Numerous studies in patients with cancer have demonstrated the presence of tumor-specific DNA, RNA and protein in the peripheral blood.1, 2, 3, 4 Peripheral blood (PB) cell free DNA (cfDNA) can be used for the diagnosis and monitoring of cancer,2, 3, 4 and is particularly useful in solid tumors in which tissue biopsy may be hazardous or not possible.1, 2, 3, 4 The success of cfDNA for genomic screening depends on multiple factors, including disease stage and tumor size, vascularity and biology.4 In contrast, in hematologic neoplasms, neoplastic cells are immersed in blood and peripheral blood cfDNA has been reported to be as reliable as bone marrow (BM) DNA in detecting molecular abnormalities.4, 5, 6, 7, 8, 9, 10, 11, 12 In theory, peripheral blood cfDNA might be more representative of the entire bone marrow and less influenced to sampling error than is BM DNA.4
The disease aplastic anemia (AA) is, in most cases, the result of immune-mediated destruction of hematopoietic stem cells.13, 14 However, recent studies have suggested that this inflammatory environment is conducive to emergence of abnormal hematopoietic clones, carrying mutations that are typically detected in patients with myelodysplastic syndrome (MDS).15 Most of these clones are detected at very low frequency, at low variant allele frequency (VAF).15 Therefore, patients with AA might be ideal to evaluate the sensitivity and reliability of cfDNA in the evaluation of mutations in the hematopoietic compartment. Furthermore, in general, patients with AA have a very low number of circulating neutrophils making results obtained from testing peripheral blood cells questionable. In addition, we have previously reported that in patients with MDS, due to the increased bone marrow apoptosis and perhaps due to inability to differentiate and circulate, these subclones are not detected when peripheral blood cells are tested.8
We tested the mutation profile in peripheral blood cfDNA in direct comparison to bone marrow aspiration samples. A panel of 54 gene (TruSight Myeloid Sequencing Panel, Illumina; San Diego, CA, USA) and next generation sequencing (NGS) were utilized to assess 120 paired bone marrow and peripheral blood cfDNA samples collected from 96 patients who had been diagnosed with AA. Paired samples were collected at the same time. All patients in the studied group had a very low neutrophils count (median 275, range 0–1380 neutrophils/ul). Twenty six of these patients had absolute neutrophils count <200/μL.
Total nucleic acid was extracted from PB plasma via the NucliSenS EasyMAG automated platform (BioMerieux; Marcy-l'Étoile, France). DNA from whole bone marrow cells was extracted QIAamp DNA Mini Kit (Qiagen; Venlo, The Netherlands) in both manual and automated (QIAcube) extractions according to manufacturer’s instruction. Molecular abnormalities were called using the Illumina-developed Somatic Variant Caller. RefSeq (NCBI; Bethesda, MD, USA) annotations were applied and molecular abnormalities were called in Illumina Variant Studio then individually verified with the Integrated Genome Viewer (Broad Institute; Cambridge, MA, USA). NGS testing for mutations in CALR, FLT3-ITD and ASXL1 were complemented by using fragment length analysis to avoid missing large indels that can be missed by NGS. cfDNA from all samples was obtained and analyzed irrespective of the severity of the disease. The efficiency of sequencing of the cfDNA was similar to that of BM cellular DNA. As a quality control, with the exception of few exons of the CEBPA gene, all amplicons in the 54 genes must meet a depth of >6000, otherwise, the sequencing was repeated. Buccal mucosa samples were tested for any mutations with VAF around between 40 and 60%. As quality control, normal plasma cfDNA was tested with every run as well positive control.
Of the 96 patients, 33 (34%) had one or more mutation in either cfDNA or BM DNA. Of the 120 samples, 48 (40%) showed one or more mutations and the total number of mutations was 64. Of the 48 samples, 26 (54%) had one mutation, 15 (31%) had two mutations and 7 (15%) had three mutations. Overall, 45 unique mutations (Table 1) were detected in the following genes: ASXL1, BCOR, BCORL1, CBLC, CSF3R, DNMT3A, EZH2, IDH1, JAK2, NPM1, NRAS, PTEN, PTPN11, RUNX1, SETBP1, SF3B1, SRSF2, STAG2, TET2, U2AF1 and ZRSR2.
Table 1. List of detected mutations.
| Gene | Nucleotide | Amino acid |
|---|---|---|
| DNMT3A | NM_022552.4:c.2470delA | NP_072046.2:p.Ile824Ter |
| ASXL1 | NM_015338.5:c.2287delC | NP_056153.2:p.Leu764TyrfsTer8 |
| ASXL1 | NM_015338.5:c.1926_1927insG | NP_056153.2:p.Gly646TrpfsTer12 |
| ASXL1 | NM_015338.5:c.1771_1772insA | NP_056153.2:p.Tyr591Ter |
| TET2 | NM_001127208.2:c.1147C>T | NP_001120680.1:p.Gln383Ter |
| JAK2 | NM_004972.3:c.1849G>T | NP_004963.1:p.Val617Phe |
| ASXL1 | NM_015338.5:c.2222A>T | NP_056153.2:p.Asp741Val |
| U2AF1 | NM_001025203.1:c.101C>T | NP_001020374.1:p.Ser34Phe |
| DNMT3A | NM_022552.4:c.1913C>A | NP_072046.2:p.Ser638Tyr |
| ASXL1 | NM_015338.5:c.2197C>T | NP_056153.2:p.Gln733Ter |
| TET2 | NM_001127208.2:c.3763_3764insA | NP_001120680.1:p.Tyr1255Ter |
| EZH2 | NM_004456.4:c.630dupA | NP_004447.2:p.Glu211ArgfsTer11 |
| RUNX1 | NM_001754.4:c.965C>G | NP_001745.2:p.Ser322Ter |
| STAG2 | NM_001042749.1:c.1027G>T | NP_001036214.1:p.Val343Leu |
| PTEN | NM_000314.4:c.674A>G | NP_000305.3:p.Tyr225Cys |
| ASXL1 | NM_015338.5:c.3110G>A | NP_056153.2:p.Trp1037Ter |
| ASXL1 | NM_015338.5:c.2276_2280delGCCAG | NP_056153.2:p.Gln760LeufsTer12 |
| BCORL1 | NM_021946.4:c.3332C>T | NP_068765.3:p.Thr1111Met |
| ASXL1 | NM_015338.5:c.2513A>G | NP_056153.2:p.Lys838Arg |
| ZRSR2 | NM_005089.3:c.1314_1315insAGCCGG | NP_005080.1:p.Gly438_Ser439insSerArg |
| TET2 | NM_001127208.2:c.3662G>T | NP_001120680.1:p.Cys1221Phe |
| TET2 | NM_001127208.2:c.3332T>A | NP_001120680.1:p.Leu1111Ter |
| RUNX1 | NM_001754.4:c.1440C>A | NP_001745.2:p.Tyr480Ter |
| BCOR | NM_001123385.1:c.4988_4989delGG | NP_001116857.1:p.Trp1663SerfsTer8 |
| SF3B1 | NM_012433.2:c.1973G>C | NP_036565.2:p.Trp658Ser |
| TET2 | NM_001127208.2:c.5636A>T | NP_001120680.1:p.Glu1879Val |
| CALR | NM_004343.3:c.1192_1194delGAG | NP_004334.1:p.Glu398del |
| IDH1 | NM_005896.2:c.394C>T | NP_005887.2:p.Arg132Cys |
| CBLC | NM_012116.3:c.1303C>T | NP_036248.3:p.Pro435Ser |
| TET2 | NM_001127208.2:c.575_576insAAT | NP_001120680.1:p.Tyr192delinsTer |
| TET2 | NM_001127208.2:c.1118_1122delAAAAT | NP_001120680.1:p.Gln373ArgfsTer15 |
| EZH2 | NM_004456.4:c.2109delA | NP_004447.2:p.Val704LeufsTer2 |
| TET2 | NM_001127208.2:c.5167C>T | NP_001120680.1:p.Pro1723Ser |
| BCOR | NM_001123385.1:c.756C>A | NP_001116857.1:p.Tyr252Ter |
| SRSF2 | NM_001195427.1:c.284C>G | NP_001182356.1:p.Pro95Arg |
| NRAS | NM_002524.4:c.35G>C | NP_002515.1:p.Gly12Ala |
| NRAS | NM_002524.4:c.37G>C | NP_002515.1:p.Gly13Arg |
| SETBP1 | NM_015559.2:c.2602G>A | NP_056374.2:p.Asp868Asn |
| PTPN11 | NM_002834.3:c.178G>C | NP_002825.3:p.Gly60Arg |
| PTPN11 | NM_002834.3:c.226G>C | NP_002825.3:p.Glu76Gln |
| RUNX1 | NM_001754.4:c.276dupC | NP_001745.2:p.Asp93ArgfsTer45 |
| NPM1 | NM_002520.6:c.863_864insCCGC | NP_002511.1:p.Trp288CysfsTer12 |
| ZRSR2 | NM_005089.3:c.1346_1360delGGAGCCGCCGCAGCC | NP_005080.1:p.Ser450_Arg454del |
| SF3B1 | NM_012433.2:c.1998G>T | NP_036565.2:p.Lys666Asn |
| DNMT3A | NM_022552.4:c.1634delA | NP_072046.2:p.Glu545GlyfsTer106 |
| DNMT3A | NM_022552.4:c.976C>T | NP_072046.2:p.Arg326Cys |
| BCORL1 | NM_021946.4:c.1942_1943insC | NP_068765.3:p.Val650ArgfsTer15 |
| TET2 | NM_001127208.2:c.2715_2716insA | NP_001120680.1:p.Met906AsnfsTer18 |
| CSF3R | NM_156039.3:c.2326C>T | NP_724781.1:p.Gln776Ter |
| TET2 | NM_001127208.2:c.2771A>G | NP_001120680.1:p.His924Arg |
| BCOR | NM_001123385.1:c.4973_4974delAG | NP_001116857.1:p.Gln1658ArgfsTer13 |
| TET2 | NM_001127208.2:c.1648C>T | NP_001120680.1:p.Arg550Ter |
| ATRX | NM_000489.3:c.5579A>G | NP_000480.2:p.Asn1860Ser |
| BCOR | NM_001123385.1:c.3809G>A | NP_001116857.1:p.Trp1270Ter |
| DNMT3A | NM_022552.4:c.2578T>C | NP_072046.2:p.Trp860Arg |
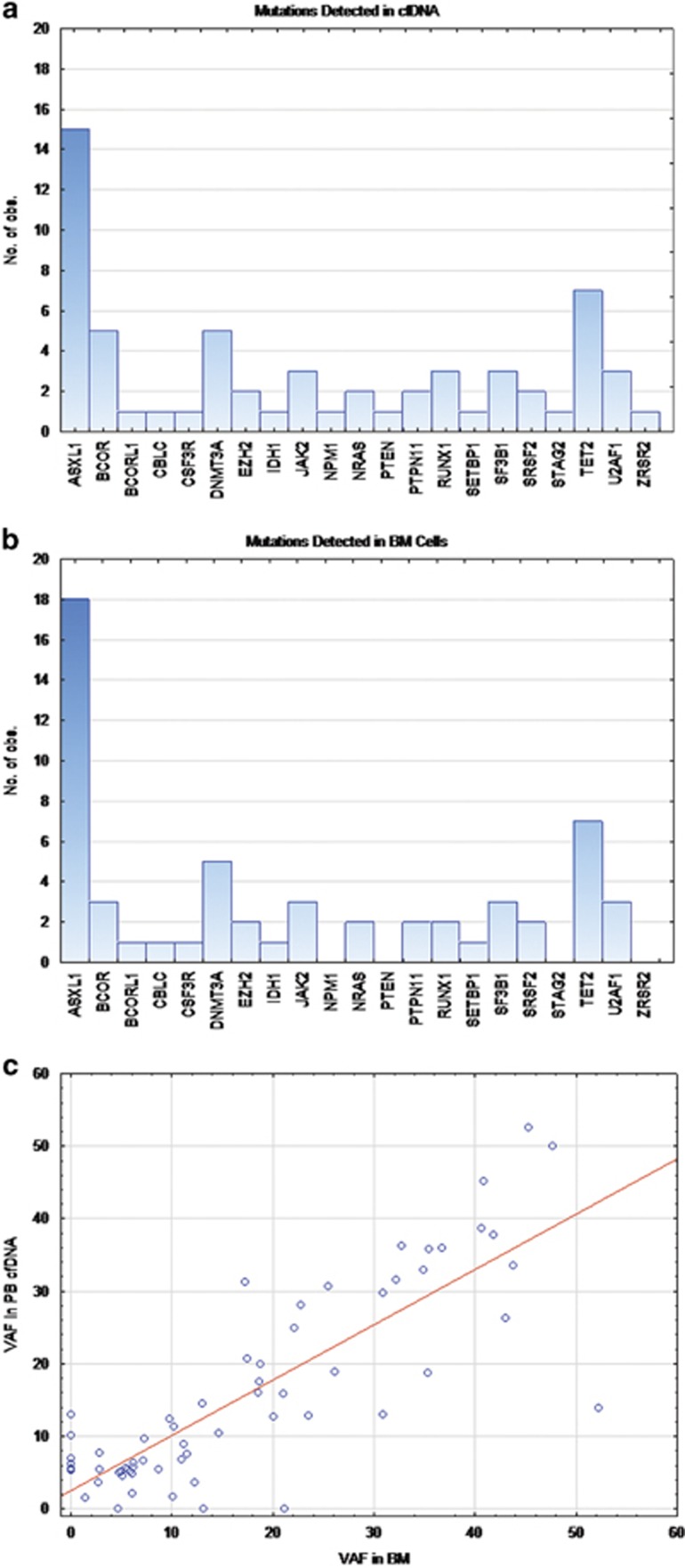
More mutations were detected in cfDNA (N=64) (Figure 1a) than in cellular BM DNA (N=57) (Figure 1b), and 6 of 33 patients with somatic mutations (18%) showed mutations in plasma cfDNA but not in BM; two patients (6%) had mutations in BM cells that were not present in cfDNA (P=0.002) (Figure 1c). Mutations detected in cfDNA and not in BM DNA were in the following genes: BCOR, NPM1, PTEN, RUNX1, STAG2, and ZRSR2. Mutations in ASXL1 were detected in the two patients who had mutations in BM but not in plasma. One of these two patients was tested twice, few months apart, and at both time points showed an ASXL1 (Tyr591Ter) mutation in bone marrow and not in cfDNA, but in the later sample, a second mutation in ASXL1 (Gly646TrpfsTer12) was detected in both BM DNA and in cfDNA. The variant allele frequency (VAF) for the Tyr591 was at 21% and 13%, respectively. The second patient with a mutation in BM DNA and not in cfDNA had an ASXL1 Gln733Ter mutation detected at VAF of 4%. The seven mutations detected in the cfDNA and not in the BM DNA had VAF of 6, 7, 10, 6, 13, 6 and 5% in STAG2, PTEN, RUNX1, BCOR, NPM1, ZRSR2 and BCOR, respectively. The most common mutation was ASXL1 (22% of cfDNA and 27% of BM cells), followed by TET2 (19% of cfDNA and 21% of BM cells), DNMT3A (7% in both cfDNA and BM cells), then BCOR (7% of cfDNA and 4% of BM cells).
Figure 1.
Comparison of cfDNA with bone marrow DNA: Frequency of mutations detected in each gene as detected in cfDNA are shown in (a) and as detected in BM cells are shown in (b). (c) shows variant allele frequency (VAF) in cfDNA as correlated with the VAF detected in the bone marrow cellular DNA (r=0.77; P-value <0.0001).
Upon comparing the VAF of various mutations as detected by BM DNA and cfDAN, there was significant correlation (r=0.77; P-value <0.0001) and overall no significant difference in VAF between the two sample types (P=0.071, Sign test). The median VAF in cfDNA was 12.6 and 10.9 in BM DNA.
These data confirm that cfDNA is as reliable as BM cells for detecting mutations, even when these mutations are present at very low frequency in in hypocellular bone marrows. If cfDNA testing proved more reliable, it might be preferred for multiple reasons to BM sampling for the purpose of serial monitoring of neoplastic processes in bone marrow, especially at early, premalignant stages. Minimal residual disease in patients with leukemia can be monitored using cfDNA, sparing the patient the need for bone marrow aspiration and biopsy. Furthermore cfDNA can be used for early diagnosis, especially in elderly patients and when a patient refuses biopsy. cfDNA may be an especially valuable source of mutation detection in marrow failure, in which marrow aspirates may not contain sufficient cells for accurate mutation analysis.
Acknowledgments
Author contributions
Concept and design: AA, MA, DT and NSY. Conduct of Laboratory work: AA, WM, IDD, VF and MA. Acquisition of data and samples: AA, DT, WM, IDD, VF, NSY and MA. Analysis and interpretation of data: AA, WM, VF, NSY and MA. Writing, review and/or revision of manuscript: all authors. Study supervision: MA.
Footnotes
AA, WM, IDD, VF and MA are employed by a diagnostic company offering cfDNA testing. All other authors declare no competing interest.
References
- Salvi S, Gurioli G, De Giorgi U, Conteduca V, Tedaldi G, Calistri D et al. Cell-free DNA as a diagnostic marker for cancer: current insights. Oncol Targets Ther 2016; 25: 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malapelle U, Pisapia P, Rocco D, Smeraglio R, di Spirito M, Bellevicine C et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res 2016; 5: 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang DH, Ensor JE, Liu ZB, Patel A, Patel TA, Chang JC et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat 2016; 155: 139–149. [DOI] [PubMed] [Google Scholar]
- Giles FJ, Albitar M. Plasma-based testing as a new paradigm for clinical testing in hematologic diseases. Expert Rev Mol Diagn 2007; 7: 615–623. [DOI] [PubMed] [Google Scholar]
- Hyman DM, Diamond EL, Vibat CR, Hassaine L, Poole JC, Patel M et al. Prospective blinded study of BRAFV600E mutation detection in cell-free DNA of patients with systemic histiocytic disorders. Cancer Discov 2015; 5: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljurf M, Abalkhail H, Alseraihy A, Mohamed SY, Ayas M, Alsharif F et al. Chimerism analysis of cell-free DNA in patients treated with hematopoietic stem cell transplantation may predict early relapse in patients with hematologic malignancies. Biotechnol Res Int 2016; 2016: 8589270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Tomita A, Nakamura F, Iriyama C, Shirahata-Adachi M, Shimada K et al. Peripheral blood cell-free DNA is an alternative tumor DNA source reflecting disease status in myelodysplastic syndromes. Cancer Sci 2016; 107: 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albitar F, Ma W, Diep K, De Dios I, Agersborg S, Thangavelu M et al. Deep sequencing of cell-free peripheral blood DNA as a reliable method for confirming the diagnosis of myelodysplastic syndrome. Genet Test Mol Biomark 2016; 20: 341–345. [DOI] [PubMed] [Google Scholar]
- Ma W, Kantarjian H, Zhang X, Yeh CH, Zhang ZJ, Verstovsek S et al. Plasma levels of JAK2 mRNA in patients with chronic myeloproliferative diseases with and without V617F mutation: implications for prognosis and disease biology. Int J Lab Hematol 2010; 32 (1 Pt 2): 95–102. [DOI] [PubMed] [Google Scholar]
- Ma W, Kantarjian H, Zhang X, Jilani I, Sheikholeslami MR, Donahue AC et al. Detection of nucleophosmin gene mutations in plasma from patients with acute myeloid leukemia: clinical significance and implications. Cancer Biomark 2009; 5: 51–58. [DOI] [PubMed] [Google Scholar]
- Ma W, Tseng R, Gorre M, Jilani I, Keating M, Kantarjian H et al. Plasma RNA as an alternative to cells for monitoring molecular response in patients with chronic myeloid leukemia. Haematologica 2007; 92: 170–175. [DOI] [PubMed] [Google Scholar]
- Jilani I, Estey E, Manshuri T, Caligiuri M, Keating M, Giles F et al. Better detection of FLT3 internal tandem duplication using peripheral blood plasma DNA. Leukemia 2003; 17: 114–119. [DOI] [PubMed] [Google Scholar]
- Rovó A, Tichelli A, Dufour C. Diagnosis of acquired aplastic anemia. Bone Marrow Transplant 2013; 48: 162–167. [DOI] [PubMed] [Google Scholar]
- Young NS. Current concepts in the pathophysiology and treatment of aplastic anemia. Hematol Am Soc Hematol Educ Program 2013; 2013: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med 2015; 373: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]