Abstract
Accumulative studies revealed that E3 ubiquitin ligases have important roles in colorectal carcinogenesis. The pathogenic mechanisms of colorectal cancer (CRC) initiation and progression are complex and heterogeneous, involving somatic mutations, abnormal gene fusion, deletion or amplification and epigenetic alteration, which may cause aberrant expression or altered function of E3 ligases in CRC. Defects of E3 ligases have been reported to be involved in the molecular etiology and pathogenesis of CRC. The aberrant expressed E3 ligases can function as either oncogenes or tumor suppressors depending on ubiquiting target substrates in CRC. Recently, considerable progress has been made in our understanding of the potential roles of E3 ligase-mediated ubiquitylation in colorectal carcinogenesis. There are mainly two subtypes of E3 ubiquitin ligases in humans, as defined by the presence of either a HECT domain or a RING finger domain on the basis of structural similitude. Most cancer-associated E3 ligases participate in regulating the cell cycle, apoptosis, gene transcription, cell signaling and DNA repair, the critical parts of CRC tumorigenesis. In this review, we have provided a comprehensive summary of abnormally expressed E3 ligases and their related pivotal mechanistic effects in CRC. In particular, we have highlighted the function of RING-type E3 ubiquitin enzymes in modulating cancer signaling pathways, immunity and tumor microenvironment in CRC development and progression; their mechanism(s) of action in CRC involving both ubiquitylation-dependent and ubiquitylation-independent effects; and the potential of RING E3 ligases as molecular biomarkers for predicting patient prognosis and as therapeutic targets in CRC. A better understanding of E3 ligase-mediated substrates' ubiquitylation involved in the development of CRC will provide new insights into the pathophysiology mechanisms of CRC, and unravel novel prognostic markers and therapeutic strategies for CRC.
Introduction
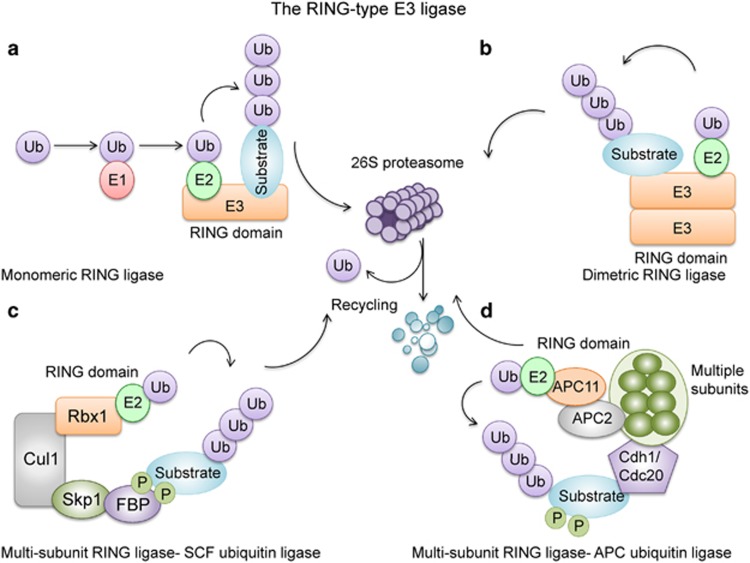
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and a major cause of cancer-related deaths in the world.1 Pathogenetic mechanisms of CRC initiation and progression are complex and heterogeneous, involving somatic gene mutations, abnormal gene fusion, gene copy number variations and epigenetic changes.2, 3, 4 The ubiquitin–proteasome system has a vital role in regulating and maintaining cellular homeostasis, including cell survival, cell differentiation, and innate and acquired immunity.5, 6, 7 Recently, considerable progress has been made in our understanding of the potential roles of E3 ligase-mediated ubiquitylation in colorectal carcinogenesis.8 There are mainly two subtypes of E3 ubiquitin ligases in humans, defined as containing either a HECT domain or a RING finger domain on the basis of structural similitude.9 Of these, the majority are RING finger and RING finger-related E3 ubiquitin ligases (Figure 1).
Figure 1.
The ubiquitin–proteasome system and the structures of RING-type E3 ligases. RING-type E3 ligases are classified into monomeric RING ligases (a), dimeric RING ligases (b) and multisubunit RING ligases, which includes SCF (c) and anaphase-promoting complex/cyclosome (APC/C) type ubiquitin ligases (d). Ubiquitylation of proteins is achieved through an enzymatic cascade involving ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin-ligating enzyme (E3). E3 enzymes provide platforms for binding E2 enzymes and specific substrates, thereby coordinating ubiquitylation of the selected substrate (a, b). Substrates of SCF and APC/C ubiquitin ligases must be modified by phosphorylation before they can bind to E3 and be subjected to ubiquitination (c, d).
In humans, over 600 RING-based ubiquitin ligases have been described and they comprised one of the biggest family of enzymes.10 These RING E3 ubiquitin ligases have been connected to the regulation of important cellular biological processes and their deregulation have been described to contribute to the pathogenesis of multiple human diseases, including malignancy.11, 12, 13, 14, 15 Both tumor-suppressive and tumor-promotive pathways are regulated by RING E3 ligase-mediated ubiquitination, and could directly suppress or exacerbate carcinogenesis. As a consequence, RING E3 ligases might function as oncogenes or tumor suppressors depending on the nature of their target substrates.16 Even a single Ring E3 ligase could have opposing functions in malignancy depending on context or the type of cancer involved.17
Many studies have described the important roles played by different types of RING finger E3 ligases in the development of CRC, including Ring Finger Proteins (RNFs), the murine double minute 2/4 (MDM2/MDMX), Tripartite motifs (TRIMs), the inhibitor of apoptotic proteins (IAPs), the S phase kinase-associated protein 1 (SKP1)–cullin1–F-box protein (SCF) E3 ligase and anaphase-promoting complex/cyclosome-type E3 ligase.18 RING-type E3 ligases have been discovered to target signaling molecules in p53 and Wnt/β-catenin pathways to modulate their activation in CRC.19, 20 Moreover, deregulation of RING finger E3 ligases involved in maintenance of genomic integrity and cellular homeostasis also contributes to tumorigenesis.10, 21 In this review, we highlight (1) the function of RING E3 ubiquitin ligases in modulating cancer growth and the tumor microenvironment in CRC development and progression; (2) their underlying mechanism(s) of action in CRC involving both ubiquitylation-dependent and ubiquitylation-independent effects; and (3) the potential of RING E3 ligases as molecular biomarkers for predicting patient prognosis and as therapeutic targets in CRC (Tables 1 and 2).
Table 1. RING-type E3 ligases in human colorectal cancer.
| Gene | Expression | Pathway | Protein partner | Biomarker | Refs |
|---|---|---|---|---|---|
| RNF4 | Upregulation | Wnt and Notch signaling pathways | β-catenin, c-Myc, c-Jun and NICD | / | 31, 32 |
| RNF14 | Upregulation | Wnt/β-catenin signaling pathway | TCF4 | / | 39, 40 |
| RNF43/ZNRF3 | Downregulation/mutation | Wnt/β-catenin signaling pathway | Fzd1/2/3/4/5/8 and LRP5/6 | Prognosis | 42, 43 |
| RNF20/40 | Downregulation | NF-κB signaling pathway | H2Bub1 | / | 46 |
| RNF85/TARF6 | Upregulation | NF-κB signaling pathway | NF-κB kinase (IKK) | / | 49, 51 |
| TRIM3 | / | p53 signaling pathway | / | / | 53 |
| TRIM15 | Downregulation | Actin cytoskeleton dynamics | Filamin-binding LIM protein 1 and VASP | / | 54 |
| TRIM40 | Upregulation | Inhibits NF-κB signaling | IKK-γ | / | 55 |
| MDM2 | Upregulation | p53 signaling pathway | p53 | / | 19, 58, 59 |
| c-IAP | Upregulation | TNF and NF-κB signaling pathways | RIP1, TRAF2 and NIK | Prognosis | 64, 65, 66 |
| XIAP | Upregulation | TNF and Wnt/β-catenin signaling pathways | Caspase-3, TLE | / | 69, 70, 71 |
| β-TRCP (SCF) | Upregulation | Wnt/β-catenin and NF-κB signaling pathways | β-catenin, IkB, p53, Yap, IKB, KRAS, Cyclin D1, WEE1, CDC25, MCL-1 and pro-caspase-3, Snail, FOXO3 and CHD1 | Prognosis | 75, 76, 77, 86, 94, 95, 96, 97, 98, 99 |
| FBW7 (SCF) | Mutation or loss | JNK and Notch signaling pathways | MYC, Cyclin E, Notch, JUN, Aurora kinase A and Mcl-1 | Prognosis | 101, 102, 103, 104, 107, 135, 136 |
| SKP2 (SCF) | Amplification/upregulation | Cell cycle progression | p27kip1 and p21 | Prognosis | 113, 115, 116 |
| CDC20 (APC) | Upregulation | Wnt/β-catenin signaling pathway | Axin2 | Prognosis | 118, 119, 120 |
| CDH1 (APC) | Mutation or loss | Cell cycle | SKP2, Cyclin B, Aurora A, Aurora B, Tpx2, Cdc6 and Cdc20 | Prognosis | 74, 122, 123, 124 |
| TRIM29 | Upregulation | / | / | Diagnosis | 126, 127 |
| Survivin | Upregulation | Anti-apoptotic pathways | / | Diagnosis | 128, 129 |
Table 2. Approaches to target E3 ubiquitin ligase activity in CRC.
| Compound | Class | Target | Proposed working mechanism | Refs |
|---|---|---|---|---|
| R-spondin | Protein/peptide | RNF43/ZNRF3 | Neutralizes receptors and upregulates Wnt signaling | 41 |
| C59 | porcupine inhibitor | Mutant RNF43/ZNRF3 | Blocks Wnt niche function | 131, 132 |
| LGK974 | Small molecule Porcupine inhibitor | RNF43/ZNRF3 | Prevents palmitylation of Wnt proteins and inhibits Wnt signaling | 131, 132 |
| HLI98 | Small molecule | MDM2 | Inhibits MDM2 ubiquitin, activates p53 pathway and induces apoptosis | 141 |
| MX69 | Small molecule | MDM2/XIAP | Degrades MDM2, activates p53 and inhibits XIAP | 142 |
| Ceramide/LCL85 | Small molecule | cIAP1/XIAP | Induces proteasomal degradation of cIAP1 and XIAP | 143 |
| MLN4924 | Small molecule | SCF E3 ubiquitin ligases | Induces G2/M arrest, apoptosis and DNA damage; inhibition of cullin neddylation | 146, 147 |
| CC0651 | Small molecule | E2 enzyme hCdc34 and SCF subunits Skp1, Cul1 and Rbx1 | Causes the SCFSkp2 substrate p27Kip1 accumulation | 149, 150 |
| Pro-TAME | Small molecule | APC/Cdc20 | Disruption of the APC/Cdc20 interaction leading to lower APC/C activity on substrates | 151, 152, 153 |
Abbreviations: APC/C, anaphase-promoting complex/cyclosome; CRC, colorectal cancer; SCF, S phase kinase-associated protein 1–cullin1–F-box protein; XIAP, X-chromosome-linked apoptotic protein.
General features and functions of the ring-type E3 ligases
RING-type E3 ligases are a large family of enzymes that participate in regulating of the stability and activity of numerous interaction substrates. Ubiquitylation of proteins is achieved through an enzymatic cascade that involves ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin-ligating enzyme (E3).22 Ubiquitylation generates when an E3 ligase enzyme binds with both the target substrate and an E2 enzyme, and then E3 ligase catalyzes the transfer of ubiquitin from E2 enzyme to the lysine residue on its target protein, leading to monoubiquitylation or polyubiquitylation (Figure 1).5, 10, 23 RING finger E3 ligases recognize their substrates via diverse protein–protein interactions that are dependent on different post-translational modifications including phosphorylation, sumoylation and glycosylation.17, 24, 25 Although the binding sites for E3 ligases on target substrates are highly specific for ubiquitylation, a single protein may be targeted by various E3 ligases and each E3 ligase has multiple substrates, including themselves. The proteins tagged by polyubiquitylation subsequently undergo degradation in the proteasome. This ubiquitin–proteasome system controls a vast array of cellular programs, such as cell division, growth and differentiation.5 Apart from proteolytic roles, ubiquitin modifications have diverse nonproteolytic effects on the substrates, span a wide spectrum that includes receptor internalization, intracellular trafficking, transcriptional activity, assembly of multiprotein complexes and regulation of enzymatic activity.26, 27
RING-type E3 ligases can be divided into monomeric, homodimeric/heterodimeric and multisubunit RING E3 ligases.28 Multisubunit RING-type E3 ligases include the cullin RING ligase superfamily and the anaphase-promoting complex/cyclosome (APC/C) complex. The largest member of cullin RING ligases known is the S phase kinase-associated protein 1 (SKP1)–cullin1 (CUL1)–F-box protein (SCF) complex; in this complex F-box protein is in charge of targeting specific phosphorylated substrate(s) for degradation (Figure 1).10, 18 Deregulation of the ubiquitin-mediated proteolysis could result in uncontrolled cell growth, genome instability and carcinogenesis. Some members of the RING family are known to be engaged in the regulation of cell proliferation, cell cycle, cell apoptosis and epithelial–mesenchymal transition as oncogenes or tumor suppressors, depending on the cellular contexts, level of enzyme expression and their target substrates.13, 14, 15, 17 Cancers including CRC often harbor promoter methylation, copy number variations or functional mutations in E3 ligases that mediate their deregulation to disturb cancer-related pathways and promote tumorigenesis.2
Mechanisms of ring-type E3 ligases in CRC
Monomeric RING domain E3 ligase
RNF family
Ring Finger Protein 4 (RNF4) is a small ubiquitin-like modifier (SUMO)-targeting RING domain ubiquitin ligase that directly connects the SUMO and the ubiquitin pathways, resulting in the proteasomal degradation of poly-SUMO-modified substrates.29 SUMO-targeting RING-type E3 ligase-mediated ubiquitylation does not only target substrates for proteolysis, but also affects protein–protein interactions, subcellular localization and transcriptional activation.30 RNF4s are increased in 30% of colon adenocarcinoma patients as compared with normal colon tissues and adenomas, indicating a potential tumorigenic role of RNF4 in CRC.31 RNF4 has been reported to target several oncoproteins including β-catenin, c-Myc, c-Jun and Notch intracellular-domain proteins for ubiquitylation in a phosphorylation-dependent manner.31, 32 Paradoxically, RNF4-mediated ubiquitylation of these proteins does not induce proteasomal degradation. Instead, RNF4 activates these oncoproteins by protein stabilization, thereby enhancing the transcriptional activities of Wnt and Notch signaling pathways. Functionally, RNF4 augments the tumorigenic signaling cascades and is crucial for cancer cell survival.
RNF4 has a particularly important role in CRC by regulating Wnt signaling. Tight regulation of Wnt signaling is important for cell proliferation, maintenance of pluripotency and the differentiation of stem cells of the intestine.33, 34 Aberrant activation of the canonical Wnt/β-catenin signaling is a major hallmark of cancer that presents in >90% of CRC, induced by either inactivating mutations in the adenomatous polyposis coli (APC) gene or activating mutations in β-catenin.4, 35, 36 Defects in APC disrupt the β-catenin degradation complex, resulting in the stabilization and activation of β-catenin, a key transcription factor for Wnt signaling. Consequently, active β-catenin translocates to the nucleus where it interacts with the lymphoid enhancer factor/T-cell factor (LEF/TCF) complex to excite a Wnt transcriptional cascade that is essential for CRC development.37, 38 RNF4 stabilized β-catenin and c-Myc by ubiquitylation, leading to enhanced and prolonged Wnt/β-catenin signal activation. RNF4-mediated stabilization of these Wnt key regulators may therefore function as an enhancer to exacerbate Wnt/β-catenin signaling and drive colorectal carcinogenesis.
Recently, another member of the RNF family protein, RNF14, was also found to activate Wnt signaling in CRC.39 RNF14 binds to LEF/TCF transcription factors and activates Wnt signaling in CRC cells and zebrafish models.39 RNF14 functions as a key interacting partner for the TCF/β–catenin complex through facilitating the recruitment of β-catenin and stabilizing its interaction with LEF/TCF, thus ensuring a high transcriptional activity even at low levels of nuclear β-catenin.39, 40 RNF14 is crucial for CRC cell survival, and its depletion suppressed colorectal tumorigenesis.
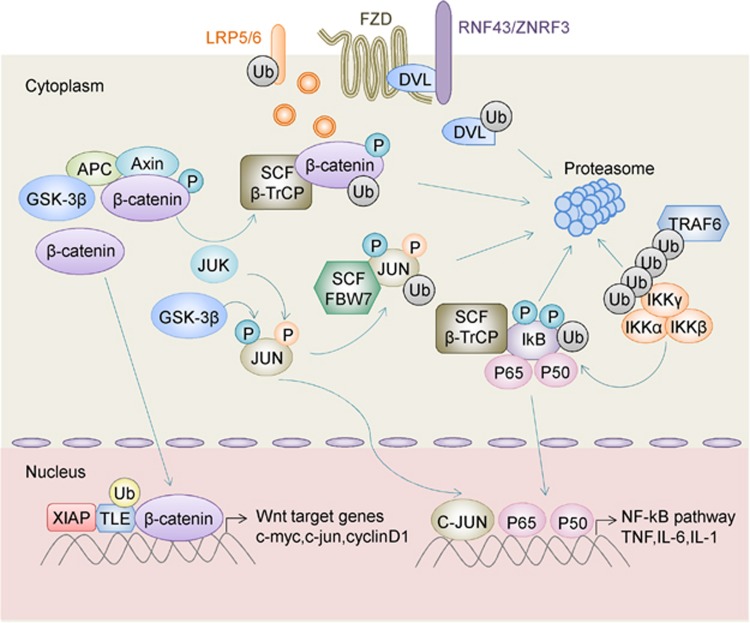
RNF43 and its homolog zinc and ring finger 3 (ZNRF3) are highly related RING finger proteins. Both ZNRF3 and RNF43 are localized to the cell plasma membrane and are defined as cell-surface transmembrane E3 ubiquitin ligases.41 The constructions of these two proteins consist of a signal peptide, an extracellular domain, a transmembrane domain as well as an intracellular RING domain. RNF43 is often mutated in CRC. One large-scale genomic analysis reported that over 18% of colorectal adenocarcinomas possess RNF43 somatic mutations.42 RNF43 together with ZNRF3 utilize Disheveled Segment Polarity Protein as an adaptor to recognize specific Wnt frizzled receptors including Fzd1-5/8 and low-density lipoprotein receptor-related protein 5/6.43 RNF43 and ZNRF3 mediated the ubiquitylation of seven transmembrane domains of frizzled receptors specifically in the cytoplasmic loops. Ubiquitinated Wnt receptors subsequently undergo endocytosis and degradation in the lysosomes, thereby suppressing Wnt signals44, 45 (Figure 2). In light of these findings, RNF43/ZNRF3 is a negative regulator of Wnt signaling via endocytosis and degradation of Wnt receptors, leading to suppressed intracellular Wnt cascade and tumor suppression. Interestingly, mutations of RNF43/ZNRF3 were mutually exclusive with APC truncation mutations in CRC.42 The mutually exclusive nature of RNF43/ZNRF3 or APC mutations suggests that the inactivation of either of these genes is sufficient to drive Wnt/β-catenin signaling and CRC, and that these mutations are functionally redundant.
Figure 2.
Regulation of Wnt/β-catenin signaling and oncogenic NF-κB pathway by ubiquitylating enzymes in CRC. Transmembrane E3 ligases RNF43 and ZNRF3 utilize disheveled segment polarity protein (DVL) as an adaptor to induce the endocytosis of Frizzled–LRP5/6 complex and abrogation of Wnt signaling. Binding of Wnt proteins to Wnt receptors results in the inactivation of GSK3β. β-catenin remains unphosphorylated, and it translocates to the nucleus where it binds to the LEF/TCF transcription complex, activating Wnt/β-catenin downstream target gene transcription. In the absence of Wnt, the scaffolding protein AXIN assembles a multiprotein destruction complex that consists of β-catenin, APC and GSK3β. β-catenin is phosphorylated by GSK3β and recognized by SCF-β-TrCP, leading to ubiquitylation and degradation of β-catenin. XIAP induces monoubiquitylation of TLE and releases TLE from the TCF/LEF complex, thereby initiating a canonical Wnt transcriptional program in CRC. K63-linked ubiquitylation of TRAF6 leads to the recruitment of the IKK (IκB kinase) complex (α, β, γ) and activation of IKK through TRAF6-mediated polyubiquitination. Activation of IKK kinase complex results in SCF-β-TrCP-mediated ubiquitination and proteasomal degradation of IκB (inhibitor of NF-κB kinase), thereby enabling the translocation of p50 and p65 to the nucleus and induction of NF-κB target genes, leading to positive regulation of NF-κB signaling. FBW7 preferentially targets phosphorylated JUN both by GSK3β or JNK and promotes JUN degradation in the proteasome. Accordingly, FBW7 depletion results in accumulation of phosphorylated JUN and stimulation of AP-1 activity. AP-1, activator protein 1; AXIN, axis inhibition protein; DVL, disheveled segment polarity protein; FBW7, F-box and WD repeat domain-containing 7; GSK3β, glycogen synthase kinase-3 beta; JUN, AP-1 transcription factor subunit; LEF/TCF, lymphoid enhancer factor/T-cell factor; LRP5/6, low-density lipoprotein receptor-related protein 5/6; TRAF6, tumor necrosis factor receptor-associated factor; XIAP, X-chromosome-linked inhibitor of apoptotic proteins.
Similar to RNF43/ZNRF3, RNF20 and RNF40 also function as tumor suppressor genes in CRC, but with distinct subcellular localization and mechanism of action. RNF20/40s are nuclear-localized E3 ubiquitin ligases that monoubiquitylate histone H2B (H2Bub1) to modulate the chromatin binding of NF-κB.46 NF-κB is a key regulator of inflammation and tumor immunity, which has a crucial role in inflammation-associated CRC.47 Subsequently, the downregulation of RNF20 and H2Bub1 augments NF-κB activity and attenuates the antitumoral T-cell response to induce chronic inflammation and inflammation-associated CRC. Moreover, RNF20/40 and H2Bub1 were frequently downregulated in ulcerative colitis and colitis-associated CRC in human, further confirming their clinical relevance.46 Blockade of NF-κB alone or in combination with antitumor drugs may be used to target CRC with inactivating mutations in RNF20/RNF40.
Another RING E3 ligase protein participating in the NF-κB pathway is tumor necrosis factor receptor-associated factor 6 (TRAF6/RNF85), which contains a RING domain interspersed with four zinc-finger motifs, a central coiled-coil domain and a highly conserved TRAF-C domain.48 TRAF6 is upregulated in CRC with positive expression in 66.7% of 135 CRC patients as determined by immunohistochemistry.49 TRAF6 functions as a signal transducer by activating NF-κB kinase (IKK), a key enzyme complex of the NF-κB pathway in response to pro-inflammatory cytokines in colon cancer cells.50 Mechanistically, TRAF6 collaborates with Ubc13/Uev1A catalyze the assembly of K63-linked polyubiquitin chains, which, in turn, activates IKK.51
TRIM family
The TRIM family is the largest subfamily of RING-type E3 ligases, which is defined as comprising an N-terminal RING domain, one or two B-boxes and a coiled-coil domain.52 Approximately 70 TRIM genes have been recognized in humans and have been subclassified on the basis of the differences in their C-terminal domain.21 Most of the TRIM proteins function as E3 ubiquitin ligases, and they are engaged in various processes such as cell proliferation, apoptosis and transcriptional regulation.7, 21, 52 Depending on modification type, ubiquitylation of their substrates could lead to activation, inhibition, proteasomal or lysosomal degradation.
TRIM3 exerts a tumor-suppressive effect in CRC progression through the regulation of p53.53 TRIM15 and TRIM40 are other tumor-suppressive TRIM proteins specifically downregulated in CRC.54 TRIM15 colocalizes with focal adhesions and it suppresses cell migration. TRIM15 interacts with actin cytoskeleton dynamics-related proteins, such as coronin 1B, cortactin, filamin-binding LIM protein 1 and vasodilator-stimulated phosphoprotein to regulate cell migration.54 TRIM40 negatively regulates NF-κB activity via ubiquitin-like modification of inhibitor of NF-κB kinase subunit gamma (IKKγ), thus preventing inflammation-associated carcinogenesis in the gut.55
Dimeric RING domain E3 ligase
MDM2/MDMX ligase
The tumor suppressor p53 has a critical role in regulating cell cycle, cell apoptosis, DNA repair and senescence.56 TP53 is one of the most frequently mutated genes in CRC. Approximately 50–60% of CRCs harbor TP53 mutations, resulting in p53 inactivation or loss.57 The dimeric RING-type E3 ligase MDM2 is one of the major regulators of p53. MDM2 directly binds with p53 and forms a protein complex, mediating p53 degradation via the ubiquitin–proteasome pathway.58, 59 Overexpression of MDM2 is an early event in CRC progression and is positively correlated with p53 loss.19 In vivo mouse models clearly demonstrated that the inactivation or loss of p53 stimulates tumorigenesis, whereas established tumors are disappeared or diminished after restoration of p53.60 These data indicate that recovery of wild-type p53 function is a valid therapeutic approach, and the specific targeting of MDM2/MDMX to reactivate p53 is a viable anticancer strategy.61 On the basis of this notion, structure-based drug design has led to the discovery of several MDM2 or MDMX antagonists that block the interactions between murine double minute (MDM) and p53, leading to p53 stabilization and activation for cancer treatment.62
IAP Family/XIAP
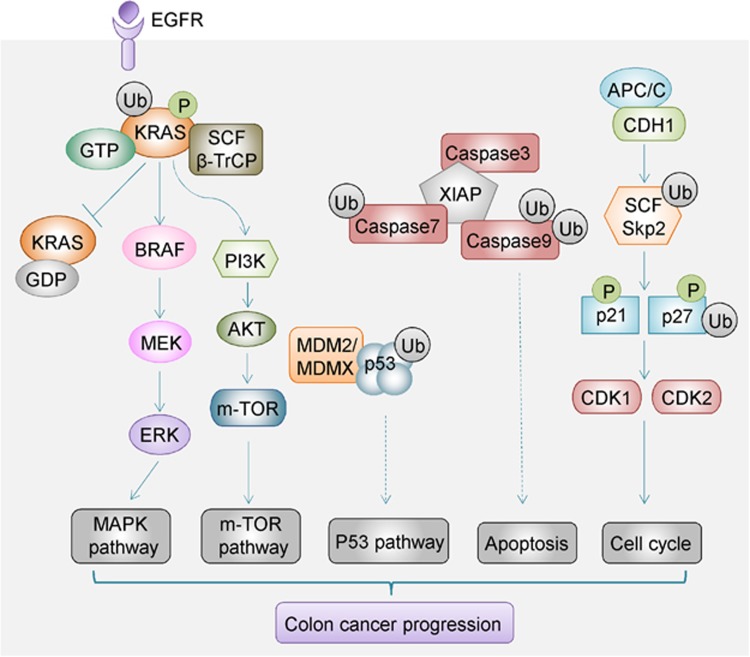
Human inhibitor of apoptotic protein (IAP) family comprised X-chromosome-linked IAP (XIAP), cellular IAP1/2, melanoma IAP and IAP-like protein 2.63 A growing amount of evidence establishes that IAP proteins target substrates in the ubiquitin–proteasome system degradation machinery. Targeting of proteins for degradation could occur not only by their own RING domains, but also through forming protein complexes with other IAPs containing the RING domains. IAPs monoubiquitinate or polyubiquitinate their binding partners, including activated caspases, tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2), receptor-interacting protein 1 (RIP1) and NF-κB-inducing kinase (NIK), proteins that mainly participate in apoptosis and TNF and NF-κB signaling cascade64, 65 (Figure 3). Both TNF and NF-κB are correlated with colitis-associated CRC. Several IAP family proteins such as XIAP, cIAP1 and cIAP2 are upregulated in CRC. Notably, increased expression of apoptosis regulator cIAP2 was significantly associated with shorter survival of CRC patients.66
Figure 3.
Regulation of important signaling pathways by E3 ubiquitin ligases in CRC. β-TrCP mediates polyubiquitination and degradation of phosphorylated KRAS. Loss of β-TrCP causes accumulation of active KRAS and the activation of downstream MAPK and m-TOR signaling to promote CRC progression. MDM2/MDMX directly binds to p53, and it mediates p53 degradation in the ubiquitin–proteasome system to inhibit the p53 pathway in CRC. XIAP promotes ubiquitylation and degradation of a number of caspase proteins to inhibit the caspase-dependent apoptosis pathway in CRC development. APC-type E3 ligase CDH1 cooperates with SCF-type E3 ligase SKP2 in governing cell cycle progression in CRC. β-TrCP, β-transducin repeat-containing protein; CDH1, Cadherin 1; MAPK, mitogen-activated protein kinase; m-TOR, the mechanistic target of rapamycin; SKP2, S phase kinase-associated protein 2; XIAP, X-chromosome-linked IAP.
XIAP is an important factor in the regulation of apoptosis induction as it acts as a potent caspase inhibitor.63, 67 XIAP contains three structural domains, the baculoviral IAP repeat domain, the UBA domain and the RING domain.68 The RING domain of XIAP processes E3 ubiquitin enzyme activity in the C-terminal and it promotes autoubiquitylation as well as transubiquitylation of their binding partners. In CRC, XIAP inhibits caspase-3 activity by ubiquitining active p17 and mediating its degradation via proteasome in HCT-116 cells harboring PIK3CA mutations.69 XIAP is thus a potential druggable target (TRAIL) to reverse TNF-related apoptosis inducing ligand resistance phenotype in PIK3CA-mutant CRC. TNF-related apoptosis-inducing ligand-targeted drugs in combination with XIAP-targeting molecules may expand the limited therapeutic probabilities for this subtype of CRC patients. Another study revealed an alternative oncogenic effect of XIAP, which involved the monoubiquitylation of TLE. TLE is a potent repressor of the TCF/LEF complex. Ubiquitylation of TLE releases TLE from the TCF/LEF complex, thereby allowing β-catenin to bind to LEF/TCF and initiate a canonical Wnt transcriptional program in CRC.70 Notably, XIAP-mediated ubiquitylation mainly alters TLE protein subcellular location, rather than promoting its degradation.71
Multisubunit RING domain E3 ligase
SCF-type E3 ligase
One large family of RING-based E3 ligase is known as SCF. SCFs utilize Cullin1 as the scaffold protein, F-box as receptors of substrates and RBX1/2 as E3 ubiquitin ligases.72 This superfamily of E3 ligases has vital roles in cancer biology. The RING domain coordinating with zinc ions is essential for SCF-type E3 ligase activity. A number of studies have demonstrated the roles of SCF-type E3 ubiquitin ligases in controlling cell size, proliferation and growth; their deregulation has been participated in abnormal cell growth and tumorigenesis.73, 74 Three well-characterized SCF complex family members are β-transducin repeat-containing protein (β-TrCP), F-box and WD repeat domain-containing 7, and SKP2 in CRC.
β-TrCP
β-TrCP is an F-box protein that acts as the substrate-recognition subunit for the SCFβ-TrCP E3 ubiquitin ligase, which mediates the ubiquitylation of various crucial substrates and post-translationally marks substrates for degradation, including β-catenin, p53, Yap and NF-κB inhibitor (IKB).75, 76, 77 Two paralogs of this F-box protein exist in humans, β-TrCP1 and β-TrCP2, which have uniform functions and appear to be superfluous in their biological effects. Among the SCF-type E3 ligase, the expression of β-TrCP1 was found to be upregulated in CRC, particularly in the metastases in CRC patients.78
As mentioned previously, activated Wnt signaling contributes to CRC development. The level of active β-catenin is normally under control of the β-catenin destruction complex, consisting of APC, glycogen synthase kinase-3 beta, casein kinase I a and Axin2.79 Without or due to lack of Wnt signal stimulation, these proteins act in collaboration to phosphorylate β-catenin.80, 81, 82 Phosphorylated β-catenin is then targeted by β-TrCP1/2 for ubiquitin-mediated proteasomal degradation (Figure 2),83, 84 thereby suppressing the Wnt transcriptional cascade. Hence, β-TrCP1/2 functions as a tumor suppressor, negatively regulating Wnt signaling.
KRAS mutation is an early and common event in CRC. Mutant KRAS is a key driver oncogene in CRC and it predicts poor outcome in CRC patients.85 Experimental evidence indicates that β-TrCP mediates the ubiquitylation and degradation of activated KRAS (GTP-bound). Loss of β-TrCP impairs KRAS degradation, thus causing the accumulation of active KRAS and the activation of its downstream effectors (Figure 3).86, 87 This constitutes an alternative mechanism by which β-TrCP exerts a tumor-suppressive effect in CRC development. However, Baker et al. established recently that Ras protein monoubiquitination suppresses GAP-mediated hydrolysis and increases the proportion of activated (GTP-bound) Ras.88 Monoubiquitination modulates Ras activity and enhances affinity of downstream effectors such as Raf and PI3 kinase to promote tumorigenesis.89 By changing the activity of oncoproteins via ubiquitination, this ubiquitination machinery may provide a new therapeutic target for CRC treatment.
Despite these in vitro findings, β-TrCP1 is paradoxically upregulated in 56% of the CRC as compared with normal tissue, and its overexpression was correlated with reduced apoptosis and worse prognosis.78 Elevated activity of β-TRCP has been widely detected in CRC cells, and it has been suggested that β-TrCP is enhanced as part of a negative feedback in response to β-catenin/TCF activation.90 However, such upregulation might be futile in CRC because of mutations in APC, Axin or β-catenin, which impair β-catenin phosphorylation and hence weaken its degradation by β-TrCP.91, 92, 93 In such a case, the tumor-suppressive effect of β-TrCP is attenuated. On the other hand, β-TrCP might exert an oncogenic effect by modulating the NF-κB pathway. β-TrCP has been found to bind with phosphorylated IKBs, inhibitors of NF-κB, and to mediate their proteasomal degradation.78, 90 Decreased IKB expression in turn enables the nuclear translocation and transcriptional activation of NF-κB, which contributes to the inhibition of apoptosis and promotion of tumor metastasis.78, 90 Thus, the upregulation of β-TrCP may exert a positive or negative effect on CRC, depending on the presence of co-occurring mutations and the level of β-TrCP expression.
Many key regulatory proteins and substrates are regulated by β-TrCP in a tissue-specific manner. Some of the known β-TrCP substrates like Cyclin D1, WEE1 G2 checkpoint kinase and cell division cycle 25A are cell cycle regulators.94 Some proteins, such as myeloid cell leukemia 1 and pro-caspase-3, targeted for degradation by β-TrCP are implicated in cell apoptosis.95, 96 Furthermore, β-TrCP also functions in cell adhesion and migration through regulation of Snail, the suppressor of E-cadherin in a phosphorylation-dependent manner.97 Recent work has also shown that β-TrCP1 targets the FOXO3 tumor suppressor,98 and epigenetic regulators including chromodomain helicase DNA-binding protein 1 for phosphorylation-dependent ubiquitylation degradation.99 Given the promiscuity of β-TrCP, it is perhaps not surprising that β-TrCP has contradictory roles in tumorigenesis in a context-dependent manner.
FBW7
FBW7 is another F-box protein that functions as a substrate receptor for SCF-type E3 ubiquitin ligase to promote polyubiquitylation.100 FBW7 degrades several key oncoproteins including Cyclin E, JUN, Mcl-1, MYC, Notch and aurora kinase A in a phosphorylation-dependent manner.101, 102 Most of these targeted substrates are key transcription regulators, allowing FBW7 to regulate multiple pathways with oncogenic signature that are in charge of gene expression clusters, thus extending the impact of FBW7 far beyond its direct substrates. Previous studies discovered that 6–10% of CRC contains FBW7 mutations.103 Somatic FBW7 mutation was also detected in 9% of family adenomatous polyposis carcinomas or hereditary non-polyposis CRC.104 Given that FBW7 targets oncogenic proteins for degradation, it possesses a tumor-suppressive character and functions in cell division, growth and differentiation pathways.105
Mutation of Fbw7 combined with Apcmin has been shown to promote CRC in mice.106, 107 Transforming growth factor-beta-induced factor and kruppel-like factor (KLF5) were elevated in ApcminFbw7ARG/+ tumors, and these proteins were strong candidates for mediating the oncogenic effect and may cause a similar consequence in Fbw7-mutant human CRC.108 In another transgenic mouse model of CRC, co-deletion of p53 and Fbw7 induced malignant adenocarcinomas that exert strong invasiveness and metastatic abilities.108 These tumors exhibited a chromosomal unstable feature, and dual p53 and Fbw7 knockout represents a novel mouse model of metastatic and chromosomally unstable CRC.109
SKP2
SKP2 primarily regulates cell cycle regulators via the ubiquitin–proteasome pathway.110 SKP2 protein exerts a remarkable impact on the progression of CRC. Overexpression of SKP2 promotes cell proliferation in normal colon cells, whereas knockdown of SKP2 inhibited cell proliferation and induced the apoptosis of SW620 colon cancer cells.111, 112 SKP2, as an F-box protein, recognizes cyclin-dependent kinase (CDK) inhibitors p27kip1 and p21 and mediates their ubiquitin-mediated proteasomal degradation.113 Degradation of p21 and p27 by SKP2 activates CDK1 and CDK2 and promotes S and G2 cell cycle phase progression, thereby positively regulating the cell cycle114 (Figure 3). In keeping with this possibility, drugs that target SKP2 could block cell cycle progression and inhibit tumor growth. The upregulation of SKP2 in colon cancer patients is correlated with a decrease in its target p27, and this phenotype is correlated with malignant tumor behavior and poor clinical outcomes.115, 116 SKP2 may be utilized as a novel independent prognostic marker for CRC.
APC/C-type E3 Ligase
Besides SCF ubiquitin ligases, the APC/C is another major multisubunit RING-type E3 ligase. Among the APC core constituents, co-activators cell division cycle 20 (Cdc20) and Cadherin 1 (Cdh1) have been well characterized. Intriguingly, even though both Cdc20 and Cdh1 have the ability to catalyze the APC-type E3 ligase complex, they may exert opposing roles in tumorigenesis because different substrates are recognized and targeted for degradation.117 APC-type E3 ligases are primarily involved in governing cell cycle progression, especially in the mitosis phase.6
APCCdc20
Emerging evidence indicates that Cdc20 is elevated in CRC cell lines and in primary CRC tissues compared with normal colon cells and adjacent non-tumor tissues.118 Cdc20 expression was correlated with advanced tumor stage, tumor metastasis and shorter overall survival of patients, suggesting that Cdc20 could be utilized as a promising predictor for human CRC. Another study also revealed that Cdc20 expression was increased markedly in 77% of CRC cases in further support of this finding.119 In line with these findings, Cdc20 depletion suppressed Wnt signaling through the upregulation of conductin. In addition, conductin with Cdc20-resistant CRC cells manifested Wnt signaling inhibition and weakened colony formation vability.120 Therefore, Cdc20 overexpression exerts an oncogenic role in colorectal tumorigenesis.
APCCdh1
In contrast to Cdc20, Cdh1 (E-cadherin) might display a tumor-suppressive effect in human colorectal carcinogenesis.121 Loss function of E-cadherin has been well demonstrated in the pathogenesis of sporadic CRC, and Cdh1 gene mutation also predisposes to early-onset CRC.122 APCCdh1 may function as a tumor suppressor through proteolysis of SKP2 protein in colorectal tumorigenesis.123 Several APCCdh1 targets, which include Aurora A, Aurora B, Cyclin B, microtubule nucleation factor (Tpx2), Cdc6 and Cdc20, are correlated with cell cycle progression, and their degradation via ubiquitylation suppressed tumorigenesis.74 Correspondingly, the downregulation of E-cadherin is frequently detected in CRC, and it thus can be used as a prognostic marker in CRC patients.124
Ring-type E3 ligases as promising biomarkers in CRC
RING-type E3 ligases are often deregulated at the early stage of tumorigenesis, and hence might serve as biomarkers for cancer diagnosis (Table 1). RNF43 is one of the most frequently mutated genes in CRCs, and somatic mutations of RNF43 are present in ~17.6–18.9% of CRC patients in different cohorts.125 Truncated mutations of RNF43 are mutually exclusive with loss of function mutations of APC in colorectal tumors. RNF43 mutations are predominately caused by small insertions/deletions in homopolymeric tracts, with a high proportion in the microsatellite instability CRC types. Overall, RNF43 is mutated in 79.7% of high microsatellite instability colorectal adenocarcinomas.42
TRIM29 expression is increased in human precancerous aberrant crypt foci biopsy tissues compared with normal colon mucosa biopsies by colonoscopy.126 That indicates that TRIM29 may be a suitable marker for the early diagnosis of CRC. Survivin is one of the IAP protein family members related to anti-apoptotic pathways.127 Overwhelming evidence indicates that survivin expression is elevated in precancerous lesions such as tubular adenomas and low-grade dysplasia, suggesting that survivin upregulation is an early event in colorectal tumor formation.128, 129 Thus, survivin could be a promising biomarker for early screening of CRC. In addition, high expression of survivin is maintained throughout the progression from normal mucosa–adenoma to carcinoma, suggesting that survivin may be a suitable biomarker throughout different stages of carcinogenesis.
Alterations in RING-type E3 ligases are also useful biomarkers for predicting prognosis and therapeutic response. To date, therapeutic targeting of the Wnt pathway has only yielded meagre clinical benefit.130 This could be attributed to a lack of patient selection. E3 ligases RNF43 and ZNRF3 are two transmembrane enzymes that remove Wnt receptors from cell surface and represent the first upstream Wnt pathway component mutated in CRC.41 Patients with RNF43 and ZNRF3 gene deletion or inactivation mutation are thus predicted to benefit from the administration of Wnt antagonist or Wnt pathway inhibitors. Indeed, Wnt inhibitors are highly effective in suppressing the growth of CRC-harboring RNF43/ZNRF3 mutations. Both Wnt-specific acyltransferase porcupine (PORCN) inhibitors, C59 and LGK974, are effective in suppressing growth of RNF43/ZNRF3-mutant intestinal tumors in vivo.131, 132 The cultured organoids biopsy from CRC patients that possesses RNF43 mutations respond sensitively to the FZD5 antibody treatment, revealing mutant RNF43 as a potential tractable target for therapeutic antibodies.133
FBW7 is the fourth most commonly mutated gene in CRC.107 CRC-harboring FBW7 mutations are insensitive to blockade of mitogen-activated protein kinase/ERK signaling by targeted therapy of sorafenib or regorafenib.134 Mechanistically, CRC cells carrying FBW7 mutations are deficient in apoptosis and demonstrate attenuated degradation of Mcl-1, which will cause resistance to regorafenib.135 Besides FBW7 mutations, mRNA level of FBW7 was significantly lower in CRC tissues compared with adjacent normal tissues. Low expression of FBW7 indicates a worse survival in CRC patients. Multivariate analysis indicated that FBW7 expression is an independent predictor in CRC patients.136 Thus, FBW7 expression may be a suitable prognostic factor for patients with CRC.
Ring-type E3 ligases as therapeutic targets
E3 ligases are attractive drug targets for cancer therapy, given the pivotal roles exhibited by E3 ligases in the regulation of diverse and important cellular pathways, including Wnt, p53, K-ras and oncogenic NF-κB in CRC. Compared with bortezomib, the general proteasome inhibitor, which non-selectively blocks the whole system protein degradation,137 compounds that target a particular E3 enzyme are expected to more selectively modulate the levels of proteins, thereby rendering improved specificity with minimal toxicity. Therefore, targeting RING-type E3 ubiquitin ligases to anticancer has harvested growing attention. Targeting E3 ubiquitin ligases using small molecular weight compounds or neutralizing antibodies is currently being designed and tested in different preclinical cancer models as well as in clinical trials (Table 2).
One of the most efficient therapeutic agent targeting E3 ligases is arsenic trioxide, which is used clinically to treat APL patients. Arsenic trioxide exhibits its chemotherapeutic effect though promoting the degradation of PML zinc-finger protein and retinoic acid receptor alpha (PML-RAR-α), a driver oncogenic fusion gene present in APL.138 Arsenic trioxide induces the oligomerization of PML-RAR-α and strengthens its interaction with SUMO-conjugating enzyme UBC9, leading to elevated SUMOylation. SUMOylation of PML-RAR-α triggers the recruitment of RNF4 into PML-nuclear bodies to ubiquitylate the fusion oncoprotein PML-RAR-α and promote its proteasomal degradation.139, 140 It would be of great interest to measure whether some RNF proteins can be activated in a similar manner in CRC to suppress oncogenic signaling.
Many experimental agents targeting RING-type E3 ligases are also entering the cancer drug pipeline. Specific targeting of either MDM2 or MDMX is a promising approach to recover the activity of wild-type p53 for cancer therapy.62 Targeting MDM2 ubiquitin ligase with small molecules or compounds has been recently developed and is being tested in cancer models. HLI98, for example, specifically inhibits Hdm2 to activate p53 signaling pathway and induce apoptosis in cancer.141 Structure-based drug design has also obtained (MDM) antagonists that disturb the interaction between (MDM) and p53, resulting in p53 reactivation for cancer treatment. Gu et al. utilized chemical screening to identify novel inhibitors of MDM2 protein–XIAP mRNA interaction, which suppresses MDM2 stability and blocks XIAP translation, leading to both antiproliferative and pro-apoptotic effects in cancer models. In p53 wild-type cancers, MDM2/XIAP inhibitors can degrade MDM2 to activate p53 and induce apoptosis, whereas in p53-deficient cancers inhibition of XIAP expression can also activate caspase cascade to induce apoptosis. Importantly, one of the MDM2/XIAP inhibitors, MX69, showed no obvious effect on normal human hematopoiesis inhibition and with a good tolerance in animal models.142 Ceramide and its analog LCL85 induce the protein degradation of cIAP1 and XIAP in the proteasome and sensitize primary and metastatic CRC cells to Fas-mediated apoptosis.143, 144 In vivo LCL85 is an effective sensitizer for Fas-mediated apoptosis, and it inhibits lung metastasis in preclinical mouse tumor models at a non-cytotoxic dose.143 By targeting these two ligases, ceramide might enhance the efficacy of Fas ligand-based CRC therapy, particularly cytotoxic T-lymphocyte-based immunotherapy.145
Compounds such as MLN4924, CC0651 and pro-TAME have been developed to target key subunits of the SCF or APC/C complex, which have shown promising efficacy in CRC with some of them entering clinical trials.146 One recent study revealed that treatment of HT-29 and HCT-116 cells with the SCF E3 ubiquitin ligase inhibitor MLN4924 efficiently sensitized CRC cells to irradiation. MLN4924 enhanced radiation sensitivity by inducing cell cycle arrest at the G2/M phase, cell apoptosis and DNA damage, and this effect was dependent on p27.147 Further preclinical work will be needed to verify the effectiveness and safety of MLN4924 as a valid radiosensitizing agent against CRC. MLN4924-mediated inhibition of cullin neddylation has shown prospect in the preclinical cancer models.146, 148
CC0651 selectively inhibits E2 enzyme human Cdc34 and evidently influences the core components Skp2, Cul1 and Rbx1 of SCF, thus potently inhibiting the ubiquitination of p27Kip1 by E3 ligase SCFSkp2.149, 150 CC0651 and its analogs are able to inhibit proliferation of CRC cells and cause the accumulation of p27Kip1, a SCFSkp2 substrate.149 Pro-TAME inhibits APC-Cdh1 activation and induces mitotic arrest in cancer cells.151 Reducing Cdc20 expression sensitizes cells to the effect of pro-TAME, consistent with the APC–Cdc20 interaction as the relevant target of this compound.151 Moreover, experimental evidence indicates that Apcin and a few apcin synthetic derivatives synergize with pro-TAME to inhibit mitotic exit in human CRC cells.152, 153
The major concerns of E3 ligase for clinical application are lack of specificity for the intended E3 ligase, which might result in unwanted side effects and give rise to drug resistance. Several high-throughput screening assays are now used for the fast filtrating of small molecular compounds of E3 ligases; specific antagonists of E3 ubiquitin enzymes will continue to be investigated as a promising kind of anticancer drugs.154 Progress in elucidating the structural aspects of RING-type E3 ligases will provide clues for rational drug design. Chemical crosslinkers are beneficial to study the structures of intermediates generated by E3-catalyzed reactions, allowing to anatomize the distinct catalytic processes during the E3 enzymes catalyzing substrate ubiquitylation. Apart from targeting E3 ligases' activity directly, another strategy is to develop homologs of tumor-suppressive RING-type E3 ligases that aim to antagonize their target proteins. In addition, recent work has revealed that ubiquitylation is an invertible process involving ubiquitylation as well as deubiquitylation. Restrain of deubiquitylation enzymes may provide a conceivable cancer therapeutic modality in the future.
Conclusions and future perspectives
In recent years, we have gained deeper insights into the regulation of E3 ligases and the mechanisms underlying E3 ligase-mediated ubiquitin–proteasome degradation. Moreover, large-scale genomic studies have uncovered genomic amplification, deletions, mutations and aberrant epigenetic regulation that lead to the deregulation of RING-type E3 ligases in cancers.154 Further work should aim to unveil the function(s) of individual RING-type E3 ligases in tumorigenesis and comprehensively elucidate their mechanisms of action. In the past few years we have seen much progress in the development of small molecule drugs that target E3 ligase receptors or adaptors to block aberrant signaling cascades in CRC. With accumulating structural information regarding individual RING finger E3 ligases and their role in protein complexes, there is great potential for development of targeted therapeutics. Future work should aim to unveil the functions of the distinct E3 enzymes in the different cellular contexts, and elucidate the crystal structure of key regulators with therapeutic potential to facilitate rational drug development. E3 ligase-based cancer biomarkers will be verified and used in clinical practice as diagnostic approaches or prognostic indexes for the benefit of CRC patients.
Acknowledgments
This project was supported by RGC-GRF Hong Kong (766613, 14106415, 14114615, 14111216), HMRF Hong Kong (03140856), The National Key Technology R&D Program (2014BAI09B05) and 135 program project (2016YFC1303200).
Author contributions
LL drafted the manuscript. CCW revised the manuscript. BG commented on the manuscript. JY commented on and revised the manuscript.
Glossary
- CRC
colorectal cancer
- UPS
the ubiquitin–proteasome system
- APC
adenomatous polyposis coli
- TCF
T-cell factor
- LEF
lymphoid enhancer factor
- RNF
Ring Finger Protein
- SUMO
small ubiquitin-like modifier
- ZNRF3
zinc and ring finger 3
- DVL
disheveled segment polarity protein
- LRP
low-density lipoprotein receptor-related protein
- TRAF6
tumor necrosis factor receptor-associated factor
- IκB
inhibitor of NF-κB kinase
- TRIM
tripartite motif
- MDM2/MDMX
the murine double minute 2/4
- IAP
inhibitor of apoptotic proteins
- XIAP
X-chromosome-linked IAP
- CRL
cullin RING ubiquitin ligases
- SCF
SKP1–cullin1–F-box protein
- β-TrCP
β-Transducin repeat-containing protein
- GSK3β
glycogen synthase kinase-3 beta
- AXIN
axis inhibition protein
- FBW7
F-box and WD repeat domain-containing 7
- CDK
Cyclin-dependent kinase
- SKP2
S phase kinase-associated protein 2
- Cdh1
Cadherin 1
- APC/C
anaphase-promoting complex/cyclosome.
Footnotes
The authors declare no conflict of interest.
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet 2013; 14: 703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol 2011; 8: 686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol 2011; 6: 479–507. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67: 425–479. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 2006; 6: 369–381. [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 2008; 8: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med 2014; 20: 1242–1253. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci 2012; 125: 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78: 399–434. [DOI] [PubMed] [Google Scholar]
- Wang XW, Wei W, Wang WQ, Zhao XY, Guo H, Fang DC. RING finger proteins are involved in the progression of barrett esophagus to esophageal adenocarcinoma: a preliminary study. Gut Liver 2014; 8: 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Schluter A, Peters MA, Viehweger B, Ostendorff HP, Soosairajah J et al. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev 2005; 19: 2307–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon YJ, Khelifa S, Ratnikov B, Scott DA, Feng Y, Parisi F et al. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell 2015; 27: 354–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Shimelis H, Linn DE, Jiang R, Yang X, Sun F et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 2009; 15: 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhao C, Zhuang T, Jonsson P, Sinha I, Williams C et al. RING finger protein 31 promotes p53 degradation in breast cancer cells. Oncogene 2016; 35: 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature 2009; 458: 438–444. [DOI] [PubMed] [Google Scholar]
- Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 2011; 11: 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK et al. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 2000; 10: 429–439. [DOI] [PubMed] [Google Scholar]
- Abdel-Fattah G, Yoffe B, Krishnan B, Khaoustov V, Itani K. MDM2/p53 protein expression in the development of colorectal adenocarcinoma. J Gastrointest Surg 2000; 4: 109–114. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009; 17: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer 2011; 11: 792–804. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995; 373: 81. [DOI] [PubMed] [Google Scholar]
- Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol 2016; 17: 626–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, Suzuki T et al. E3 ubiquitin ligase that recognizes sugar chains. Nature 2002; 418: 438–442. [DOI] [PubMed] [Google Scholar]
- Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta 2014; 1843: 75–85. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 2009; 78: 363–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 2011; 12: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 2014; 21: 301–307. [DOI] [PubMed] [Google Scholar]
- Sun H, Hunter T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J Biol Chem 2012; 287: 42071–42083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denuc A, Marfany G. SUMO and ubiquitin paths converge. Portland Press Limited 2010; 38: 34–39. [DOI] [PubMed] [Google Scholar]
- Thomas JJ, Abed M, Heuberger J, Novak R, Zohar Y, Beltran Lopez AP et al. RNF4-dependent oncogene activation by protein stabilization. Cell Rep 2016; 16: 3388–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Prieto R, Cuijpers SA, Kumar R, Hendriks IA, Vertegaal AC. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 2015; 14: 1859–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev 2014; 28: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring A, Kim Y-M, Kahn M. Wnt/catenin signaling in adult stem cell physiology and disease. Stem Cell Rev Rep 2014; 10: 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev 2007; 17: 45–51. [DOI] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. β-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 2005; 12: 364–371. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009; 17: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Piloto S, Zeng W, Hoverter NP, Schilling TF, Waterman ML. Ring Finger Protein 14 is a new regulator of TCF/β‐catenin‐mediated transcription and colon cancer cell survival. EMBO Rep 2013; 14: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantù C, Valenta T, Basler K. A RING finger to wed TCF and β‐catenin. EMBO Rep 2013; 14: 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H-X, Xie Y, Zhang Y, Charlat O, Oster E, Avello M et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012; 485: 195–200. [DOI] [PubMed] [Google Scholar]
- Giannakis M, Hodis E, Mu XJ, Yamauchi M, Rosenbluh J, Cibulskis K et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 2014; 46: 1264–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Spence JR. Cancer biology: a new RING to Wnt signaling. Curr Biol 2012; 22: R849–R851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Charlat O, Zamponi R, Yang Y, Cong F. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol Cell 2015; 58: 522–533. [DOI] [PubMed] [Google Scholar]
- Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012; 488: 665–669. [DOI] [PubMed] [Google Scholar]
- Tarcic O, Pateras IS, Cooks T, Shema E, Kanterman J, Ashkenazi H et al. RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep 2016; 14: 1462–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfer L. Cytokines, inflammation and colon cancer. Curr Cancer Drug Targets 2011; 11: 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson KM, Bhakar AL, Barker PA. TRAF6‐dependent NF‐kB transcriptional activity during mouse development. Dev Dyn 2004; 231: 122–127. [DOI] [PubMed] [Google Scholar]
- Zhang T, Wang H, Han L. Expression and clinical significance of tumor necrosis factor receptor-associated factor 6 in patients with colon cancer. Iran Red Crescent Med J 2016; 18: e23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Li XB, Meng Y, Fan L, Li M, Fang J. TRAF6 upregulates expression of HIF-1alpha and promotes tumor angiogenesis. Cancer Res 2013; 73: 4950–4959. [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 2000; 103: 351–361. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Hatakeyama S. TRIM proteins and diseases. J Biochem 2017; 161: 135–144. [DOI] [PubMed] [Google Scholar]
- Piao MY, Cao HL, He NN, Xu MQ, Dong WX, Wang WQ et al. Potential role of TRIM3 as a novel tumour suppressor in colorectal cancer (CRC) development. Scand J Gastroenterol 2016; 51: 572–582. [DOI] [PubMed] [Google Scholar]
- Lee OH, Lee J, Lee KH, Woo YM, Kang JH, Yoon HG et al. Role of the focal adhesion protein TRIM15 in colon cancer development. Biochim Biophys Acta 2015; 1853: 409–421. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Okumura F, Takahashi N, Kataoka A, Kamiyama T, Todo S et al. TRIM40 promotes neddylation of IKKgamma and is downregulated in gastrointestinal cancers. Carcinogenesis 2011; 32: 995–1004. [DOI] [PubMed] [Google Scholar]
- Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 2009; 1: a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlin A, Smith G, Carey FA, Wolf CR, Steele RJ. The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut 2005; 54: 1283–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387: 296. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997; 387: 299. [DOI] [PubMed] [Google Scholar]
- Zhang X, Pageon L, Post SM. Impact of the Mdm2(SNP309-G) allele on a murine model of colorectal cancer. Oncogene 2015; 34: 4412–4420. [DOI] [PubMed] [Google Scholar]
- Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer 2003; 3: 102–109. [DOI] [PubMed] [Google Scholar]
- Wade M, Li Y-C, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep 2006; 7: 988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 2010; 29: 4198–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J, Vucic D. IAP family of cell death and signaling regulators. Methods Enzymol 2014; 545: 35–65. [DOI] [PubMed] [Google Scholar]
- Krajewska M, Kim H, Kim C, Kang H, Welsh K, Matsuzawa S-i et al. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin Cancer Res 2005; 11: 5451–5461. [DOI] [PubMed] [Google Scholar]
- Schile AJ, García-Fernández M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev 2008; 22: 2256–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galban S, Duckett CS. XIAP as a ubiquitin ligase in cellular signaling. Cell Death Differ 2010; 17: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenschwender M, Bittner S, Seibold K, Wajant H. XIAP-targeting drugs re-sensitize PIK3CA-mutated colorectal cancer cells for death receptor-induced apoptosis. Cell Death Dis 2014; 5: e1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AJ, Wallace HA, Freeman TJ, Beauchamp RD, Lee LA, Lee E. XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Mol Cell 2012; 45: 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardavaccaro D, Pagano M. Oncogenic aberrations of cullin-dependent ubiquitin ligases. Oncogene 2004; 23: 2037–2049. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 2002; 416: 703–709. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004; 5: 739–751. [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Wei W. SCF and APC E3 Ubiquitin Ligases in Tumorigenesis. Springer International Publishing: Cham, Switzerland, 2014. [Google Scholar]
- Hart M, Concordet J, Lassot I, Albert I, Del los Santos R, Durand H et al. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr Biol 1999; 9: 207–211. [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang C-Y, Guan K-L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ-TRCP. Genes Dev 2010; 24: 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack P, Caligiuri M, Pelletier M, Boisclair M, Theodoras A, Beer-Romero P et al. SCF [beta]-TRCP and phosphorylation dependent ubiquitination of I [kappa] B [alpha] catalyzed by Ubc3 and Ubc4. Oncogene 2000; 19: 3529. [DOI] [PubMed] [Google Scholar]
- Ougolkov A, Zhang B, Yamashita K, Bilim V, Mai M, Fuchs SY et al. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J Natl Cancer Inst 2004; 96: 1161–1170. [DOI] [PubMed] [Google Scholar]
- Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol 2013; 5: a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Hamada F, Ishidate T, Ki Anai, Kawahara K, Toyoshima K et al. Axin, an inhibitor of the Wnt signalling pathway, interacts with β‐catenin, GSK‐3β and APC and reduces the β‐catenin level. Genes Cells 1998; 3: 395–403. [DOI] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg G-H, Tan Y et al. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 2002; 108: 837–847. [DOI] [PubMed] [Google Scholar]
- Peters JM, McKay RM, McKay JP, Graff JM. Casein kinase I transduces Wnt signals. Nature 1999; 401: 345–350. [DOI] [PubMed] [Google Scholar]
- Liu C, Kato Y, Zhang Z, Do VM, Yankner BA, He X. β-Trcp couples β-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci USA 1999; 96: 6273–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K et al. An F‐box protein, FWD1, mediates ubiquitin‐dependent proteolysis of β‐catenin. EMBO J 1999; 18: 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps AI, Buchanan DD, Makar KW, Win AK, Baron JA, Lindor NM et al. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br J Cancer 2013; 108: 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Allam US, Ahsan A, Chen G, Krishnamurthy PM, Marsh K et al. KRAS protein stability is regulated through SMURF2: UBCH5 complex-mediated beta-TrCP1 degradation. Neoplasia 2014; 16: 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong W-J, Yoon J, Park J-C, Lee S-H, Lee S-H, Kaduwal S et al. Ras stabilization through aberrant activation of Wnt/β-catenin signaling promotes intestinal tumorigenesis. Sci Signal 2012; 5: ra30. [DOI] [PubMed] [Google Scholar]
- Baker R, Lewis SM, Sasaki AT, Wilkerson EM, Locasale JW, Cantley LC et al. Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat Struct Mol Biol 2013; 20: 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER et al. Ubiquitination of Ras enhances activation and facilitates binding to select downstream effectors. Sci Signal 2011; 4: ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman VS, Slaga TJ, Pagano M, Minamoto T, Ronai Ze, Fuchs SY. Wnt/β-catenin signaling induces the expression and activity of βTrCP ubiquitin ligase receptor. Mol Cell 2000; 5: 877–882. [DOI] [PubMed] [Google Scholar]
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β‐catenin is a target for the ubiquitin–proteasome pathway. EMBO J 1997; 16: 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017; 36: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde R, Smits R, Clevers H. APC signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer 2001; 1: 55–67. [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer 2008; 8: 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, He X, Hsu J-M, Xia W, Chen C-T, Li L-Y et al. Degradation of Mcl-1 by β-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol 2007; 27: 4006–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Gallegos JR, Gu Q, Huang Y, Li J, Jin Y et al. SAG/ROC-SCFβ-TrCP E3 ubiquitin ligase promotes pro-caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia 2006; 8: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 2004; 6: 931–940. [DOI] [PubMed] [Google Scholar]
- Tsai WB, Chung YM, Zou Y, Park SH, Xu Z, Nakayama K et al. Inhibition of FOXO3 tumor suppressor function by betaTrCP1 through ubiquitin-mediated degradation in a tumor mouse model. PLoS One 2010; 5: e11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Lu X, Wang G, Lan Z, Liao W, Li J et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 2017; 542: 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 2013; 14: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 2008; 8: 83–93. [DOI] [PubMed] [Google Scholar]
- Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 2004; 303: 1374–1378. [DOI] [PubMed] [Google Scholar]
- Al-Shamsi HO, Jones J, Fahmawi Y, Dahbour I, Tabash A, Abdel-Wahab R et al. Molecular spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53, and APC somatic gene mutations in Arab patients with colorectal cancer: determination of frequency and distribution pattern. J Gastrointest Oncol 2016; 7: 882–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki M, Yamaguchi T, Iijima T, Takahashi K, Matsumoto H, Mori T. Somatic mutations of the CDC4 (FBXW7) gene in hereditary colorectal tumors. Oncology 2009; 76: 430–434. [DOI] [PubMed] [Google Scholar]
- Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, Sarkar FH et al. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett 2012; 586: 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis H, Lewis A, Behrens A, Tomlinson I. Investigation of the atypical FBXW7 mutation spectrum in human tumours by conditional expression of a heterozygous propellor tip missense allele in the mouse intestines. Gut 2014; 63: 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology 2010; 139: 929–941. [DOI] [PubMed] [Google Scholar]
- Grim JE, Knoblaugh SE, Guthrie KA, Hagar A, Swanger J, Hespelt J et al. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol Cell Biol 2012; 32: 2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minella AC, Grim JE, Welcker M, Clurman BE. p53 and SCFFbw7 cooperatively restrain cyclin E-associated genome instability. Oncogene 2007; 26: 6948–6953. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J 2000; 19: 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Mo X, Yu J, Huang S, Huang Z, Gao L. Interference of Skp2 effectively inhibits the development and metastasis of colon carcinoma. Mol Med Rep 2014; 10: 1129–1135. [DOI] [PubMed] [Google Scholar]
- Uddin S, Ahmed M, Bavi P, El-Sayed R, Al-Sanea N, AbdulJabbar A et al. Bortezomib (Velcade) induces p27Kip1 expression through S-phase kinase protein 2 degradation in colorectal cancer. Cancer Res 2008; 68: 3379–3388. [DOI] [PubMed] [Google Scholar]
- Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M et al. The cell-cycle regulatory protein Cks1 is required for SCFSkp2-mediated ubiquitinylation of p27. Nat Cell Biol 2001; 3: 321–324. [DOI] [PubMed] [Google Scholar]
- Bashir T, Pagano M. Cdk1: the dominant sibling of Cdk2. Nat Cell Biol 2005; 7: 779–781. [DOI] [PubMed] [Google Scholar]
- Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer 2005; 103: 1336–1346. [DOI] [PubMed] [Google Scholar]
- Bochis OV, Irimie A, Pichler M, Berindan-Neagoe I. The role of Skp2 and its substrate CDKN1B (p27) in colorectal cancer. J Gastrointest Liver Dis 2015; 24: 225–234. [DOI] [PubMed] [Google Scholar]
- Peters J-M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 2006; 7: 644–656. [DOI] [PubMed] [Google Scholar]
- Wu W-j, Hu K-s, Wang D-s, Zeng Z-l, Zhang D-s, Chen D-l et al. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J Transl Med 2013; 11: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro T, Tanikawa C, Furukawa Y, Katagiri T, Nakamura Y, Matsuda K. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene 2008; 27: 1562–1571. [DOI] [PubMed] [Google Scholar]
- Hadjihannas MV, Bernkopf DB, Bruckner M, Behrens J. Cell cycle control of Wnt/beta-catenin signalling by conductin/axin2 through CDC20. EMBO Rep 2012; 13: 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pećina-Šlaus N. Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int 2003; 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards FM, McKee SA, Rajpar MH, Cole TR, Evans DGR, Jankowski JA et al. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum Mol Genet 1999; 8: 607–610. [DOI] [PubMed] [Google Scholar]
- Fujita T, Liu W, Doihara H, Wan Y. Regulation of Skp2-p27 axis by the Cdh1/anaphase-promoting complex pathway in colorectal tumorigenesis. Am J Pathol 2008; 173: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli A, Zlobec I, Minoo P, Baker K, Tornillo L, Terracciano L et al. Prognostic significance of the wnt signalling pathway molecules APC, β‐catenin and E‐cadherin in colorectal cancer-a tissue microarray‐based analysis. Histopathology 2007; 50: 453–464. [DOI] [PubMed] [Google Scholar]
- Kedrin D, Gala MK. Genetics of the serrated pathway to colorectal cancer. Clin Transl Gastroenterol 2015; 6: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebov OK, Rodriguez LM, Soballe P, DeNobile J, Cliatt J, Nakahara K et al. Gene expression patterns distinguish colonoscopically isolated human aberrant crypt foci from normal colonic mucosa. Cancer Epidemiol Biomarkers Prev 2006; 15: 2253–2262. [DOI] [PubMed] [Google Scholar]
- Sah N, Khan Z, Khan G, Bisen P. Structural, functional and therapeutic biology of survivin. Cancer Lett 2006; 244: 164–171. [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Farma JM, Coppola D, Hakam A, Fulp WJ, Chen DT et al. Expression of the antiapoptotic protein survivin in colon cancer. Clin Colorectal Cancer 2011; 10: 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-Y, Yu C-P, Lin C-K, Nieh S, Hsu K-F, Chiang H et al. Expression of survivin and cortactin in colorectal adenocarcinoma: association with clinicopathological parameters. Dis Markers 2009; 26: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov 2014; 13: 513–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Hao H-X, Growney JD, Woolfenden S, Bottiglio C, Ng N et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA 2013; 110: 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, van Es JH, van den Born M, Clevers H. Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43;Znrf3-mutant neoplasia. Proc Natl Acad Sci USA 2015; 112: 7548–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhart Z, Pavlovic Z, Chandrashekhar M, Hart T, Wang X, Zhang X et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat Med 2017; 23: 60–68. [DOI] [PubMed] [Google Scholar]
- Schütte M, Risch T, Abdavi-Azar N, Boehnke K, Schumacher D, Keil M et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat Commun 2017; 8: 14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, Tan S, Zou F, Yu J, Zhang L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene 2017; 36: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki M, Mimori K, Ishii H, Yokobori T, Takatsuno Y, Sato T et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. Int J Cancer 2010; 126: 1828–1837. [DOI] [PubMed] [Google Scholar]
- Kozuch PS, Rocha-Lima CM, Dragovich T, Hochster H, O'Neil BH, Atiq OT et al. Bortezomib with or without irinotecan in relapsed or refractory colorectal cancer: results from a randomized phase II study. J Clin Oncol 2008; 26: 2320–2326. [DOI] [PubMed] [Google Scholar]
- Zhang X-W, Yan X-J, Zhou Z-R, Yang F-F, Wu Z-Y, Sun H-B et al. Arsenic trioxide controls the fate of the PML-RARα oncoprotein by directly binding PML. Science 2010; 328: 240–243. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 2008; 10: 538–546. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 2008; 10: 547–555. [DOI] [PubMed] [Google Scholar]
- Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med 2007; 13: 23–31. [DOI] [PubMed] [Google Scholar]
- Gu L, Zhang H, Liu T, Zhou S, Du Y, Xiong J et al. Discovery of dual inhibitors of MDM2 and XIAP for cancer treatment. Cancer Cell 2016; 30: 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschall AV, Zimmerman MA, Torres CM, Yang D, Chen MR, Li X et al. Ceramide targets xIAP and cIAP1 to sensitize metastatic colon and breast cancer cells to apoptosis induction to suppress tumor progression. BMC Cancer 2014; 14: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe GL, Redd PS, Paschall AV, Lu C, Gu L, Cai H et al. Ceramide mediates FasL-induced caspase 8 activation in colon carcinoma cells to enhance FasL-induced cytotoxicity by tumor-specific cytotoxic T lymphocytes. Sci Rep 2016; 6: 30816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Chen J, Ouyang N, Su F, Wang M, Heemann U. Soluble Fas ligand released by colon adenocarcinoma cells induces host lymphocyte apoptosis: an active mode of immune evasion in colon cancer. Br J cancer 2001; 85: 1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res 2011; 71: 3042–3051. [DOI] [PubMed] [Google Scholar]
- Wan J, Zhu J, Li G, Zhang Z. Radiosensitization of human colorectal cancer cells by MLN4924: an inhibitor of NEDD8-activating enzyme. Technol Cancer Res Treat 2016; 15: 527–534. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009; 458: 732–736. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 2011; 145: 1075–1087. [DOI] [PubMed] [Google Scholar]
- Kotz J. Celgene skips SKP2. SciBX 2011; 4: 1–2. [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 2010; 18: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidou M, Curtis NL, Bolanos-Garcia VM. Cdc20: at the crossroads between chromosome segregation and mitotic exit. Trends Biochem Sci 2017; 42: 193–205. [DOI] [PubMed] [Google Scholar]
- Sackton KL, Dimova N, Zeng X, Tian W, Zhang M, Sackton TB et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 2014; 514: 646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street I, Lackovic K, Haupt Y, Monahan B, Wolyniec K, Haupt S et al. 1039 A high throughput screening platform for the identification of small molecule inhibitors of the E3 ligase E6AP. Eur J Cancer 2012; 48: S250. [Google Scholar]