, as being linear, curved, or kinked. A helix is classified as being kinked if at least one local bending angle in the middle of the helix is >20°. Based on this alternative analysis, the residues in the middle of the helix (106–108) were found to have a higher local bending angle for the ApoE4 isoform (109.5 (11.9)), in accordance with our other analysis.
Figure 4.
Bending of helix 3 in ApoE4. (a) Helix 3 (residues 89–125) shown in red in a cartoon representation of the ApoE3/E4 isoform attains a bent conformation during MD run of ApoE4. (b) Given here is the distribution of the bending angle of helix 3 in ApoE3 and ApoE4. The bending in ApoE4 is indicated by a bending angle of ∼158° (red color in frequency histogram plot). ApoE3 has this helix in comparatively straight/elongated form. (c) The two peaks for bending angle of helix 3 calculated from MD run of only N-terminal domains of ApoE3 (black) and ApoE4 (red) are in the same region and >160°, indicating almost no bending in either isoforms. To see this figure in color, go online.
To investigate the possible role of C-terminal domain in this bending (and later on domain interaction), we performed independent control-simulations with only N-terminal domain (residues 1–200) of both the isoforms. The results obtained from these simulations of only the N-terminal domain clearly indicates that the bending in helix 3 of ApoE4 is much less pronounced in absence of the C-terminal domain (Fig. 4 c). Hence, it can be inferred that in ApoE4, the conformations of N-terminal domain helices may be dependent on the C-terminal domain. It is to be mentioned here that this analysis is performed on part of the trajectories considered after removing the first 50 ns as equilibration time, and then averaging over all the MD runs. We believe that the observation of bent structure in the case of ApoE4 in our simulation potentially contributes to the so-called “domain interaction”, as will be discussed in the following sections.
Interactions between N- and C-terminal domains are prominent in ApoE4
The ApoE3/ApoE4 isoform is composed of an N-terminal domain and a large C-terminal domain. Multiple related studies have explored potential interactions between the two independently folded domains, i.e., domain interactions, and these has been suggested to play a unique role in the characterization of ApoE4 (66, 67, 68).
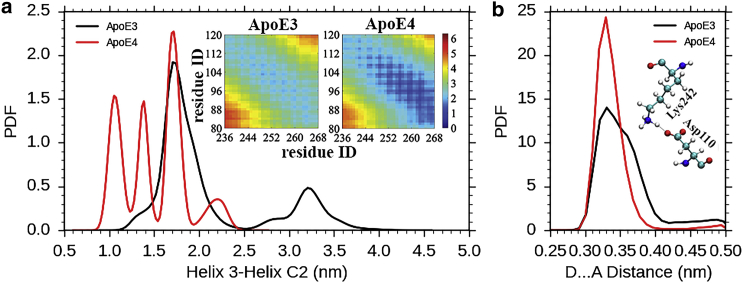
To investigate how different the domain interaction is in the two isoforms, we compare the distance of proximity between the center of mass of regions of N- and C-terminal domains of ApoE isoforms, as obtained from trajectories of MD simulation. Toward this end, we have considered helix 2 (residues 55–79), helix 3 (residues 89–125), and helix 4 (residues 131–164) as the representatives of N-terminal domain and helix C2 (residues 278–299) as a representative of the entire C-terminal domain (because apart from being the longest helical stretch of C-terminal domain, helix C2 also completely retains its helicity in both ApoE3 and ApoE4 during the entire period of all the simulations). As depicted in the distribution of Fig. 5 a, the helix 3-helix C2 distance is significantly smaller (∼1.51 nm), and occasionally even reaches <1.0 nm in ApoE4, as compared to that in ApoE3 (∼2.10 nm). However, we find that helix 2-helix C2 distance is of the same range for both the isoforms, and helix 4-helix C2 distance never reaches below 1.2 nm in either of the isoforms (Fig. S6). Thus, we expect the domain interaction to be prominent in ApoE4 due to the relatively closer proximity of helix 3 (which contains the mutation point) with helix C2.
Figure 5.
Domain interactions. (a) The helix 3-helix C2 distance in ApoE4 reaches <1 nm. (Inset) The contact map plot also shows that the regions helix 3 and helix C2 are closer in ApoE4 as compared to ApoE3. (b) The DA distance of Asp110-Lys242 salt bridge is lesser in ApoE4, indicating stronger interactions. (Inset) Shown here is the salt bridge, Asp110-Lys242, which is expected to be responsible for domain interaction. To see this figure in color, go online.
Similarly, on comparing the time-averaged contact maps of ApoE3 and ApoE4, we find the regions helix C2 (residues 236–266) and helix 3 (residues 89–125) come in close proximity during the MD run of ApoE4, although these remain far apart in ApoE3 (Fig. 5 a), indicative of domain interaction prevailing in ApoE4 involving the helix containing the mutation site. We have also observed that hinge helix 2 (residues 190–199) is closer to helix 4 (residues 131–164) and helix C2 (residues 236–266) in the average ApoE3 structure obtained from our MD runs, but are far apart in ApoE4 (Fig. S7). This observation may be attributed to the fact that during the MD runs of ApoE4, hinge helix 2 is completely unstructured.
Domain interaction was one of the first isoform-specific differences identified between ApoE3 and ApoE4. This concept was introduced to explain the preferential binding of ApoE4 to low density lipoprotein and of ApoE3 to HDL (7). The isoform-specific domain interaction as observed in our simulation studies might point toward isoform-specific differential ApoE-lipid interaction.
Salt bridge plays a crucial role in domain interactions
Many buried hydrophilic residues that are located between ApoE domains point outside and are potentially available for domain interaction by forming buried H-bonds and salt bridges between domains (25). Among all such possible interactions, H-bond between Arg61 and Thr194, and salt-bridge Glu255-Lys95 are suggested to play significant roles in domain interaction in the solution NMR structure of ApoE3 (25). In our current simulation, we did not find the H-bond between Arg61 and Thr194 to be stable during our MD runs. It was reported by Dong and co-workers (67, 68) that the interaction between the N- and C-terminal domains is a unique property of ApoE4 characterized by salt-bridge formation between Arg61 and Glu255, as Arg112 causes the Arg61 to extend away from the four-helix bundle and interact with Glu255, making the structure more compact. On the contrary, in ApoE3, Cys112 causes the Arg61 side chain to move in-between two helices, reducing the Arg61-Glu255 contact, resulting in an overall less compact structure (67). And the study by Williams et al. (61) in 2015, completely disagrees with the experiments as they did not observe these residues in contact during their replica exchange/discrete MD simulations of ApoE4. During our simulation of ApoE4 too, these residues remain far apart (3.03 nm (2.1)), rendering the formation of this salt bridge impossible.
We have focused on determining if any other salt bridges, not previously reported, might be responsible for the structural difference between ApoE3 and ApoE4. Toward this quest, we could identify the salt-bridge Arg112-Glu109 in ApoE4, whereas in ApoE3 no ionic interaction involving Cys112 and Glu109 was observed (Fig. S8). The substitution of Arg112 in ApoE4 leads to the ionic interaction with Glu109, which excludes the Asp110 side chain from its usual position, causing it to be more exposed and allowing it to become available for interaction with residues in the C-terminal domain. We have also found quite a few salt bridges involving residues of helix 3 and helix C2, but only a few were stable during the entire trajectory. Specifically, we observed two salt bridges, Glu255-Lys95 and Asp110-Lys242 (in ApoE3 and ApoE4), both involving residues of helix 3 (residues 89–125) and helix C2 (residues 236–266) that might play a role in domain interaction. Although the solution NMR structure of ApoE3 indicates probable interactions between the N-terminal and C-terminal domains by the Glu255-Lys95 salt bridge (25), we found Asp110-Lys242 (Fig. 5 b) to be the most prominent and stable in ApoE4, with the distance between the two residues reaching < 0.28 nm during the course of MD runs of ApoE4 (Fig. 5 b). Also, we see that the donor-acceptor distance in salt-bridge Asp110-Lys242 is lower in ApoE4 as compared to ApoE3, whereas the DA distance in salt-bridge GLu255-Lys95 is similar for the two isoforms (Fig. S9). Thus, we can state that Asp110-Lys242 is the unique salt bridge that promotes domain interactions only in ApoE4.
Tertiary fold of ApoE3 and ApoE4 is significantly different
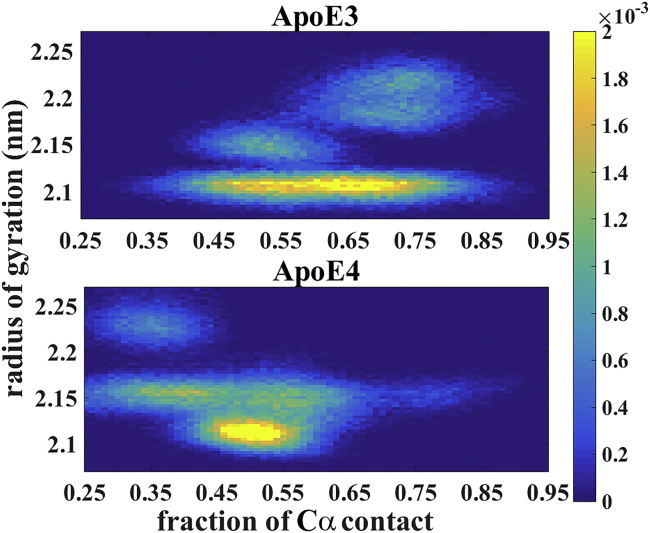
The single-residue mutation and the concomitant change in the secondary structure between ApoE3 and ApoE4 can potentially give rise to significant differences in the overall topology or the tertiary structure between the two isoforms. Toward this end, we investigate the overall compactness and the retention of Cα contacts (defined earlier in the text) of both the ApoE isoforms. We consider that a loss in fraction of contacts along with simultaneous increase in radius of gyration is a clear indication of loss in compactness or rigidity of the protein. Fig. 6 delineates the joint probability distribution of radius of gyration and fraction of contacts for both the isoforms based on our multi-microsecond simulation data. The plots clearly indicate that in ApoE4, with an increase in radius of gyration there is a significant loss in contacts, but in ApoE3 the contacts are well maintained despite increase in the radius of gyration. Thus, ApoE3 is topologically more rigid as compared to ApoE4 and maintains its structure during the MD run. We also note that although the domain interaction is prevalent in ApoE4, one should have in mind that this isoform has far less compactness accounting for its lower stability with respect to ApoE3. As we will see in later sections, the lack of rigidity in ApoE4 potentially gives rise to structural heterogeneity and multiple partially unfolded states.
Figure 6.
Joint probability distribution of the radius of gyration and fraction of Cα contact. ApoE3 has a higher fraction of contacts at a larger radius of gyration, indicating that it retains its structure. On the other hand, ApoE4 has a less compact structure, as indicated by a lesser fraction of contact at higher radius of gyration. Color bar gives the probability density at each point. To see this figure in color, go online.
ApoE4 is thermodynamically less stable than ApoE3
Our free energy calculations show that the mutation C112R destabilizes the ApoE4 structure by an amount of 1.76 ± 0.15 kcal/mol relative to ApoE3, which is consistent with the results reported in the past (61). Experimental studies and guanidium-HCl denaturation confirmed that the two domains of ApoE unfold independently in all three isoforms (69). However, the difference in stability of the N-terminal domain of ApoE isoforms runs in the order ApoE4 < ApoE3 < ApoE2 (69). ApoE4 is known to unfold even less cooperatively than does ApoE3 (21), suggesting that stable intermediate conformations are formed more readily for ApoE4 than for ApoE3. We note that the free energy perturbation techniques used in this work provide estimates of free energy changes due to local change only and global structural effects are not generally captured in these techniques. Nonetheless, the free energy estimate validates that ApoE is thermodynamically less stable than ApoE3, which is consistent with the underlying themes of our current work.
On the other hand, Fig. S10 compares the free energy profile, obtained via subsequent umbrella sampling simulations, and corresponding probability distribution as a function of helical fraction of hinge helix 2 (residues 190–199) regions. The free energy profiles of the hinge helix 2 region depict a free energy minima at 50% helicity in ApoE3, whereas for ApoE4 the free energy minima appears at nearly 0% helicity—suggesting a complete lack of α-helicity, in accordance with equilibrium MD simulation results.
Discussion
The two residues cysteine and arginine at position 112 of ApoE3 and ApoE4, respectively, are structurally different and thus, as a result of C112R mutation, we can expect that the mutant ApoE4 has a structure that is different from the wild-type ApoE3. The major question that we aimed to address in this work is: what is the consequence of this single-residue difference on the key structural differences between ApoE3 and ApoE4?
The noteworthy difference in time-averaged secondary structure of the two isoforms is that hinge helix 2 (residues 190–199) is completely unstructured in ApoE4, whereas it has a helical conformation in ApoE3. Consistent with our unbiased MD simulation results, the free energy profile as a function of the helical fraction of hinge helix 2 (residues 190–199) supports the completely unstructured conformation of this region in ApoE4. To understand why this region is not retaining its helical form in ApoE4, we have looked into the sequence of both hinge helix 2 (residues 190–199) and helix 3 (residue 89–125) (Fig. S11), as helix 3 is the mutation-containing helix and hinge helix 2 lies closest to the mutation site, Arg112. As mentioned earlier, Arg112 in ApoE4 forms a salt bridge with Glu109, but the cysteine at position 112 of ApoE3 is free to have interactions with other nearby residues. We calculated the distance between 112th residue (of both ApoE3 and ApoE4) and all other amino acid residues of hinge helix 2 (residues 190–199 and considering center of mass of each residue) independently, and observed that two alanine residues (Ala192 and Ala193) of hinge helix 2 (residues 190–199) are closer to Cys112 compared to Arg112 with the distance between Ala192/193 and Cys112 being as low as 0.5 nm in ApoE3—i.e., the nonbonded interactions between residues are effective in ApoE3 and not in ApoE4 (Fig. S12). We may assume that the absence of such close interaction between residues of hinge helix 2 and helix 3 (more importantly Cys112) in ApoE4 causes the hinge helix 2 to become unstructured during the MD run even though this region was helical when the MD run was initiated.
We have found hinge helix 2 to be unstructured in ApoE4 in both the presence and the absence of C-terminal domain, indicating that this structural difference between the two isoforms is independent of domain interaction. However, we found helix 3 (residues 89–125) to always attain a bent conformation in ApoE4 in the presence of the C-terminal domain. This can be explained again by the fact that Arg112 in ApoE4 is involved in an ionic interaction with Glu109 that excludes the Asp110 side chain from its usual position, causing it to be more exposed and allowing it to become available for interaction with Lys242 of its C-terminal domain. This Asp110-Lys242 salt bridge in ApoE4 somewhat puts a strain on helix 3, causing it to bend at the region around the 110th residue. Scrutinizing the sequence of helix 3 (Fig. S11) indicated that there are two residues with very low helical propensity—Asp107 (0.69 kcal/mol) and Gly105 (1 kcal/mol) (70)—around which the helix is broken and bent and the region is almost at the middle of the entire helix. In ApoE3 too, only a very small amount of bending of helix 3 is observed (Fig. 4 b), probably due to poor helical propensity of the same Asp107 and Gly105 residues. However, unlike in ApoE4, due to absence of any domain interactions the helix bending is not retained during the MD-runs of ApoE3 isoform, and as a result, the helix is majorly straightened.
During the MD runs, ApoE3 was found to be topologically more rigid as compared to ApoE4 while maintaining its initial structure. The domain interaction is found to be prevalent in ApoE4, which may be considered as a cause for the decrease in its contacts or topological rigidity with respect to the initial structure, thereby accounting for the lower stability of ApoE4 with respect to ApoE3.
The Markov state models constructed based on the simulation trajectories provided only one solely populated macrostate for ApoE3, which has highly stable hinge helix 2 (residues 190–199). The overall secondary structure of ApoE3 as obtained from MSM is the same as the time-averaged secondary structure obtained from the simulation trajectories. In contrast to this, for ApoE4, the MSM identified an ensemble of three metastable partially unfolded macrostates of significant population containing unstructured regions near hinge helix 2. Morrow et al. (64) have reported that ApoE4 unfolds even less cooperatively than does ApoE3, suggesting that stable intermediate conformations are formed more readily for ApoE4 than for ApoE3. The observation of the partially unfolded conformations as obtained in our simulation in the case of ApoE4 are in qualitative agreement with a previous experimental report of molten globule formation by ApoE4 (64). Molten globules have often been implicated in a wide variety of physiological processes. The ApoE4 molten globule could exert a number of effects that might contribute to a disease phenotype. For example, the instability of the N-terminal domain of ApoE4 may explain its increased ability to bind and remodel phospholipid micelles (69). As a result, lipoprotein particles containing ApoE4 could differ from ApoE3 particles in their composition. In AD, the lipid-binding properties of ApoE4 could also influence cellular function, resulting in pathological effects (7).
Our free energy-based simulation finds that ApoE4 is thermodynamically less stable than ApoE3 and specifically the hinge-helix 2 region (residue 190–199) has very low helical propensity. For ApoE, these differences in conformational stability and folding behavior of the N-terminal domain among the isoforms are important in lipid binding (67). In particular, variation in the stability of the N-terminal domains of these two isoforms might contribute to their differences in their lipoprotein-binding preferences and other biological functions in vivo. Moreover, the recent work by Frieden et al. (71) suggests that the first step in lipid binding might be domain-domain separation between the N- and C-terminal domains caused by breaking-up of salt bridges by the charged lipid molecules. Certainly, ApoE undergoes large conformational changes on lipid binding and the presence of partially folded states could facilitate conformational change in case of ApoE4.
It is well known that ApoE is majorly implicated in the transport of lipid (9). In vivo, ApoE is almost always bound to lipids and cholesterol, and the actual amount of lipid-free ApoE is expected to be small (22). On lipidation, the protein undergoes large structural changes (72, 73), and although there is no high-resolution structure of lipid-bound ApoE, the structural differences between the ApoE isoforms potentially lead to their differential association with lipids and lipoproteins (74). To understand if the structural differences observed here in the lipid-free ApoE isoforms are retained in the lipidated ApoE, this work can be extended by studying the differences in association of 1,2-dimyristoyl-sn-glycero-3-phosphocholine-ApoE3 or 1,2-dimyristoyl-sn-glycero-3-phosphocholine-ApoE4 with ligands, such as Aβ.
Conclusions
In conclusion, we find that compared to isoform ApoE3, ApoE4 is less structured, less thermodynamically stable, topologically less compact, and structurally more heterogeneous with significant populations of multiple metastable states. Our simulation results also point to a bent architecture of the long-mutation-containing helix 3 in ApoE4, with strong prevalence of interaction between the N- and C- terminal domains. Our simulation analysis also insinuates that the salt bridge, Asp110-Lys242, is critical for domain interactions in ApoE4. The domain interaction can be considered as the major cause for bending of helix 3 (residues 89–125) and the salt bridge Arg112-Glu109 as the reason for unstructured hinge helix 2 (residues 190–199). These two structural differences between ApoE3 and ApoE4 can be considered as the underlying cause for ApoE4 to be thermodynamically less stable than ApoE3.
We believe that the potential biological implications of the structural differences between ApoE3 and ApoE4 isoforms proposed in this work will serve as a topic of future exploration. It is generally considered that binding of ApoE isoforms with the Aβ oligomers have a potentially significant role in the onset of Alzheimer’s disease (72, 73). ApoE3 and ApoE4 are speculated to have a differential binding rate with Aβ oligomers, with ApoE4 rated to be a better stabilizer of Aβ (74) than ApoE3. It will be worth exploring whether the origin of differential interaction of ApoE isoforms with Aβ oligomers is rooted in the structural differences noted in this work.
Author Contributions
A.R. performed research, analyzed data, and cowrote the manuscript. N.A. analyzed data and cowrote the manuscript. J.M. designed research, performed research, analyzed data, and cowrote the manuscript.
Acknowledgments
J.M. is grateful to Dr. Kanchan Garai for introducing us to the topic of Apolipoprotein E and useful experimental insights from his laboratory.
This work was supported by startup funds for the Tata Institute of Fundamental Research Hyderabad (CIS/TIFR-H) (to J.M.), with early career research funds provided by the Department of Science and Technology (DST) Science and Engineering Research Board (ECR/2016/000672) (to J.M.). A.R. thanks the Science and Engineering Research Board (SERB) for grant PDF/2016/002712. A.R. and N.A. thank the Tata Institute of Fundamental Research for the fellowship.
Editor: Francesca Marassi.
Footnotes
Twelve figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31093-7.
Supporting Material
References
- 1.Kyrtsos C.R., Baras J.S. Studying the role of ApoE in Alzheimer’s disease pathogenesis using a systems biology model. J. Bioinform. Comput. Biol. 2013;11:1342003. doi: 10.1142/S0219720013420031. [DOI] [PubMed] [Google Scholar]
- 2.Weisgraber K.H., Mahley R.W. Human apolipoprotein E: the Alzheimer’s disease connection. FASEB J. 1996;10:1485–1494. doi: 10.1096/fasebj.10.13.8940294. [DOI] [PubMed] [Google Scholar]
- 3.Hauser P.S., Ryan R.O. Impact of apolipoprotein E on Alzheimer’s disease. Curr. Alzheimer Res. 2013;10:809–817. doi: 10.2174/15672050113109990156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahley R.W., Weisgraber K.H., Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olaisen B., Teisberg P., Gedde-Dahl T., Jr. The locus for apolipoprotein E (apoE) is linked to the complement component C3 (C3) locus on chromosome 19 in man. Hum. Genet. 1982;62:233–236. doi: 10.1007/BF00333526. [DOI] [PubMed] [Google Scholar]
- 6.Mahley R.W. Central nervous system lipoproteins: ApoE and regulation of cholesterol metabolism. Arterioscler. Thromb. Vasc. Biol. 2016;36:1305–1315. doi: 10.1161/ATVBAHA.116.307023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong N., Weisgraber K.H. Understanding the association of apolipoprotein E4 with Alzheimer disease: clues from its structure. J. Biol. Chem. 2009;284:6027–6031. doi: 10.1074/jbc.R800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perugini M.A., Schuck P., Howlett G.J. Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J. Biol. Chem. 2000;275:36758–36765. doi: 10.1074/jbc.M005565200. [DOI] [PubMed] [Google Scholar]
- 9.Hatters D.M., Peters-Libeu C.A., Weisgraber K.H. Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Weisgraber K.H. Apolipoprotein E distribution among human plasma lipoproteins: role of the cysteine-arginine interchange at residue 112. J. Lipid Res. 1990;31:1503–1511. [PubMed] [Google Scholar]
- 11.Garai K., Verghese P.B., Frieden C. The binding of apolipoprotein E to oligomers and fibrils of amyloid-β alters the kinetics of amyloid aggregation. Biochemistry. 2014;53:6323–6331. doi: 10.1021/bi5008172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acharya P., Segall M.L., Snow J. Comparison of the stabilities and unfolding pathways of human apolipoprotein E isoforms by differential scanning calorimetry and circular dichroism. Biochim. Biophys. Acta. 2002;1584:9–19. doi: 10.1016/s1388-1981(02)00263-9. [DOI] [PubMed] [Google Scholar]
- 13.Fisher C.A., Ryan R.O. Lipid binding-induced conformational changes in the N-terminal domain of human apolipoprotein E. J. Lipid Res. 1999;40:93–99. [PubMed] [Google Scholar]
- 14.Corder E.H., Saunders A.M., Schmader K.E. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 15.Laws S.M., Hone E., Martins R.N. Expanding the association between the APOE gene and the risk of Alzheimer’s disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J. Neurochem. 2003;84:1215–1236. doi: 10.1046/j.1471-4159.2003.01615.x. [DOI] [PubMed] [Google Scholar]
- 16.Strittmatter W.J., Weisgraber K.H., Roses A.D. Binding of human apolipoprotein E to synthetic amyloid β peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Dhanasekaran P., Lund-Katz S. Molecular mechanisms responsible for the differential effects of apoE3 and apoE4 on plasma lipoprotein-cholesterol levels. Arterioscler. Thromb. Vasc. Biol. 2013;33:687–693. doi: 10.1161/ATVBAHA.112.301193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davignon J., Gregg R.E., Sing C.F. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Hauser P.S., Narayanaswami V., Ryan R.O. Apolipoprotein E: from lipid transport to neurobiology. Prog. Lipid Res. 2011;50:62–74. doi: 10.1016/j.plipres.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi P.K., Fischer K., Wilson J.F. Variants near CHRNA3/5 and APOE have age- and sex-related effects on human lifespan. Nat. Commun. 2016;7:11174. doi: 10.1038/ncomms11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisgraber K.H. Apolipoprotein E: structure-function relationships. Adv. Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 22.Frieden C., Garai K. Structural differences between apoE3 and apoE4 may be useful in developing therapeutic agents for Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2012;109:8913–8918. doi: 10.1073/pnas.1207022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson C., Wardell M.R., Agard D.A. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 24.Segrest J.P., Jones M.K., Anantharamaiah G.M. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 25.Chen J., Li Q., Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. USA. 2011;108:14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivashanmugam A., Wang J. A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein binding and for receptor binding activity. J. Biol. Chem. 2009;284:14657–14666. doi: 10.1074/jbc.M901012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatters D.M., Zhong N., Weisgraber K.H. Amino-terminal domain stability mediates apolipoprotein E aggregation into neurotoxic fibrils. J. Mol. Biol. 2006;361:932–944. doi: 10.1016/j.jmb.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 28.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 29.Brooks B.R., Brooks C.L., 3rd, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, openMM, and CHARMM/openMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abraham M.J., Murtola T., Lindah E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 32.Páll S., Abraham M.J., Lindahl E. Lecture Notes in Computer Science. Springer; Berlin, Germany: 2015. Tackling exascale software challenges in molecular dynamics simulations with GROMACS; pp. 3–27. [Google Scholar]
- 33.Best R.B., Zhu X., MacKerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sagui C., Darden T.A. Molecular dynamics simulations of biomolecules: long-range electrostatic effects. Annu. Rev. Biophys. Biomol. Struct. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- 35.Evans D.J., Holian B.L. The Nosé-Hoover thermostat. J. Chem. Phys. 1985;83:4069–4074. [Google Scholar]
- 36.Parrinello M., Rahman A. Crystal structure and pair potentials: a molecular-dynamics study. Phys. Rev. Lett. 1980;45:1196–1199. [Google Scholar]
- 37.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 38.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 39.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 40.Gonzalez T.F. Clustering to minimize the maximum intercluster distance. Theor. Comput. Sci. 1985;38:293–306. [Google Scholar]
- 41.Prinz J.-H., Wu H., Noé F. Markov models of molecular kinetics: generation and validation. J. Chem. Phys. 2011;134:174105. doi: 10.1063/1.3565032. [DOI] [PubMed] [Google Scholar]
- 42.Chodera J.D., Noé F. Markov state models of biomolecular conformational dynamics. Curr. Opin. Struct. Biol. 2014;25:135–144. doi: 10.1016/j.sbi.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwantes C.R., McGibbon R.T., Pande V.S. Perspective: Markov models for long-timescale biomolecular dynamics. J. Chem. Phys. 2014;141:090901. doi: 10.1063/1.4895044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez-Hernández G., Paul F., Noé F. Identification of slow molecular order parameters for Markov model construction. J. Chem. Phys. 2013;139:015102. doi: 10.1063/1.4811489. [DOI] [PubMed] [Google Scholar]
- 45.Lloyd S. Least squares quantization in PCM. IEEE Trans. Inf. Theory. 1982;28:129–137. [Google Scholar]
- 46.Deuflhard P., Weber M. Robust Perron cluster analysis in conformation dynamics. Linear Algebra Appl. 2005;398:161–184. [Google Scholar]
- 47.Scherer M.K., Trendelkamp-Schroer B., Noé F. PyEMMA 2: a software package for estimation, validation, and analysis of Markov models. J. Chem. Theory Comput. 2015;11:5525–5542. doi: 10.1021/acs.jctc.5b00743. [DOI] [PubMed] [Google Scholar]
- 48.Tribello G.A., Bonomi M., Bussi G. PLUMED 2: new feathers for an old bird. Comput. Phys. Commun. 2014;185:604–613. [Google Scholar]
- 49.Pietrucci F., Laio A. A collective variable for the efficient exploration of protein beta-sheet structures: application to SH3 and GB1. J. Chem. Theory Comput. 2009;5:2197–2201. doi: 10.1021/ct900202f. [DOI] [PubMed] [Google Scholar]
- 50.Bonomi M., Branduardi D., Parrinello M. PLUMED: a portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 2009;180:1961–1972. [Google Scholar]
- 51.Kumar S., Rosenberg J.M., Kollman P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 52.Grossfield, A. WHAM: the weighted histogram analysis method, version 2.0.9. http://membrane.urmc.rochester.edu/content/version-209.
- 53.Loeffler H.H., Michel J., Woods C. FESetup: automating setup for alchemical free energy simulations. J. Chem. Inf. Model. 2015;55:2485–2490. doi: 10.1021/acs.jcim.5b00368. [DOI] [PubMed] [Google Scholar]
- 54.Shirts M.R., Chodera J.D. Statistically optimal analysis of samples from multiple equilibrium states. J. Chem. Phys. 2008;129:124105. doi: 10.1063/1.2978177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen Y., Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wetterau J.R., Aggerbeck L.P., Weisgraber K.H. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J. Biol. Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 57.Aggerbeck L.P., Wetterau J.R., Lindgren F.T. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J. Biol. Chem. 1988;263:6249–6258. [PubMed] [Google Scholar]
- 58.Zhang Y., Vasudevan S., Wang J. A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 59.Huang R.Y.C., Garai K., Gross M.L. Hydrogen/deuterium exchange and electron-transfer dissociation mass spectrometry determine the interface and dynamics of apolipoprotein E oligomerization. Biochemistry. 2011;50:9273–9282. doi: 10.1021/bi2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 61.Williams B., 2nd, Convertino M., Dokholyan N.V. ApoE4-specific misfolded intermediate identified by molecular dynamics simulations. PLOS Comput. Biol. 2015;11:e1004359. doi: 10.1371/journal.pcbi.1004359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chetty P.S., Mayne L., Phillips M.C. Helical structure, stability, and dynamics in human apolipoprotein E3 and E4 by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2017;114:968–973. doi: 10.1073/pnas.1617523114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoel P.G., Port S.C., Stone C.J. Houghton Mifflin Harcourt; Boston, MA: 1972. Introduction to Stochastic Processes. [Google Scholar]
- 64.Morrow J.A., Hatters D.M., Weisgraber K.H. Apolipoprotein E4 forms a molten globule. A potential basis for its association with disease. J. Biol. Chem. 2002;277:50380–50385. doi: 10.1074/jbc.M204898200. [DOI] [PubMed] [Google Scholar]
- 65.Bansal M., Kumar S., Velavan R. HELANAL: a program to characterize helix geometry in proteins. J. Biomol. Struct. Dyn. 2000;17:811–819. doi: 10.1080/07391102.2000.10506570. [DOI] [PubMed] [Google Scholar]
- 66.Dong L.M., Wilson C., Agard D.A. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J. Biol. Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 67.Dong L.M., Weisgraber K.H. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 68.Mahley R.W., Weisgraber K.H., Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J. Lipid Res. 2009;50(Suppl):S183–S188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morrow J.A., Segall M.L., Weisgraber K.H. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 70.Pace C.N., Scholtz J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frieden C., Wang H., Ho C.M.W. A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain-domain interactions. Proc. Natl. Acad. Sci. USA. 2017;114:6292–6297. doi: 10.1073/pnas.1705080114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peters-Libeu C.A., Newhouse Y., Weisgraber K.H. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J. Biol. Chem. 2006;281:1073–1079. doi: 10.1074/jbc.M510851200. [DOI] [PubMed] [Google Scholar]
- 73.Peters-Libeu C.A., Newhouse Y., Weisgraber K.H. Apolipoprotein E∗dipalmitoylphosphatidylcholine particles are ellipsoidal in solution. J. Lipid Res. 2007;48:1035–1044. doi: 10.1194/jlr.M600545-JLR200. [DOI] [PubMed] [Google Scholar]
- 74.Nguyen D., Dhanasekaran P., Lund-Katz S. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry. 2010;49:10881–10889. doi: 10.1021/bi1017655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.