Abstract
Flavopiridol is a synthetically produced flavonoid that potently inhibits the proliferation of human tumor cell lines. Flavopiridol exerts strong antitumor activity via several mechanisms, including the induction of cell cycle arrest and apoptosis, and the modulation of transcriptional regulation. The aim of the present study was to determine the effect of flavopiridol on a subpopulation of cluster of differentiation (CD)44+/CD24− human breast cancer MCF7 stem cells. The CD44+/CD24− cells were isolated from the MCF7 cell line by fluorescence-activated cell sorting and treated with 100, 300, 500, 750 and 1,000 nM flavopiridol for 24, 48 and 72 h. Cell viability and proliferation assays were performed to determine the inhibitory effect of flavopiridol. Gene expression profiling was analyzed using Illumina Human HT-12 v4 Expression BeadChip microarray. According to the results, the half maximal inhibitory concentration (IC50) value of flavopiridol was 500 nM in monolayer cells. Flavopiridol induced growth inhibition and cytotoxicity in breast cancer stem cells (BCSCs) at the IC50 dose. The present study revealed several differentially regulated genes between flavopiridol-treated and untreated cells. The result of the pathway analysis revealed that flavopiridol serves an important role in translation, the ribosome biogenesis pathway, oxidative phosphorylation, the electron transport chain pathway, carbon metabolism and cell cycle. A notable result from the present study is that ribosome-associated gene expression is significantly affected by flavopiridol treatment. The data of the present study indicate that flavopiridol exhibits antitumor activity against CD44+/CD24− MCF7 BCSCs through different mechanisms, mainly by inhibiting translation and the ribosome biogenesis pathway, and could be an effective chemotherapeutic molecule to target and kill BCSCs.
Keywords: flavopiridol, ribosome biogenesis, breast cancer stem cell, microarray
Introduction
Breast cancer is a common type of malignancy in the world and a major cause of mortality in females between 30 and 59 years of age (1). Breast cancer is a heterogeneous disease in terms of histology, pathology, and genetic and molecular profiles (2). Despite diagnostic and therapeutic advances, breast cancer patients still often exhibit relapse or metastasis subsequent to therapy (3).
Tumors are morphologically heterogeneous, composed of undifferentiated and differentiated cells (4). Cancer stem cells (CSCs) have been identified as a subpopulation within the tumor possessing the ability to self-renew and differentiate into non-tumorigenic cell populations that constitute the bulk of the tumor (5). CSCs have been associated with tumor initiation, therapy resistance and tumor recurrence. CSCs are a major problem for cancer therapy, and the elimination of CSCs is required for an effective treatment (6). The presence of CSC population in breast cancer has been demonstrated in several studies (7,8). Breast cancer stem cells (BCSCs) were first isolated by Al-Hajj et al (9) in 2003 from the pleural effusions of a patient. Specific cell surface markers and biomarkers are used to identify and isolate BCSCs. The adhesion molecule cluster of differentiation (CD) 44 is a multifunctional cell surface transmembrane glycoprotein that serves a role in cell adhesion, proliferation, differentiation, motility and migration (10). In breast cancer, CD44+/CD24− expression was demonstrated as prospective phenotype to isolate BCSCs. Al-Hajj et al (9) reported that breast cancer cells exhibiting an increased expression of CD44+/CD24− were able to form tumors when injected into immunodeficient mice.
Cyclin-dependent kinases (CDKs) serve an essential role in the control of the cell cycle, and are associated with cytoskeletal dynamics, epigenetic regulation, controlling stem cell self-renewal, regulating metabolism, cell migration, regulation of transcription, DNA damage, and genomic and chromosomal instability (11). The dysregulation of CDK expression contributes to the loss of normal cell cycle control, which leads to the formation and progression of cancer (12). Therefore, the inhibition of CDKs by small-molecule CDK inhibitors may be an effective treatment of cancer. The dysregulation of cyclin D and the CDK pathway in cancer cells may inhibit senescence and promote cellular proliferation (13). By using various different mechanisms, malignant cells may increase cyclin D-dependent activity. The cyclin D-CDK4/6-retinoblastoma pathway controls the cell cycle restriction point, and is commonly dysregulated in breast cancer, making it a possible target for anticancer therapy (14).
Flavopiridol is a semisynthetic flavonoid that was the first CDK inhibitor used in clinical trials (15). Flavopiridol exhibits an antitumor effect against a variety of tumor types, including several solid tumors, through cytostatic activity, and induces cell cycle arrest and apoptosis (16). This flavonoid is a promising anticancer drug that is undergoing phase I and II clinical trials for chronic myeloid leukemia and pancreatic cancer (17,18). Our previous study demonstrated that flavopiridol induced growth inhibition and apoptosis in CD133+/CD44+ prostate CSCs (19).
BCSCs have been proposed to be responsible for numerous properties of breast cancer such as resistance, metastatic properties and recurrence (20). Conventional anticancer therapies may kill the majority of the cancer cells, but CSCs are not affected by these therapies (21). For a more effective treatment of breast cancer, it may be necessary to target CSCs. Genome-wide gene expression profiling based on microarray analysis is a powerful tool to elucidate the possible mechanisms of cancer drugs. The present study aimed to investigate the cytotoxic effects and underlying mechanism of action of flavopiridol against human breast CSCs.
Materials and methods
Cell culture conditions and reagents
Human breast cancer MCF7 cells were obtained from Interlab Cell Line Collection (Genova, Italy) and were grown in monolayer cell culture in RPMI 1640 culture medium (Lonza Group AG, Basel, Switzerland) containing 10% heat-inactivated fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 1% penicillin and 1% streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The cells were cultured in 25-cm2 polystyrene flasks (Corning Life Sciences, Corning, NY, USA) and incubated for 48 h at 37°C in a humidified atmosphere of 5% CO2. Flavopiridol (Sigma-Aldrich; Merck KGaA) was prepared as 10 mM stock solution in dimethyl sulfoxide (DMSO), and the final volume of DMSO did not exceed 0.1% of the total incubation volume and was not cytotoxic to the tumor cells at these concentrations (data not shown).
Fluorescence-activated cell sorting (FACS)
To sort the CSCs subpopulations in the human breast cancer MCF7 cell line, the antibodies of expressed surface markers CD44+/CD24−, anti-CD44 conjugated to fluorescein isothiocyanate (10 µl/106 cell; FITC; cat. no. 555478; BD Biosciences, Franklin Lakes, NJ, USA) and anti-CD24 conjugated to phycoerythrin (10 µl/106 cell; PE; clone 32D12; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) were used. The MCF7 cells were seeded and grown to 80% confluence. The cells were detached using a non-enzymatic cell dissociation solution (Sigma-Aldrich; Merck KGaA) and resuspended in RPMI 1640 culture medium. A total of ~5×104 cells were incubated with anti-CD44-fluorescein isothiocyanate (FITC; clone G44 26; BD Biosciences, Franklin Lakes, NJ, USA) and anti-CD24-phycoerythrin (PE; clone 32D12; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) in FACS stain buffer (cat. no. 554657; BD Pharmingen, Franklin Lakes, NJ, USA) for 15 min at 4°C. After 15 min, the cells were washed with the above FACS wash buffer and resuspended in FACS stain buffer (cat. no 554657, BD Pharmingen, Franklin Lakes, NJ, USA) to a density of 107 cells/ml. The cells were sorted into a CD44+/CD24− population (sorted cells) using a FACSAria flow cytometer (BD Biosciences).
Analysis of cell viability
The viability of the cells following treatment was determined using the Muse® Count & Viability kit (Muse Cell Analyzer; EMD Millipore, Billerica, MA, USA) according to the protocol of the manufacturer. The cells were seeded in triplicate in 6-well plates at a density of 1×104 cells/well. Subsequent to a 24-h incubation, the cells were exposed to 500, 750 and 1,000 nM flavopiridol. The plates were then incubated at 37°C in a 5% CO2 incubator for 24, 48 and 72 h. Subsequent to incubation, all cells were collected and diluted with PBS. In total, 50 µl of the cell suspension was then added to 450 µl Muse® Count & Viability reagent (dilution, 10X), incubated for 5 min at room temperature and analyzed using the Muse Cell Analyzer. Data were presented as proportional viability (%) by comparing the treated group with the untreated cells.
RNA isolation and microarray analysis
The BCSCs were treated with a dose of flavopiridol equivalent to its half maximal inhibitory concentration (IC50). Total RNA was extracted from the treated and untreated cells using the RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) according to the protocol of the manufacturer. Biotin-labeled RNA samples for hybridization on Illumina Human HT-12 v4 Expression BeadChip (Illumina, Inc., San Diego, CA, USA) were prepared according to the recommended sample labeling procedure of Illumina, Inc. A total of 250 ng total RNA was used for cDNA synthesis, followed by an amplification/labeling step to synthesize biotin-labeled cRNA. The quality of the cRNA was controlled using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). Hybridization was performed at 58°C in GEX-HCB buffer (Illumina, Inc.) at a concentration of 150 ng cRNA/µl. The BeadChips were subsequently washed, blocked and conjugated with cyanine 3-streptavidin (Thermo Fisher Scientific, Inc.). The microarrays were scanned in the iScan System (Illumina, Inc.). The obtained amplification data (fold-changes in the quantification cycle values of all the genes) were processed in Agilent GeneSpring Data Analysing Software (Agilent Technologies, Inc.) and >2 fold-change was used for filtering criteria.
Statistical analysis
The statistical software package SPSS version 20.0 for Windows (IBM Corp., Armonk, NY USA) was used for all statistical analysis. All experiments were performed independently three times. Statistical analysis was tested by one-way analysis of variance, followed by Tukey's or Dunnett's post hoc tests. All data are presented as mean ± standard deviation from 3 independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
Sorting breast cancer MCF7 cells and purity of the CD44+/CD24− sorted subpopulations
Human breast cancer MCF7 cells were separated with FACS, yielding a CD44+/CD24− population (Fig. 1). The present study obtained MCF7 CSC and non-CSC subpopulations. According to the results, the mean percentage of MCF7 CSCs and non-CSCs were 1.6 and 98.4%, respectively. The purity of the CSCs samples was tested with anti-CD44 and anti-CD24 antibodies. The sorting rate analysis and purity of the cells were evaluated sequentially, and the rate was 96.7±5.4% for the sorted cells. To confirm the flow cytometry analyses, the cells were re-evaluated following sorting, and the analyses were repeated subsequent to one passage. The results revealed that the cell purity following sorting was >90%.
Figure 1.
Flow cytometry analysis of CD44+/CD24− subpopulations in MCF7 cell lines. CD44+/CD24− populations are presented in P1. CD, cluster of differentiation; SSC-A, side scatter area; FSC-A, forward scatter area; FITC-A, fluorescein isothiocyanate area; PE-A, phycoerythrin area.
Increasing cytotoxicity of CD44+/CD24− BCSCs with flavopiridol
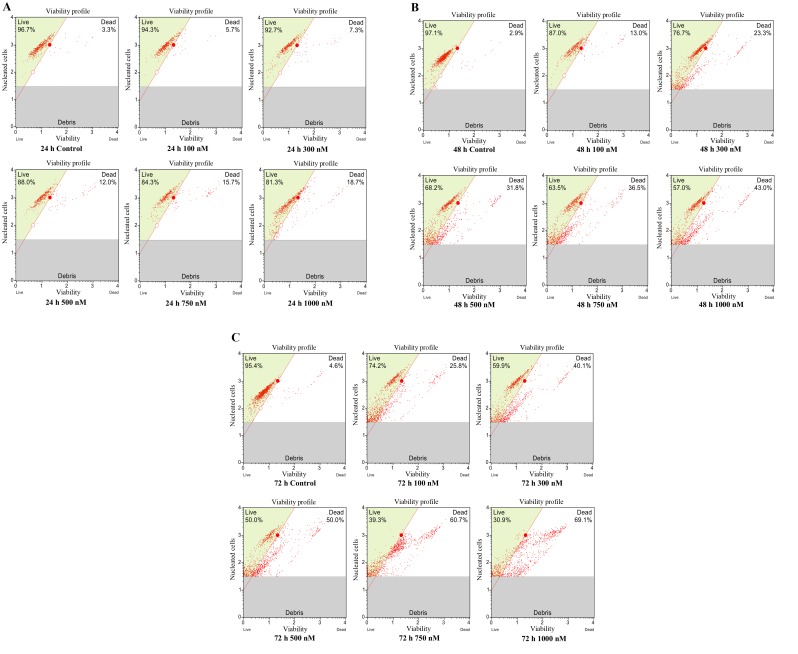
Cytotoxicity assays were performed to determine the therapeutic effect of flavopiridol. MCF7 CSCs were exposed to 100–1,000 nM flavopiridol for 24, 48 and 72 h, and the percentage of viable cells in the samples was determined by a cell viability assay. Flavopiridol reduced the cell viability of CSCs in a time- and concentration-dependent manner (Fig. 2A-C). According to the data, there were no significant decreases in cell viability at the low doses (100 and 300 nM) of flavopiridol treatment for 24 h compared with that of the untreated cells (P=0.642). After 48 h of treatment, flavopiridol significantly reduced the cell viability of BCSCs at 500, 750 and 1,000 nM compared with that of the untreated cells (P=0.000). After 72 h of treatment with flavopiridol, the IC50 was calculated as 500 nM.
Figure 2.
Representative cell viability profile of CD44+/CD24− breast cancer stem cells non-treated or treated with 100, 300, 500, 750 and 1,000 nM flavopiridol subsequent to (A) 24, (B) 48 and (C) 72 h of incubation. Each concentration was studied as three replicates.
Microarray analysis for the identification of differentially expressed genes in MCF7 CD44+/CD24− cells treated with flavopiridol
To analyze the molecular mechanisms underlying the anticancer effect of flavopiridol in BCSCs, the MCF7 CD44+/CD24− cells were treated with 500 nM flavopiridol for 72 h. To identify flavopiridol-regulated genes and determine the possible mechanism underlying the differential role of flavopiridol on the growth of MCF7 CD44+/CD24− cells, global gene expression profiling was undertaken following treatment with flavopiridol using the Illumina Human HT-12 v4 Expression BeadChip. According to the results of microarray analysis, 65 genes were identified as significantly affected subsequent to treatment with flavopiridol, since the expression of 57 genes decreased and the expression of 8 genes increased compared with that in untreated cells at 72 h (Table I).
Table I.
Changes in the expression of upregulated and downregulated genes following treatment with flavopiridol.
| A, Translation pathway and ribosome biogenesis pathway | ||||
|---|---|---|---|---|
| Probe ID | Symbol | Fold-changea | Regulation | Definition |
| 4920193 | RPL27A | −2.7026234 | Down | RPL27a |
| 6060356 | RPL13A | −2.1516730 | Down | RPL13a |
| 3360228 | RPS20 | −2.0229893 | Down | RPS20 |
| 5290082 | RPLP1 | −2.1037197 | Down | RiP, large, P1 |
| 1710369 | RPL3 | −2.5006313 | Down | RPL3, transcript variant 2 |
| 620754 | RPS5 | −2.3889322 | Down | RPS5 |
| 7040095 | RPL17 | −2.0128388 | Down | RPL17, transcript variant 2 |
| 990273 | RPL37A | −2.2212677 | Down | RP L37a |
| 3060477 | RPL8 | −2.4217634 | Down | RPL8, transcript variant 2 |
| 3610241 | RPL19 | −3.2778310 | Down | RPL19 |
| 3800332 | RPS25 | −2.3831854 | Down | RPS25 |
| 5260682 | RPS14 | −2.5092149 | Down | RPS14, transcript variant 2 |
| 5220037 | RPS2 | −3.9913297 | Down | RPS2 |
| 6960181 | RPS12 | −2.9727620 | Down | RPS12 |
| 7510482 | RPS4X | −2.1651378 | Down | RPS4, X-linked |
| 20021 | RPS15 | −2.4493800 | Down | RPS15 |
| 5560349 | RPS11 | −2.6253710 | Down | RRPS11 |
| 5890730 | RPS26L | −2.8872151 | Down | Predicted: Homo sapiens 40S RPS26-like |
| 510195 | RPL27 | −2.4229383 | Down | RPL27 |
| 840647 | RPL36 | −2.7411752 | Down | RPL36, transcript variant 1 |
| 4250445 | RPL4 | −2.0928760 | Down | RPL4 |
| 6250097 | RPS9 | −2.3448272 | Down | RPS9 |
| 1410537 | RPSA | −2.1424713 | Down | RPSA, transcript variant 1 |
| 6270546 | RPS6 | −2.3560820 | Down | RPS6 |
| 6590377 | RPS26 | −2.1171474 | Down | RPS26 |
| 4250445 | RPL4 | −2.0012200 | Down | RPL4 |
| 3610309 | LOC653881 | −2.6014566 | Down | Predicted: Similar to RPL3 |
| 2490450 | LOC91561 | −2.2661705 | Down | Predicted: Similar to RPS2, transcript variant 3 |
| 3440670 | LOC402251 | −2.3879724 | Down | Predicted: Similar to eukaryotic translation elongation factor 1 α 2 |
| 4060446 | LOC649150 | −3.2092447 | Down | Predicted: Similar to eukaryotic translation elongation factor 1 α 2 |
| 1440398 | LOC644511 | −2.2366867 | Down | Predicted: Similar to RPL13a, transcript variant 1 |
| 1570491 | LOC648000 | −2.3058624 | Down | Predicted: Similar to 60S RPL7, transcript variant 1 |
| 2320494 | LOC653314 | −3.1144562 | Down | Homo sapiens similar to RPL19 |
| 6280021 | LOC441876 | −2.8330393 | Down | Predicted: Similar to 40S RPS16, |
| 870593 | LOC285053 | −2.3860030 | Down | Predicted: Similar to RPL18a, transcript variant 1 |
| 5720747 | LOC441775 | −2.6068625 | Down | Predicted: Similar to 60S RPL18 |
| 2190546 | LOC388654 | −2.2629400 | Down | Predicted: Similar to laminin receptor 1 (RPSA) |
| 5720747 | LOC441775 | −2.6068625 | Down | Predicted: Similar to 60S RPL18 |
| 6330373 | EEF1B2 | −2.3026142 | Down | Eukaryotic translation elongation factor 1 β 2, transcript variant 1 |
| 3850121 | EEF1A1 | −3.1777650 | Down | Eukaryotic translation elongation factor 1 α 1 |
| B, Oxidative phosphorylation and electron transport chain pathway | ||||
| 3850110 | COX6A1 | −2.1455740 | Down | Cytochrome c oxidase subunit Vıa polypeptide 1 |
| 4490259 | COX8A | −2.9969997 | Down | Cytochrome c oxidase subunit 8A (ubiquitous) |
| C, Carbon metabolism | ||||
| 2760358 | NME1-2 | −2.3299380 | Down | NME1-NME2 readthrough |
| 1940360 | TPI1 | −2.8388138 | Down | Triosephosphate isomerase 1 |
| 6590253 | ALDOA | −2.1273860 | Down | ALDOA |
| 6520128 | GPX4 | −2.1577030 | Down | Glutathione peroxidase |
| D, Mammary gland development pathway | ||||
| 5860138 | RIPK4 | −2.1919790 | Down | Receptor-interacting serine-threonine kinase 4 |
| E, G protein-mediated signaling pathway via Gα12/Gα13 family | ||||
| 2850402 | PFN1 | −2.4898353 | Down | Profilin 1 |
| F, Tumor necrosis factor-mediated signaling pathway | ||||
| 670673 | BCL2L1 | −2.0002713 | Down | BCL2-like 1, nuclear gene encoding mitochondrial |
| G, Signaling pathway pertinent to immunity | ||||
| 1980594 | FTHL8 | −3.2119188 | Down | Ferritin, heavy polypeptide-like 8 |
| 2970431 | FTHL7 | −4.2565985 | Down | FTHL7 |
| H, Toll-like receptor signaling pathway | ||||
| 3840154 | SPP1 | −3.0712519 | Down | SPP1, transcript variant 1 |
| I, Signaling by TGF-β receptor complex | ||||
| 1430239 | UBC | −2.8056865 | Down | UBC |
| J, Regulatory and cell adhesion signaling pathways | ||||
| 5570132 | ACTB | −3.4210854 | Down | Actin, β |
| K, NRF2 pathway | ||||
| 4920767 | FTL | −3.1391878 | Down | Ferritin, light polypeptide |
| L, Folate-alcohol and cancer pathway | ||||
| 6510754 | ALDH1A1 | −2.9203625 | Down | Aldehyde dehydrogenase 1 familyer A1 |
| M, Cell adhesion signaling pathway | ||||
| 610437 | CD24 | 2.9155455 | Up | CD24 molecule |
| N, Calcium/calcium-mediated signaling pathway | ||||
| 7100711 | CALM2 | 2.7985630 | Up | Calmodulin 2 (phosphorylase kinase, delta) |
| O, Amino acid metabolism | ||||
| 450161 | FAHD1 | 2.1592160 | Up | Fumarylacetoacetate hydrolase domain containing 1 |
| P, Protein modification pathway | ||||
| Probe ID | Symbol | Fold-changea | Regulation | Definition |
| 4590110 | SEPT9 | 2.2080840 | Up | Septin 9 |
| Q, Cell cycle | ||||
| 870491 | BUB3 | 2.1889267 | Up | BUB3 budding uninhibited by benzimidazoles 3 |
| R, Regulation of actin cytoskeleton | ||||
| 2760292 | PPP1CC | 2.6673288 | Up | Protein phosphatase 1, catalytic subunit |
| S, Vasopressin-regulated water reabsorption | ||||
| 4230520 | DNCL1 | 2.0563870 | Up | Dynein, cytoplasmic, light polypeptide 1 |
| T, Transport pathway | ||||
| 1740136 | SLC38A2 | 2.1263490 | Up | Solute carrier family 38, member 2 |
>2 fold-change was considered to be significant (P<0.05). NME, nucleoside diphosphate kinase; ALDOA, aldolase A, fructose-bisphosphate; BCL-2, B-cell lymphoma 2; RP, ribosomal protein; FTHL7, ferritin, heavy polypeptide-like 7; SPP1, secreted phosphoprotein 1; UBC, ubiquitin C; CD, cluster of differentiation; TGF, transforming growth factor; ACTB, actin, beta; FTL, ferritin, light polypeptide; ALDH1A1, aldehyde dehydrogenase 1 family, member A1; CD24, CD24 molecule; CALM2, calmodulin 2; FAHD1, fumarylacetoacetate hydrolase domain containing 1; BUB3, BUB3 budding uninhibited by benzimidazoles 3; PPP1CC, protein phosphatase 1, catalytic subunit; DNCL1, dynein, cytoplasmic, light polypeptide 1; SLC38A2, solute carrier family 38, member 2.
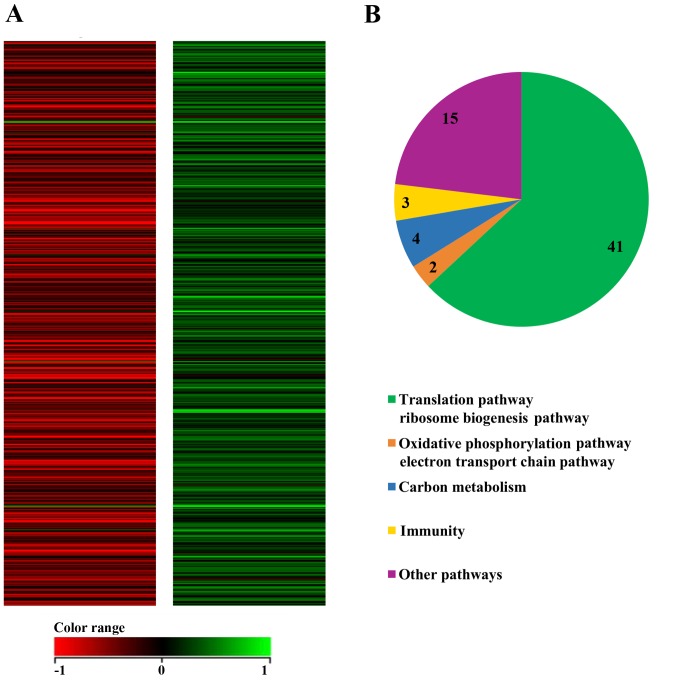
To investigate the mechanism involved in the flavopiridol-induced antiproliferative effect on MCF7 CD44+/CD24− CSCs, pathway analysis was performed using the WikiPathways database (www.wikipathways.org). Specifically, these pathways are involved in the translation pathway, ribosome biogenesis, oxidative phosphorylation, the electron transport chain pathway, carbon metabolism, mammary gland development, protein modification and the cell cycle (Fig. 3A and B).
Figure 3.
(A) Heat map showing the normalized expression of differentially regulated genes, filtering criteria >2-fold change in flavopiridol-treated and untreated MCF7 CD44+/CD24− cancer stem cells. Red color indicates high expression, while green color indicates expression. (B) Pie chart representing the proportion of genes associated with various pathways. CD, cluster of differentiation.
Discussion
BCSCs have been identified as subpopulations of cells within breast tumors that possess tumor-initiating potential in addition to the ability to self-renew and differentiate into a diverse range of progeny cells that make up the tumor (22) These cells are resistant to traditional therapies against cancer, including chemotherapy and radiation therapy (5). Although treatments associated with cancer therapy kill the majority of tumor cells, CSCs are not killed (23). Therefore, a more effective strategy for the treatment of breast cancer may target CSCs. The present study investigated the effect and underlying mechanism of flavopiridol on BCSCs with respect to antitumor properties. The results demonstrated that flavopiridol dose-dependently induced the growth inhibition of BCSCs.
To isolate populations of BCSCs within tumors, the phenotypic definition of a CSC must first be established. CSCs have been identified using cell surface markers in the majority of cancer types. The present study isolated BCSCs based on the CD44+/CD24− phenotype from the breast cancer MCF7 cell line. Al-Hajj et al (9) revealed that breast cancer tumorigenic cells exhibit a CD44+/CD24−/low phenotype. Several studies have used the CD44+/CD24−and/or the aldehyde dehydrogenase (ALDH)+ phenotype for BCSC isolation (9,24). Ginestier et al (25) isolated stem-like cells from primary breast xenografts using CD44+/CD24− and ALDH activity, revealing that these cells displayed the greatest tumor-initiating capacity, generating tumors in non-obese diabetic/severe combined immunodeficiency mice from as little as 20 cells.
Cyclins and CDK inhibitors are involved in cell morphogenesis, adhesion, migration, DNA repair, transcription, cytoskeleton dynamics and cell motility. Flavopiridol is the first CDK inhibitor that exhibits an antitumor effect against a variety of tumor types in several solid tumors (26) The results of the present study revealed that flavopiridol reduced the level of cell viability of BCSCs in a dose- and time-dependent manner, and that flavopiridol appears to possess multiple targets within tumor cells. The number of publications involving the effect of flavopiridol on CSC is quite limited. Soner et al (19) demonstrated that flavopiridol induced growth inhibition and apoptosis by the upregulation of p53 and caspases 3 and 8 in CD133+/CD44+ prostrate CSCs.
The translation and ribosome biogenesis pathways serve important roles in numerous cellular processes and are more active in cancer cells compared with those in normal cells. The inhibition of translation and ribosome biogenesis have been reported to be associated with alterations in the cell cycle and the regulation of cell growth (27). The present study demonstrated that flavopiridol induced the downregulation of translation and ribosome biogenesis genes in CSCs. According to previous studies, flavopiridol induced G1/S-phase cell cycle arrest (28,29). The mechanism of flavopiridol on the cell cycle may be associated with ribosome biogenesis. Cancer cells have been suggested to exhibit a higher rate of ribosome biogenesis compared with that in normal cells. Changes of proto-oncogenes and tumor-suppressor genes activate the mechanisms that stimulate cell growth and proliferation, and initiate certain pathways that enhance ribosome biogenesis (30,31). Derenzini et al (32) demonstrated that the inhibition of ribosomal RNA synthesis caused an accelerated or delayed G1/S-phase progression in rat hepatoma cells.
The present study demonstrated the effects of flavopiridol on BCSCs, suggesting that flavopiridol induced growth inhibition in CD44+/CD24− BCSCs and inhibited the translation and ribosome biogenesis pathways. Flavopiridol is one of the most promising chemotherapy drug candidates for the treatment of cancer. However, the data on the effects of flavopiridol on cancer remain limited. An increased understanding of the mechanisms responsible for the effects of the drug is required to improve novel therapeutic strategies for breast cancer. Combination drug therapies targeting CSCs may be an effective method to prevent relapse and resistance in cancer therapies.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- 3.Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: Markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 4.Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28:4006–4012. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.Bhat-Nakshatri P, Goswami CP, Badve S, Sledge GW, Jr, Nakshatri H. Identification of FDA-approved drugs targeting breast cancer stem cells along with biomarkers of sensitivity. Sci Rep. 2013;3:2530. doi: 10.1038/srep02530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, Mazumdar M, Chakraborty S, Manna A, Saha S, Khan P, Bhattacharjee P, Guha D, Adhikary A, Mukhjerjee S, Das T. Curcumin inhibits breast cancer stem cell migration by amplifying the E-cadherin/β-catenin negative feedback loop. Stem Cell Res Ther. 2014;5:116. doi: 10.1186/scrt506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells; Proc Natl Acad Sci USA; 2003; pp. 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams K, Motiani K, Giridhar PV, Kasper S. CD44 integrates signaling in normal stem cell, cancer stem cell and (pre)metastatic niches. Exp Biol Med (Maywood) 2013;238:324–338. doi: 10.1177/1535370213480714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: Cell cycle regulators and beyond. Dev Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Casimiro MC, Crosariol M, Loro E, Li Z, Pestell RG. Cyclins and cell cycle control in cancer and disease. Genes Cancer. 2012;3:649–657. doi: 10.1177/1947601913479022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Sullivan CC. Overcoming endocrine resistance in hormone-receptor positive advanced breast cancer-the emerging role of CDK4/6 inhibitors. Int J cancer Clin Res. 2015;2:pii: 029. doi: 10.23937/2378-3419/2/4/1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spring L, Bardia A, Modi S. Targeting the cyclin D-cyclin-dependent kinase (CDK) 4/6-retinoblastoma pathway with selective CDK 4/6 inhibitors in hormone receptor-positive breast cancer: Rationale, current status, and future directions. Discov Med. 2016;21:65–74. [PMC free article] [PubMed] [Google Scholar]
- 15.DiPippo AJ, Patel NK, Barnett CM. Cyclin-dependent kinase inhibitors for the treatment of breast cancer: Past, present, and future. Pharmacotherapy. 2016;36:652–667. doi: 10.1002/phar.1756. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro GI. Preclinical and clinical development of the cyclin-dependent kinase inhibitor flavopiridol. Clin Cancer Res. 2004;10:4270s–4275s. doi: 10.1158/1078-0432.CCR-040020. [DOI] [PubMed] [Google Scholar]
- 17.Lin TS, Ruppert AS, Johnson AJ, Fischer B, Heerema NA, Andritsos LA, Blum KA, Flynn JM, Jones JA, Hu W, et al. Phase II study of flavopiridol in relapsed chronic lymphocytic leukemia demonstrating high response rates in genetically high-risk disease. J Clin Oncol. 2009;27:6012–6018. doi: 10.1200/JCO.2009.22.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvajal RD, Tse A, Shah MA, Lefkowitz RA, Gonen M, Gilman-Rosen L, Kortmansky J, Kelsen DP, Schwartz GK, O'Reilly EM. A phase II study of flavopiridol (alvocidib) in combination with docetaxel in refractory, metastatic pancreatic cancer. Pancreatology. 2009;9:404–409. doi: 10.1159/000187135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soner BC, Aktug H, Acikgoz E, Duzagac F, Guven U, Ayla S, Cal C, Oktem G. Induced growth inhibition, cell cycle arrest and apoptosis in CD133+/CD44+ prostate cancer stem cells by flavopiridol. Int J Mol Med. 2014;34:1249–1256. doi: 10.3892/ijmm.2014.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velasco-Velázquez MA, Homsi N, De La Fuente M, Pestell RG. Breast cancer stem cells. Int J Biochem Cell Biol. 2012;44:573–577. doi: 10.1016/j.biocel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skvortsova I, Debbage P, Kumar V, Skvortsov S. Radiation resistance: Cancer stem cells (CSCs) and their enigmatic pro-survival signaling. Semin Cancer Biol. 2015;35:39–44. doi: 10.1016/j.semcancer.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Lawson JC, Blatch GL, Edkins AL. Cancer stem cells in breast cancer and metastasis. Breast Cancer Res Treat. 2009;118:241–254. doi: 10.1007/s10549-009-0524-9. [DOI] [PubMed] [Google Scholar]
- 23.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 24.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senderowicz AM. Flavopiridol: The first cyclin-dependent kinase inhibitor in human clinical trials. Invest New Drugs. 1999;17:313–320. doi: 10.1023/A:1006353008903. [DOI] [PubMed] [Google Scholar]
- 27.Brighenti E, Treré D, Derenzini M. Targeted cancer therapy with ribosome biogenesis inhibitors: A real possibility? Oncotarget. 2015;6:38617–38627. doi: 10.18632/oncotarget.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newcomb EW, Tamasdan C, Entzminger Y, Alonso J, Friedlander D, Crisan D, Miller DC, Zagzag D. Flavopiridol induces mitochondrial-mediated apoptosis in murine glioma GL261 cells via release of cytochrome c and apoptosis inducing factor. Cell Cycle. 2003;2:243–250. doi: 10.4161/cc.2.3.357. [DOI] [PubMed] [Google Scholar]
- 29.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, Van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 30.Sherr CJ. The pezcoller lecture: Cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 31.Kusnadi EP, Hannan KM, Hicks RJ, Hannan RD, Pearson RB, Kang J. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene. 2015;556:27–34. doi: 10.1016/j.gene.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Derenzini M, Montanaro L, Chillà A, Tosti E, Vici M, Barbieri S, Govoni M, Mazzini G, Treré D. Key role of the achievement of an appropriate ribosomal RNA complement for G1-S phase transition in H4-II-E-C3 rat hepatoma cells. J Cell Physiol. 2005;202:483–491. doi: 10.1002/jcp.20144. [DOI] [PubMed] [Google Scholar]