Abstract
Most anticancer therapies to date focus on druggable features of tumor epithelia. Despite the increasing repertoire of treatment options, patient responses remain varied. Moreover, tumor resistance and relapse remain persistent clinical challenges. These observations imply an incomplete understanding of tumor heterogeneity. The tumor microenvironment is a major determinant of disease progression and therapy outcome. Cancer-associated fibroblasts (CAFs) are the dominant cell type within the reactive stroma of tumors. They orchestrate paracrine pro-tumorigenic signaling with adjacent tumor cells, thus exacerbating the hallmarks of cancer and accelerating tumor malignancy. Although CAF-derived soluble factors have been investigated for tumor stroma-directed therapy, the underlying transcriptional programs that enable the oncogenic functions of CAFs remain poorly understood. Nuclear receptors (NRs), a large family of ligand-responsive transcription factors, are pharmacologically viable targets for the suppression of CAF-facilitated oncogenesis. In this study, we defined the expression profiles of NRs in CAFs from clinical cutaneous squamous cell carcinoma (SCC) biopsies. We further identified a cluster of driver NRs in CAFs as important modifiers of CAF function with profound influence on cancer cell invasiveness, proliferation, drug resistance, energy metabolism and oxidative stress status. Importantly, guided by the NR profile of CAFs, retinoic acid receptor β and androgen receptor antagonists were identified for concurrent therapy with cisplatin, resulting in the inhibition of chemoresistance in recurred SCC:CAF xenografts. Our work demonstrates that treatments targeting both the tumor epithelia and the surrounding CAFs can extend the efficacy of conventional chemotherapy.
Introduction
The tumor microenvironment consists of a variety of stromal cells and a fibrotic matrix that surround and support malignant epithelia.1, 2 The interactions among the various components of the tumor microenvironment, mediated largely by cytokines and growth factors, are significant. Tumor epithelia can change the nature of the microenvironment, and conversely, the microenvironment can affect how a tumor grows and spreads.3, 4 Furthermore, tumor stroma co-evolution further disrupts tissue organization,5, 6 and the resultant loss of organ homeostasis creates a feed-forward reaction permissive to tumor aggressiveness and malignancy.4, 7 Despite this, many conventional cancer treatments are designed around druggable features of tumor epithelia, ignoring the supportive role of stromal cells. The diversity of patient outcomes from such treatments not only suggests that rapid resistance occurs, but also highlights an incomplete understanding of the tumor microenvironment.8, 9
As the most abundant cell population in the tumor stroma, cancer-associated fibroblasts (CAFs) are a potent source of growth factors, extracellular matrix components, matrix remodeling enzymes, inflammatory cytokines and reactive oxygen species (ROS). Hence, CAFs create a microenvironment that promotes proliferation, invasiveness, oxidative stress, aberrant metabolism, immune evasion and therapy resistance of tumors. Although CAFs have been well characterized by their expression of alpha-smooth muscle actin,10 fibroblast (FIB) activation protein,11 platelet-derived growth factor receptors,12 asporin13 and collagen 11α1,14 the underlying transcriptional programs enabling the pro-oncogenic functions of CAFs remain poorly understood. Moreover, whereas transcription factor signaling nodes control many cellular behaviors, most transcription factors cannot be directly modulated by chemical drugs, and are considered poor pharmacological targets.15, 16
Nuclear hormone receptors (NRs) represent a unique class of transcription factors that regulate gene expression under the strict control of endogenous or synthetic ligands.3, 17 In humans, the 48 known NRs play numerous roles in development, physiology and pathology. Thus, ligands of NRs have the potential to modulate the cytokine profile of CAFs, leading to tumor suppression or tumor sensitization to conventional chemotherapy. However, the expression of NRs in CAFs from squamous cell carcinoma (SCC) tumors is unknown, and their non-redundant roles in SCC progression and chemoresistance is unclear.
As the primary experimental system to explore CAF NR-directed therapy, we defined an NR profile for CAFs from patients diagnosed with cutaneous SCC. Guided by this expression profile, the genetic and pharmacological targeting of specific driver NRs in CAFs diminished SCC invasiveness, proliferation, drug resistance, energy metabolism and oxidative stress status. Furthermore, primary and recurred xenograft tumor growth was attenuated by a combination treatment with NR ligands and cisplatin, in part due to reduced chemoresistance. Our findings suggest that NR-directed ligands that have successfully treated other pathologies such as inflammation, dyslipidemia and diabetes, may be repurposed as concurrent treatments to conventional anticancer chemotherapeutics.
Results
NRs are differentially expressed between CAFs and normal FIBs
Paired samples of CAFs and peri-tumoral FIBs from archived SCC biopsies (n=10) were microdissected (Figure 1a). RNA was then extracted and profiled for the expression of NR transcripts. Among the 26 NR transcripts detected, 18 were upregulated (more than twofold), five remained unchanged and three were downregulated (more than twofold) in CAFs compared with FIBs. Thirteen of the 21 differentially expressed NRs were druggable targets (Figure 1b).
Figure 1.
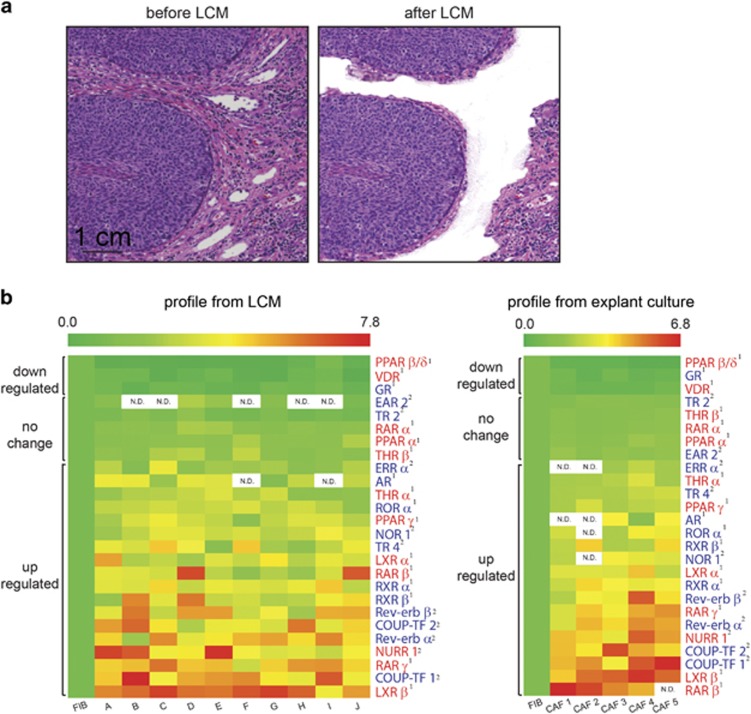
NRs are differentially expressed between CAFs and normal FIBs. (a) Representative images of hematoxylin- and eosin-stained sections of archived patient SCC biopsies before (left) and after (right) laser capture microdissection. CAFs within the tumor stroma directly bordering tumor epithelia were selectively excised while excluding obvious immune or vascular structures. Scale bar: 1 cm. (b) Relative expression of detectable NR transcripts in CAFs derived from metastatic and non-metastatic patient SCC biopsies (n=10), compared against expression levels in matched pairs of peri-normal FIBs by RT-qPCR (left). Profiling was repeated using CAFs explanted from human SCC tumors (n=5) and cultured in vitro (right). Expression values in CAFs are relative to that in normal FIBs, The first column in the heatmap represents the expression of NRs from five different FIB controls. NRs that form heterodimers with retinoid X receptors (RXRs) are labeled in red, while those that form homodimers are labeled in blue. Superscript numbers distinguish NRs with known ligands (1) from orphan NRs (2). Color scales: green=downregulated, red=upregulated. ‘N.D.’ denotes that the gene was not detected by RT-qPCR.
Peroxisome proliferator-activated receptor (PPAR) β expression was downregulated to the greatest extent in CAFs, followed by the vitamin D receptor (VDR) and the glucocorticoid receptor (GR). Notably, RXRα and RXRβ were upregulated in CAFs compared with FIBs. As RXRs form heterodimeric partners with many NRs, we stratified the 21 NRs according to whether they heterodimerized with the RXRs. Nine of these 21 NRs were heterodimeric partners of RXRs (excluding the RXRs themselves) (Figure 1b).
To further study CAFs in vitro, we first confirmed whether the NR signature of SCC CAFs remains stable when CAFs are removed from their native environment for culture. To this end, we performed NR profiling on patient CAFs explanted from human SCC tumors. Explanted CAFs were subjected to FACS analysis to verify their expression of vimentin and alpha-smooth muscle actin (Supplementary Figure S1). A highly similar NR profile was observed between the microdissected CAFs and the explanted CAFs (Figure 1b), indicating that the NR profile of SCC CAFs is retained during in vitro culture.
The SCC transcriptome is modified by changes in the CAF NR profile
We hypothesized that by disrupting the NR status in CAFs, their paracrine interaction with cancer cells becomes compromised, leading to changes in the cancer cell transcriptome that inhibit SCC aggressiveness. Guided by the NR profile of SCC CAFs, the individual knockdown or overexpression of specific NRs in CAFs was performed. Briefly, if the expression of an NR was downregulated in CAFs relative to FIBs, that NR was overexpressed (NR-OE). Conversely, an NR that was highly expressed in CAFs relative to FIBs was subjected to siRNA knockdown (NR-KD). NR-OE or NR-KD was verified with RT-qPCR and immunoblot (Supplementary Fig S2). NR-OE or NR-KD did not affect CAF viability (Supplementary Figure S3a). NR-OE/KD-CAFs were subsequently co-cultured with the malignant A-5RT3 SCC cells for 48 h. The A-5RT3 cells were then isolated for RNA extraction and microarray analysis (Figure 2a).
Figure 2.
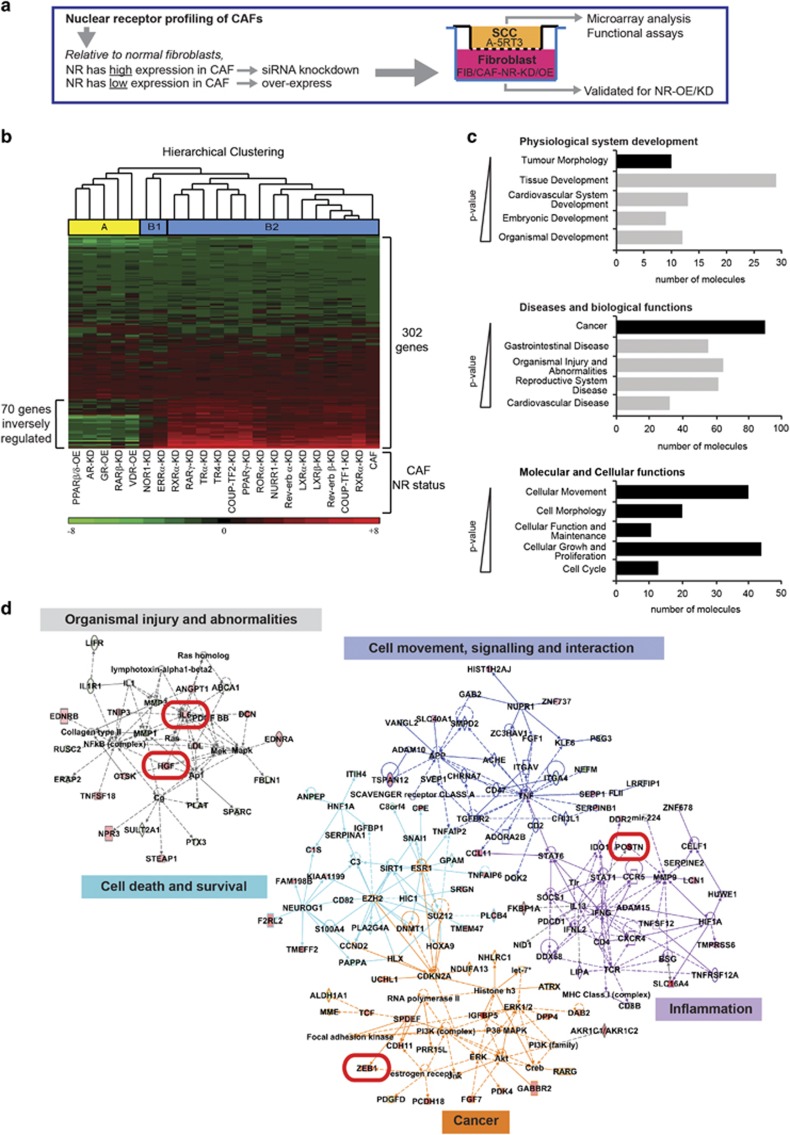
The SCC transcriptome is modified by changes in the CAF NR profile. (a) Experimental approach for the overexpression or knockdown of specific NRs in CAFs. FIBs, CAFs and NR-KD/OE-CAFs were co-cultured with A-5RT3 cells in a transwell setup for 48 h. A-5RT3 cells were isolated for microarray and functional analyses. NR-KD/OE-CAFs were isolated for validation of NR knockdown or overexpression. (b) Heatmap and hierarchical clustering of 302 genes differentially expressed by >1.5-fold between A-5RT3:CAF and A-5RT3:FIB co-culture samples. Total RNA from A-5RT3 cells after co-culture with FIBs, CAFs or NR-KD/OE-CAFs was amplified using the Applause WT-Amp System (Nugen, UK) and labeled with the Encore Biotin Module (Nugen, UK) for loading onto GeneChip Human Gene 1.0 ST Array (Affymetrix) chips. Raw microarray files were imported into Partek Genomic Suite (v6.6) and normalized using robust multiarray averaging (RMA). ANOVA was performed for all A-5RT3:CAF co-culture samples with contrast set against the A-5RT3:FIB sample. Color scales: green=downregulated; red=upregulated. Cluster A (yellow) identifies NR-KD/OE-CAFs that distinctly modified A-5RT3 gene expression. Seven genes that were upregulated in the A-5RT3 cells by co-culture with CAFs were instead downregulated (that is, inversely regulated) when the A-5RT3 cells were co-cultured with the NR-KD/OE-CAFs belonging to cluster A. (c) Ingenuity Pathway Core Analysis of gene ontologies enriched by the 302 gene set. Gene ontologies were ranked in decreasing order of the strength of association based on their P-value. A higher association is indicated by a smaller P-value. Only the top five ontologies with P-values <0.05 were included. (d) Top biological networks associated with the 302 gene set. Relationships between genes are color-coded according to their assigned biological functions. Key mediators of tumor malignancy, namely, HGF, ZEB, IL6 and POSTN, are boxed in red.
The microarray analysis highlighted 302 genes differentially expressed by >1.5-fold between the A-5RT3 cells from the A-5RT3:CAF and A-5RT3:FIB co-cultures (Figure 2b). Using the Ingenuity Pathway Analysis software for deeper analysis, the ontology term ‘Tumor morphology’ was enriched (P<0.024) and ‘Cancer’ was identified as the topmost-enriched pathology (P<0.030).
Next, we identified molecular and cellular functions in A-5RT3 cells that were altered by co-culture with CAFs. ‘Cellular movement’ (P<0.029), ‘Cell morphology’ (P<0.024), ‘Cellular Function and Maintenance’ (P<0.024), ‘Cellular Growth and Proliferation’ (P<0.027) and ‘Cell Cycle’ (P<0.024) were the most strongly associated gene functions (Figure 2c). The top networks in our analysis included ‘Organismal injury and abnormalities’ (score=37), ‘Cancer’ (score=34), ‘Cell death and survival’ (score=22), ‘Cell movement, signaling and interaction’ (score=18) and ‘Inflammation’ (score=15) (Figure 2d), reflecting a wound-like environment created by co-culture with CAFs.
To understand how the individual KD/OE of NRs in CAFs affected the A-5RT3 transcriptome, the expression of these 302 genes was examined in all A-5RT3:NR-KD/OE-CAF co-cultures. Hierarchical clustering revealed two major clusters; Cluster A: PPARβ/δ-OE-CAFs, androgen receptor (AR)-KD-CAFs, GR-OE-CAFs, retinoic acid receptor (RARβ)-KD-CAFs, VDR-OE-CAFs, and Cluster B containing two sub-clusters, B1 and B2. Cluster B2 gene signatures were highly similar to that of A-5RT3:CAF co-culture, indicating a functional redundancy among these CAF NRs, that is, the disruption of their individual expression was mutually compensated, thus limiting their impact on the SCC transcriptome. Although Cluster B1 gene signatures differed from the A-5RT3:CAF gene signature, these NRs are orphan receptors, making them poor pharmacological targets. In contrast, Cluster A alterations to the NR status of CAFs clearly modified the transcriptome of A-5RT3 cells in co-culture; 70 genes were inversely regulated (that is, downregulated in A-5RT3 from Cluster A co-cultures as opposed to upregulated in A-5RT3:CAF co-culture) (Figure 2b). Importantly, genes associated with tumor malignancy, such as hepatocyte growth factor (HGF), zinc-finger E-box-binding 1 (ZEB1), periostin (POSTN) and interleukin (IL)-6, were strongly upregulated in A-5RT3:CAF co-culture, whereas in A-5RT3 cells co-cultured with Cluster A NR-KD/OE-CAFs, these same genes were downregulated (Figures 2c and d). Thus, the KD/OE of cluster A NRs in CAFs are likely to attenuate SCC malignancy.
Genetic modulation of the NR expression of CAFs inhibits SCC malignancy and reduces chemoresistance
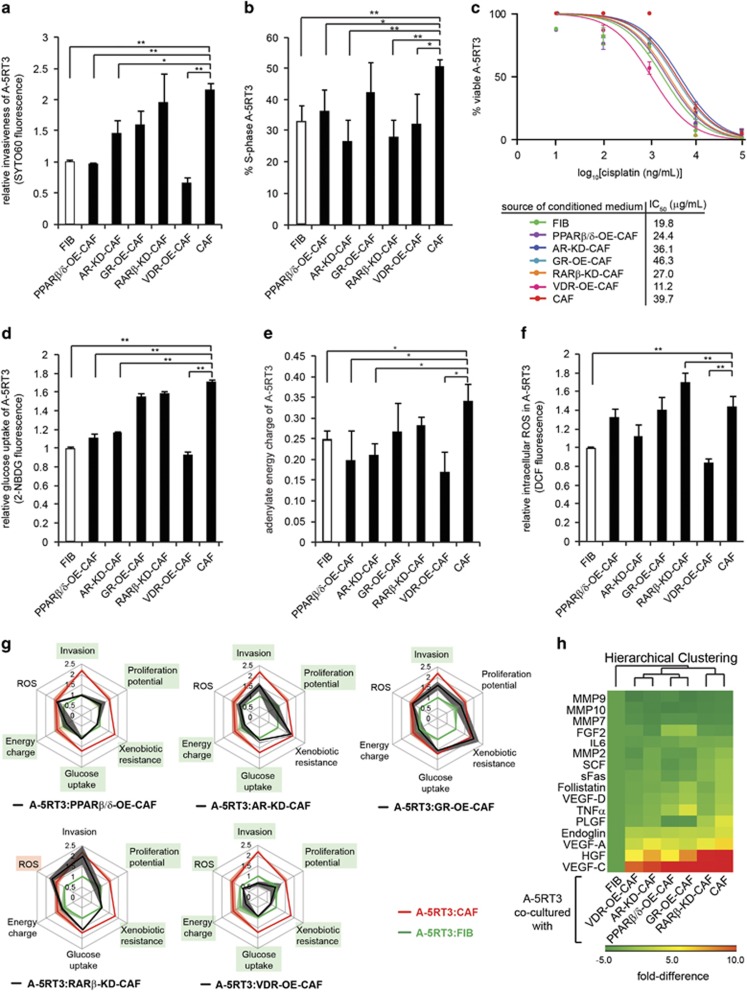
Based on the microarray analysis, the malignant SCC phenotypes of invasion, proliferation, drug resistance, energy and ROS metabolism were influenced by the CAF NR status, especially NRs belonging to Cluster A. We examined the invasiveness of A-5RT3 cells using the transwell invasion assay. FIBs, CAFs or NR-OE/KD-CAFs seeded in the lower chamber provided the chemotactic gradient. CAFs potentiated the invasiveness of A-5RT3 cells by twofold compared with FIBs. While RARβ-KD-CAFs did not significantly reduce A-5RT3 invasiveness, PPARβ/δ-OE-CAFs and VDR-OE-CAFs reduced A-5RT3 invasiveness by 75% and 50%, respectively, compared with A-5RT3:CAF co-culture. AR-KD-CAFs and GR-OE-CAFs also inhibited A-5RT3 cell invasion by 27.5% and 23.0%, respectively (Figure 3a).
Figure 3.
Genetic modulation of CAF NR expression inhibits SCC malignancy and reduces chemoresistance. (a) Relative measurements of A-5RT3 cell invasiveness when co-cultured with FIBs, CAFs or NR-KD/OE-CAFs. A total of 4 × 104 A-5RT3 cells were seeded on 6.5 mm Boyden Chamber inserts with 8 μm pores coated with 50 μg/ml type I rat-tail collagen in acetic acid. A total of 1.2 × 105 FIBs, CAFs or NR-KD/OE-CAFs were seeded in the lower chamber. Complete medium was replaced by serum-free defined medium at the start of the invasion assay, with cells in the lower chamber conditioning the medium over 48 h to provide the chemotactic gradient. Invaded A-5RT3 cells on the bottom of the inserts were stained with the infrared dye, SYTO60 (Life Technologies, USA). SYTO60 is cell permeant and fluoresces at 680 nm only upon binding to cellular DNA. SYTO60 signal was read using the Odyssey CLx infrared imaging system (LI-COR Biosciences, USA). (b) A-5RT3 cells (4 × 104 per 6.5 mm insert) were co-cultured with FIBs, CAFs or NR-KD/OE-CAFs (1.2 × 105 per 24 well) for 48 h. The percentage of A-5RT3 cells in the S-phase of the cell cycle was identified by BrdU incorporation into DNA, followed by FACS analysis. (c) Dose–response curves for the apoptosis of A-5RT3 cells cultured in CM from FIBs, CAFs or NR-KD/OE- CAFs and subjected to the indicated concentrations of cisplatin for 16 h. The 48 h CM of FIBs, CAFs and NR-KD/OE-CAFs was collected and passed through a 0.2 μm filter to remove debris before being added to A-5RT3 cells. A total of 10, 100, 1000, 10000 and 100000 ng/ml of cisplatin was added to A-5RT3 in CM for 16 h of treatment. Thereafter, apoptosis was measured using Annexin-V and propidium iodide staining. IC50 values were calculated using the graphing software, Prism. (d) Relative glucose uptake by A-5RT3 co-cultured with FIB, CAF or NR-KD/OE-CAFs for 48 h. A total of 10 μM of the fluorescent glucose analog, 2-NBDG, was added to cells in glucose-, serum- and phenol red-free DMEM and incubated at 37 °C, 5% CO2 for 30 min. A-5RT3 cells were subsequently rinsed and isolated for FACS analysis. (e) Relative adenylate energy charge of A-5RT3 cells co-cultured with FIB, CAF or NR-KD/OE-CAFs for 48 h. Nucleotides standards and samples (Sigma Aldrich, USA) were prepared in milli-Q water and resolved on a Shimadzu LC-20AD series Quaternary Gradient HPLC system with a photodiode array detector using a Hypersil ODS 150 mm × 4.6 mm I.D. C18 reverse-phase column with 3 μm particle size and 5 μm Hypersil ODS C18 guard column (Thermo Scientific, USA). Separation of nucleotides was carried out with a mobile phase containing 60 mmol/l of KH2PO4, 0.45 mmol/l of tetrabutylammonium bromide (TBAB) and 1.26 mol/l of acetonitrile, pH 3.20. The retention times of the individual nucleotides were matched to those of the respective nucleotide standards. The areas under the peaks that corresponded to the respective nucleotides were tabulated, and energy charge was calculated using the following formula: Energy charge=([ATP]+0.5[ADP])/([ATP]+[ADP] + [AMP]). (f) Relative intracellular ROS production by A-5RT3 co-cultured with FIBs, CAFs or NR-KD/OE-CAFs for 48 h, measured using 10 μM 5-(and 6-) chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester fluorophores (CM-H2DCFDA) (Molecular Probes, USA) and FACS analysis. (g) Multi-parametric evaluation of CAF NR effects on SCC. Measured parameters from A-5RT3 cells co-cultured with FIBs (green line), CAFs (red line) and NR-KD/OE-CAFs (black line) are superimposed. Shading represents the standard error of each parameter. A-5RT3 parameters significantly altered by co-culture with NR-KD/OE-CAFs are boxed in green. (h) Heatmap showing the relative quantities of analytes detected in the CM from A-5RT3:FIB, A-5RT3:CAF or A-5RT3:NR-KD/OE-CAF co-cultures. Color scales: green=downregulated; red=upregulated. Values represent means±s.d. from n=3 independent experiments. *P<0.05, **P<0.01.
Next, we examined the proliferation of A-5RT3 cells co-cultured with FIBs, CAFs and NR-OE/KD-CAFs. Bromodeoxyuridine incubation followed by FACS analysis revealed that A-5RT3:CAFs co-culture had increased proliferation when compared with A-5RT3:FIB co-culture (53.4±2.4% vs 33.5±5.1% in S-phase, respectively) (Figure 3b). Except for GR-OE-CAFs, all other NR-OE/KD-CAFs inhibited A-5RT3 cell proliferation by ⩾14% (Figure 3b).
Although cisplatin is common in SCC chemotherapy, drug resistance arises quickly. Our microarray analysis suggested that chemoresistance is reinforced by the tumor stroma. Conditioned medium (CM) from CAFs conferred chemoprotective effects on A-5RT3 tumor cells treated with cisplatin, demonstrated by a twofold increase in the IC50 dose of cisplatin compared with A-5RT3 cells in FIB CM (FIB CM IC50: 19.8 μg/ml vs, CAF CM IC50: 39.7 μg/ml). IC50 is defined as the cisplatin dose required to reduce A-5RT3 survival to 50% of the total cell population. GR-OE-CAF CM increased the IC50 compared with CAF CM, whereas AR-KD-CAF CM did not significantly change the IC50. In contrast, VDR-OE-CAF CM abrogated CAF-mediated chemoresistance to the greatest extent (IC50: 11.2 μg/ml), followed by PPARβ/δ-OE-CAF CM (24.4 μg/ml) and RARβ-KD-CAF CM (27 μg/ml) (Figure 3c).
Altered energy metabolism is another cancer hallmark. We observed that A-5RT3:CAF co-culture displayed a 60% increase in glucose uptake with a concomitant 40% elevation of adenylate energy charge compared with A-5RT3:FIB co-culture (Figures 3d and e). Although GR-OE-CAFs and RAR-KD-CAFs did not significantly reduce A-5RT3 cell glucose uptake and energy charge, PPARβ/δ-OE-, VDR-OE- and AR-KD-CAFs inhibited glucose uptake by up to 50% (Figure 3e) and lowered the energy charge of the co-cultured A-5RT3 cells (Figure 3e). We also assessed ROS levels in our co-culture systems. A-5RT3:CAFs co-culture displayed a 40% increase in intracellular ROS production relative to A-5RT3:FIB co-culture, evidence of exacerbated oxidative stress. ROS production in A-5RT3 cells was only attenuated by co-culture with VDR-OE-CAFs (Figure 3f).
To consolidate the antitumor efficacy of targeting specific CAF NRs, we overlaid the multi-parametric data from A-5RT3:NR-OE/KD-CAF co-cultures onto the data from A-5RT3:CAF and A-5RT3:FIB co-cultures (Figure 3g). We observed that CAFs potentiate all six aggressive behaviors of the co-cultured A-5RT3 cells (red line) compared with FIBs (green line). The attenuation of any of these six SCC behaviors by NR-OE/KD-CAFs is indicated by the black line approaching the green line. Our analysis suggests that PPARβ/δ, VDR and, to a lesser extent, AR and RARβ expression in CAFs elicit antitumor effects and reduce chemoresistance (Figure 3g). Interestingly, PPARβ/δ-OE-CAFs and VDR-OE-CAFs both inhibited mitochondrial dysfunction in co-cultured A-5RT3 (Supplementary Figure S3c), indicating that energy metabolism is important in CAF-mediated oncogenesis.
Finally, CM from the co-cultures was subjected to a multiplexed protein assay screening for 16 secreted pro-tumor factors. The paracrine signaling network between CAFs and SCC cells was indeed disrupted by the altered CAF NR status (Figure 3h). HGF and VEGF-C levels in the CM were attenuated to the greatest extents by NR-OE/KD-CAFs compared with CAFs.
Collectively, these results demonstrate that the transcriptome of SCC cancer cells is sensitive to the NR status of CAFs and suggest that SCC hallmarks are inhibited because oncogenic tumor stroma crosstalk is compromised.
Pharmacological modulation of CAF NRs inhibits SCC malignancy and reduces chemoresistance in vitro
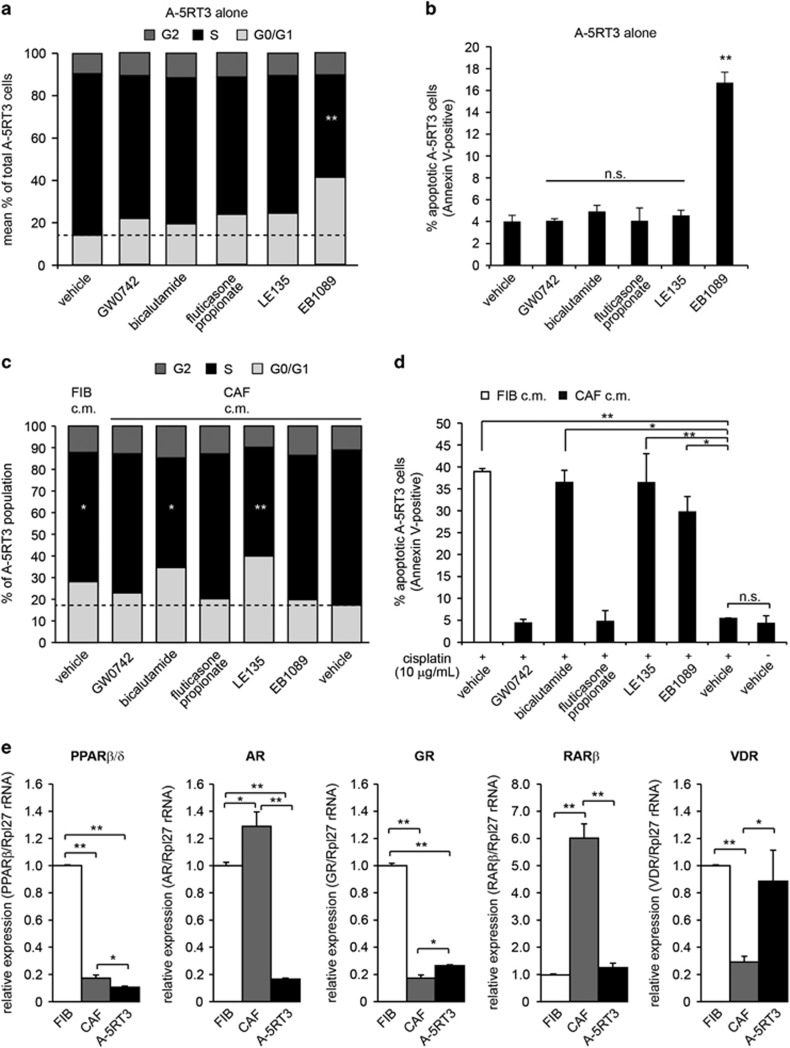
The KD/OE of specific NRs in CAFs resulted in the simultaneous attenuation of several cancer hallmarks. To test CAF NR-directed therapy in an experimental preclinical setting, we selected small molecule agonists of PPARβ/δ (GW0742), VDR (EB1089) and GR (fluticasone propionate), or antagonists of RARβ (LE135) and AR (bicalutamide) for evaluation. We first assessed whether these ligands had intrinsic anticancer activity. A-5RT3 cells were treated with each ligand at 5 μM for 24 h and analyzed by FACS for bromodeoxyuridine uptake and Annexin-PI staining. The results showed that only EB1089, the VDR agonist, significantly reduced A-5RT3 proliferation and increased A-5RT3 apoptosis (Figures 4a and b). Using A431 cells, another cell line representative of cutaneous SCC, we observed that only EB1089 treatment inhibited A431 cell proliferation, while only LE135 treatment promoted apoptosis of A431 cells (Supplementary Figures S4a, b).
Figure 4.
Pharmacological modulation of CAF NRs inhibits SCC malignancy and reduces chemoresistance in vitro. (a) Percentages of A-5RT3 cells in the G0/G1-, S- or G2-phases of the cell cycle when subjected to NR ligand treatments. Agonists of PPARβ/δ (GW0742), VDR (EB1089) and GR (fluticasone propionate), and antagonists of AR (bicalutamide) and RARβ (LE135) were used at a final concentration of 5 μM. (b) Percentages of A-5RT3 cells positive for Annexin-V staining when subjected to the indicated treatments. (c) Percentages of A-5RT3 cells in the G0/G1-, S- or G2-phases of the cell cycle after being cultured for 48 h in the CM from NR ligand-treated CAFs. (d) Percentages of A-5RT3 cells positive for Annexin-V staining. A total of 10 μg/ml cisplatin was added to A-5RT3 cells cultured in the CM of NR ligand-treated CAFs. After 16 h of cisplatin treatment, A-5RT3 cells were isolated for FACS analysis. (e) Relative mRNA expression of the indicated NRs in FIBs, CAFs and A-5RT3 cells. The ribosomal RNA gene Rpl27 was used as a housekeeping gene. Values are means±s.d. from n=3 independent experiments. n.s.: not significant, *P<0.05, **P<0.01.
To explore whether these NR ligands demonstrate anticancer properties via CAFs, we pre-treated CAFs with the ligands for 48 h. The CAFs were rinsed thoroughly to remove excess ligand, and fresh serum-free medium was added for conditioning. The 48-hour CM from pre-treated CAFs was collected and added to A-5RT3 cells. CAF CM potentiated A-5RT3 cell proliferation compared with FIBs CM. CM from GW0742 and fluticasone propionate pre-treated CAFs did not significantly alter A-5RT3 proliferation. Interestingly, A-5RT3 cells cultured in LE135 or bicalutamide pre-treated CAF CM showed reduced proliferation (Figure 4c). This was not observed when A-5RT3 cells were directly exposed to the ligands, suggesting a CAF-dependent effect. In contrast, CM from EB1089 pre-treated CAFs did not have any anti-proliferative effect, indicating that EB1089 acts directly on A-5RT3 cells to inhibit proliferation (Figure 4c). For A431 cells, the CM of GW0742 treated CAFs promoted cell proliferation (Supplementary Figure S4c). Notably, when the CM from the ligand-treated co-cultures was subjected to a multiplexed protein assay screening for 8 secreted pro-tumor factors, the co-cultures treated with a combination of LE135 and bicalutamide displayed the strongest suppression of pro-tumor paracrine activity as compared with vehicle-treated controls (Supplementary Figure S5).
Next, we explored whether our selected NR ligands could sensitize A-5RT3 cells to cisplatin. A-5RT3 cells were cultured in pre-treated CAF CM in the presence of 10 μg/ml cisplatin. Nearly 40% of A-5RT3 cells underwent apoptosis when cultured in FIB CM, compared with only 5% when cultured in CAF CM. Pre-treating CAFs with LE135, bicalutamide or EB1089 abrogated the CAF-enabled resistant phenotype in A-5RT3 cells (Figure 4d). On the other hand, we observed that the CM of CAFs pre-treated with the five NR ligands abrogated CAF-mediated chemoresistance to 5-fluorouracil in A431 cells, with LE135 pretreatment of CAFs exhibiting the most potent paracrine effect (Supplementary Figure S4d).
In the earlier NR-KD/OE-CAF experiments, VDR and PPARβ/δ demonstrated potent anticancer properties. However, RARβ and AR appeared as better pharmacological targets for CAF-directed therapy. Therefore, we tested whether this discrepancy could be explained by the relative expression of the NRs in A-5RT3 cells and CAFs. RT-qPCR revealed that VDR expression was higher in A-5RT3 cells than in CAFs (Figure 4e). Thus, A-5RT3 cells are more responsive to EB1089. Unlike VDR-OE-CAFs that demonstrated anticancer effects, pre-treating CAFs with EB1089 had limited impact because of the low endogenous expression of VDR. Conversely, RARβ and AR expression was much higher in CAFs than A-5RT3 cells (Figure 4e). In A431 cells, AR and RARβ expression is also virtually absent (Supplementary Figure S4e). Hence, the pretreatment of CAFs with RARβ and AR antagonists, which bind to the abundant levels of their cognate NRs in CAFs, resulted in a non-cell autonomous inhibition of SCC aggressiveness and chemoresistance (Figures 4c and d). Collectively, our in vitro data suggest that LE135 and bicalutamide, and thus RARβ and AR, are potential NR candidates for CAF-directed therapy.
Cisplatin with LE135 and bicalutamide concurrent therapy inhibits SCC malignancy and reduces chemoresistance in vivo
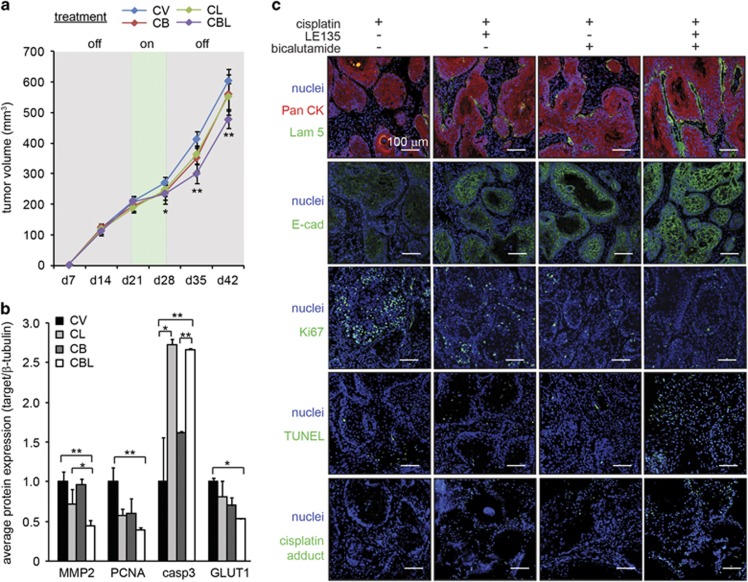
Based on in vitro data, a concurrent treatment using cisplatin with LE135 and/or bicalutamide was effective against SCC via CAFs. To test their efficacy in vivo, we first titrated the dose of cisplatin in immunodeficient NSG (NOD SCID gamma) mice bearing subcutaneous A-5RT3:CAF xenografts. An intraperitoneal dose of 2 mg/kg cisplatin administered twice (on weeks 3 and 4) was required to impede tumor growth (Supplementary Figure S5a). Although 10 mg/kg cisplatin reduced the tumor volumes, it was accompanied by proteinuria (Supplementary Figure S6a, b) which indicates nephrotoxicity, a common side effect of cisplatin chemotherapy. Therefore, 2 mg/kg cisplatin in combination with 5 mg/kg LE13518, 19 and 10 mg/kg bicalutamide20, 21 was intraperitoneally administered twice on weeks 3 and 4. Cisplatin in combination with LE135 (CL) or bicalutamide (CB) did not significantly improve treatment outcome compared with cisplatin alone treatment (CV). Significant delay in tumor growth was observed only when both LE135 and bicalutamide were used concurrently with cisplatin (CBL). However, tumor remission, defined as a reduction in tumor volume, was not achieved (Figure 5a).
Figure 5.
Concurrent therapies of cisplatin with LE135 and/or bicalutamide inhibit SCC malignancy and reduces chemoresistance in vivo. (a) Tumor volume measurements in mice implanted with A-5RT3:CAF xenograft tumors (n=6 per experiment condition, two tumors per mouse) receiving intraperitoneal injections of 2 mg/kg cisplatin, alone (CV) or in combination with 5 mg/kg LE135 (CL), 10 mg/kg bicalutamide (CB) or both (CBL). The treatments were administered on two consecutive weeks (d21 and d28, treatment: ‘on’). (b) Densitometry measurements from zymogram for MMP2 and immunoblots of PCNA, caspase 3 and GLUT1 in mice receiving intraperitoneal injections of NR ligands and cisplatin. β-Tubulin from the same samples was used as a loading and transfer control. Values are means±s.d. from n=3 independent experiments. *P<0.05, **P<0.01. (c) Immunofluorescence staining for pan-cytokeratin (PanCK), laminin 5 (Lam 5), E-cadherin (E-cad), Ki67, TUNEL and cisplatin adducts in the xenograft tumors of mice receiving intraperitoneal injections of NR ligands and cisplatin. Scale bar: 100 μm.
Gelatin zymography and immunoblot analysis of tumor lysates revealed that the properties of the A-5RT3:CAF tumor xenografts were altered by the treatments. MMP2, PCNA and GLUT1 expression was significantly reduced only by CBL therapy, suggesting that the full combination of drugs was required to impair tumor invasiveness, proliferation and energy metabolism, respectively. Caspase 3 expression was also strongly elevated by the CL and CBL treatments (Figure 5b, Supplementary Figure S6c). Immunofluorescence staining further showed that laminin 5 expression, which is inversely associated with tumor invasiveness, was only restored by CBL treatment (Figure 5c). The staining of the epithelial marker, E-cad (E-cadherin), was more intense and defined at cell:cell boundaries upon CL, CB and CBL treatments compared with CV. The proliferation marker Ki67 was reduced by CL, CB and CBL treatment compared with CV. Moreover, terminal deoxynucleotidyltransferase dUTP nick end labeling assay (TUNEL) showed that the number of apoptotic tumor cells was increased only by CBL therapy, corresponding with increased intratumoral cisplatin:DNA adducts (Figure 5c).
Despite the attenuation of tumor aggressiveness, tumor remission did not occur, likely owing to insufficient intratumoral accumulation of cisplatin. Furthermore, the inflection point at week 4 in the tumor growth curve suggested that cisplatin resistance was developing (Figure 5a). Hence, the injection of cisplatin directly into tumors could improve treatment outcomes. As SCC tumors are located superficially, the intratumoral injection of cisplatin is feasible.
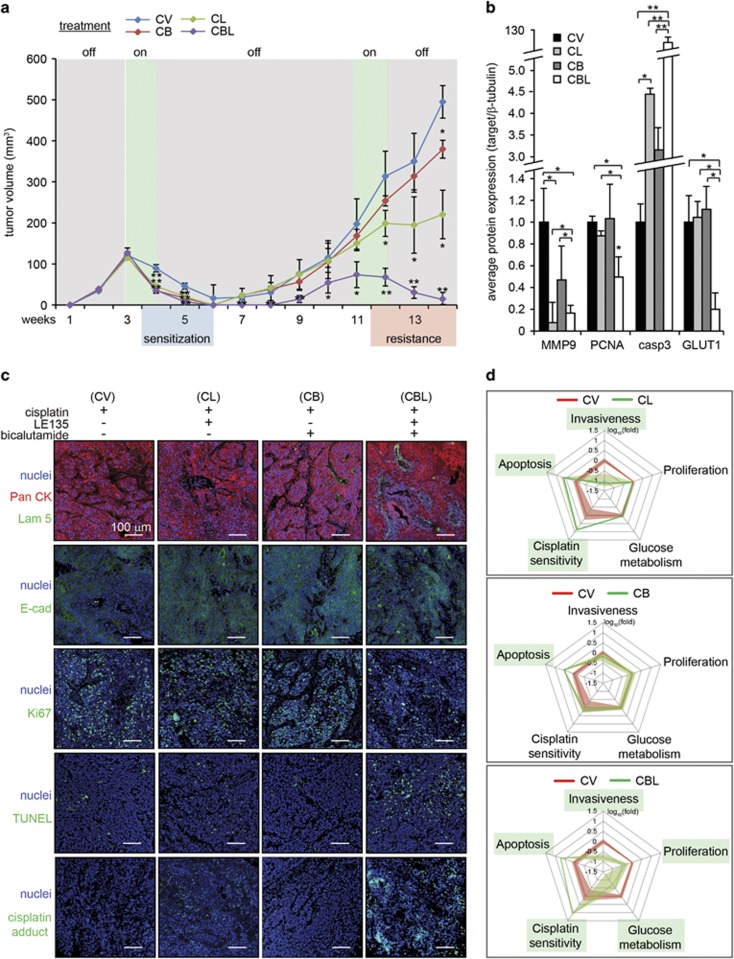
To assess the efficacy of this treatment route, we established tumor xenografts in another group of NSG mice. On weeks 3 and 4, when xenograft volumes approached 150 mm3, 2 mg/kg cisplatin was injected directly into the tumors, with 5 mg/kg LE135 and/or 10 mg/kg bicalutamide injected intraperitoneally as before, given their effect in earlier experiments. CL, CB and CBL treatments significantly reduced tumor volumes compared with CV treatment. Although treatment was discontinued beyond week 4, tumor remission was sustained until week 6. This suggested that the combined therapies had sensitized tumors to cisplatin.
To observe whether tumors would recur, the same mice were maintained even after tumors became impalpable (Figure 6a). Tumors reappeared on week 7 for mice that received CV, CL and CB therapies. Tumor recurrence was also observed in CBL-treated mice on week 9, suggesting the presence of residual disease after the first round of combinatorial therapy. These results presented an opportunity to study whether the surviving A-5RT3:CAF tumors had developed resistance. The tumors were allowed to grow until tumor volumes approached 150 mm3 before treatment was resumed (mice were treated on weeks 11 and 12). Recurred tumor xenografts resisted second-round CV and CB treatments; CL treatment impeded tumor growth without causing remission (Figure 6a). Notably, tumor xenografts responded optimally to second-round CBL treatment, becoming barely palpable by week 14 (Figure 6a).
Figure 6.
Intratumoral injection of cisplatin increases the efficacy of combination therapies. (a) Tumor volume measurements in mice implanted with A-5RT3:CAF xenograft tumors (n=6 per experiment condition, two tumors per mouse) receiving intratumoral injections of 2 mg/kg cisplatin, alone (CV) or in combination with intraperitoneal injections of 5 mg/kg LE135 (CL), or 10 mg/kg bicalutamide (CB) or both (CBL). The treatments were administered on weeks 3 and 4, withdrawn from weeks 5–10 and resumed on weeks 11 and 12. (b) Densitometry measurements from zymogram for MMP2 and immunoblots of PCNA, caspase 3 and GLUT1 in mice that received intraperitoneal injections of NR ligands and intratumoral injections of cisplatin. β-Tubulin from the same samples was used as a loading and transfer control. Values are means±s.d. from n=3 independent experiments. *P<0.05, **P<0.01. (c) Immunofluorescence staining for pan-cytokeratin (PanCK), laminin 5 (Lam 5), E-cadherin (E-cad), Ki67, TUNEL and cisplatin adducts in xenograft tumors of mice receiving intraperitoneal injections of NR ligands and intratumoral injections of cisplatin. Scale bar: 100 μm. (d) Multi-parametric evaluation of CAF NRs effect on SCC. Measured parameters from CV-treated tumors (red line) and combination therapies (CL, CB, CBL; green line) are superimposed. Shading represents the standard error of each parameter. Parameters significantly altered by combination therapies are boxed in green.
Gelatin zymography analysis of tumor lysates revealed that MMP9 expression was strongly repressed by CL and CBL treatments. PCNA and GLUT1 protein expression was only reduced by CBL treatment. Meanwhile, caspase 3 protein expression was upregulated fourfold, threefold and >100-fold by CL, CB and CBL treatments, respectively, compared with CV treatment (Figure 6b, Supplementary Figure S6d). Immunofluorescence staining also showed that CBL therapy was most effective in restoring laminin 5 expression, accompanied by intense and well-defined E-cad staining, reduced Ki67 staining and increased number of TUNEL-positive nuclei (Figure 6c). Importantly, the numerous cisplatin:DNA adducts in CBL-treated tumors suggested that CBL therapy played a role in reducing chemoresistance.
Finally, gelatin zymography, immunoblot (PCNA, GLUT1 and caspase 3) and immunofluorescence (cisplatin adducts) data representing SCC tumor invasiveness, proliferation, energy metabolism, apoptosis and cisplatin sensitivity, respectively, were consolidated into a multi-parametric chart of treatment efficacy. The red line represents tumor response to CV treatment which forms the baseline for comparison (Figure 6d). The green line represents tumor response to CL, CB or CBL therapy. Our data suggest that RARβ and AR antagonists are effective when concurrently administered with cisplatin chemotherapy.
To validate the efficacy of NR antagonists as an adjunctive treatment alongside another chemotherapy drug commonly used in SCC, we treated mice bearing A431:CAF tumor xenografts with 10mg/kg of 5-fluorouracil alone (FV) or in combination with bicalutamide (FB), LE135 (FL) or both NR antagonists (FBL). Stratified responses were observed after the first round of treatment, with combinatorial treatments eliciting modest inhibition of tumor growth. A second round of treatment was administered on day 35, and FBL treatment exhibited the largest treatment response in terms of tumor growth inhibition (Supplementary Figure S7a). Immunostaining of A431:CAF tumor sections revealed that the combinatorial treatments FL, FB and FBL reduced tumor invasiveness and increased apoptosis, further verified by MMP zymography and immunoblot. FB and FBL treatments inhibited tumor proliferation. (Supplementary Figure S7b-d). These findings further demonstrate that CAF-mediated resistance to chemotherapy in SCC can be abrogated by combinatorial treatment with RARβ and AR antagonists, contingent on the profiling of NR expression in CAFs.
Discussion
CAFs are the major cell population within the tumor stroma and are involved in cancer progression.22, 23, 24 The transcription-based programs underlying the pro-tumor functions of CAFs are not fully understood, and transcription factors are generally considered undruggable.16, 25 NRs have surfaced as ideal drug targets primarily because their transcriptional activity is precisely modulated by the binding of small lipophilic molecules.26
The complete NR profile of skin SCC CAFs has not been explored in previous studies. Herein, we showed that among the 48 known human NRs, 18 were upregulated and three were downregulated in SCC CAFs relative to FIBs. To test the functional relevance of individual CAF NRs in SCC malignancy, NRs that were upregulated in SCC CAFs were subjected to siRNA knockdown, whereas downregulated NRs were overexpressed (NR-KD/OE-CAFs). When NR-KD/OE-CAFs were co-cultured with malignant A-5RT3 SCC cells, the A-5RT3 transcriptome responded to genetic disruption of the CAF NR signature. This highlights a non-cell autonomous effect mediated by an altered paracrine signaling network when CAF NR expression is modified. In particular, we identified RARβ, PPARβ/δ, VDR, GR and AR in CAFs as key players in the attenuation of SCC invasiveness, proliferation, energy metabolism, ROS production and response to chemotherapy. Intriguingly, these same NRs perform critical functions in skin homeostasis, especially during wound healing.27, 28 As cancers have been described as ‘wounds that do not heal’,29, 30 our work strongly suggests that the deregulation of these NRs in FIBs is in part responsible for the manifestation of many pathological skin conditions, including cutaneous SCC.17 This underscores their tremendous potential as therapeutic targets for a spectrum of skin disorders.
Although the range of chemotherapy options for SCC is increasing, the rapid onset of tumor resistance remains a key challenge for sustained treatment efficacy, especially for conventional chemotherapeutics such as cisplatin.31 In our in vitro experiments, SCC CAFs doubled the IC50 of cisplatin in A-5RT3 cells. Chemoresistance in A-5RT3 cells was abrogated by the knockdown of PPARβ/δ, RARβ and VDR in SCC CAF, and CM from CAFs treated with the RARβ antagonist, LE135, and the AR antagonist, bicalutamide, could no longer induce cisplatin resistance in A-5RT3. Notably, our microarray results identified a strong upregulation of HGF, ZEB1, IL6 and POSTN in A-5RT3 cells co-cultured with SCC CAFs. The expression of each of these molecules is associated with the acquisition of resistant phenotypes in various cancers. HGF binding to its receptor, c-MET, triggers the PI3K/Akt and MAPK pathways, which inhibit apoptosis upon chemotherapy challenge.32 IL6 enacts multidrug resistance via similar intracellular cascades.33 ZEB1 is a driver of EMT and an inducer of the cancer stem cell phenotype. Post-EMT cells and cancer stem cells are chemorefractory and notoriously difficult to destroy.34 Finally, POSTN is elevated in the cancer stem cell niche, promotes cancer cell survival and facilitates metastatic colonization.35 Compatible with the above findings, when RARβ, PPARβ/δ, VDR, GR or AR expression in SCC CAFs was disrupted, the expression of HGF, ZEB1, IL6 and POSTN in A-5RT3 cells was significantly reduced, indicating less chemoresistance.
In vivo, concurrent treatments using low-dose cisplatin with LE135 and/or bicalutamide (CL, CB or CBL therapy) resulted in significant A-5RT3:CAF xenograft sensitization to cisplatin compared with cisplatin alone (CV). Tumor relapse upon withdrawal of treatments occurred for all combination therapies tested. However, when treatments were resumed, CV-treated tumors continued to grow. CL therapy inhibited tumor growth and CBL therapy resulted in a near-complete remission of tumors. Importantly, the sustained accumulation of cisplatin adducts in CBL-treated tumors indicated that cisplatin resistance was reduced. Our data illustrate that guided by NR profiling in CAFs, additive or synergistic effects can be achieved when NR ligands are used concurrently with cytotoxic chemotherapy. Nonetheless, we note that our in vivo xenograft experiments involved immune compromised NSG mice, which precludes the influence of tumor-associated immune responses. As immune cells also have an important role in the tumor microenvironment, further studies using immunocompromised mouse models repopulated with matched patient immune cells and patient-derived xenografts are warranted.
Previous CAF-directed treatments have targeted either fibrotic signaling pathways or FIB activation protein activity, with varying success rates. Because CAFs are potent sources of fibroblast growth factor, platelet-derived growth factor and vascular endothelial growth factor, several Phase I/II trials using receptor tyrosine kinase inhibitors that block these signaling pathways are also being conducted.22 Tranilast, an anti-allergy and antifibrotic drug, is well known to inhibit FIB proliferation and FIB secretion of TGFβ.36 An in vitro study of mice lymphoma and Lewis lung carcinoma showed that tranilast inhibited CAF-mediated immunosuppression.37 Another study involving melanoma A375 tumor xenografts in mice showed that tranilast abrogated CAF-mediated resistance to radiotherapy.38 In humans, tranilast yielded promising results in two preliminary clinical studies of prostate cancer.39, 40 On the other hand, sibrotuzumab, a FIB activation protein-neutralizing antibody, failed to achieve even one complete or partial remission in a phase II trial involving 25 patients with metastatic colon cancer.41 Another phase II trial with talabostat, a small molecule inhibitor of FIB activation protein, also yielded disappointing patient outcomes.42 Interestingly, a preclinical study revealed that activation of stromal VDR using calcipotriol increased the efficacy of gemcitabine chemotherapy in a mouse model of pancreatic ductal adenocarcinoma.43 These studies and ours underscore the importance of developing complementary and combination therapies based on a deeper understanding of CAFs.
Materials and methods
Cell culture
Primary FIBs (ATCC, Manassas, VA, USA) were cultured in FibroGRO-LS (Millipore, Billerica, MA, USA) and used at passage 3–5. SCC CAFs explanted from patient SCC tumors were purchased from Asterand Biosciences (Detroit, MI, USA) and verified for their CAF expression of vimentin and alpha-smooth muscle actin. CAFs were maintained in DMEM supplemented with 5% FBS (Hyclone Laboratories, South Logan, UT, USA). A-5RT3 cells (Heidelberg, German Cancer Research Center) were maintained as previously described.44 Cell lines were tested to exclude mycoplasma contamination and authenticated against the International Cell Line Authentication Committee (ICLAC) database.
Laser capture microdissection of CAFs
Formalin-fixed, paraffin-embedded skin SCC biopsies from 10 male patients (A-J) were obtained from the National Skin Centre, Singapore. The study was approved by the National Healthcare Group Domain-Specific Review Boards (NHG-DSRB). Microdissection of SCC CAFs and peri-tumoral FIBs was performed using the PALM MicroBeam (Carl Zeiss, Oberkochen, Germany).
Transfection of CAFs
CAFs were transfected with Dharmafect SMARTpool siRNAs targeting NR transcripts, using DharmaFECT1 (Dharmacon, Lafayette, CO, USA). For NR overexpression, NR genes (Addgene, Cambridge, MA, USA) were delivered to CAFs with the FugeneHD reagent (Promega, Fitchburg, WI, USA).
RNA isolation, RT-qPCR and microarray
RNA isolation for RT-qPCR was performed as previously described.45 ProbeLibrary (Roche Applied Science, Mannheim, Germany) was used to design primers and probes for RT-qPCR (Table 1). RNA isolation and processing for microarray was performed using the RecoverAll kit (Ambion, Foster City, CA, USA), the Full Spectrum Complete Transcriptome RNA Amplification Kit (System Bioscience, Palo Alto, CA, USA), the Applause WT-Amp System (Nugen, San Carlos, CA, USA) and the Encore Biotin Module (Nugen) for loading onto GeneChip Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA, USA) chips, according to manufacturer's protocol.
Table 1. List of primers and probes.
| Gene | Nomenclature | Abbreviation | Sequence (5’ –3’) | Probe no. |
|---|---|---|---|---|
| Thyroid hormone receptor | NR1A1 | TRα | F: ccgctgtcacttgtgagg | 58 |
| R: gctgtcatatttgcaggaatagg | ||||
| NR1A2 | TRβ | F: ttggcatggcaacagattt | 42 | |
| R: ccggttctcctctatcagctt | ||||
| Retinoic acid receptor | NR1B1 | RARα | F: aggctaccactatggggtca | 89 |
| R: gttgatgatgcagttcttgtcc | ||||
| NR1B2 | RARβ | F: ttgtgttcacctttgccaac | 51 | |
| R: cggttcctcaaggtcctg | ||||
| NR1B3 | RARγ | F: cgctccatccaagagactg | 70 | |
| R: gagggaactgggccgtag | ||||
| Peroxisome proliferator-activated receptor | NR1C1 | PPARα | F: agattcaggagaggcaagaaaa | 56 |
| R: gcatcagtcttcatgggtga | ||||
| NR1C2 | PPARβ/δ | F: ctcaatgggggaccacag | 11 | |
| R: agctccgggagaggtctg | ||||
| NR1C3 | PPARγ | F: gacctgaaacttcaagagtaccaaa | 39 | |
| R: tgaggcttattgtagagctgagtc | ||||
| Reverse erbA | NR1D1 | Rev-erbα | F: cctggactccaacaacaaca | 52 |
| R: tagagggattcagggctggt | ||||
| NR1D2 | Rev-erbβ | F: agaacaagaatagttacctgtgcaac | 69 | |
| R: ggagacttactcattggacaaacc | ||||
| RAR-related orphan receptor | NR1F1 | RORα | F: tacatgtgaaggctgcaagg | 2 |
| R: ggagtaggtggcattgcttt | ||||
| NR1F2 | RORβ | F: gatcactcacgccatccaat | 66 | |
| R: aaccacttccaagcaacctg | ||||
| NR1F3 | RORγ | F: gcgctccaacatcttctccc | 6 | |
| R: cacgttcccacatctcc | ||||
| Liver X receptor | NR1H2 | LXRβ | F: acagcaaggacgacttccac | 39 |
| R:actcgaagatggggttgatg | ||||
| NR1H3 | LXRα | F:agtgtcggcttcgcaaat | 52 | |
| R:ggcggatctgttcttctgac | ||||
| Farnesoid X receptor | NR1H4 | FXR | F:acctgtgaggggtgtaaaggt | 2 |
| R:gccccgttttacacttg | ||||
| Vitamin D receptor | NR1I1 | VDR | F: atcggcatgatgaaggagtt | 6 |
| R:ttccgcttcaggatcatctc | ||||
| Pregane X receptor | NR1I2 | PXR | F:tcgccattactctgaagtcctac | 57 |
| R:catgatcttcaggaacaagaacc | ||||
| Constitutive androstane receptor | NR1I3 | CAR | F:gctggcatgaggaaagaca | 29 |
| R:tgcacaggtgtttgctgtg | ||||
| Human nuclear factor 4 | NR2A1 | HNFα | F:gagatccatggtgttcaagga | 68 |
| R:gtgccgagggacaatgtagt | ||||
| NR2A2 | HNFγ | F:tcttggtggaatgggctaaa | 8 | |
| R:agctctcaacagtgccacct | ||||
| Retinoid X receptor | NR2B1 | RXRα | F:acatgcagatggacaagacg | 26 |
| R:gagagccccttggagtcag | ||||
| NR2B2 | RXRβ | F:agctcccccaggattctc | 66 | |
| R:cagggagtgacactgttgagtt | ||||
| NR2B3 | RXRγ | F:tgtcatgggcatgaagagg | 82 | |
| R:cctcactctcagctcgctct | ||||
| Testis receptor | NR2C1 | TR2 | F:cctgatctgtctgcacaacac | 17 |
| R:tggtccttggtctggagaat | ||||
| NR2C2 | TR4 | F:ggcgcaaatcctgaggta | 80 | |
| R:tcggtggagattatctggatg | ||||
| Tailess-related receptor | NR2E2 | TLX | F:ggacctatgtctgcaaatctgg | 75 |
| R:tttctgtgcgtcttgtccac | ||||
| Photoreceptor-specific nuclear receptor | NR2E3 | PNR | F:ggtggcctgtggagtcag | 25 |
| R:caatctgtccaaacttcagttccatcgtgctgttcacgtcaga | ||||
| Chicken ovalbumin upstream promoter-transcription factor | NR2F1 | COUP-TFI | F:atcgtgctgttcacgtcaga | 2 |
| R:gctcctcacgtactcctcca | ||||
| NR2F2 | COUP-TFII | F:gccatagtcctgttcacctca | 36 | |
| R:aatctcgtcggctggttg | ||||
| ErbA2-related gene-2 | NR2F6 | EAR2 | F:ccaggagcaggtggacaa | 46 |
| Estrogen receptor | NR3A1 | ERα | F:aaccagtgcaccattgataaaa | 69 |
| R:tcctcttcggtcttttcgtatc | ||||
| NR3A2 | ERβ | F:cgagagttaaaactccaacacaaa | 15 | |
| R:tcgctgtgaccagagggta | ||||
| Estrogen receptor-related receptor | NR3B1 | ERRα | F:ggcggcagaagtacaagc | 3 |
| R:attcactggggctgctgt | ||||
| NR3B2 | ERRβ | F:agagaggcaggcagatctca | 79 | |
| R:gtgagccagagatgctttcc | ||||
| NR3B3 | ERRγ | F:ccagccaaaaagccattg | 83 | |
| R:gacagtagggtcaggcatgg | ||||
| Glucocorticoid receptor | NR3C1 | GR | F:agttttcttcaaaagagcagtgg | 56 |
| R:caatcattccttccagcaca | ||||
| Mineralocorticord receptor | NR3C2 | MR | F:acaggtagacggcgagagac | 58 |
| R:tcagggagactgtggtagcc | ||||
| Progesterone receptor | NR3C3 | PR | F:ggatgagcttaatggtgtttgg | 58 |
| R:gattctttcatccgctgttca | ||||
| Androgen receptor | NR3C4 | AR | F:gccttgctctctagcctcaa | 14 |
| R:gtcgtccacgtgtaagttgc | ||||
| NGF-induced factor B | NR4A1 | NGFIB | F:acagcttgcttgtcgatgtc | 34 |
| R:ggttctgcagctcctccac | ||||
| Nur related factor 1 | NR4A2 | NURR1 | F:gcccatgtcgactccaac | 64 |
| R:actcatttgatagtcagggttcg | ||||
| Neuron-derived orphan receptor 1 | NR4A3 | NOR1 | F:catcagcagccatccattc | 19 |
| R:ctgcttgaagtacatggaggtg | ||||
| Steroidogenic factor 1 | NR5A1 | SF-1 | F:gcaggtgcatggtcttcaa | 10 |
| R:agttctgcagcagcgtcat | ||||
| Liver receptor homologous protein 1 | NR5A2 | LRH-1 | F:tgtctgaaattcttggtgctctt | 11 |
| R:tgacttgttcctggacacctt | ||||
| Germ cell nuclear factor | NR6A1 | GCNF | F:aagtactcgccctccgatg | 58 |
| R:tggaacttgtgatagaggtagatga | ||||
| DSS-AHC critical region on the X chromosome, gene 1 | NR0B1 | DAX1 | F:ccatcaagtgctttctttcca | 67 |
| R:cctgaatgtacttcacgcactg | ||||
| Short heterodimeric partner | NR0B2 | SHP | F:tgctgtctggagtccttctg | 1 |
| R:ccagtgagcctcctgctg | ||||
| Controls | ||||
| 18S ribosomal RNA | 18S rRNA | F:gtaacccgttgaaccccatt | 48 | |
| R:ccatccaatcggtagtagcg | ||||
| TATA Box promoter | TBP | F:gctggttatcgggagttgg | 30 | |
| R:actggcctggtgtcctagag |
Microarray data analysis
Raw microarray files were imported into Partek Genomic Suite (v6.6) and normalized using robust multiarray averaging. Analysis of variance was performed for all samples with contrast set against the A-5RT3:FIB sample. The 302 genes differentially expressed by >1.5-fold between the A-5RT3:CAF and A-5RT3:FIB samples were examined in all A-5RT3:NR-KD/OE-CAF samples and hierarchically clustered. Ingenuity Pathway Analysis software provided functional annotation of focus molecules. Gene ontology enrichment and top associated networks were assessed based on P-values calculated by the analysis platform.
Invasion, proliferation, glucose uptake and ROS assays
A total of 4 × 104 A-5RT3 cells and 1.2 × 105 FIBs, CAFs or NR-KD/OE-CAFs were seeded in the upper and lower chambers, respectively. The FIBs provided the chemotactic gradient. Invasion assay was performed as previously described,46 with modification. Invaded A-5RT3 cells on the bottom of the inserts were stained with the infrared dye, SYTO60 (Life Technologies, Camarillo, CA, USA) and quantified with Odyssey CLx (LI-COR Biosciences, Lincoln, NE, USA). Proliferation, glucose uptake and ROS in A-5RT3 cells were assessed as previously described.47, 48, 49 FACS was performed with the Accuri C6 Flow Cytometer (BD Biosciences, San Diego, CA, USA) and analyzed using FlowJo (Treestar, Ashland, OR, USA).
Energy charge determination
Energy charge in A-5RT3 cells was determined as previously described.50
Multiple analyte detection
MILLIPLEX MAP Human MMP, Human Angiogenesis/Growth Factor and Human Circulating Cancer Biomarker kits were used for multiplexed immunoassay detection of secreted factors in CM (Merck Millipore, Billerica, MA USA). The MAGPIX system was used for data acquisition and analysis was performed with the onboard software (Luminex Corporation, Austin, TX, USA). The concentration of pro-tumor secreted factors in the conditioned media of A-5RT3:CAF and A431:CAF co-cultures treated with LE135, bicalutamide or both were measured by multiplexed immunoassay performed by Eve Technologies (Calgary, Alberta, Canada).
Drug treatments and apoptosis measurements
The 48 h CM (sterile filtered) of FIBs, CAFs and NR-KD/OE-CAFs was added to A-5RT3 cells. In total, 10, 100, 1000, 10000 and 100000 ng/ml of cisplatin (Sigma Aldrich, St Louis, MO, USA) was added to A-5RT3 in CM for 16 h. Apoptosis was measured with Annexin-V and propidium iodide (BioLegend, San Diego, CA, USA) and FACS. IC50 values were calculated using the graphing software, Prism (GraphPad, CA, USA). Agonists of PPARβ/δ (GW0742), VDR (EB1089) and GR (fluticasone propionate), and antagonists of AR (bicalutamide) and RARβ (LE135) were used at a final concentration of 5 μM (Tocris Bioscience, Bristol, UK). Terminal deoxynucleotidyltransferase dUTP nick end labeling assay (Roche Applied Science) was used for apoptosis measurements in tissue sections.
Animal experiments
Six-week-old male NSG mice (n=6 per treatment) (Jackson Laboratories, Sacramento, CA, USA) were subcutaneously injected with 2 × 105 A-5RT3 cells admixed with 6 × 105 CAFs in 50 μl of growth factor-reduced matrigel. Treatments started 3 weeks after tumor xenografts were established (tumor volumes ⩾150 mm3). For cisplatin treatment, mice received intraperitoneal or intratumoral injections of 2 mg/kg cisplatin for 2 consecutive weeks. For combination treatments, intraperitoneal injection of 5 mg/kg LE135 and/or 10 mg/kg bicalutamide was administered each time cisplatin was injected. Power analysis was used to determine sample size. Double-blind randomization was used for allocation of the experimental groups. All animal experiments were carried out in accordance to the guidelines of the Institutional Animal Care and Use Committee (ARF-SBS/NIE-A0250AZ, -A0324 and -A0321) of Nanyang Technological University, Singapore.
Immunoblotting and immunostaining
Primary antibodies against mouse and human NRs, PCNA, GLUT1 and β-tubulin (Santa Cruz Biotechnology, Dallas, TX, USA), Ki67 (Dako, Produktionsvej, Glostrup, Denmark), E-cad (Cell Signaling, Danvers, MA, USA) were used. Dye-conjugated secondary antibodies were from LI-COR and Life Technologies. Protein bands were detected using Odyssey CLx (LI-COR Bioscience). Microscopy images were obtained using the LSM710 (Carl Zeiss).
Statistical analysis
Statistical differences were evaluated with two-tailed Mann–Whitney U-test or one-way analysis of variance test with SPSS software where appropriate. P-values <0.05 indicate statistical significance.
Acknowledgments
The work was supported by grants from the Singapore Ministry of Education (MOE2014-T2-1-012 and AcRF Tier 1 RG134/15) to NST. JSKC is a recipient of the Nanyang President Graduate Scholarship.
Author contributions
JSKC, MKS and NST were involved in conception, design and development of methodology. JSKC, MKS, ZQT, HCC and JST were involved in data acquisition and analysis. JSKC and NST wrote and reviewed the manuscript.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Shi Y, Du L, Lin L, Wang Y. Tumour-associated mesenchymal stem/stromal cells: emerging therapeutic targets. Nat Rev Drug Discov 2016; 16: 35–52. [DOI] [PubMed] [Google Scholar]
- Spaw M, Anant S, Thomas SM. Stromal contributions to the carcinogenic process. Mol Carcinog 2016; 56: 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AS, Vakoc CR. Targeting transcription factors in cancer. Trends Cancer 2015; 1: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzarri M, Cucina A. Tumor and the microenvironment: a chance to reframe the paradigm of carcinogenesis? Biomed Res Int 2014; 2014: 934038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti MP, Martinez-Outschoorn UE, Chiavarina B, Pavlides S, Whitaker-Menezes D, Tsirigos A et al. Understanding the 'lethal' drivers of tumor-stroma co-evolution: emerging role(s) for hypoxia, oxidative stress and autophagy/mitophagy in the tumor micro-environment. Cancer Biol Ther 2010; 10: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C et al. Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: a new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle 2010; 9: 3256–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenschein C, Soto AM. Carcinogenesis explained within the context of a theory of organisms. Prog Biophys Mol Biol 2016; 122: 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer 2009; 9: 665–674. [DOI] [PubMed] [Google Scholar]
- Togo S, Polanska UM, Horimoto Y, Orimo A. Carcinoma-associated fibroblasts are a promising therapeutic target. Cancers (Basel) 2013; 5: 149–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby IA, Zakuan N, Billet F, Desmouliere A. The myofibroblast, a key cell in normal and pathological tissue repair. Cell Mol Life Sci 2016; 73: 1145–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AM, Jung J, Aziz N, Kissil JL, Pure E. Targeting fibroblast activation protein inhibits tumor stromagenesis and growth in mice. J Clin Invest 2009; 119: 3613–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J, Ehnman M, Ostman A. PDGF receptors in tumor biology: prognostic and predictive potential. Future Oncol 2014; 10: 1695–1708. [DOI] [PubMed] [Google Scholar]
- Simkova D, Kharaishvili G, Slabakova E, Murray PG, Bouchal J. Glycoprotein asporin as a novel player in tumour microenvironment and cancer progression. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2016; 160: 467–473. [DOI] [PubMed] [Google Scholar]
- Vazquez-Villa F, Garcia-Ocana M, Galvan JA, Garcia-Martinez J, Garcia-Pravia C, Menendez-Rodriguez P et al. COL11A1/(pro)collagen 11A1 expression is a remarkable biomarker of human invasive carcinoma-associated stromal cells and carcinoma progression. Tumour Biol 2015; 36: 2213–2222. [DOI] [PubMed] [Google Scholar]
- Redmond AM, Carroll JS. Defining and targeting transcription factors in cancer. Genome Biol 2009; 10: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Higgins PJ. Drugging the undruggable: transcription therapy for cancer. Biochim Biophys Acta 2013; 1835: 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K, Smith AG. Nuclear receptor function in skin health and disease: therapeutic opportunities in the orphan and adopted receptor classes. Cell Mol Life Sci 2016; 73: 3789–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Zhang Y, Guo Y, Xie J, Wang J, He X et al. All-trans retinoic acid attenuates experimental colitis through inhibition of NF-kappaB signaling. Immunol Lett 2014; 162: 34–40. [DOI] [PubMed] [Google Scholar]
- van de Pavert SA, Olivier BJ, Goverse G, Vondenhoff MF, Greuter M, Beke P et al. Chemokine CXCL13 is essential for lymph node initiation and is induced by retinoic acid and neuronal stimulation. Nat Immunol 2009; 10: 1193–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesei A, Leonetti C, Di Donato M, Gabucci E, Porru M, Varchi G et al. Effect of small molecules modulating androgen receptor (SARMs) in human prostate cancer models. PLoS ONE 2013; 8: e62657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Lamoureux F, Crafter C, Davies BR, Beraldi E, Fazli L et al. Synergistic targeting of PI3K/AKT pathway and androgen receptor axis significantly delays castration-resistant prostate cancer progression in vivo. Mol Cancer Ther 2013; 12: 2342–2355. [DOI] [PubMed] [Google Scholar]
- Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: orchestrating the composition of malignancy. Genes Dev 2016; 30: 1002–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer 2006; 6: 392–401. [DOI] [PubMed] [Google Scholar]
- Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016; 16: 582–598. [DOI] [PubMed] [Google Scholar]
- Darnell JE Jr.. Transcription factors as targets for cancer therapy. Nat Rev Cancer 2002; 2: 740–749. [DOI] [PubMed] [Google Scholar]
- Sladek FM. Nuclear receptors as drug targets: new developments in coregulators, orphan receptors and major therapeutic areas. Expert Opin Ther Targets 2003; 7: 679–684. [DOI] [PubMed] [Google Scholar]
- Rieger S, Zhao H, Martin P, Abe K, Lisse TS. The role of nuclear hormone receptors in cutaneous wound repair. Cell Biochem Funct 2015; 33: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan NS, Vazquez-Carrera M, Montagner A, Sng MK, Guillou H, Wahli W. Transcriptional control of physiological and pathological processes by the nuclear receptor PPARbeta/delta. Prog Lipid Res 2016; 64: 98–122. [DOI] [PubMed] [Google Scholar]
- Byun JS, Gardner K. Wounds that will not heal: pervasive cellular reprogramming in cancer. Am J Pathol 2013; 182: 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak HF. Tumors: wounds that do not heal-redux. Cancer Immunol Res 2015; 3: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res 2001; 478: 23–43. [DOI] [PubMed] [Google Scholar]
- Gao CF, Vande Woude GF. HGF/SF-Met signaling in tumor progression. Cell Res 2005; 15: 49–51. [DOI] [PubMed] [Google Scholar]
- Ghandadi M, Sahebkar A. Interleukin-6: a critical cytokine in cancer multidrug resistance. Curr Pharm Des 2016; 22: 518–526. [DOI] [PubMed] [Google Scholar]
- Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta 2009; 1796: 75–90. [DOI] [PubMed] [Google Scholar]
- Hong LZ, Wei XW, Chen JF, Shi Y. Overexpression of periostin predicts poor prognosis in non-small cell lung cancer. Oncol Lett 2013; 6: 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzawa H, Kikuchi S, Arai N, Koda A. The mechanism involved in the inhibitory action of tranilast on collagen biosynthesis of keloid fibroblasts. Jpn J Pharmacol 1992; 60: 91–96. [DOI] [PubMed] [Google Scholar]
- Ohshio Y, Hanaoka J, Kontani K, Teramoto K. Tranilast inhibits the function of cancer-associated fibroblasts responsible for the induction of immune suppressor cell types. Scand J Immunol 2014; 80: 408–416. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gan G, Wang B, Wu J, Cao Y, Zhu D et al. Cancer-associated fibroblasts promote irradiated cancer cell recovery through autophagy. EBioMedicine 2017; 17: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K, Mizokami A, Li YQ, Narimoto K, Sugimoto K, Kadono Y et al. Tranilast inhibits hormone refractory prostate cancer cell proliferation and suppresses transforming growth factor beta1-associated osteoblastic changes. Prostate 2009; 69: 1222–1234. [DOI] [PubMed] [Google Scholar]
- Izumi K, Mizokami A, Shima T, Narimoto K, Sugimoto K, Kobori Y et al. Preliminary results of tranilast treatment for patients with advanced castration-resistant prostate cancer. Anticancer Res 2010; 30: 3077–3081. [PubMed] [Google Scholar]
- Hofheinz RD, al-Batran SE, Hartmann F, Hartung G, Jager D, Renner C et al. Stromal antigen targeting by a humanised monoclonal antibody: an early phase II trial of sibrotuzumab in patients with metastatic colorectal cancer. Onkologie 2003; 26: 44–48. [DOI] [PubMed] [Google Scholar]
- Narra K, Mullins SR, Lee HO, Strzemkowski-Brun B, Magalong K, Christiansen VJ et al. Phase II trial of single agent Val-boroPro (Talabostat) inhibiting fibroblast activation protein in patients with metastatic colorectal cancer. Cancer Biol Ther 2007; 6: 1691–1699. [DOI] [PubMed] [Google Scholar]
- Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 2014; 159: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CR, Tan C, Teo Z, Tay CY, Phua T, Wu YL et al. Loss of TAK1 increases cell traction force in a ROS-dependent manner to drive epithelial-mesenchymal transition of cancer cells. Cell Death Dis 2013; 4: e848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio DC, Ares M Jr., Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010; 2010: pdb prot5439. [DOI] [PubMed] [Google Scholar]
- Marshall J. Transwell((R)) invasion assays. Methods Mol Biol 2011; 769: 97–110. [DOI] [PubMed] [Google Scholar]
- Eruslanov E, Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol Biol 2010; 594: 57–72. [DOI] [PubMed] [Google Scholar]
- Rothaeusler K, Baumgarth N. Assessment of cell proliferation by 5-bromodeoxyuridine (BrdU) labeling for multicolor flow cytometry. Curr Protoc Cytom 2007; Chapter 7: Unit 7; 31. [DOI] [PubMed]
- Zou C, Wang Y, Shen Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J Biochem Biophys Methods 2005; 64: 207–215. [DOI] [PubMed] [Google Scholar]
- Chen Y, Xing D, Wang W, Ding Y, Du L. Development of an ion-pair HPLC method for investigation of energy charge changes in cerebral ischemia of mice and hypoxia of Neuro-2a cell line. Biomed Chromatogr 2007; 21: 628–634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.