Abstract
As a protease complex involved in the cleavage of amyloid precursor proteins that lead to the formation of amyloid β fibrils implicated in Alzheimer's disease, γ-secretase is an important target for developing therapeutics against Alzheimer's disease. γ-secretase is composed of four subunits: nicastrin (NCT) in the extracellular (EC) domain, presenilin-1 (PS1), anterior pharynx defective 1, and presenilin enhancer 2 in the transmembrane (TM) domain. NCT and PS1 play important roles in binding amyloid β precursor proteins and modulating PS1 catalytic activity. Yet, the molecular mechanisms of coupling between substrate/modulator binding and catalytic activity remain to be elucidated. Recent determination of intact human γ-secretase cryo-electron microscopy structure has opened the way for a detailed investigation of the structural dynamics of this complex. Our analysis, based on a membrane-coupled anisotropic network model, reveals two types of NCT motions, bending and twisting, with respect to PS1. These underlie the fluctuations between the “open” and “closed” states of the lid-like NCT with respect to a hydrophilic loop 1 (HL1) on PS1, thus allowing or blocking access of the substrate peptide (EC portion) to HL1 and to the neighboring helix TM2. In addition to this alternating access mechanism, fluctuations in the volume of the PS1 central cavity facilitate the exposure of the catalytic site for substrate cleavage. Druggability simulations show that γ-secretase presents several hot spots for either orthosteric or allosteric inhibition of catalytic activity, consistent with experimental data. In particular, a hinge region at the interface between the EC and TM domains, near the interlobe groove of NCT, emerges as an allo-targeting site that would impact the coupling between HL1/TM2 and the catalytic pocket, opening, to our knowledge, new avenues for structure-based design of novel allosteric modulators of γ-secretase protease activity.
Introduction
The γ-secretase complex is a membrane protease involved in the proteolytic cleavage of a range of substrates (1, 2, 3), including the amyloid precursor protein (APP) and the Notch protein (2). Amyloidogenic APP cleavages by β- and γ-secretases generate amyloid β (Aβ) peptides, which oligomerize and form the Aβ plaques leading to Alzheimer's disease (AD) pathology (1). Because of this pathogenic effect, γ-secretase has been a target for the development of drugs against AD (4).
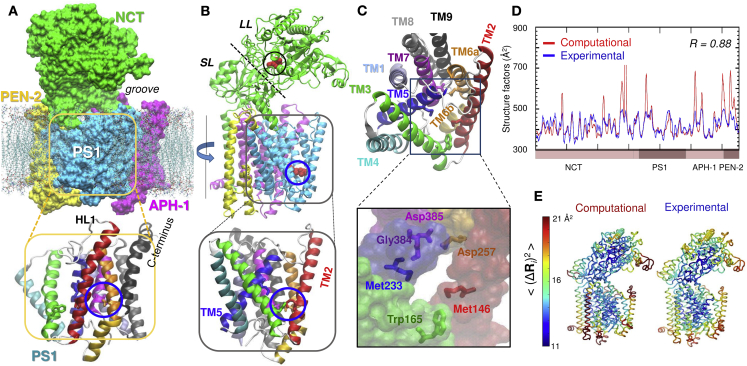
The γ-secretase complex is composed of four subunits/proteins: presenilin-1 (PS1) (5), nicastrin (NCT), anterior pharynx defective 1 (APH-1), and presenilin enhancer 2 (PEN-2) (6, 7) (Fig. 1, A and B). PS1 serves as the catalytic core (8, 9, 10). Fig. 1 C displays the catalytic residues Asp257 and Asp385, along with the nearby residues (Met146, Trp165, Met233, and Gly384) that coordinate the inhibitor, DAPT, observed in cryo-electron microscopy (11). Cleavage of substrate (99 residue C-terminal fragment of the APP) by PS1 proceeds in two steps (12): ε-cleavage (endopeptidase activity), followed by γ-cleavage (carboxypeptidase activity) to release the Aβ peptide to the extracellular (EC) medium. PS1 is composed of nine transmembrane (TM) helices, TM1–TM9 (Fig. 1 C). Many mutations implicated in the formation of Aβ, including 90% of familial AD mutations, are located in PS1 (2, 13, 14, 15, 16). NCT forms the EC domain, except for its C-terminal helix, which inserts into the membrane. It is composed of two lobes, large and small (LL and SL). Glu333 near the interlobe interface, and the nearby DYIGS motif D336–S340, have been pointed out to be involved in substrate binding (17, 18, 19) (Fig. 1 B). PEN-2 helps stabilize the complex (20). APH-1 has seven TM helices; it plays an essential role in Notch signaling and aids in the assembly of premature components (21).
Figure 1.
Structure of γ-secretase, its catalytic cavity, and equilibrium fluctuations. (A) γ-secretase in a lipid bilayer, constructed using the coordinate data in PDB: 5FN2 (11) and the lipid molecules from the Orientations of Proteins in Membranes database. The complex is composed of four subunits shown in different colors (PS1, NCT, APH-1, and PEN-2). NCT forms the ectodomain (except for its C-terminal helix, residues 665–698); the other three subunits form the TMD. The lipid bilayer is shown as gray sticks, with the polar heads in red. The lower diagram shows the nine TM helices of PS1 color-coded as in (C), with the catalytic site enclosed in a blue circle. The hydrophilic loop HL1 connecting TM1 and TM2 is labeled. (B) Ribbon diagram of the complex shown upon rotating the structure in (A) by 90°. Glu333, near the substrate-binding site of NCT, is shown in red, enclosed in a black circle; the catalytic site of PS1 is enclosed in a blue circle. The dashed line on NCT separates the large lobe (LL) and small lobe (SL), which form a large surface groove. (C) Top view of PS1. The lower diagram shows the catalytic residues Asp257 and Asp385, and neighboring Met146, Trp165, Met233, and Gly384 (TM helices colored as in top ribbon diagram). (D) Comparison of experimentally observed (blue) and computationally predicted (red) structure factors for the complex resolved by cryo-electron microscopy. The correlation coefficient between the two sets of data is 0.88. (E) Same comparison as in D, in terms of ribbon diagrams color-coded by residue MSFs from experiments and ANM computations.
Targeting γ-secretase for AD therapy requires the development of a compound that would inhibit the production of Aβ (especially Aβ42), which can also maintain the interactions with other substrates (2). Although a number of γ-secretase inhibitors have been developed and tested in humans with both AD and cancer, harmful side effects associated with blocking the Notch signaling pathway have been reported (2, 22). This motivated the development of γ-secretase modulators (GSMs) for the treatment of AD (23). Earlier studies have also pointed to the roles of γ-secretase-associated proteins (24), and to allosteric sites located within PS1 (25, 26, 27). Clinical trials have been initiated for a GSM, E2012, which reduces the production of Aβ without interfering with Notch signaling (23, 28).
The active site of PS1 is located at the interior of its TM horseshoe-like fold (29) (Fig. 1). TM2 and TM6 serve as “doors” for substrate entry (30), as also observed in recent molecular dynamics (MD) simulations of the PS1 subunit (31). Recent coarse-grained (CG) and atomic simulations also showed that an extended surface covering TM2, TM6, TM9, and the C-terminal PAL motif, was involved in binding the APP at TM domain (TMD) (32). The similar microsecond simulations of Aguayo-Ortiz et al. (33) also showed that the same three helices, plus TM7, were concertedly involved in the transition of PS1 between active and inactive states (coupled to the protonation of Asp257 and Asp385), but these movements were not correlated with those involved in substrate recognition. Earlier studies have drawn attention to the EC-exposed parts of hydrophilic loop 1 (HL1) (S104–T124), to TM5 (L219–L241), and to the C-terminal fragment of PS1 near HL1/TM2 as sites involved in APP binding (12, 34, 35), and substrate binding to these regions has been proposed to induce structural changes that enable PS1 catalytic activity (12). Thus, conflicting findings on the type of, or even existence of, couplings between the conformational changes of PS1 involved in substrate binding and those involved in catalytic activity have been reported. The mechanism of modulation of activity by GSMs has also not been resolved. HL1/TM2 with TM5 were reported to form a pocket for phenylimidazole-type GSM binding (35), but how modulator binding would interfere with the activity at a catalytic pocket > 30 Å away remains unclear. Bai et al. (29) suggested that the distance between the binding sites on NCT and PS1 could be shortened upon rotation of NCT, and Xie et al. (36) suggested that rotation of NCT LL relative to SL might expose a substrate-binding site in NCT that is otherwise buried by an exposed loop (C140–L167). Other studies point to the role of NCT conformational dynamics (37, 38), or its role as a gatekeeper for substrate binding or excluding larger substrates (39), whereas earlier studies questioned the NCT-substrate-binding ability (40) and the role of Glu333 in NCT (41).
Here, we examine the structural dynamics of intact γ-secretase using the anisotropic network model (ANM) (42, 43), with the goal of elucidating the collective mechanisms of motions intrinsically favored by the intact quaternary structure, and identifying potential sites for allosteric modulation of its APP binding properties. Allosteric modulation, or allo-targeting, has emerged as a rational strategy for selectively interfering with specific interactions involved in particular pathways, although retaining their catalytic activity (44, 45, 46). Even though the γ-secretase complex has been resolved at relatively low resolutions by cryo-electron microscopy (4.5 Å in 2014 (47), 4.0–4.3 Å (11), and 3.4 Å in 2015 (29)), ANM analysis can advantageously use the low-resolution data to generate a unique and robust solution for the global dynamics of the quaternary structure, as proven in numerous applications (to other systems) and comparisons with experimental data (48, 49, 50, 51). Here, we first characterize the mechanism of couplings between the subunits (and surrounding membrane) and identify key sites (e.g., global hinges) that control the overall mechanics of the complex using the ANM. The validity of ANM results is verified by CG MD simulations, where applicable. Then, we perform druggability simulations (52), which are particularly useful for identifying allosteric sites (53, 54), and determine the hot spots, either orthosteric or allosteric, that can potentially bind modulators of allosteric dynamics. The integrated analysis of the two sets of data using the ProDy interface (55, 56) opens, to our knowledge, new avenues for the rational design of allosteric modulators of γ-secretase.
Methods
ANM for membrane proteins
The ANM (42, 43) represents the protein as an elastic network where each Cα atom is a node, and node pairs within a cutoff distance (of 15 Å) are connected by elastic springs of uniform force constant γ. The overall potential V of the system is a summation over the harmonic potentials of all springs. The second derivative of V with respect to residue movements yields a closed form expression for the ANM Hessian matrix H. Eigenvalue decomposition of H yields 3N-6 nonzero eigenvalues λk and corresponding eigenvectors uk, which define the frequencies and shapes of the ANM (normal) modes uniquely accessible to the structure. The size of motion along mode k is proportional to (1/λk)1/2, i.e., lower modes, also called softer modes, make larger contributions to overall structural change (see Fig. S1), e.g., the contributions of ANM modes 1 and 2 were 0.12 and 0.06, respectively, summing up to a total of 0.18. The extension to membrane proteins (membrANM) (57) permits the incorporation of the effect of the lipid bilayer (environment) on the system (the γ-secretase complex) dynamics. The TM horseshoe-like structure of the TMD creates a hollow region exposed to membrane, hence the importance of using membrANM. The membrane is constructed using a face-centered cubic lattice with a circular shape, the thickness and position of which are obtained from the Orientations of Proteins in Membranes database (58). Computations were performed using the ProDy application programming interface (55, 56). ANM predicts the direction of motion along different modes, whereas the size of the motion is arbitrary (dependent on γ). Here, ANM conformers along each mode were illustrated/visualized by adopting an root-mean-square-deviation (RMSD) of 4 Å with respect to the PDB structure. The corresponding interresidue distance changes were comparable to those observed in CG MD simulations. ANM conformers were mapped into full atomic representations using PULCHRA (59), succeeded by energy-minimization using NAMD (60).
CG MD simulations were performed using GROMACS v5.1.4 (61) with MARTINI 2.2 force field (62). The CG MD set up (protein embedded in the phosphatidylcholine bilayer) was generated using GHARMM-GUI MARTINI bilayer maker (63). The trajectories (or 10,000 frames per run) were subjected to principal component analysis (PCA) (55, 64) using ProDy application programming interface (55, 56). The contribution of principal components 1 and 2 (also called PCA modes 1 and 2) were 0.59 (0.51) and 0.14 (0.15), respectively, summing up to a total of 0.73 (0.66) for the first (second) run (see Fig. S1).
Druggability simulations, also called mixed-probe (65), cosolvent-based (66, 67), or mixMD (54) simulations, were performed using the method described in our previous study (52), implemented in the DruGUI module of ProDy (55, 56). The method has been benchmarked against several well-studied systems (52, 68), such as murine double mutant-2, protein tyrosine phosphatase 1B, lymphocyte function-associated antigen 1, vertebrate kinesin-5, p38 mitogen-activated protein kinase, and cytochrome c. Mainly, a grid-based method is used and the binding free energy of probe molecules at each grid i is evaluated as ΔGi = −RT ln(ni/n0). Here, ni/n0 is the ratio of the observed density of probes in MD simulations (ni) to the density in protein-free system (n0), R is the gas constant, and T is the absolute temperature (K). More details on this and other methods are presented in the Supporting Materials and Methods.
The PDB structure of γ-secretase (11) and its ANM-generated conformers were used as targets for docking of several inhibitors (DAPT, BMS-708163, CHEMBL2159511, CHEMBL2159687, and CHEMBL21596910) and GSMs (E2012 and ST1120) using Surflex-Dock (69), implemented in SYBYL-X 1.3, with the same protocol as reported in our previous studies (70, 71, 72, 73).
Visualization of data was performed using ProDy NMWiz (55, 56), VMD v1.9.1 (74), PyMOL v1.8.4.2 (75), and GnuPlot v4.6 (76).
Results
NCT undergoes two types of en bloc movements with respect to the TMD: global bending and twisting
Fig. 1, D and E illustrate the comparison of the ANM-predicted mean-square fluctuations (MSFs) of residues, <(ΔRi)2>, with the structure factors reported experimentally (11). ΔRi denotes the change in the spatial position (Ri) of residue i. The correlation coefficient between the two sets is 0.88. Panel E provides an alternative comparison, with color-coded diagrams. These results show the excellent agreement between the ANM-predicted intrinsic dynamics of the γ-secretase complex in the absence of membrane and that observed in the cryo-electron microscopy environment.
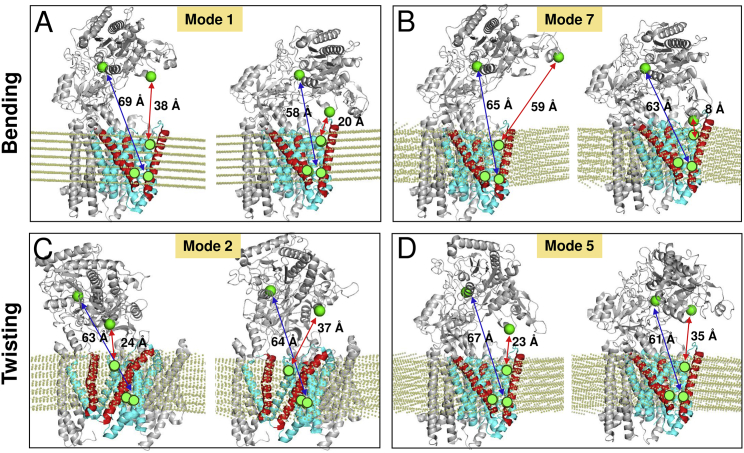
Next, we examined the collective movements of the complex in the lipid bilayer. We focused on the collective movements driven by the lowest frequency modes, which usually provide information on cooperative events relevant to functional mechanisms (48, 49, 50, 51). Our membrANM analysis clearly showed that a major mechanism of collective motion encoded by the structure is the reorientation of NCT with respect to PS1, via two types of reconfiguration: bending and twisting, illustrated in Fig. 2. Each panel in the figure displays a pair of alternative conformers differing by an RMSD of 4 Å with respect to the original structure, sampled during the fluctuations along the indicated soft (most easily accessible) modes. See also Movies S1 and S2 for the collective motions driven by the two softest modes. During bending movements (Fig. 2, A and B; Movie S1), the NCT LL moves up and down above the EC-facing vestibule of PS1, and during the twisting motion (Fig. 2, C and D; Movie S2) it cooperatively rotates back and forth about the normal to the plane of the membrane. NCT thus acts as a lid on the EC-facing vestibule of PS1; its movements give rise to open or closed conformers of the large surface groove, which alternately expose or cover the EC-exposed vestibule of PS1, as indicated by distance changes between selected residues.
Figure 2.
Bending and twisting motions of NCT with respect to TMD. (A–D) Pairs of conformers sampled during bending (modes 1 and 7) and twisting (modes 2 and 5) of γ-secretase. In each case, two ANM conformers are shown to illustrate the type of conformational fluctuations driven by the indicated mode. We display the membrane in yellow dots, PS1 in cyan, and PS1 TM2, TM3, and TM7 helices in red. The position of Glu333 in NCT, and Asp257 and Asp385 in PS1, are indicated by green spheres. Distances between Asp257 and Glu333 (blue arrow), Asp541 (NCT) and Tyr115 (PS1) (red arrows in A, C, and D), and Glu584 (NCT) and Tyr115 (PS1) (red arrows in B) are shown. Note that the distances depend on the size of ANM motions, which scale with the force constant γ. Here, the distances corresponding to an RMSD of 4 Å with respect to the original (PDB) structure are shown, for each mode direction. MD simulations (Fig 3) indicated that such distance changes take place within microseconds. See Movies S1 and S2 for the animations of the respective modes 1 and 5.
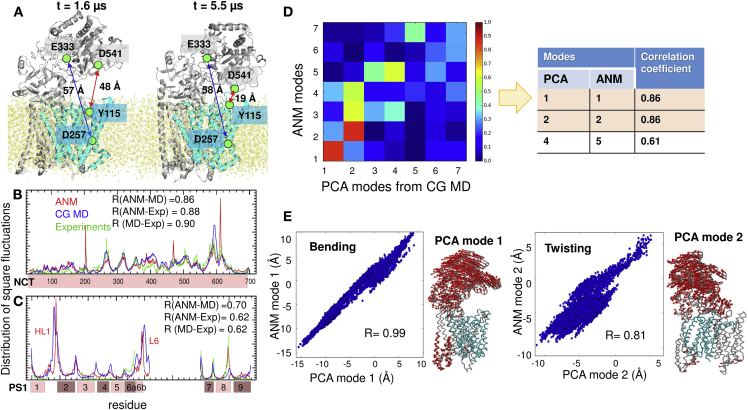
The validity of these two types of motion was further verified by two independent CG MD simulations of 10 μs each. The simulations yielded MSF profiles in excellent agreement with ANM predictions (Figs. 3, B and C; S2, A and B). The distance changes between NCT and PS1 observed in CG MD (Fig. 3 A) confirmed that those listed for ANM conformers in Fig. 2, based on an RMSD of 4 Å from the initial structure, were reasonable approximations. Fig. S3 shows that the CG MD trajectories exhibited RMSDs of 8–11 Å during 10 μs simulations. PCA of CG MD trajectories further confirmed that global bending and twisting of NCT were the most dominant modes of motion of the complex, as also reported in a recent study by Aguayo-Ortiz et al. (33). As shown in the mode-mode correlation map in Fig. 3 D, PCA modes 1 and 2 (PCA1 and PCA2) exhibited a correlation coefficient of 0.86 each, with the respective ANM modes 1 and 2. Snapshots from the trajectories aligned along PCA1 yielded a correlation of 0.99 with their alignment along ANM mode 1; and that along PCA2, yielded a correlation of 0.81 with ANM mode 2 (Fig. 3 E). See similar results from CG MD run 2 in Fig. S2. Finally, we also analyzed five different structures resolved (11, 29) for NCT and PS1. The MSF profile based on their structural covariance was found to be consistent with both ANM and CG MD profiles (see Figs. 3, B and C; S2, A and B).
Figure 3.
Results from CG MD simulations of γ-secretase. (A) Motions of NCT with respect to PS1 (cyan). Two snapshots at t = 1.6 and 5.5 μs are displayed (from run 1). The distances between Asp257 and Glu333 (blue arrows) and Tyr115 and Asp541 (red arrows) are indicated. See Fig. S3 for more details. (B) Comparison of the MSF profile of NCT residues predicted by the ANM (red curve), observed in CG MD (blue), and deduced from ensemble analysis of five PDB structures (green). Correlations between the three pairs vary in the range 0.86–0.90, as indicated. (C) Same as (B), for PS1. (D) Correlations between the softest (seven) modes predicted by the ANM and those obtained from the PCA of 10 μs MD trajectory (run 1). High correlations are shown in dark red and weak correlations in dark blue. The table lists the pairs that exhibit the highest correlations. (E) Projections of 10,000 frames from 10 μs trajectory onto the ANM mode 1 and PCA mode 1 directions (left), and ANM mode 2 and PCA mode 2 directions (right). PCA mode 1 is equivalent to the bending (mode 1) predicted by ANM, as illustrated in the ribbon diagram, and PCA mode 2 is equivalent to the twisting mode (ANM mode 2). Similar results obtained in CG MD run 2 are presented in Fig. S2.
NCT lid-like movements accompanied by PS1 HL1 rearrangements modulate the exposure/covering of the PS1 vestibule
The lid-like motions of NCT suggest that this subunit may modulate the access or binding of ligands or drugs to the groove, and thereby to PS1. The distance changes between NCT and PS1 during these cooperative motions were indicated in Figs. 2 and 3. Two residue pairs were selected as references: 1) Glu333 in NCT near the groove, which has been proposed to serve as substrate recruitment pocket (19), and the PS1 catalytic residues Asp257/Asp385 (29); and 2) NCT Asp541 or Glu584, at the entrance of the groove, distinguished by their high mobility (see Fig. 4 A), and Tyr115 in PS1, which has been reported to bind a GSM (35). The former pair provides a measure of the overall distance between functional sites in the two subunits; the latter emphasizes the ability of selected functional sites to come into close proximity, thus allowing for a physical interaction between the two subunits near the known substrate- or GSM-binding regions. The accessibility of an ensemble of intersubunit distances favored by the intrinsic dynamics of the complex is consistent with the ability of γ-secretase to catalyze the cleavage of substrates of various lengths (1, 2, 3, 77, 78), and the shifts in the sizes of Aβ peptides caused by different modulators (22). Overall, these results suggest that these modes of motion favored by the quaternary architecture can be exploited for binding or accommodating various peptides or cleavage sites, and they modulate the protease activity by providing or hindering access to binding sites.
Figure 4.
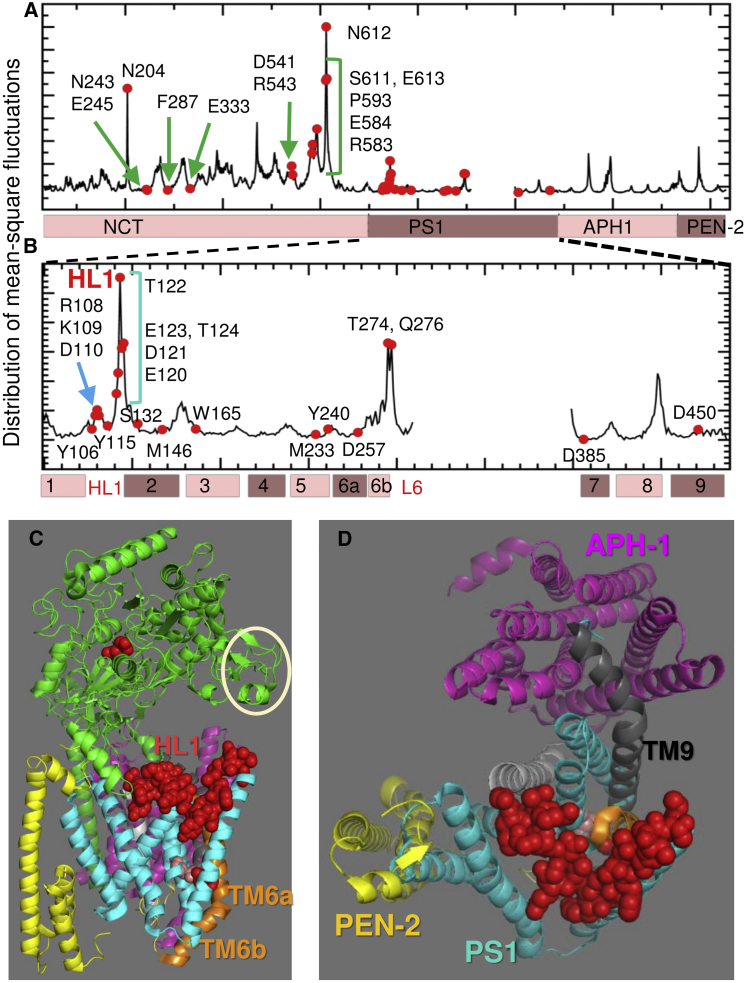
Mobility profile of γ-secretase and the critical position and dynamics of HL1. (A) Distribution of residue MSFs for the intact γ-secretase in the membrane (NCT portion identical to the ANM curve is shown in red in Fig. 3B). Highest peaks in NCT are labeled, as well as sites serving as anchors or hinges (e.g., E245, F287, and E333), which exhibit small fluctuations in space. (B) Close-up view of key residues along PS1 MSF profile. PS1 loop HL1 (S104–T124) central portion exhibits large movements. The catalytic residues (D257 and D385) are highly stable (minima). (C) Location of HL1 loop residues (shown as red space-filling) and the broken TM6 (orange) of PS1 where D257 is located. All other structural elements are colored as in Fig. 1A. The location of NCT peak residues is indicated by the white circle. (D) Top view of the TMD (colored as in Fig. 1; NCT is removed for visual clarity). HL1 covers a large portion of the EC-facing vestibule of PS1. See Figs. 5 and S4 for more details.
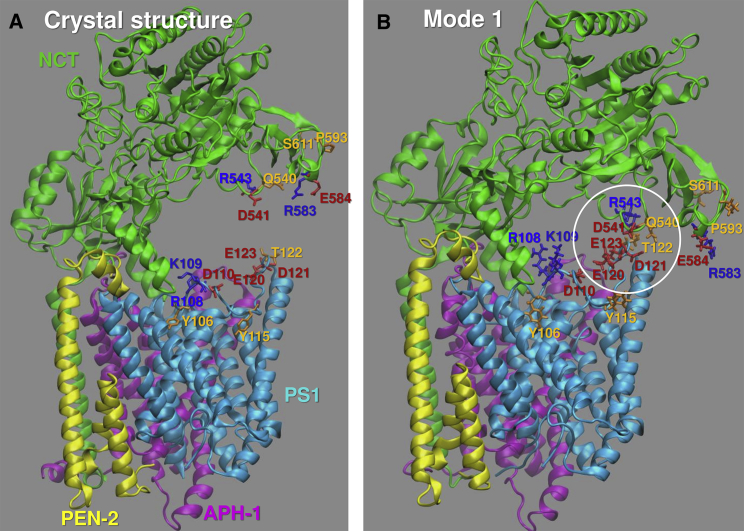
Minima in the MSF profile of the complex predicted in the presence of the lipid bilayer (Fig. 4 A) indicate the key regions that potentially act as hinges controlling the bending/torsion of NCT. Among minima, we note Phe287, which was suggested (29, 36) to act as a hinge. Glu333 also exhibits small fluctuations, consistent with its stable positioning at the interface between anticorrelated lobes of NCT. In contrast, the NCT residues His610-T614 (peak at N612) show large movements distinguished by a sharp peak. These, and spatially neighboring residues (e.g., D541, R543, R583, E584, and P593) are all located at the “mouth” of the surface groove, enclosed by the white circle in Fig. 4 C. Fig. 4 B displays the MSF profile of PS1 residues. The highest peak in this case is at E120-T124. These, along with the charged residues R108, K109, and D110, take part in HL1 (S104–T124; shown by red spheres in panels C and D), which connects TM1 and TM2. This loop covers the EC-facing vestibule of PS1 (as can be seen from the top view of the three TM subunits in Fig. 4 D). Its high mobility has been noted in an earlier study (31). Notably, several charged residues distinguished by their high mobility make intersubunit contacts (Figs. 5 and S4) during the cooperative bending of NCT to interact with the PS1 HL1 (see also Movie S1). Table S1 provides the list of intersubunit contacts made during these movements, including Q540 (NCT)-T122 (PS1), which can form a hydrogen bond driven by mode 1 (Fig. 5), or D541 (NCT)-R108 (PS1), which can form a salt bridge driven by mode 7 (Fig. S4).
Figure 5.
Intersubunit contacts between NCT LL and PS1 HL1 facilitated by the global bending mode. (A) Initial structure of the complex (PDB: 5FN2). (B) Closed form enabled by ANM mode 1, based on an RMSD of 4 Å from the initial structure. Q540, D541, R543, R583, E584, P593, and S611 in NCT, and Y106, R108-D110, Y115, and E120–E123 in PS1 are shown as red (negatively charged), blue (positively charged), or orange (polar) sticks. We note a cluster of interactions involving D121–E123 on HL1 and Q540–R543 at the NCT LL surface groove mouth (encircled). See also Movie S1 and Table S1 for close intersubunit interactions in the closed form favored by mode 1.
The ability of HL1 to make conformational rearrangements and intersubunit interactions that can interfere with the access of the substrate (APP N-terminal segment) suggests that this region may serve as a target site for binding allosteric modulators of γ-secretase function. Y106/Y115 have indeed been reported to be GSM-binding residues (35). Druggability simulations below will show that this loop harbors a druggable site, in support of their significance for potential alteration of the γ-secretase substrate-binding properties.
Another PS1 region that shows large movements is the broken C-terminal half of TM6 (TM6b; see peak at T274-Q276 in Fig. 4 B) and the adjoining loop (L6) that connects to TM7. A large part of this intracellular-exposed loop was not resolved, consistent with its conformational flexibility, also noted earlier (11, 31, 33). The kink in TM6, which apparently underlies the high mobility of TM6b, is very close to the catalytic pocket, and the flexibility of TM6b (and L6) is likely to facilitate the exposure of the catalytic pocket to the lipid environment.
The anticorrelated motions of NCT are enhanced by coupled membrane undulations, whereas the catalytic site stably maintains its local geometry
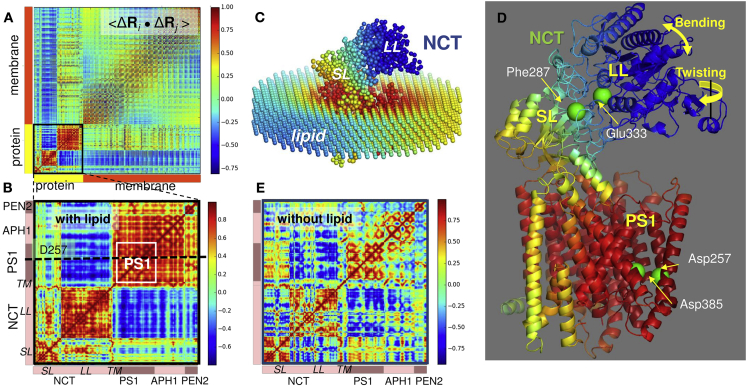
Fig. 6 A shows the orientational cross correlations, Cij = < ΔRi · ΔRj >/ [<(ΔRi)2> <(ΔRi)2>]1/2, between the movements of network nodes, each node representing an amino acid or a site on the lipid bilayer (Fig. 6 C). We focus here on the effect of the 10 softest modes, which usually represent the most easily accessible and cooperative movements of the system. <ΔRi · ΔRj > is the cross correlation between the movements of residues i and j. By definition, Cij varies in the range −1 ≤ Cij ≤ 1, with the lower and upper limits corresponding to fully correlated (+1) or fully anticorrelated (−1) pairs. Anticorrelated pairs undergo coupled, but opposite-direction, movements (e.g., opening/closing of a cleft). Fig. 6 A reveals the preferential intra- and intersubunit couplings within γ-secretase, as well as the type of coupling to the surrounding lipid bilayer. The two red blocks along the diagonal of Fig. 6 A show the strong intrasubunit coupling within NCT and within the TM subunits, as can be seen more clearly in the enlarged portion of the map in Fig. 6 B. The latter shows that the two NCT lobes, SL and LL, form two internally coherent entities when they are dynamically decoupled with respect to each other, separated by a hinge region near F287; the C-terminal helix of NCT (labeled TM along the axes) is instead correlated with PS1, indicating the role of this helix in maintaining the coupling between NCT and PS1.
Figure 6.
Cross correlations between the motions of γ-secretase residues in the membrane environment. (A) Cross correlation map for γ-secretase complex and surrounding lipid bilayer. The entries in the map represent the orientational cross correlations (−1 ≤ Cij ≤ 1; see the scale on the color-coded bar) between all pairs of nodes (residues or lipid sites) i and j. Red blocks along the diagonal indicate the strongly correlated substructures, and blue regions indicate the anticorrelated (moving in opposite direction) pairs of substructures. Results are based on 10 softest ANM modes. (B) Close-up view of cross correlations within γ-secretase. (C) Network representation of the complex and membrane, colored by the type and strength of correlation with respect to Asp257 (indicated by dashed line in B). Blue and red regions exhibit anticorrelated and correlated movements, respectively, with respect to Asp257; yellow/green regions are uncorrelated. (D) γ-secretase color-coded by cross correlations of residues with Asp257. Strongest anticorrelations are observed at the NCT LL (blue), whereas PS1 and APH-1 form a highly correlated block (red). The NCT SL (residues 34–248 and 651–664) and NCT terminal TM helix (residues 665–698) include hinge sites (yellow/green). (E) Same as in (B), in the absence of membrane. An overall weakening in the cooperativity of subunit movements is observed.
Fig. 6 A further shows that the NCT LL undergoes anticorrelated movements with respect to the membrane. To further assess the role of the lipid in the collective dynamics of γ-secretase, we repeated the ANM analysis in the absence of membrane. The map in Fig. 6 E is the counterpart of that in Fig. 6 B computed in the absence of the lipid bilayer. It clearly shows that the strong intra- or intersubunit couplings are significantly weakened, and that the blocks that exhibited coherent dynamics became fragmented, in the absence of membrane. Toward assessing the effect of adopting a more detailed network model for the lipid bilayer, we repeated the membrANM computations with a higher-density representation of the membrane, which confirmed that the softest modes were robustly retained (see Fig. S5). Overall, our analysis suggests that the membrane consolidates the strength of correlations within subunits, and anticorrelations between NCT and TM subunits.
To understand better the physical implications of these cross correlations, we focused on residue D257 as a representative site for the catalytic region in PS1. Fig. 6 D shows the structure color-coded by the strength and types of the cross correlations of D257 with all other residues, deduced from the row corresponding to D257 in the map Fig. 6 B, i.e., the ith node/residue in Fig. 6 D is colored by the Cij value in the row corresponding to j = D257 (indicated by dashed line) in Fig. 6 B. The structural elements that move in the same direction are colored red (strong coupling) or orange (moderate coupling); those coupled but moving in the opposite directions are blue. Green regions indicate the sites that are uncorrelated, which often act as anchors (to maintain stability) or hinges (between anticorrelated substructures on both sides). The diagram clearly shows that the catalytic residues D257 and D385 take part in a tightly correlated block, i.e., their relative positions, as well as those of the near neighbors M146, W165, and M233 (see Fig. 1 C), remain almost fixed. This is consistent with the precise positioning of a catalytic residue required for chemical specificity, as noted earlier (79) for a series of enzymes. Yet, this does not mean that PS1 is entirely rigid. A closer examination of a selected mode (Figs. 7 and S6) shows that the PS1 HL1 and TM2 N-terminal part (both exposed to the EC region) exhibit anticorrelated (blue; opposite directions) motions with respect to the catalytic region, as will be further detailed below, even though the two catalytic residues D257 and D385 stably maintain their relative positions.
Figure 7.
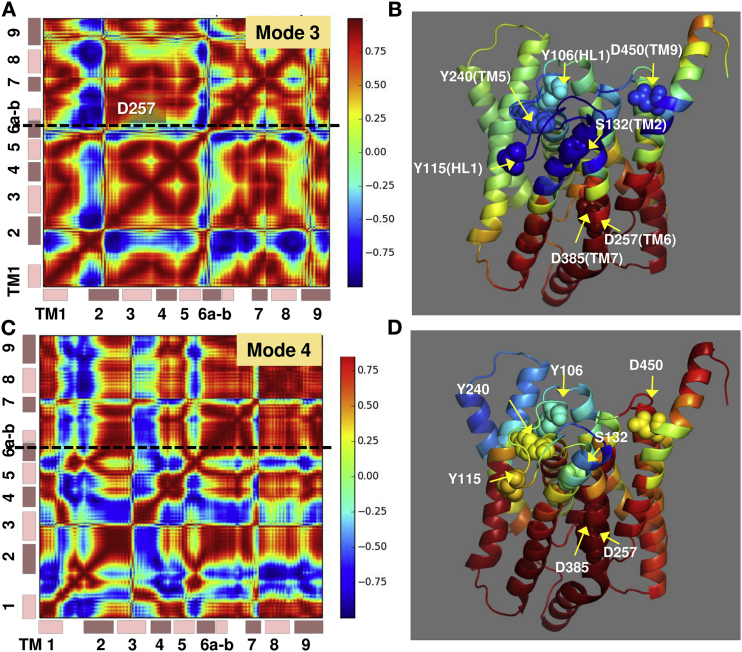
Anticorrelations between EC-exposed ends and the catalytic site observed in ANM modes 3 and 4 accessible to the complex. Cross correlations driven by ANM modes 3 (A) and 4 (C) are displayed, and corresponding respective diagrams (B and D) color-coded by the correlations of all PS1 residues with respect to Asp257. The two catalytic residues (D257 in TM6 and D385 in TM7) are shown in spheres as well as selected residues (e.g., Y106 (HL1), Y115 (HL1), Y240 (TM5), S132 (TM2), and D450 (TM9)) that either exhibit strong anticorrelations (blue) or act as hinge centers (yellow). Y106, Y115, and Y240 are involved in GSM binding, and S132 and D450 in substrate binding.
Within PS1, HL1- and EC-exposed ends of TM2, TM5, and TM9 exhibit anticorrelated movements with respect to the catalytic site
Among the residues that show strong anticorrelations with respect to the catalytic region, we note Y115 and Y106 in HL1, Y240 on TM5, D450 on TM9, and S132 on TM2, shown in spheres in Fig. 7, B and D. S132 and D450 have been pointed out to play a key role in substrate binding (12), and Y106/Y115 and Y240 have been reported to bind a GSM (35). These coupled residues identified here occupy minima positions in Fig. 4 B, which means that they are severely constrained in the global dynamics of PS1 (and the overall complex). Therefore, structural changes at those sites would be accompanied by cooperative compensating rearrangements, or allosteric responses, which would mutually engage the strongly anticorrelated regions.
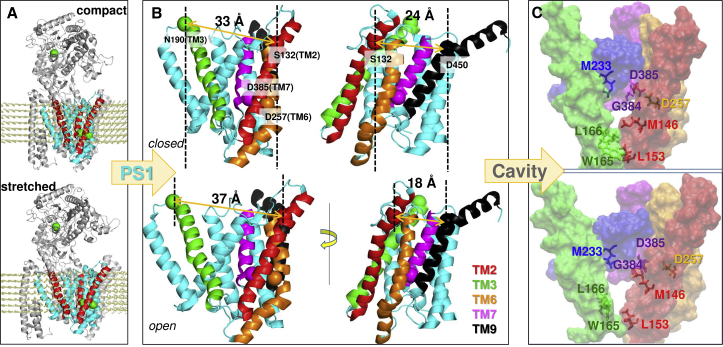
We also distinguished another mode that cooperatively involved NCT LL tip motions and PS1 collective “breathing”: ANM mode 14 (Fig. 8; Movie S3). During this motion, the cross-sectional area of PS1 (viewed from the EC side) changed from an approximately circular to an elliptical shape and vice versa. Fig. 8 A shows the end conformers alternately accessed, referred to as compact and stretched conformers. Precisely, the EC-facing vestibule lined by TM2-TM3-TM7 stretches into an open form (evidenced by the N190 (TM3)-S132 (TM2) distance increase), whereas TM2-TM6-TM9 contracts (S132-D450 distance decreases), and vice versa (Fig. 8 B).
Figure 8.
Asymmetric breathing motion of PS1. (A) A pair of conformers, compact and stretched, sampled during reconfiguration of PS1 along ANM mode 14 of the protease complex-lipid system is shown. The conformers are obtained based on an RMSD of 4 Å each with respect to the initial (PDB) structure. TM2, TM3, and TM7 are colored red. Glu333 in NCT1, and Asp257 and Asp385 in PS1, are shown in green spheres. (B) Close-up view of interhelical distance changes in the two conformers. TM2, TM3, TM6, TM7, and TM9 are colored as in Fig. 1C; other TM helices are cyan. The left diagrams display PS1 from the same perspective as in (A); the right diagrams are rotated around the normal to the membrane plane, to facilitate the visualization of the TM2–TM9 distance. Asp257, Asp385, Ser132 (TM2), Asn190 (TM3), and Asp450 (TM9) are shown in spheres colored by the corresponding TMs. S132–N190 (left) and S132–D450 (right) distances show opposite changes, i.e., an increase in the former is accompanied by a decrease in the latter and vice versa. (C) Catalytic cavity in the respective conformers, with TM2–TM7 shown in space-filling. Side chain positions obtained by energy minimization are shown for selected residues. See also Movie S3.
Our CG MD simulations confirmed the same behavior: N190 and S132/D450 exhibited even larger distance changes (up to 10/17 Å) in CG MD simulations, and a decrease in S132-N190 distance was often accompanied by an increase in S132-D450 distance (Fig. S7 A). To further understand the origin of this particular mode, we examined the PS1 subunit behavior in MD simulations. PCA of PS1 alone showed that a principal mode (PCA2) intrinsically favored by the PS1 tertiary structure strongly correlated with the stretching/contraction mode observed in ANM mode 14 (Fig. S7 B).
Remarkably, the net effect of this cooperative motion is a change in interhelical packing near the catalytic site, as can be seen in Fig. 8 C. In the closed PS1, Met146 (TM2) is making close hydrophobic contacts with Leu166 (TM3); likewise, Trp165 (TM3) is interacting with Leu153 (TM2). These tight interactions might restrict the recruitment of a new molecule, whereas ligands can enter/exit more easily in the stretched state. It is interesting to note that when the PS1 sample is of the stretched state, the NCT LL tip (His610-T614) opens-up simultaneously, and vice versa (see Movie S3). Thus, synchronous movements of NCT and PS1 may help ligand insertion into the cavity.
Conclusion from ANM analysis
Overall, this analysis shows that despite the lack of correlation between the global bending and twisting of NCT and the local conformational changes in the catalytic site, the complex has access to mechanisms of motions that couple the NCT ligand-binding and PS1 catalytic sites. This is achieved through intermediate regions such as the NCT LL mouth/tip R583-E613, the PS1 HL1, and the EC-exposed ends of TM5, TM9, and TM2 helices, whose motions are mediated by global hinges (e.g., at the NCT-TMD interface or the NCT LL-SL groove). Thus, our analysis identified sites that may play a key mechanical role in substrate binding and proper positioning for cleavage. The next question is then: are these sites druggable? We now proceed to druggability simulations to explore whether the regions distinguished here by correlated events, or those key sites controlling these correlated movements, can be effectively targeted by drug-like molecules.
Druggability simulations reveal multiple druggable sites on PS1, including both orthosteric and allosteric sites
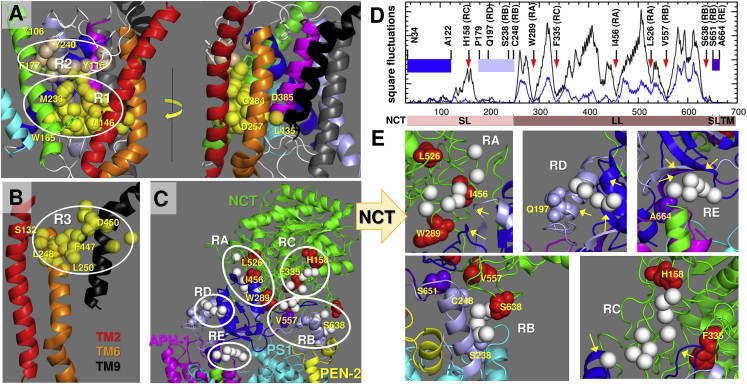
We first performed a series of druggability simulations for PS1 alone and for PS1 in the contact of the intact protease (Table S2). Simulations for the intact γ-secretase yielded six druggable sites on PS1, labeled R1–R6 in Figs. 9, A and B and S8. Druggable sites are indicated by circles enclosing the clusters of probe molecules (shown as spheres) that were found to consistently bind those regions, called hot spots. Among them, R1 was invariably detected in all runs performed for the intact secretase as well as isolated PS1 to be a high-affinity druggable site, with an estimated binding affinity of −14.4 to −11.6 kcal/mol (Table S2). R1 coincides with the central catalytic cavity of PS1 (Fig. 9 A) and therefore represents an orthosteric binding site. Probes bound therein were observed to make contacts with the catalytic residues Asp257 and Asp385, as well as the spatial neighbors Met146, Trp165, Met233, and Gly384, reported to possibly bind the inhibitor DAPT (11). Even though this region is buried, it easily attracted probes (originally located in the solution).
Figure 9.
High-affinity sites identified by druggability simulations of the intact complex. (A and B) Druggable regions of PS1. TM helices are colored as in Fig. 1A. Three druggable sites are observed, labeled as R1–R3, composed each of a cluster of hot spots (in cream/yellow spheres). Y106 and Y115 (HL1); S132 and M146 (TM2); W165 and F177 (TM3); M233 and Y240 (TM5); L248, L250, and D257 (TM6); G384 and D385 (TM7); and L435, F447, and D450 (TM9) are shown as sticks and labeled. (C) Druggable regions on NCT, labeled RA-RE. Druggable hotspots are shown in white. Sites colored blue, ice blue, and purple participate in the hinge regions, and red spheres act as hinge centers. (D) Square fluctuations driven by ANM modes 1 (black line) and 2 (blue line). The hinge regions and residues are marked by bands (blue, ice blue, and purple) and red arrows. Residues of interest at druggable sites are indicated. (E) Zoom-in views of each druggable region in NCT. Hinge residues close to druggable sites are labeled and indicated by yellow arrows. See Table S2 for estimated binding affinities at those sites, and Fig. S8 for additional druggable sites.
Notably, the R2 site, adjacent to R1, included the HL1 residues Y106 and Y115 as well as the spatially neighboring F177 and Y240, which coincides with a GSM-binding site (35). These are highly stable residues (minima in Fig. 4 B) that communicate directly with HL1 residues affected by the global bending (Figs. 2, A and B, 5, and S4) and twisting (Fig. 2, C and D), in addition to undergoing anticorrelated motions with respect to the catalytic site (Fig. 7), hence the significance of this site allosteric modulation of activity.
The druggable site R3 was located between TM2 and TM9, near the EC region, close to S132 and D450 (Fig. 9 B). These two residues were reported to be involved in substrate gating (12). The same site also includes L250, whose distance from L435 was reported (35) to change upon GSM binding.
Site R4 was widely distributed along TM1 and TM8, at the interface with APH-1 (see Fig. S8). It was detected in all three runs conducted for the intact protein and confirmed in two performed for the isolated subunit PS1 (Table S2). Likewise, R6 emerged in the simulations of the intact protein, at the interface between TM8–TM9 and subunit APH-1. R5 was near the C-terminal end of TM9.
Druggable sites on NCT are localized at hinge sites that control the opening/closure of the substrate-binding groove
Figs. 9, C and E and S8 give an overview of druggable sites on NCT and APH-1. No druggable site was detected on PEN2. A new TM region, R7, at the interface between the NCT C-terminal TM helix and APH-1, was found in all three simulations to yield high binding energies (≤ −13.0 kcal/mol) (see Table S2). As for the EC region, five sites RA-RE (Fig. 9, C and E) were detected. Although corresponding probe-binding affinities were relatively lower compared to the other hot spots on PS1 and APH-1, these are evidently more accessible, being in the EC region, and interestingly most were located near the hinge region between NCT and TM subunits, or around the substrate-binding groove whose opening/closure with respect to the PS1 HL1 was noted above (Figs. 5 and S4), suggesting that RA-RE could serve as allo-targeting sites.
Fig. 9, D and E present a more detailed analysis. Fig. 9 D shows square fluctuations in NCT residues driven by the softest ANM modes (modes 1 and 2 that control the opening/closure of the groove mouth; see Fig. 2, A and C). NCT sequence spans the regions SL and LL, and then back to SL and TM (C-terminal helix). Many SL residues are severely constrained, participating in two hinge-bending centers: one with the TM subunits APH-1 and PS1, and another with the NCT LL. The constrained regions (minima in Fig. 9 D) include N34-A122 (shown in blue band), P179-C248 (ice blue band), and S651-A664 (purple band). These are colored in the same way in the ribbon diagrams displayed in Fig. 9, C and E. All the druggable sites on NCT either overlap or make close contacts with these constrained regions. The hinge centers therein are indicated by the red spheres in the ribbon diagrams (Fig. 9, C and E) and red arrows in the fluctuation profile (Fig. 9 D). Among the druggable sites, we note that RB is located at the hinge near the LL-SL interface as well as PS1 HL1, making it a highly susceptible site for binding a GSM.
Overall, this analysis shows that NCT presents several druggable sites, including those closely neighboring hinge centers that mediate the cooperative rearrangements of the EC part of the complex with respect to its TMD or the movements of NCT LL with respect to SL. These movements directly affect the exposure or closure of the surface groove over HL1. Targeting those sites thus opens, to our knowledge, new avenues for allosterically controlling the access of substrate to PS1 EC-facing vestibule and potentially obstructing access to the γ-secretase active site.
PS1 inhibitors and modulators bind orthosteric and allosteric sites consistent with druggability simulations
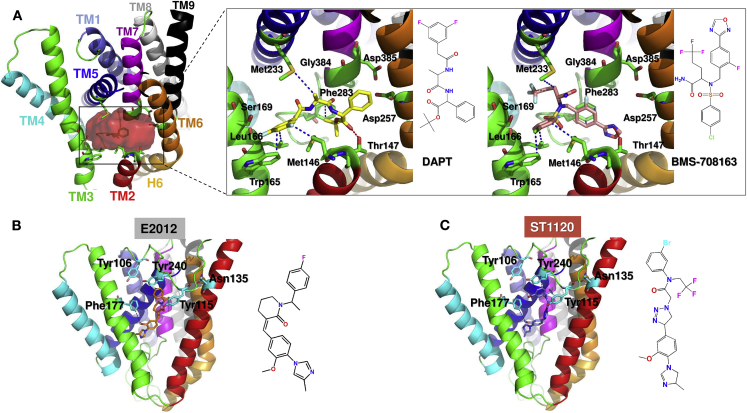
We first examined the binding properties of two reported γ-secretase inhibitors (DAPT and BMS-708163, with the IC50 values of 120 and 0.3 nM, respectively) onto the R1 site. As depicted in Fig. 10 A, high-affinity binding poses were attained for these drugs, although R1 was relatively buried in the TMD. The binding pocket is lined by TM2, TM3, TM5, and TM7, and involves Met146, Thr147, Trp165, Leu166, Ser169, Met233, Phe283, and Gly384, in close agreement with the hot spot and coordinating residues reported above (see Fig. 9 A). Met146, Trp165, and Leu166 make hydrophobic contacts with the chlorobenzene group on DAPT and m-difluorobenzene group on BMS-708163; the methyl group on DAPT and the methylene group on BMS-708163 are both positioned to interact with the methyl thio group on Met233; and Thr147 forms a hydrogen bond with one of the carbonyl groups on DAPT. Interestingly, Phe283 exhibits a π-π conjugation with a fluorobenzene group on BMS-708163, which may explain the stronger activity of BMS-708163 compared to that of DAPT. Overall, these docking results suggest that Met146, Thr147, Trp165, and Met233 play a role in binding orthosteric ligands consistent with the R1 region deduced from druggability simulations to serve as hot spots.
Figure 10.
Binding of inhibitors (DAPT and BMS-708163) and modulators (E2012 and ST1120) to PS1. (A) Orthosteric site near D257 and D385, also identified as the top-ranking druggable site R1 (in Fig. 9A), binds both inhibitors. Binding pose of each inhibitor is shown in the magnified diagrams on the right. The same set of residues (labeled) coordinate both drugs. (B and C) Binding of modulators E2012 and ST1120 to the allosteric site R2 identified in Fig. 9A. Coordinating residues include Y106 and Y115 (on the EC loop HL1), N135 (TM2), Phe177(TM3), and Y240 (TM5).
As a further test, we examined the binding pose of three additional γ-secretase inhibitors from the ChEMBL database, CHEMBL2159511 (IC50: 1.3 nM), CHEMBL2159691 (IC50: 37 nM), and CHEMBL2159687 (IC50: 20 nM). These compounds share the same scaffold but have different functional groups ((methylsulfonyl)ethane, acetonitrile, and ethanol). As shown in Fig. S9, Trp165, Leu166, and Met233 interact with the fluorine atom that connects to the benzene. Strong hydrophobic interactions were observed between Phe283 and the aromatic groups of the ligands. Thr147 formed a hydrogen bond with each of the functional groups as indicated by the interatomic distance of 2.0–2.1 Å. However, we found that (methylsulfonyl)ethane formed strong hydrophobic interactions with Leu268 (2.7 Å, shown by dashed line) and the backbone of Lys265 (2.2 Å), which might make CHEMBL2159511 more potent.
Docking simulations were also performed for two GSMs: E2012 and ST1120. These simulations confirmed the high affinity of site R2 (Fig. 10, B and C), which involved Y106 and Y115 (on HL1), N135 (TM2), Phe177 (TM3), and Y240 (TM5). The two modulators exhibited almost the same interactions upon slight rearrangements of the conformations of these key residues. For example, both formed a hydrogen bond with Tyr115, and made π-π interaction with Tyr240 and hydrophobic interaction with Phe177. ST1120 has been reported to bind to the extended HL1 at the EC vestibule of PS1 (35), and E2012 has been reported to bind to the N-terminal fragment TM1–TM6 of PS1 (80).
We also performed docking simulations, using conformers sampled along ANM 14, to examine the effect of volume expansion near the catalytic site. Both inhibitors, DAPT and BMS-708163, exhibited higher affinity to bind the expanded state of PS1, compared to binding the PDB structure. As illustrated in Fig. S10, BMS-708163 interacts with Met146 and Trp165 in the expanded, but not in the contracted, conformation of PS1. That is, PS1 volume increase enhances orthosteric inhibitor binding. Computations repeated with modes 1 and 7 had smaller effects on inhibitor binding, but affected the modulator binding affinity. Interestingly, the modulators E2012 and ST1120 both exhibited the highest docking score in the original (PDB) structure and the lowest in mode in the closed/compact states driven by modes 1 and 7, indicating that the NCT lid closure interfered with modulator binding.
Discussion
Anticorrelated movement of NCT with respect to PS1 HL1 emerges as a structure-encoded mechanism for modulating substrate recruitment and positioning
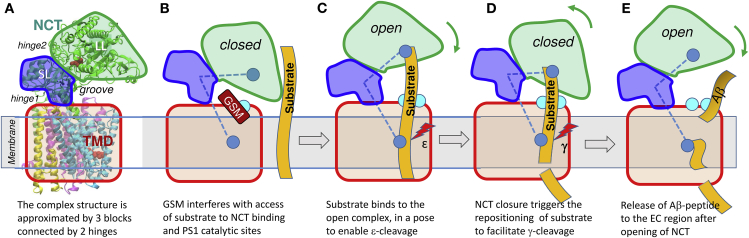
Our analysis suggests a simple mechanistic model (Fig. 11) for facilitating substrate binding, successive cleavage at two sites, and release of peptide (Aβ) to the EC region, mediated by the structure-encoded global dynamics of the intact γ-secretase complex. We have shown that the γ-secretase ectodomain (NCT) is highly dynamic, as indicated by previous studies (36, 37, 38). It enjoys large bending and twisting motions, allowing for changes in the size of a major groove that has been reported to bind substrate (12) and modulators (35). Based on the hinge regions observed in our ANM analysis, γ-secretase can be divided into three substructures: the TMD (composed of PS1, AHP-1, PEN-2, and the C-terminal helix of NCT; red), the NCT SL (blue), and the NCT LL (green). Two hinge regions separate these three substructures, labeled hinge 1 and hinge 2 in Fig. 11 A. NCT fluctuates between open and closed forms; due to the rotational flexibility endowed by these hinges (and thermal fluctuations), either form may be stabilized upon ligand binding. For example, binding of a modulator near HL1 can potentially stabilize the closed form as shown in Figs. 2, 5 B, and S4. NCT harbors substrate-binding sites (17, 18, 19) and the TMD contains the catalytic site, schematically shown by blue spheres in Fig. 11, B–E. The structure-encoded bending and twisting of NCT gives rise to dramatic changes in the distance between NCT LL and PS1 at the groove; if NCT is in the open state (Fig. 11 C), the substrate can insert into the groove and bind to NCT or PS1, but if NCT is closed (as in Fig. 11 B), it cannot. NCT motions are thus proposed to play a critical role in allowing or blocking substrate recognition and/or binding.
Figure 11.
Schematic description of a model for peptide binding, repositioning, cleavage, and release modulated by the opening/closure of NCT with respect to the TMD. (A) Schematic diagram of γ-secretase. NCT is composed of two lobes, LL and SL, shown in green and blue separated by hinge 2, and the transmembrane domain (TMD) is in red, with interfacial hinge 1 enabling the relative movement of NCT SL with respect to TMD. NCT fluctuates between open and closed states. (B) Binding of modulator (GSM, dark red) to the allosteric site (cyan) near HL1/TM2 (on PS1) or the interlobe groove (on NCT) stabilizes the closed form and obstructs the access of the substrate peptide (orange, extended from EC to TM region) to its binding site and to the PS1 catalytic site. Blue ball in NCT is substrate-binding site and blue ball in TMD is catalytic site. (C) Suggested binding pose of substrate, attached on top to NCT, and exposed at the bottom to the catalytic site. (D) Sliding of the substrate to expose a new cleavage site to the catalytic region, facilitated by the closure of NCT. Open and closed states may thus help proper positioning of the successive cleavage sites: ε cleavage occurs in open state (C) and γ cleavage in closed state (D). (E) Subsequent opening of NCT permits the release of the Aβ peptide to the EC region.
Fig. 11, C and D schematically describes the successive ε- and γ-cleavages of the substrate. According to the two-step substrate-recognition process proposed in a recent study (12): 1) a first binding site (for ε-cleavage) forms upon reconfiguration of PS1 residues S132 and D450 to expose a hydrophobic site for substrate binding and 2) the C-terminal fragment on PS1, which closely neighbors the HL1/TM2 N-terminal end, serves as a second site for γ-cleavage of the substrate (APP). We note that both sites were identified by our simulations to be druggable (Fig. 9), and the latter was shown to bind GSMs (Fig. 10). We propose here that the anticorrelated movements of NCT, to which one end of the substrate is bound, may assist in the repositioning of the substrate for γ-cleavage. Finally, the release of the resulting Aβ peptide to the EC region can also be facilitated by the opening of NCT (Fig. 11 E).
The intact γ-secretase offers multiple sites for allo-targeting
Enzymatic activity is often inhibited by targeting the catalytic site with orthosteric inhibitors. However, in recent years, allosteric inhibition, especially in the case of multidomain/multisubunit proteins that interact with multiple substrates, has emerged as a more specific approach to interfere with selected pathways upon blocking selected interactions (81, 82). γ-secretase is such a target. There has been a growing interest in developing allosteric modulators of γ-secretase, which would be selective for Aβ modulation, but would not impair the processing of other γ-secretase substrates such as Notch, EphB2, or EphA4. This motivated the original discovery of nonsteroidal anti-inflammatory drugs (R-flurbiprofen and sulindac sulfide) by AstraZeneca, and more recently, a second generation of GSMs (E2012) by Eisai (23). The two groups of compounds were anticipated to have a different mechanism of action. Recent studies have shown that the photoaffinity probe E2012-BPyne specifically labels the N-terminal fragment of PS1 (80) when PS1 is in the active state (but not inactive state), indicating that (the parent imidazole) E2012 binds the specific location that becomes accessible upon PS1 activation. Notably, this binding site is different from those of other GSMs (cid GSM-1, allosteric GSI BMS-708163, or substrate docking site peptide inhibitor pep11). Johnson et al. concluded that “multiple binding sites within the γ-secretase complex exist, each of which may contribute to different modes of modulatory action” (80). Our study demonstrates that such allosteric sites indeed exist in the complex, and many of these were also shown to play a key role in controlling the collective dynamics, and thereby substrate-binding properties.
A recent study of the mode of actions of phenylimidazole-type GSMs using a chemical biology approach and systematic mutagenesis revealed that an increase in the proteolytic activity of γ-secretase and a reduction in Aβ toxicity takes place in a selective way (35), as a result of a conformational change at the catalytic center induced upon binding of a modulator (ST1120) to the extended HL1 at the EC vestibule of PS1. Although this study was based on a structural model derived from the PS1 ortholog from the archaea Methanoculleus marisnigri (83), we had the opportunity to use the cryo-electron microscopy structure resolved for the human γ-secretase complex and draw conclusions for PS1 conformational dynamics based on the intact structure embedded in the lipid bilayer. Photoaffinity labeling experiments and alanine substitutions suggested that the EC pocket formed by HL1/TM2/TM5—and in particular the PS1 residues Y106 and Y115 (on the EC loop HL1), N135 (TM2), and Y240 (TM5)—were involved in the interaction of PS1 with the phenylimidazole-type GSM (35). Our analysis (Fig. 10) confirms the involvement of these residues in binding ST1120 and further sheds light on detailed interatomic interactions at the binding site, as well as the mechanistic basis of the GSM action.
The lipid bilayer restricts the motion of the TMD. However, the distance changes in TM2–TM3 or TM2–TM9, indicated by both ANM and CG MD (see Fig. 8; Figs. S6 and S7), support the view that TM2–TM3 (11) and TM2–TM9 (12) can serve as lateral gates for substrate entering. We suggest that TM2 plays a major role in gating motions that control the access to the catalytic site, from the horseshoe-like opening of PS1, and computational studies that take account of the adaptability of (or volume changes in) the PS1 central cavity are likely to detect new inhibitors.
NCT can serve as a new target for allosteric modulation of γ-secretase dynamics
In addition to PS1, which harbors a number of allosteric sites, our analysis suggests that NCT can also bind allosteric modulators (Fig. 9). Many residues that take part in hinge regions at NCT were found to be druggable, meaning that the equilibrium between the open and closed states of NCT can be shifted in one direction or another (depending on the modulator), or that the movements accessible to the complex could be altered. Previous studies have reported inhibitory actions originating from structural change (38) and also dynamic changes in NCT (37). Here, we attribute the effect of NCT to the twisting and bending motions inherently favored by the overall architecture of γ-secretase, which were also reported in recent CG MD simulations of the intact structure (33). Bai et al. (29) suggested that NCT rotation could shorten the distance between the sites of NCT and PS1. Here, we show how NCT can indeed have contacts with PS1 HL1 residues, such as Y115, which are involved in modulator binding (35) (Fig. S4; Table S1).
The cleft (or groove) between NCT LL and SL and PS1 HL1 is distinguished here as a region that is likely to bind a modulator. In particular, the site RB in this cleft was found to be highly druggable (Fig. 9 E). This site neighbors the hinge residues V557, S638, and S651 and the anchor helical residues S238–C248, and thus represents a mechanically constrained, yet solvent-exposed and druggable, site. Targeting this site would directly interfere with the hinge-bending mobility of the NCT SL-TMD interface. Furthermore, the same site is located only 10–15 Å from HL1 (e.g., residues K109 and N112), such that binding of a modulator is likely to interfere with the HL1 motions, which in turn may alter the interhelical interactions involving S132, Y446, F447, and D450. Note that the latter set of residues harbor another druggable site, R3, which may also serve as an allosteric modulation site.
In summary, although we focused (and demonstrated in silico) the high affinity of sites R1 and R2 on PS1 to bind (in accord with earlier experiments) inhibitors and modulators, respectively (Fig. 10; Figs. S9 and S10), two other sites for allo-targeting emerged from this analysis: R3 on PS1 (Fig. 9 B) and RB on NCT (Fig. 9 E). These results, to our knowledge, open up new paths for the development of allosteric modulators of γ-secretase APP processing activity, to be advanced by future work.
Author Contributions
J.Y.L., Z.F., X.-Q.X., and I.B. designed and performed research. J.Y.L., Z.F., and I.B. analyzed data and wrote the manuscript.
Acknowledgments
The authors acknowledge funding from the National Institutes of Health (P41 GM103712 (I.B.), P30 DA035778 (I.B. and X.-Q.X.), and R01 DA025612 (X.-Q.X.)) and the Department of Defense (W81XWH-16-1-0490 (X.-Q.X.)).
Editor: Michael Feig.
Footnotes
Ji Young Lee and Zhiwei Feng contributed equally to this work.
Supporting Materials and Methods, ten figures, two tables, and three movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31126-8.
Contributor Information
Xiang-Qun Xie, Email: xix15@pitt.edu.
Ivet Bahar, Email: bahar@pitt.edu.
Supporting Material
The extracellular NCT subunit undergoes a cooperative bending movement with respect to the TM subunits inducing significant (∼20 Å) changes in the distance between its exposed loops and the EC-facing vestibule of PS1. There are five spheres showing Asp257/Asp385 (PS1) in yellow, Tyr115 (PS1-HL1) in green, Asp541 (NCT) in orange, and Glu333 (NCT) in blue. See Fig. 2 for the conformations of the intact γ-secretase sampled during these fluctuations.
The extracellular NCT subunit undergoes a cooperative twisting movement with respect to the TM subunits that are practically rigidly embedded in the membrane. Note the accompanying membrane movements. There are five spheres showing Asp257/Asp385 (PS1) in yellow, Tyr115 (PS1-HL1) in green, Asp541 (NCT) in orange, and Glu333 (NCT) in blue. See Fig. 2 for the conformations of the intact γ-secretase sampled during these fluctuations.
The breathing motion of PS1 and cooperative movement of the NCT subunit. The catalytic sites Asp257 and Asp385 on PS1, and the central residue Glu333 on NCT are shown in green spheres. See Fig. 8 for details during these fluctuations.
References
- 1.Selkoe D.J., Wolfe M.S. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 2.De Strooper B., Iwatsubo T., Wolfe M.S. Presenilins and γ-secretase: structure, function, and role in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2012;2:a006304. doi: 10.1101/cshperspect.a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Struhl G., Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol. Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe M.S. γ-Secretase as a target for Alzheimer’s disease. Adv. Pharmacol. 2012;64:127–153. doi: 10.1016/B978-0-12-394816-8.00004-0. [DOI] [PubMed] [Google Scholar]
- 5.Sobhanifar S., Schneider B., Dötsch V. Structural investigation of the C-terminal catalytic fragment of presenilin 1. Proc. Natl. Acad. Sci. USA. 2010;107:9644–9649. doi: 10.1073/pnas.1000778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active γ-Secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 7.Kimberly W.T., LaVoie M.J., Selkoe D.J. γ-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc. Natl. Acad. Sci. USA. 2003;100:6382–6387. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfe M.S., Xia W., Selkoe D.J. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 9.De Strooper B., Saftig P., Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 10.Struhl G., Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 11.Bai X.C., Rajendra E., Scheres S.H.W. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takagi-Niidome S., Sasaki T., Tomita T. Cooperative roles of hydrophilic loop 1 and the C-terminus of presenilin 1 in the substrate-gating mechanism of γ-secretase. J. Neurosci. 2015;35:2646–2656. doi: 10.1523/JNEUROSCI.3164-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selkoe D.J. Presenilin, Notch, and the genesis and treatment of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2001;98:11039–11041. doi: 10.1073/pnas.211352598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F., Hasegawa H., Fraser P. TMP21 is a presenilin complex component that modulates γ-secretase but not ε-secretase activity. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- 15.Chávez-Gutiérrez L., Bammens L., De Strooper B. The mechanism of γ-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L., Zhou R., Shi Y. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Aβ42 and Aβ40 peptides by γ-secretase. Proc. Natl. Acad. Sci. USA. 2017;114:E476–E485. doi: 10.1073/pnas.1618657114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah S., Lee S.F., Yu G. Nicastrin functions as a γ-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Yu G., Nishimura M., St George-Hyslop P. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature. 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- 19.Dries D.R., Shah S., Yu G. Glu-333 of nicastrin directly participates in γ-secretase activity. J. Biol. Chem. 2009;284:29714–29724. doi: 10.1074/jbc.M109.038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takasugi N., Tomita T., Iwatsubo T. The role of presenilin cofactors in the γ-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 21.Lee S.F., Shah S., Yu G. A conserved GXXXG motif in APH-1 is critical for assembly and activity of the γ-secretase complex. J. Biol. Chem. 2004;279:4144–4152. doi: 10.1074/jbc.M309745200. [DOI] [PubMed] [Google Scholar]
- 22.Golde T.E., Koo E.H., Miele L. γ-Secretase inhibitors and modulators. Biochim. Biophys. Acta. 2013;1828:2898–2907. doi: 10.1016/j.bbamem.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borgegard T., Juréus A., Lundkvist J. First and second generation γ-secretase modulators (GSMs) modulate amyloid-β (Aβ) peptide production through different mechanisms. J. Biol. Chem. 2012;287:11810–11819. doi: 10.1074/jbc.M111.305227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frykman S., Teranishi Y., Tjernberg L.O. Identification of two novel synaptic γ-secretase associated proteins that affect amyloid β-peptide levels without altering Notch processing. Neurochem. Int. 2012;61:108–118. doi: 10.1016/j.neuint.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Weggen S., Eriksen J.L., Koo E.H. Evidence that nonsteroidal anti-inflammatory drugs decrease amyloid β 42 production by direct modulation of γ-secretase activity. J. Biol. Chem. 2003;278:31831–31837. doi: 10.1074/jbc.M303592200. [DOI] [PubMed] [Google Scholar]
- 26.Beher D., Clarke E.E., Shearman M.S. Selected non-steroidal anti-inflammatory drugs and their derivatives target γ-secretase at a novel site. Evidence for an allosteric mechanism. J. Biol. Chem. 2004;279:43419–43426. doi: 10.1074/jbc.M404937200. [DOI] [PubMed] [Google Scholar]
- 27.Shelton C.C., Zhu L., Li Y.M. Modulation of γ-secretase specificity using small molecule allosteric inhibitors. Proc. Natl. Acad. Sci. USA. 2009;106:20228–20233. doi: 10.1073/pnas.0910757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crump C.J., Johnson D.S., Li Y.M. Development and mechanism of γ-secretase modulators for Alzheimer’s disease. Biochemistry. 2013;52:3197–3216. doi: 10.1021/bi400377p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai X.C., Yan C., Shi Y. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe N., Image Image I.I., Image Image I. Functional analysis of the transmembrane domains of presenilin 1: participation of transmembrane domains 2 and 6 in the formation of initial substrate-binding site of γ-secretase. J. Biol. Chem. 2010;285:19738–19746. doi: 10.1074/jbc.M110.101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somavarapu A.K., Kepp K.P. The dynamic mechanism of presenilin-1 function: Sensitive gate dynamics and loop unplugging control protein access. Neurobiol. Dis. 2016;89:147–156. doi: 10.1016/j.nbd.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Li S., Zhang W., Han W. Initial substrate binding of γ-secretase: The role of substrate flexibility. ACS Chem. Neurosci. 2017;8:1279–1290. doi: 10.1021/acschemneuro.6b00425. [DOI] [PubMed] [Google Scholar]
- 33.Aguayo-Ortiz R., Chávez-García C., Dominguez L. Characterizing the structural ensemble of γ-secretase using a multiscale molecular dynamics approach. Chem. Sci. 2017;8:5576–5584. doi: 10.1039/c7sc00980a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong P., Vetrivel K.S., Thinakaran G. Mutation analysis of the presenilin 1 N-terminal domain reveals a broad spectrum of γ-secretase activity toward amyloid precursor protein and other substrates. J. Biol. Chem. 2010;285:38042–38052. doi: 10.1074/jbc.M110.132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeo K., Tanimura S., Iwatsubo T. Allosteric regulation of γ-secretase activity by a phenylimidazole-type γ-secretase modulator. Proc. Natl. Acad. Sci. USA. 2014;111:10544–10549. doi: 10.1073/pnas.1402171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie T., Yan C., Shi Y. Crystal structure of the γ-secretase component nicastrin. Proc. Natl. Acad. Sci. USA. 2014;111:13349–13354. doi: 10.1073/pnas.1414837111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elad N., De Strooper B., Chávez-Gutiérrez L. The dynamic conformational landscape of γ-secretase. J. Cell Sci. 2015;128:589–598. doi: 10.1242/jcs.164384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Lu S.H., St George-Hyslop P.H. Structural interactions between inhibitor and substrate docking sites give insight into mechanisms of human PS1 complexes. Structure. 2014;22:125–135. doi: 10.1016/j.str.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolduc D.M., Montagna D.R., Wolfe M.S. Nicastrin functions to sterically hinder γ-secretase-substrate interactions driven by substrate transmembrane domain. Proc. Natl. Acad. Sci. USA. 2016;113:E509–E518. doi: 10.1073/pnas.1512952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao G., Liu Z., Kopan R. γ-secretase composed of PS1/Pen2/Aph1a can cleave notch and amyloid precursor protein in the absence of nicastrin. J. Neurosci. 2010;30:1648–1656. doi: 10.1523/JNEUROSCI.3826-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chávez-Gutiérrez L., Tolia A., de Strooper B. Glu(332) in the Nicastrin ectodomain is essential for γ-secretase complex maturation but not for its activity. J. Biol. Chem. 2008;283:20096–20105. doi: 10.1074/jbc.M803040200. [DOI] [PubMed] [Google Scholar]
- 42.Atilgan A.R., Durell S.R., Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eyal E., Lum G., Bahar I. The anisotropic network model web server at 2015 (ANM 2.0) Bioinformatics. 2015;31:1487–1489. doi: 10.1093/bioinformatics/btu847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nussinov R., Tsai C.J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 45.Wagner J.R., Lee C.T., Amaro R.E. Emerging computational methods for the rational discovery of allosteric drugs. Chem. Rev. 2016;116:6370–6390. doi: 10.1021/acs.chemrev.5b00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dokholyan N.V. Controlling allosteric networks in proteins. Chem. Rev. 2016;116:6463–6487. doi: 10.1021/acs.chemrev.5b00544. [DOI] [PubMed] [Google Scholar]
- 47.Lu P., Bai X.C., Shi Y. Three-dimensional structure of human γ-secretase. Nature. 2014;512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahar I., Lezon T.R., Eyal E. Global dynamics of proteins: bridging between structure and function. Annu. Rev. Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bahar I. On the functional significance of soft modes predicted by coarse-grained models for membrane proteins. J. Gen. Physiol. 2010;135:563–573. doi: 10.1085/jgp.200910368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuglebakk E., Tiwari S.P., Reuter N. Comparing the intrinsic dynamics of multiple protein structures using elastic network models. Biochim. Biophys. Acta. 2015;1850:911–922. doi: 10.1016/j.bbagen.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 51.López-Blanco J.R., Chacón P. New generation of elastic network models. Curr. Opin. Struct. Biol. 2016;37:46–53. doi: 10.1016/j.sbi.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 52.Bakan A., Nevins N., Bahar I. Druggability assessment of allosteric proteins by dynamics simulations in the presence of probe molecules. J. Chem. Theory Comput. 2012;8:2435–2447. doi: 10.1021/ct300117j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghanakota P., Carlson H.A. Driving structure-based drug discovery through cosolvent molecular dynamics. J. Med. Chem. 2016;59:10383–10399. doi: 10.1021/acs.jmedchem.6b00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghanakota P., Carlson H.A. Moving beyond active-site detection: MixMD applied to allosteric systems. J. Phys. Chem. B. 2016;120:8685–8695. doi: 10.1021/acs.jpcb.6b03515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bakan A., Meireles L.M., Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakan A., Dutta A., Bahar I. Evol and ProDy for bridging protein sequence evolution and structural dynamics. Bioinformatics. 2014;30:2681–2683. doi: 10.1093/bioinformatics/btu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lezon T.R., Bahar I. Constraints imposed by the membrane selectively guide the alternating access dynamics of the glutamate transporter GltPh. Biophys. J. 2012;102:1331–1340. doi: 10.1016/j.bpj.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lomize M.A., Lomize A.L., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 59.Rotkiewicz P., Skolnick J. Fast procedure for reconstruction of full-atom protein models from reduced representations. J. Comput. Chem. 2008;29:1460–1465. doi: 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Páll S., Abraham M.J., Lindahl E. Tackling exascale software challenges in molecular dynamics simulations with GROMACS. Solving Software Challenges for Exascale. 2015;8759:3–27. [Google Scholar]
- 62.de Jong D.H., Singh G., Marrink S.J. Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 63.Qi Y., Ingólfsson H.I., Im W. CHARMM-GUI Martini maker for coarse-grained simulations with the Martini force field. J. Chem. Theory Comput. 2015;11:4486–4494. doi: 10.1021/acs.jctc.5b00513. [DOI] [PubMed] [Google Scholar]
- 64.Amadei A., Linssen A.B., Berendsen H.J. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 65.Sayyed-Ahmad A., Gorfe A.A. Mixed-probe simulation and probe-derived surface topography map analysis for ligand binding site identification. J. Chem. Theory Comput. 2017;13:1851–1861. doi: 10.1021/acs.jctc.7b00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ung P.M., Ghanakota P., Carlson H.A. Identifying binding hot spots on protein surfaces by mixed-solvent molecular dynamics: HIV-1 protease as a test case. Biopolymers. 2016;105:21–34. doi: 10.1002/bip.22742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uehara S., Tanaka S. Cosolvent-based molecular dynamics for ensemble docking: practical method for generating druggable protein conformations. J. Chem. Inf. Model. 2017;57:742–756. doi: 10.1021/acs.jcim.6b00791. [DOI] [PubMed] [Google Scholar]
- 68.Bakan A., Kapralov A.A., Bahar I. Inhibition of peroxidase activity of cytochrome c: De novo compound discovery and validation. Mol. Pharmacol. 2015;88:421–427. doi: 10.1124/mol.115.097816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cleves A.E., Jain A.N. Knowledge-guided docking: accurate prospective prediction of bound configurations of novel ligands using Surflex-Dock. J. Comput. Aided Mol. Des. 2015;29:485–509. doi: 10.1007/s10822-015-9846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng Z., Pearce L.V., Xie X.Q. Structural insight into tetrameric hTRPV1 from homology modeling, molecular docking, molecular dynamics simulation, virtual screening, and bioassay validations. J. Chem. Inf. Model. 2015;55:572–588. doi: 10.1021/ci5007189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng Z., Alqarni M.H., Xie X.Q. Modeling, molecular dynamics simulation, and mutation validation for structure of cannabinoid receptor 2 based on known crystal structures of GPCRs. J. Chem. Inf. Model. 2014;54:2483–2499. doi: 10.1021/ci5002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie X.Q., Chen J.Z., Billings E.M. 3D structural model of the G-protein-coupled cannabinoid CB2 receptor. Proteins. 2003;53:307–319. doi: 10.1002/prot.10511. [DOI] [PubMed] [Google Scholar]
- 73.Chen J.Z., Wang J., Xie X.Q. GPCR structure-based virtual screening approach for CB2 antagonist search. J. Chem. Inf. Model. 2007;47:1626–1637. doi: 10.1021/ci7000814. [DOI] [PubMed] [Google Scholar]
- 74.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 75.DeLano, W. L. 2017. PyMOL. http://www.pymol.org.
- 76.Williams, T., and C. Kelley. 2017. Gnuplot 4.5: an interactive plotting program. http://gnuplot.info.
- 77.Wolfe M.S. Toward the structure of presenilin/γ-secretase and presenilin homologs. Biochim. Biophys. Acta. 2013;1828:2886–2897. doi: 10.1016/j.bbamem.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tanzi R.E., Bertram L. Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 79.Yang L.W., Bahar I. Coupling between catalytic site and collective dynamics: a requirement for mechanochemical activity of enzymes. Structure. 2005;13:893–904. doi: 10.1016/j.str.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pozdnyakov N., Murrey H.E., Johnson D.S. γ-Secretase modulator (GSM) photoaffinity probes reveal distinct allosteric binding sites on presenilin. J. Biol. Chem. 2013;288:9710–9720. doi: 10.1074/jbc.M112.398602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Smet F., Christopoulos A., Carmeliet P. Allosteric targeting of receptor tyrosine kinases. Nat. Biotechnol. 2014;32:1113–1120. doi: 10.1038/nbt.3028. [DOI] [PubMed] [Google Scholar]
- 82.Nussinov R., Tsai C.J. The different ways through which specificity works in orthosteric and allosteric drugs. Curr. Pharm. Des. 2012;18:1311–1316. doi: 10.2174/138161212799436377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X., Dang S., Shi Y. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493:56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The extracellular NCT subunit undergoes a cooperative bending movement with respect to the TM subunits inducing significant (∼20 Å) changes in the distance between its exposed loops and the EC-facing vestibule of PS1. There are five spheres showing Asp257/Asp385 (PS1) in yellow, Tyr115 (PS1-HL1) in green, Asp541 (NCT) in orange, and Glu333 (NCT) in blue. See Fig. 2 for the conformations of the intact γ-secretase sampled during these fluctuations.
The extracellular NCT subunit undergoes a cooperative twisting movement with respect to the TM subunits that are practically rigidly embedded in the membrane. Note the accompanying membrane movements. There are five spheres showing Asp257/Asp385 (PS1) in yellow, Tyr115 (PS1-HL1) in green, Asp541 (NCT) in orange, and Glu333 (NCT) in blue. See Fig. 2 for the conformations of the intact γ-secretase sampled during these fluctuations.
The breathing motion of PS1 and cooperative movement of the NCT subunit. The catalytic sites Asp257 and Asp385 on PS1, and the central residue Glu333 on NCT are shown in green spheres. See Fig. 8 for details during these fluctuations.