Abstract
Aberrant expression of microRNA (miRNA) is important in the progression of various human cancers, however further investigation is required in order to fully elucidate mechanisms of and validate actions of endogenous non-coding RNAs. The present study demonstrated that the expression of miR-646 was downregulated in colorectal cancer tissues and cell lines. Notably, it was observed that miR-646, a tumor suppressor, inhibited colorectal cancer cell progression through directly targeting Nin one binding protein (NOB1) expression, which possesses anti-tumor properties in colorectal cancer. Furthermore, knockdown of NOB1 expression was responsible for the tumor-suppressive effect of miR-646. The findings suggest that miR-646 may act as a therapeutic target for the treatment of colorectal cancer.
Keywords: renal cell carcinoma, miRNA, proliferation, migration, NOB1
Introduction
Colorectal cancer is one of the most prevalent gastrointestinal cancers in the world, accounting for 9% of adult malignancies (1,2). Unfortunately, an astounding number of colorectal cancer patients have advanced stages diagnosed at initial diagnosis (3). Although surgical resection remains the most curative treatment for colorectal cancer and improves the prognosis of patients, its effect is still limited because patients finally develop relapse or metastasis (4,5). Therefore, the improvement of new diagnostic and treatment strategy for colorectal cancer is urgently required.
In mammals, microRNAs (miRNAs) are a class of small, non-protein-coding potential, single-stranded RNAs which suppress the expression of target genes through directly binding to the 3′-untranslated region (UTR) of mature mRNAs (6,7). Notably, miRNAs have been reported to be differently expressed in human cancers and play crucial roles in cell growth, migration, apoptosis, angiogenesis, invasion, metastasis and so forth (8–12). miRNAs serve as oncogene or tumor suppressor in various human cancers, involving colorectal cancer (13–15). Specifically, miR-646 is a novel miRNAs which was first discovered to be mammalian specific (16). Previous studies reported that miR-646 functioned as an anti-tumor miRNA which abrogated human lung cancer or renal caner progression (17,18). Importantly, one genomic deletion was encountered involving miR-646/AK309218 in copy number variants (CNVs) of colorectal cancer (19). In CpG Island Methylator Phenotype positive (CIMP+) colorectal tumors, miR-646 was downregulated compared with other tumor markers (20). However, the role of miR-646 in colorectal cancer is yet to be elucidated.
NOB1, as an essential protein encoding the Nin one binding protein by two-hybrid screening, is originally discovered in Saccharomyces cerevisiae and involved in the proteasome as well as ribosome biogenesis (21–23). The human NOB1 gene, localized in the nucleus, is mainly expressed in the prostate, colorectal and lung (24–26). NOB1 plays pivotal role of modulating some physiological and pathological functions (27). Upregulation of the NOB1 gene is identified to elicit various human cancers. In human ovarian cancer, downregulation of NOB1 inhibits cell proliferation (28). Similarly, the disregulated expression of NOB1 is identified in papillary thyroid carcinoma and involved in tumorigenesis (29). Suppressed NOB1 blocked various human cancer progressions, including colorectal cancer (30), non-small cell lung cancer (31), oral squamous cell carcinoma (31), laryngeal cancer (31) and so forth.
In the present study, we demonstrated that miR-646 serves as a tumor-suppressive miRNA in colorectal cancer, which blunted colorectal cancer cell proliferation and migration. Furthermore, miR-646 attenuated NOB1 protein level through directly binding its 3′UTR. Notably, we further investigated that downregulation of NOB1 also depressed colorectal cancer progression.
Materials and methods
Clinical sample
A total of 15 colorectal cancer tissues and normal tissues were obtained for patients who underwent surgical resections at the first Department of General Surgery, Shidong Hospital (Shanghai, China). The fresh tissues were frozen in liquid nitrogen to protect the protein or RNA away from degradation. The use of human tissues was approved by the ethics committee of Shidong Hospital.
Cell culture
The human colorectal cancer cell lines HCT116, SW480, SW620 and normal adult human intestine cell lines NCM460 were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HCT116, SW480, SW620 were cultured in serum-containing medium as recommended in a humidified 5% CO2 atmosphere at 37°C. The normal adult human intestine cell lines NCM460 was cultured in M3 base media as previously described (32).
Cell transfection and vector construction
According to manufacturer's protocol, miR-646/mock (GenePharma, Shanghai, China) and scrambled shRNA (stem-loop-stem structure) against NOB1 (sh-NOB1)/sh-con (Guangzhou RiboBio Co., Ltd., Guangzhou, China) were transiently transfected in SW480 or SW620 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) at a final concentration of 50 nM. The cells were harvested at 48 h after transfection. The effects of gene silencing were measured via western blotting and real-time PCR analysis.
The sh-NOB1 sequence was designed and synthesized (sh-NOB1: 5′-AAGGTTAAGGTGAGCTCAT-3′). The sh-con sequence was designed and synthesized: 5′-AATTCTCCGAACGTGTCACGT-3′. The full-length NOB1 (NM_014062.2) cDNA containing plasmid was ordered from IBSbio (IBS Solutions Co. Ltd, Shanghai, China). The complete NOB1 cDNA was reverse transcribed by PCR and ligated into the pcDNA3.1 mammalian expression vector (Invitrogen).
MTT assay
Five days after lentivirus infection, SW480 or SW620 cells were trypsinized, resuspended, seeded in a 96-well plate with a density of 2×103 cells/well and incubated at 37°C. The number of viable cells was measured at daily intervals (days 1–5). At each time-point, 20 µl of 5 mg/ml MTT (Dingguo Biotechnology, Beijing, China) was added and incubation was continued for 4 h. At the end of the incubation period, the medium was removed carefully and 150 µl of acidified isopropanol (in 0.01 M HCl) was added. The plates were agitated and the absorbance was measured at 490 nm on the spectrophotometer Biotek ELx800 (Beijing, China). Each data point was collected from five parallel wells.
Colony formation assay
The SW480 or SW620 cells were seeded in 6-well plates (8×102 cells/well) (in three duplicate wells) and cultured at 37°C in 5% CO2. After two weeks, the cells were washed with PBS once and fixed with paraformaldehyde for 30 min and washed with PBS and stained with Giemsa for 20 min. ddH2O was used to wash the cells three times to obtain a clean background. The number of colonies and cell number in each colony were counted and statistically analyzed.
Wound-healing assay
SW480 or SW620 cells were cultured in 25 cm2 culture flask to 80–90% confluency, collected by digestion and centrifugation, and then seeded into six-well plates at 2.5×105 cells per well. miR-646 mimic/mock or sh-NOB1/sh-con was transfected into SW480 or SW620 cells. The floating cells were removed by washing with PBS, and the width of scratch was observed at 0 and 48 h using inverted microscope (50-fold). Each experiment was repeated three times.
Quantitative real-time PCR (qRT-PCR)
Total RNA from tissues or cells were extracted using Trizol reagent (Invitrogen) according to the manufacturer's protocol. cDNAs were synthesized using a PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan). qRT-PCR was performed with KAKA SYBR FAST qPCR kit (Kapa Biosystems, Inc., Wilmington, MA, USA) using a 7900HT Fast Real-Time PCR system (Applied Biosystems Japan Ltd., Tokyo, Japan). The expression level were normalized to endogenous small nuclear RNA U6 or GAPDH. The 2−ΔΔCt method was used to analyze the expression level relative to the endogenous control. Following primers were used: NOB1-Forward, 5′-ATCTGCCCTACAAGCCTAAAC-3′; NOB1-Reverse, 5′-TCCTCCTCCTCCTCCTCAC-3′; U6-Forward, 5′CTCGCTTCGGCAGCACA3′; U6-Revers, 5′AACGCTTCACGAATTTGCGT3′; GAPDH-Forward, 5′-GAAGGTGAAGGTCGGAGTC-3′; GAPDH-Reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
Protein extracting and western blot analysis
The cells were lysed using RIPA buffer plus protease inhibitors and phosphatase inhibitors. For western blot analysis, 25 µg of protein extracts were loaded to 12% sodium dodecylsulfate-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. The membranes were incubated with a primary antibody at 4° overnight, and incubated with a secondary antibody in 1 h with room temperature. The expression of GAPDH was used as loading control. The information of antibodies were listed as follow: NOB1 (1:10,000; Abcam, Cambridge, MA, USA) or GAPDH (1:5,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Luciferase reporter assay
Luciferase reporter assay using the one step directed cloning kit (Novoprotein, Shanghai, China). The 3′untranslated regions (3′UTR) of NOB1 mRNA containing the intact miR-646 recognition sequences were PCR-amplified and subcloned into the Sac I and Sal I sites of pmirGLO vector. The wild-type plasmid was created containing the 3′-UTR of NOB1 with complementary sequence of miR-646 (NOB1 3′-UTR wt), and mutant plasmid with the mutation sequence without complementary sequence of miR-646 (NOB1 3′-UTR mut1, mut2 and mut3). Following primers were used: (Forward) NOB1-3′-UTR wild-F, 5′-CAAGCTTAGCGAGTTCCCGCAGGCAAAT-3′; (Reverse) NOB1-3′-UTR wild-R, 5′-CTCTAGACATGATCTCTGGGCACAC-3′;
Statistical analysis
All data were analyzed using the Graphpad 6.0 statistical software. Results are expressed as means ± SD from at least 3 independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-646 is downregulated in colorectal cancer tissues and cell lines
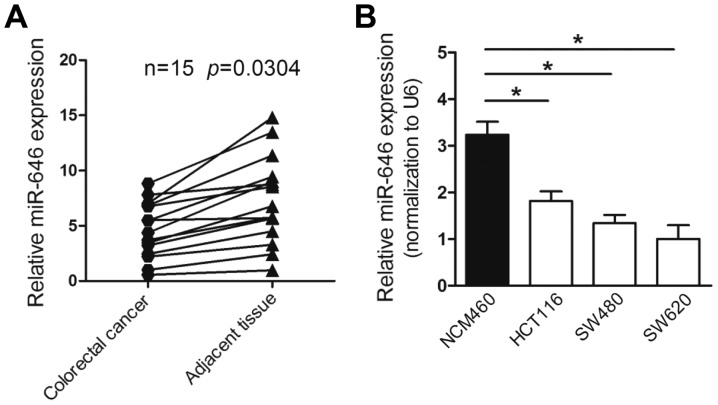
To explore the expression of miR-646 in colorectal cancer, we first performed quantitative real-time PCR (qRT-PCR) to detect miR-646 mRNA level in colorectal cancer tissues and non-tumorous tissues from 15 colorectal cancer patients. The results proved that the expression of miR-646 was remarked downregulated in colorectal cancer compared to adjacent non-cancerous tissues (P<0.05) (Fig. 1A). Furthermore, the mRNA level of miR-646 was lower-expressed in various colorectal cancer cell lines, involving HCT116, SW480 and SW620 compared to normal adult human intestine cell lines NCM460 (Fig. 1B).
Figure 1.
miR-646 is downregulated in colorectal cancer tissues and cell lines. (A) Comparison of miR-646 expression in 15 paired colorectal cancer tissues and adjacent non-cancer tissues via qRT-PCR. U6 was used as an internal control. (B) miR-646 expression in a series of the human colorectal cancer cell lines (HCT116, SW480, SW620) and normal adult human intestine cell lines NCM460. *P<0.05.
Taken together, the results above indicated that the level of miR-646 was reduced in colorectal cancer tissues and cell lines.
miR-646 attenuates colorectal cancer proliferation and migration in vitro
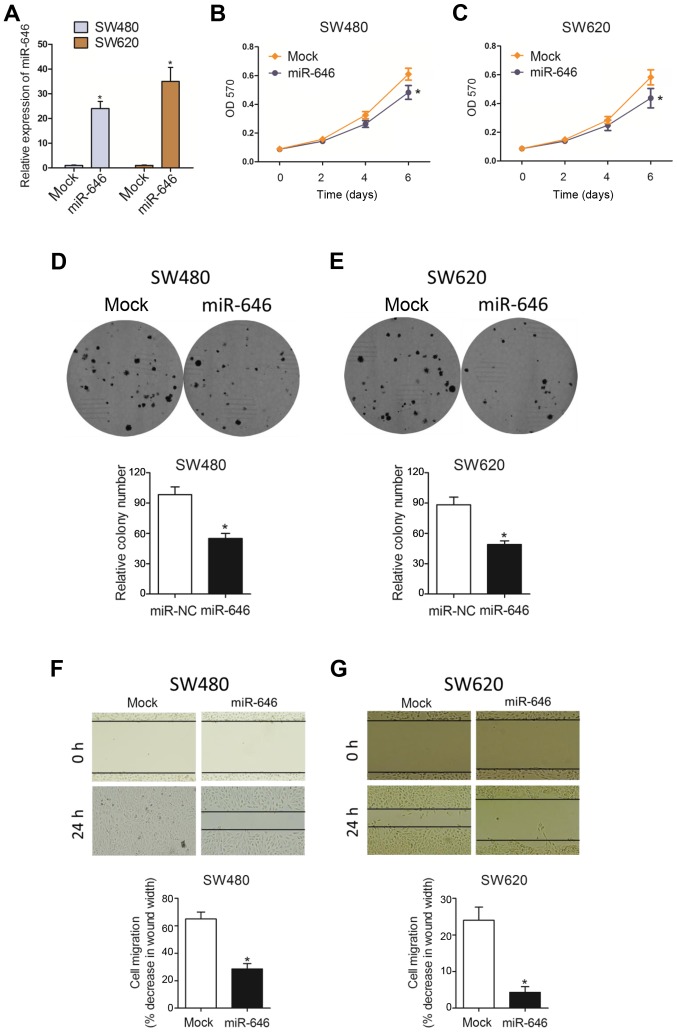
To examine the phenotypic role of miR-646 in colorectal cancer progression, we constructed colorectal cancer cell lines (SW480 and SW620) with miR-646 mimics to overexpress miR-646 expression(Fig. 2A). As Fig. 2B and C showed, MTT assay revealed that overexpressed miR-646 (miR-646) suppressed colorectal cancer cell proliferation in both SW480 and SW620 cell lines compared to the control group (mock). Similarly, we also substantiated that miR-646 attenuated cell proliferation in both cells using colony formation assay (Fig. 2D and E). In addition, compared to mock group, miR-646 mimic significantly abolished colorectal cancer cell migration through wound-healing assay (Fig. 2F and G).
Figure 2.
miR-646 attenuates colorectal cancer proliferation and migration in vitro. (A) qRT-PCR analysis after the transfection with miR-646 in SW480 and SW620 cells when compared to that of mock group. (B-C) MTT proliferation change with miR-646 comparing to mock in SW480 (B) and SW620 (C) cells. (D-E) Colony formation assay after the transfection with miR-646 in SW480 (D) and SW620 (E) cells when compared to that of mock group. (F-G) Wound healing assay after the transfection with miR-646 in SW480 (F) and SW620 (G) cells when compared to that of mock group. *P<0.05 vs. (A-C, F, G) mock or (D and E) miR-NC.
Taken together, all data above suggested that miR-646 inhibited colorectal cancer cell proliferation and migration.
miR-646 directly targets NOB1
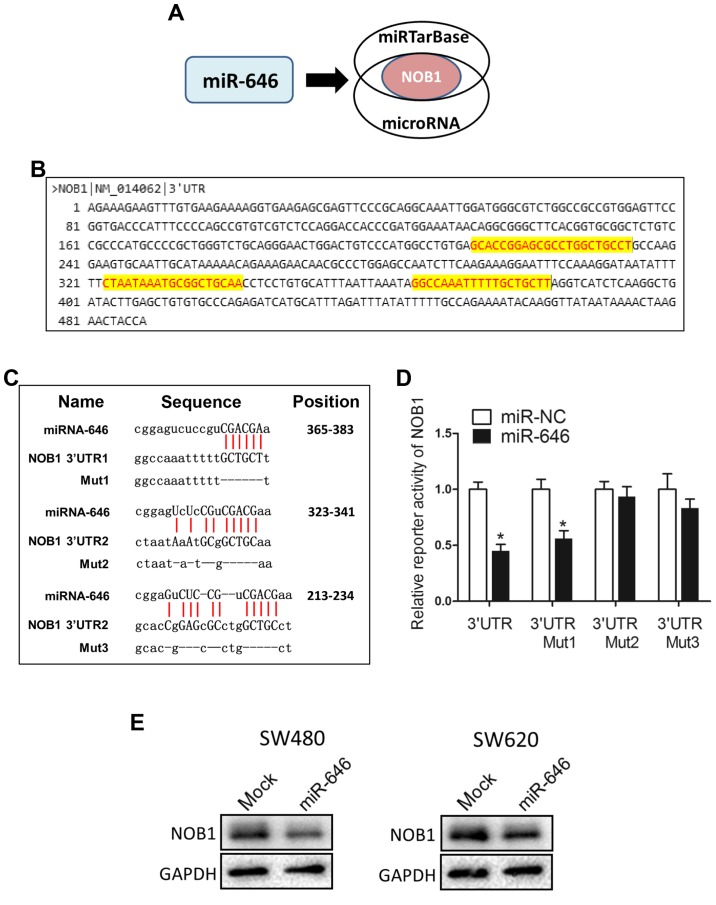
To dissect the detailed mechanism underlying miR-646-mediated depression of colorectal cancer progression, we used two different miRNA target-predicting algorithms (miRTarBase and microRNA) to select the potential target genes of miR-646 and then focused on only one candidate target genes related with cell proliferation and migration (NOB1) (Fig. 3A). Fig. 3B showed that NOB1 included three putative miR-646 binding sites in its 3′UTR region. We constructed luciferase reporters through cloning the 3′-UTR of NOB1 wild-type as well as NOB1 mutant-type via the deletion complementary sequence located at 365–383 (Mut1), 323–341 (Mut2) and 213–234 (Mut3) (Fig. 3C). Dual-luciferase reporter assays elucidated that the coexpression of miR-646 in 293T cells significantly reduced the firefly luciferase reporter activity of the NOB1 wild-type 3′-UTR but barely impacted the mutant 3′-UTR (Fig. 3D). Interestingly, for reporters with a single mutant 3′-UTR, only that of the NOB1 mut1 was also downregulated by miR-646 (Fig. 3D). This result attested that wild-type and mut2/3 3′-UTR of NOB1 were targeted by miR-646. In addition, we performed WB to confirm that overexpressed miR-646 blunted NOB1 protein level into both colorectal cancer cell lines (Fig. 3E).
Figure 3.
miR-646 directly targets NOB1. (A) The schematic illustration of the proposed model depicting of NOB1 as the potential target gene of miR-646 as mentioned above from 2 microRNA prediction databases, miRTarBase and microRNA. (B) The possible binding sites in the 3′UTR of NOB1. (C) NOB1 were predicted to be miR-646 targets, including predicted binding site in the 3′-UTRs of NOB1; sequences of the wild-type (NOB1 3′-UTR) and mutated 3′-UTR-Renilla luciferase reporters (NOB1 3′-UTR Mut1, Mut2 and Mut3). (D) NOB1 3′-UTR luciferase reporter assays in 293 T cells. (E) Western blot analysis for NOB1 protein levels of miR-646 comparing to mock transfection in SW480 and SW620 cell lines.*P<0.05 vs. miR-NC.
Taken together, these results suggested that miR-646 abrogated NOB1 protein and activity through directly targeting the 3′-UTR of NOB1.
Sh-NOB1 deletes cell proliferation and migration in colorectal cancer
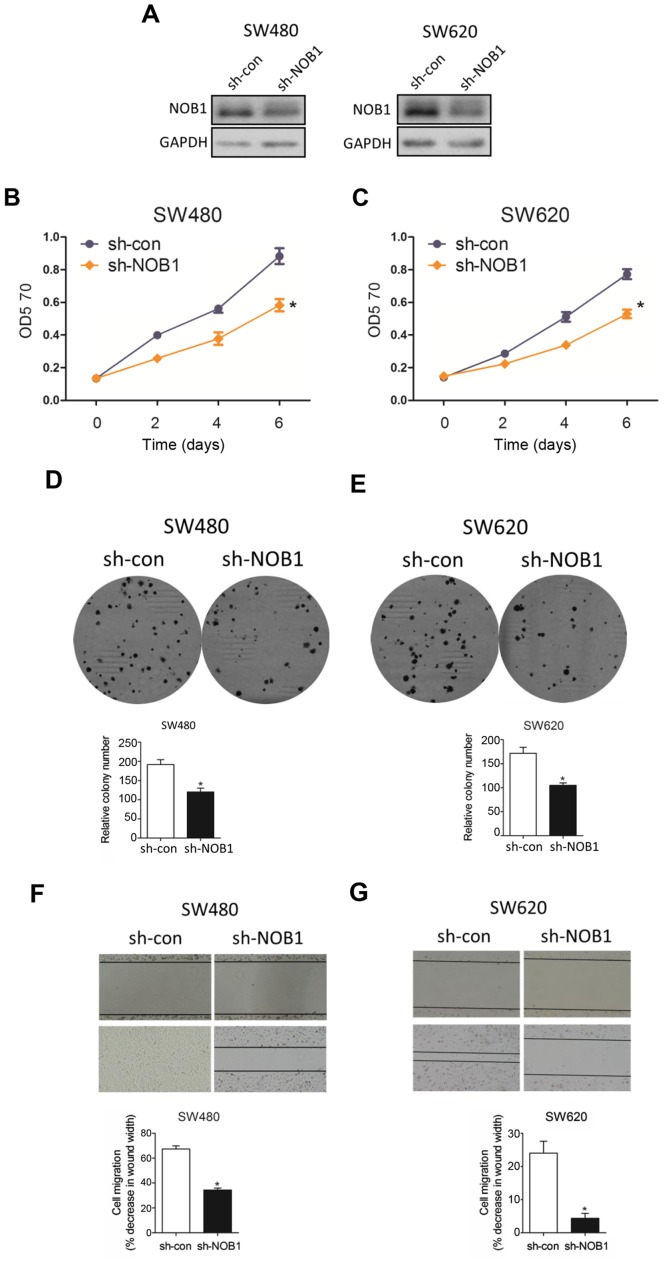
In an attempt to visualize the functional role of NOB1 in colorectal cancer, we knocked down NOB1 expression with sh-NOB1 and WB analysis substantiated that sh-NOB1 potently interfered NOB1 protein in both cell lines (Fig. 4A). As expected, sh-NOB1 minimized cell proliferation with MTT assay (Fig. 4B) and colony formation assay (Fig. 4C). In parallel, sh-NOB1 substantially depressed cell migration capability through wound-healing assay (Fig. 4D), consistent with the phenotype caused by overexpressed miR-646 in colorectal cancer.
Figure 4.
Sh-NOB1 deletes cell proliferation and migration in colorectal cancer. (A) Western blot analysis for NOB1 protein levels of sh-NOB1 comparing to sh-con transfection in SW480 and SW620 cell lines. (B-C) MTT proliferation change with sh-NOB1 comparing to sh-con in SW480 (B) and SW620 (C) cells. (D-E) Colony formation assay after the transfection with sh-NOB1 in SW480 (D) and SW620 (E) cells when compared to that of sh-con group. (F-G) Wound healing assay after the transfection with sh-NOB1 in SW480 (F) and SW620 (G) cells when compared to that of sh-con group.*P<0.05 vs.sh-con.
Taken together, all data above suggested that sh-NOB1 attenuated colorectal cancer cell progression.
Discussion
In the present work, we reported that miR-646 expression was downregulated in colorectal cancer tissues and cell lines compared to that in surrounding non-cancerous tissues as well as normal human intestine cells. Functional studies certified that overexpressed miR-646 obviously suppressed cell growth and cell migration via MTT, colorectaly formation and wound healing assay in SW480 and SW620 cell lines. Importantly, miR-646 targeted downstream gene NOB1 through directing binding a regulatory element in its coding region. Furthermore, knocking-down NOB1 expression was responsible for the tumor-suppressive effect of miR-646. Collectively, all these findings supported the conclusion that bona fide miR-646 served as a key modulator of NOB1 expression in human colorectal cancer cells, which revealed a novel mechanism of colorectal cancer progression.
Altered miRNAs expression has been reported to be associated with tumor progression and overall survival in patients with colorectal cancer. A series of miRNAs have been shown to be anti-miRNAs in colorectal cancer. Zhang et al identified that miR-600 abolished cell proliferation, migration and invasion by targeting p53 in mutant p53-expressing human colorectal cancer cell lines (33). Another study reported that miR-219-5p attenuated the proliferation and invasion of colorectal cancer cells by targeting calcyphosin (34). Recently, miR-145, acting as a classic anti-miRNA, could inhibited human colorectal cancer cell migration and invasion via PAK4-dependent pathway (35). On the contrary, an astounding number of miRNAs have been reproted to be an oncogene in colorectal cancer. Specifically, miR-141 facilitated colon cancer cell proliferation by inhibiting MAP2K4 (36). Liu et al demonstrated that miR-19a enhanced colorectal cancer proliferation and migration by targeting TIA1 (37). Lu et al miR-146a induced cell migration and invasion in human colorectal cancer via carboxypeptidase M/src-FAK pathway (38). In brief, aberrant miRNAs could function as tumor suppressors or oncogenes involving multiple biological functions in colorectal cancer development.
A bulk of evidence indicated that miR-646 acted as a tumor suppressor in various human cancers (17,18). Specially, upregulated miR-646, which was a prognosis factor for overall survival of lung cancer, not only decreased EGFR pathway, but also reduced cell proliferation and metastasis of lung cancer (17). On the other hand, miR-646 functioned as an anti-tumor miRNA to repress cell growth and migration in renal cancer (18). Consistently, our present findings also demonstrated that miR-646 was downregulated in colorectal cancer tissues and cell lines to block tumor progression through downregulating specific oncogenic targets-NOB1. Therefore, miR-646 could be considered as a vital tumor suppressor in vast majority of human cancers.
Previous studies elucidated that NOB1 was found to be upregulated in various human tumors and could be the potential target for several miRNAs (39–41). Specifically, NOB1, as the target gene of miR-326, was associated with poor prognosis in glioma and gastric cancer. In cervical cancer, NOB1 reversed the effect of miR-139-3p on inhibiting cell metastasis while inducing cell apoptosis. In our work, we reported that downregulation of NOB1, as a target of miR-646, attenuated cell growth and migration of colorectal cancer.
In conclusion, our report substantiated that miR-646 acts as a suppressive non-coding RNA to mediate colorectal cancer cell progression through targeting NOB1 expression. Our finding provided a new therapeutic approach of colorectal cancer.
References
- 1.Boyle P, Langman JS. ABC of colorectal cancer: Epidemiology. BMJ. 2000;321:805–808. doi: 10.1136/bmj.321.7264.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Walsh JM, Terdiman JP. Colorectal cancer screening: Scientific review. JAMA. 2003;289:1288–1296. doi: 10.1001/jama.289.10.1288. [DOI] [PubMed] [Google Scholar]
- 4.Kang H, O'Connell JB, Maggard MA, Sack J, Ko CY. A 10-year outcomes evaluation of mucinous and signet-ring cell carcinoma of the colon and rectum. Dis Colon Rectum. 2005;48:1161–1168. doi: 10.1007/s10350-004-0932-1. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 6.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White NM, Bao TT, Grigull J, Youssef YM, Girgis A, Diamandis M, Fatoohi E, Metias M, Honey RJ, Stewart R, et al. miRNA profiling for clear cell renal cell carcinoma: Biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. J Urol. 2011;186:1077–1083. doi: 10.1016/j.juro.2011.04.110. [DOI] [PubMed] [Google Scholar]
- 9.Nelson KM, Weiss GJ. MicroRNAs and cancer: Past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Osanto S, Qin Y, Buermans HP, Berkers J, Lerut E, Goeman JJ, van Poppel H. Genome-wide microRNA expression analysis of clear cell renal cell carcinoma by next generation deep sequencing. PLoS One. 2012;7:e38298. doi: 10.1371/journal.pone.0038298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 13.Li C, Xu N, Li YQ, Wang Y, Zhu ZT. Inhibition of SW620 human colon cancer cells by upregulating miRNA-145. World J Gastroenterol. 2016;22:2771–2778. doi: 10.3748/wjg.v22.i9.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Song Y, Liu TJ, Cui YB, Jiang Y, Xie ZS, Xie SL. miRNA-22 suppresses colon cancer cell migration and invasion by inhibiting the expression of T-cell lymphoma invasion and metastasis 1 and matrix metalloproteinases 2 and 9. Oncol Rep. 2013;29:1932–1938. doi: 10.3892/or.2013.2300. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed FE. miRNA as markers for the diagnostic screening of colon cancer. Expert Rev Anticancer Ther. 2014;14:463–485. doi: 10.1586/14737140.2014.869479. [DOI] [PubMed] [Google Scholar]
- 16.Finnerty JR, Wang WX, Hébert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: Evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Chen Y, Ma D, Ji Z, Cao F, Chen Z, Ning Y, Bai C. miR-646 is a key negative regulator of EGFR pathway in lung cancer. Exp Lung Res. 2016;42:286–295. doi: 10.1080/01902148.2016.1207726. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Liu M, Feng Y, Xu YF, Huang YF, Che JP, Wang GC, Yao XD, Zheng JH. Downregulated miR-646 in clear cell renal carcinoma correlated with tumour metastasis by targeting the nin one binding protein (NOB1) Br J Cancer. 2014;111:1188–1200. doi: 10.1038/bjc.2014.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatachalam R, Verwiel ET, Kamping EJ, Hoenselaar E, Görgens H, Schackert HK, van Krieken JH, Ligtenberg MJ, Hoogerbrugge N, van Kessel AG, Kuiper RP. Identification of candidate predisposing copy number variants in familial and early-onset colorectal cancer patients. Int J Cancer. 2011;129:1635–1642. doi: 10.1002/ijc.25821. [DOI] [PubMed] [Google Scholar]
- 20.Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: Differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011;50:196–206. doi: 10.1002/gcc.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tone Y, Tanahashi N, Tanaka K, Fujimuro M, Yokosawa H, Toh-e A. Nob1p, a new essential protein, associates with the 26S proteasome of growing saccharomyces cerevisiae cells. Gene. 2000;243:37–45. doi: 10.1016/S0378-1119(99)00566-1. [DOI] [PubMed] [Google Scholar]
- 22.Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyler E, Zimmermann M, Widmann B, Gstaiger M, Pfannstiel J, Kutay U, Zemp I. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA. 2011;17:189–200. doi: 10.1261/rna.2325911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Huang H, Yuan B, Zhuang LY, Luo TP, Zhang Q. Lentivirus-mediated knockdown of NOB1 suppresses the proliferation of colon cancer cells. Z Gastroenterol. 2014;52:429–435. doi: 10.1055/s-0033-1356338. [DOI] [PubMed] [Google Scholar]
- 25.Liu K, Chen HL, Gu MM, You QS. Relationship between NOB1 expression and prognosis of resected non-small cell lung cancer. Int J Biol Markers. 2015;30:e43–e48. doi: 10.5301/jbm.5000120. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Zhang D, Qu F, Hong Y, Cao J, Pan X, Li L, Huang Y, Huang H, Yin L, et al. Knockdown of NOB1 expression inhibits the malignant transformation of human prostate cancer cells. Mol Cell Biochem. 2014;396:1–8. doi: 10.1007/s11010-013-1920-3. [DOI] [PubMed] [Google Scholar]
- 27.Veith T, Martin R, Wurm JP, Weis BL, Duchardt-Ferner E, Safferthal C, Hennig R, Mirus O, Bohnsack MT, Wöhnert J, Schleiff E. Structural and functional analysis of the archaeal endonuclease Nob1. Nucleic Acids Res. 2012;40:3259–3274. doi: 10.1093/nar/gkr1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y, Peng S, Yu H, Teng H, Cui M. RNAi-mediated downregulation of NOB1 suppresses the growth and colony-formation ability of human ovarian cancer cells. Med Oncol. 2012;29:311–317. doi: 10.1007/s12032-010-9808-5. [DOI] [PubMed] [Google Scholar]
- 29.Lin S, Meng W, Zhang W, Liu J, Wang P, Xue S, Chen G. Expression of the NOB1 gene and its clinical significance in papillary thyroid carcinoma. J Int Med Res. 2013;41:568–572. doi: 10.1177/0300060513479862. [DOI] [PubMed] [Google Scholar]
- 30.He XW, Feng T, Yin QL, Jian YW, Liu T. NOB1 is essential for the survival of RKO colorectal cancer cells. World J Gastroenterol. 2015;21:868–877. doi: 10.3748/wjg.v21.i3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W, Zhong W, Xu J, Su B, Huang G, Du J, Liu Q. Lentivirus-mediated gene silencing of NOB1 suppresses non-small cell lung cancer cell proliferation. Oncol Rep. 2015;34:1510–1516. doi: 10.3892/or.2015.4132. [DOI] [PubMed] [Google Scholar]
- 32.Moyer MP, Manzano LA, Merriman RL, Stauffer JS, Tanzer LR. NCM460, a normal human colon mucosal epithelial cell line. In Vitro Cell Dev Biol Anim. 1996;32:315–317. doi: 10.1007/BF02722955. [DOI] [PubMed] [Google Scholar]
- 33.Zhang P, Zuo Z, Wu A, Shang W, Bi R, Jin Q, Wu J, Jiang L. miR-600 inhibits cell proliferation, migration and invasion by targeting p53 in mutant p53-expressing human colorectal cancer cell lines. Oncol Lett. 2017;13:1789–1796. doi: 10.3892/ol.2017.5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Zhu L, Jiang Y, Xu J, Wang F, He Z. miR-219-5p suppresses the proliferation and invasion of colorectal cancer cells by targeting calcyphosin. Oncol Lett. 2017;13:1319–1324. doi: 10.3892/ol.2017.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheng N, Tan G, You W, Chen H, Gong J, Chen D, Zhang H, Wang Z. MiR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med. 2017;6:1331–1340. doi: 10.1002/cam4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding L, Yu LL, Han N, Zhang BT. miR-141 promotes colon cancer cell proliferation by inhibiting MAP2K4. Oncol Lett. 2017;13:1665–1671. doi: 10.3892/ol.2017.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui S, Gu Y, Sun W, You C, Liu Z, et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16:53. doi: 10.1186/s12943-017-0625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu D, Yao Q, Zhan C, Le-Meng Z, Liu H, Cai Y, Tu C, Li X, Zou Y, Zhang S. MicroRNA-146a promote cell migration and invasion in human colorectal cancer via carboxypeptidase M/src-FAK pathway. Oncotarget. 2017;8:22674–22684. doi: 10.18632/oncotarget.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J, Xu T, Yan Y, Qin R, Wang H, Zhang X, Huang Y, Wang Y, Lu Y, Fu D, Chen J. MicroRNA-326 functions as a tumor suppressor in glioma by targeting the Nin one binding protein (NOB1) PLoS One. 2013;8:e68469. doi: 10.1371/journal.pone.0068469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang P, Xi J, Liu S. MiR-139-3p induces cell apoptosis and inhibits metastasis of cervical cancer by targeting NOB1. Biomed Pharmacother. 2016;83:850–856. doi: 10.1016/j.biopha.2016.07.050. [DOI] [PubMed] [Google Scholar]
- 41.Ji S, Zhang B, Kong Y, Ma F, Hua Y. MiR-326 inhibits gastric cancer cell growth through down regulating NOB1. Oncol Res. 2017;25:853–861. doi: 10.3727/096504016X14759582767486. [DOI] [PMC free article] [PubMed] [Google Scholar]