Abstract
The present study explored the expression of tetraspanin 1 (TSPAN1) in esophageal carcinoma (EC) and its association with clinicopathological factors. TSPAN1 small interfering RNA (siRNA) was designed to target the TSPAN1 gene in Eca-109 cells in order to explore the biological function of TSPAN1 in the proliferation and apoptosis of EC. The results demonstrated that the level of TSPAN1 expression in EC tissue was significantly increased compared with that in adjacent normal tissue (P<0.001). TSPAN1 expression was also associated with histological differentiation, depth of invasion, lymph node metastasis (all P<0.05) and Ki-67 (P<0.01). However, no association was observed between TSPAN expression and gender, age or location (P>0.05). In addition, silencing TSPAN1 markedly inhibited proliferation while increasing the apoptosis rate of Eca-109 cells, which was demonstrated by detecting the expression of the cell proliferation-associated gene Ki-67 and the apoptotic gene caspase-3 (P<0.05). Taken together, these results indicated that TSPAN1 functions as a tumor-associated gene in EC through promoting cell proliferation and suppressing apoptosis, and siRNA technology may provide an advanced alternative in the development of therapeutics for EC.
Keywords: esophageal carcinoma, tetraspanin 1, RNA interference, proliferation, apoptosis
Introduction
Esophageal carcinoma (EC) is one of the most fatal cancers of the digestive system worldwide (1). Although great improvements have been made in the diagnosis and surgical treatment of EC, the overall survival rate remains poor (2). Therefore, it is necessary to study the molecular mechanisms of the development of EC. In particular, studying the role of the new tumor-associated genes in EC may help to elucidate the mechanisms of EC initiation and progression, and provide effective targets for treatment of EC. Tetraspanin 1 (TSPAN1; GenBank accession no., AF065388; also termed NET-1), which encodes a 241 amino acid protein and is located at chromosome 1p34.1, belongs to the tetraspanin superfamily (TM4SF) and was identified by Serru et al (3) from the EST database in 2000. TM4SF, a specific family member of cell membrane glycoproteins, is widely expressed in numerous tissues and cells and mediates extracellular signal transduction and performs an important role in cell development, growth and movement through the role of the four hydrophobic regions in determining the boundaries of extracellular transmembrane regions of cysteine residues (4).
Previous studies revealed that TSPAN1 was highly expressed in a number of cancers (5,6), but the role of TSPAN1 in EC was unclear. In addition, it was also uncertain whether TSPAN1 affects the proliferation and apoptosis of EC cells. Therefore, the present study detected TSPAN1 protein in EC tissues and evaluated its association with clinicopathological factors and progression of EC. The effects of target inhibiting TSPAN1 with siRNA on the EC cells were examined, and the effects on proliferation and apoptosis of EC cells were observed.
Materials and methods
Patients
A total of 60 samples of EC tissue from resected specimens and adjacent normal tissues were obtained from the Affiliated Tumor Hospital of Nantong University (Nantong, China) between April 2013 and April 2015. The study was approved by the local medical ethics committee of Nantong University, and written prior informed consent and approval were signed by the patients.
Immunohistochemical staining
Histological tumor typing was performed on the basis of resected specimens in the Department of Pathology of the Affiliated Tumor Hospital of Nantong University. The tissues were routinely fixed in 10% natural-buffered formalin for 24 h at 20°C and embedded in paraffin. The 4 µm tissue sections were dewaxed using xylene and rehydrated in graded alcohols. To reduce non-specific background staining, endogenous peroxidase activity was blocked by 3% hydrogen peroxide for 15 min at room temperature. The sections were incubated with the following primary antibodies: Rabbit anti-human polyclonal TSPAN1 antibody (cat. no. ab221740; Abcam, Cambridge, UK; dilution, 1:200) and mouse anti-human monoclonal Ki-67 antibody (cat. no. ab8191; Abcam; dilution, 1:200) at 4°C overnight. Sections were then washed three times with PBS, followed by incubation with goat anti-rabbit horseradish peroxidase (HRP)-conjugated IgG (cat. no. ab6721; Abcam; dilution, 1:100) and goat anti-mouse HRP-conjugated IgG (cat no. ab6789; Abcam; dilution, 1:100) secondary antibodies for 1 h at room temperature. Subsequent to washing with PBS three times for 10 min each, the sections were incubated with 3,3′-diaminobenzidine dilute, lightly counterstained with hematoxylin for 1 min at room temperature, and observed under a light microscope (magnification, ×200).
Intensity and extent of TSPAN1 and Ki-67 expression levels were thoroughly evaluated by two experienced pathologists who were blinded to the clinical results and the staining results were divided into three categories: (−), <10% positive parenchyma cells and/or weakly stained; (+), ≥10% positive parenchyma cells and/or strongly stained (7).
Cell culture
The ECEca-109 cells were provided by Biomics Biotechnologies Co., Ltd. (Nantong, China) and cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a humidified incubator (5% CO2).
Small interfering RNA (siRNA) construction and cell transfection
The experiment was divided into three groups: TSPAN1 siRNA group (siRNA group), negative control group (NC group) and untreated group (Un group). According to an optimization principle of siRNA, a 19 nt sequence-specific siRNA targeting TSPAN1 was designed (TSPAN1 siRNA; Biomics Biotechnologies Co., Ltd.) for studying TSPAN1 in vitro. The TSPAN1 siRNA group sequences were 5′-CCACAAUGGCUGAGCACUUdTdT-3′ (sense) and 5′-AAGUGCUCAGCCAUUGUGGdTdT-3′ (antisense). The negative control group sequences were 5′-UUCUCCGAACGUGUCACGUdTdT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAAdTdT-3′ (antisense). According to the manufacturer's protocol, TSPAN1 siRNA or NC siRNA were transfected using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at a final concentration of 25 nmol/l when the cells reached 70% confluence. All results were repeated three times.
Immunofluorescence microscopy
Eca-109 cells were cultured on 24-well plates (1×105 cells/well), transfected within 48 h, washed twice with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. The cells were permeabilized with 0.1% Triton X-100 for 15 min, followed by blocking with 1% bovine serum albumin (Sigma-Aldrich; Merck KGaA) in PBS for 30 min at room temperature. Cells were incubated at 4°C overnight with anti-TSPAN1 polyclonal antibody (cat. no. ab221740; Abcam; dilution, 1:200), then washed extensively with PBS, followed by labeling with a secondary tetramethylrhodamine-labeled antibody (cat. no. T6778; Sigma-Aldrich; Merck KGaA, dilution, 1:100) for 2 h at room temperature. The nuclei were stained by Hoechst 33258 (5 µg/ml; Invitrogen; Thermo Fisher Scientific, Inc.) for 15 min at room temperature, and the stained cells in five different fields of view were observed by fluorescence microscopy (magnification, ×400; Nikon Corporation, Tokyo, Japan).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of TSPAN1 mRNA levels
Subsequent to being transfected with siRNA for 48 h, total RNA was extracted from cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. According to the One-step Quantace kit (Quantace, Alexandria, Australia), RT-qPCR was performed in a volume of 25 µl, containing 1 µl of each primer mix (10 µM/ml), 12 µl of 2X Master Mix, 0.5 µl of 50X SYBR-Green I, 6.5 µl ddH2O and 5 µl RNA. The mixtures were first subjected to 30 min at 42°C for reverse transcription and initially denatured for 10 min at 94°C, followed by 40 cycles of amplification with the following cycling parameters: 20 sec at 95°C, 30 sec at 55°C and 30 sec at 72°C. The primer pairs for TSPAN1 gene were designed with Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA). GAPDH served as an internal control for RT-qPCR. The primer sequences were as follows: TSPAN1 forward, 5′-GTGGCTTCACCAACTATACG-3′ and reverse, 5′-GACTGCATTAGTTCGGATGT-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTC-3′. The expression levels of each gene were determined using the 2−ΔΔCq method (8).
Western blot analysis
Total protein was extracted from all groups of cells following transfection for 48 h with a radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China), and the protein concentration was determined using the Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). A total of 25 µg of protein were loaded into each lane, separated by 10% SDS-PAGE, transferred to a polyvinylidene fluoride membrane and blocked for 2 h at room temperature with 5% non-fat milk in TBS containing 0.1% Tween-20 (TBST) to exclude non-specific binding. The membranes were incubated with anti-TSPAN1 (cat. no. ab221740; Abcam; dilution, 1:1,000), anti-Ki-67 (cat. no. ab8191; Abcam; dilution, 1:1,000), mouse anti-caspase-3 (cat. no. C5737; Sigma-Aldrich; Merck KGaA; dilution, 1:1,000), and mouse anti-β-actin (cat. no. BM0626; Boster Biological Technology, Pleasanton, CA, USA; dilution, 1:2,000) primary antibodies overnight at 4°C. The membranes were washed three times in TBST for 5 min and then incubated with goat anti-rabbit (cat. no. ab6721; Abcam; dilution, 1:5,000) and goat anti-mouse (cat. no. ab6789; Abcam; dilution, 1:5,000) secondary antibodies for 2 h at room temperature, followed by washing three times in TBST for 5 min. The enhanced chemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.) was added to the target proteins and the film was visualized. The density of the bands was semi-quantitatively analyzed by Image J densitometry (v.2.1.4.7; National Institutes of Health, Bethesda, MA, USA). The detected protein was normalized by β-actin.
Detection of cell proliferation using the MTT assay
Cells on coverslips were cultured in 96-well plates for 0, 24, 48 and 72 h, at a density of 2×103 cells/well at 37°C. At the time cells were transfected with siRNA and NC for 0, 24, 48 and 72 h, MTT solution (0.2 mg/ml; Sigma-Aldrich; Merck KGaA) was added to each well for incubating cells at 37°C for 4 h away from light. The supernatant was then removed, and 150 µl dimethylsulfoxide (Sigma-Aldrich; Merck KGaA) was added to dissolve the purple crystals in each well. The optical density was determined using a Microplate Reader (Bio-Rad 680; Bio-Rad Laboratories, Inc., Hercules, CA, USA) at a wavelength of 490 nm, and cell growth curves were constructed.
Cell apoptosis analysis using fluorescein Αnnexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) double labeling
Cellular apoptosis was determined by the Annexin V-FITC Apoptosis Detection kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Cells were cultured on 6-well plates and trypsinized at 37°C. At 48 h post-transfection, cells were collected and resuspended in 400 µl 1X binding buffer, at 800 × g for 10 min at 4°C and supernatant was removed. Cells were then resuspended in 200 µl 1X binding buffer and transferred to a sterile flow cytometry (FCM) glass tube. Subsequently, 3 µl Αnnexin V-FITC and 3 µl propidium iodide were added and then incubated at 37°C for 15 min in the dark. Cells were then analyzed by CyFlow space FCM (BD Biosciences, Franklin Lakes, NJ, USA). The distribution of cells was analyzed using Cell-Quest software (v. FCS2.0; BD Biosciences) within 1 h of staining. Data from 10,000 cells was collected for each data file. Apoptotic cells were identified as Αnnexin V-FITC-positive and PI-negative cells.
Statistical analysis
The rates of TSPAN1 expression and clinicopathological factors were compared using one-way analysis of variance with post hoc contrasts by Student-Newman-Keuls test and χ2 test analyses. The cell apoptosis analysis experiments were performed independently at least three times and comparisons between the experimental groups are expressed as the mean ± standard deviation. Statistical analysis was performed using SPSS v.20.0 statistical software (IBM SPSS, Armonk, NY, USA) and image processing was performed with GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significance difference.
Results
Clinical cases and tumor characteristics
Of the 60 EC cases, 11 (18.33%) of lesions occurred in the upper esophagus, 28 (46.67%) occurred in the median esophagus and 21 (35.00%) occurred in the lower esophagus. Among them, 50 cases were male and 10 cases were female, with a median age of 63.30 years (range, 32–75 years). None of the patients received radiotherapy or chemotherapy prior to diagnosis. According to the states of cell differentiation and dysplasia appearances, cases were divided histologically into high (30.00%, 18 cases), moderate (30.00%, 18 cases) and low differentiation (40.00%, 24 cases). Depending on the depth of tumor invasion, the cases were divided into three groups: T1 in 13 cases (21.67%), T2 in 17 cases (28.33%) and T3 in 30 cases (50.00%). There were 36 cases (60.00%) with lymph node metastasis. Immunohistochemistry results also revealed that Ki-67 was expressed in EC tissue of all 60 cases, with high expression in 53 cases (88.33%) and low expression in 7 cases (11.67%).
TSPAN1 expression is increased in EC tissues compared with normal tissues
Immunohistochemical staining was used to examine the expression of TSPAN1 protein in 60 cases of EC tissue and adjacent normal tissue. Positive granules stained yellow were mainly distributed in the cytoplasm of tumor cells, and the expression of TSPAN1 protein in EC tissue (51 cases, 85.00%) was significantly higher compared with that in adjacent normal tissue (23 cases, 38.33%), with statistical significance (χ2=27.638; P<0.001). In 85.00% of EC cases, TSPAN1 was stained brown, and there was an association between increased TSPAN1 expression and lower differentiation. Furthermore, there was an association between TSPAN1 and Ki-67 expression (χ2=11.039, P=0.001; Fig. 1). The association between TSPAN1 expression and the clinicopathological parameters in EC was analyzed using the χ2 test (Table I). A statistically significant association was observed between TSPAN1 expression and histological differentiation (χ2=9.020; P=0.011), depth of tumor invasion (χ2=8.908; P=0.012) and lymph vessel invasion (χ2=6.296; P=0.012).
Figure 1.
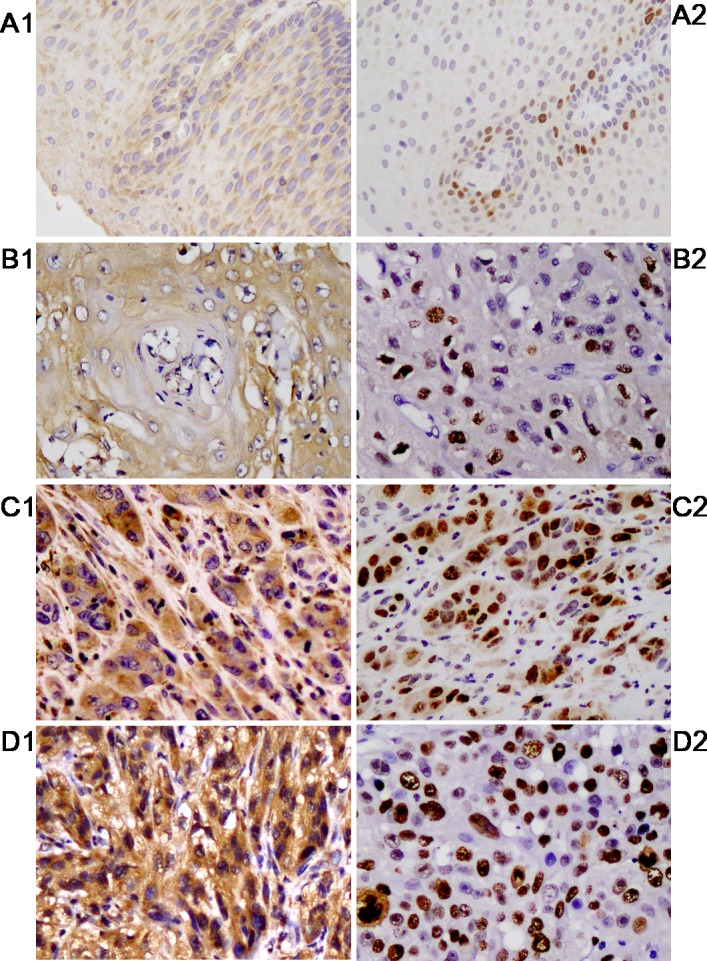
The expression of TSPAN1 and Ki-67 in EC tissue and adjacent normal tissue. Paraffin sections of EC were stained with (A) anti-TSPAN1 polyclonal antibody and (B) Ki-67 monoclonal antibody by immunocytochemistry. Brown indicates expression of the protein location and TSPAN1 located in the cytoplasm, Ki-67 located in the nuclear is indicated by yellow granulation. (A1 and B1) TSPAN1 and Ki-67 expressed negatively/or weakly in adjacent normal tissue. (A2 and B2) High differentiation, (A3 and B3) moderate differentiation and (A4 and B4) low differentiation. Magnification, ×200. EC, esophageal cancer; TSPAN1, tetraspanin 1.
Table I.
Clinicopathological characteristics of high TSPAN1 expression in 60 patients with esophageal carcinoma.
| Groups | Patients, n (%) | TSPAN1 high expression, n (%) | χ2 | P-value |
|---|---|---|---|---|
| Gender | ||||
| Male | 50 (83.33) | 43 (86.00) | 0.235 | 0.628 |
| Female | 10 (16.67) | 8 (80.00) | ||
| Age, years | ||||
| ≤60 | 23 (38.33) | 18 (78.26) | 1.329 | 0.249 |
| >60 | 37 (61.67) | 33 (89.18) | ||
| Location | ||||
| Upper esophagus | 11 (18.33) | 8 (72.72) | 1.711 | 0.425 |
| Median esophagus | 28 (46.67) | 25 (89.28) | ||
| Lower esophagus | 21 (35.00) | 18 (85.71) | ||
| Histological differentiation | ||||
| High | 18 (30.00) | 12 (66.67) | 9.020 | 0.011 |
| Moderate | 18 (30.00) | 15 (83.33) | ||
| Low | 24 (40.00) | 24 (100.00) | ||
| Depth of invasion | ||||
| T1a | 13 (21.67) | 8 (61.53) | 8.908 | 0.012 |
| T2b | 17 (28.33) | 14 (82.35) | ||
| T3c | 30 (50.00) | 29 (96.67) | ||
| Lymph node metastasis | ||||
| Positive | 36 (60.00) | 34 (94.44) | 6.296 | 0.012 |
| Negative | 24 (40.00) | 17 (70.83) | ||
| Ki-67 expression | ||||
| High | 53 (88.33) | 48 (90.57) | 11.039 | 0.001 |
| Low | 7 (11.67) | 3 (42.85) |
Depth of the tumor up to
mucous and submucosa
muscle
fibrous membranes and adjacent organs. TSPAN1, tetraspanin 1.
Immunofluorescence analysis of TSPAN1
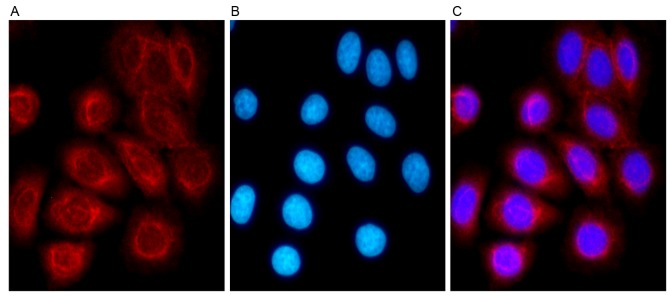
Immunofluorescence analysis was used to detect the location of TSPAN1 protein in Eca-109 cells, and a confocal microscope was used to observe fluorescence. The results indicated that TSPAN1 protein was distributed evenly in the cytoplasm around the nucleus of Eca-109 cells (Fig. 2).
Figure 2.
Immunofluorescence staining was used to detect TSPAN1 protein expression in Eca-109 cells. TSPAN1 protein was stained with anti-TSPAN1 polyclonal antibody and marked by (A) tetramethylrhodaminein red, which was located in the cytoplasm. (B) The nuclei were stained with Hoechst-33258 in blue. (C) Merged figure. Magnification, ×400. TSPAN1, tetraspanin.
TSPAN1 mRNA and protein levels
RT-qPCR and western blot analysis revealed a significant decrease in the mRNA and protein levels of the TSPAN1 siRNA group compared with the negative control group. As demonstrated in Fig. 3, TSPAN1 siRNA inhibited TSPAN1 expression at the mRNA level up to 67.7% (Fig. 3) in comparison with that of the negative control group (0.27±0.05 vs. 0.95±0.10; P<0.05). As demonstrated in Fig. 4, a silencing effect of TSPAN1 siRNA was observed at the protein level up to 62.0% compared with the negative control group (0.35±0.05 vs. 0.97±0.12; P<0.05), indicating effective silencing at the mRNA and protein levels.
Figure 3.
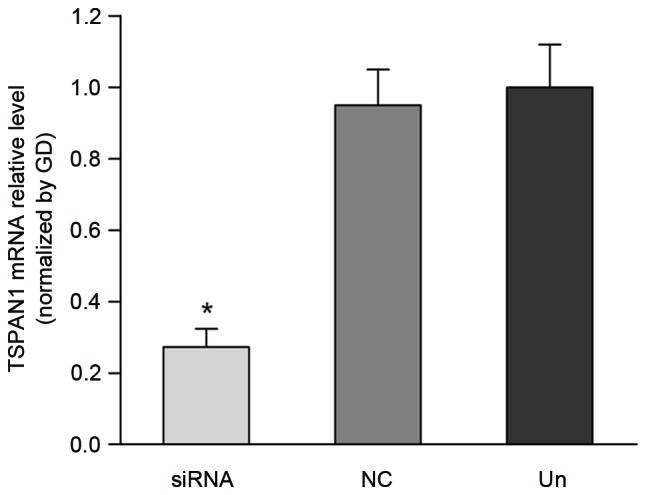
Expression of TSPAN1 mRNA in Eca-109 cells was detected by reverse transcription-quantitative polymerase chain reaction following siRNA transfection. The TSPAN1 mRNA level was significantly decreased in the TSPAN1 siRNA group compared with the negative control group (0.27±0.05 vs. 0.95±0.10; *P<0.05). TSPAN1, tetraspanin 1; siRNA, small interfering RNA; NC, negative control; Un, untreated; GD, GAPDH served as an internal control. GD, GAPDH.
Figure 4.
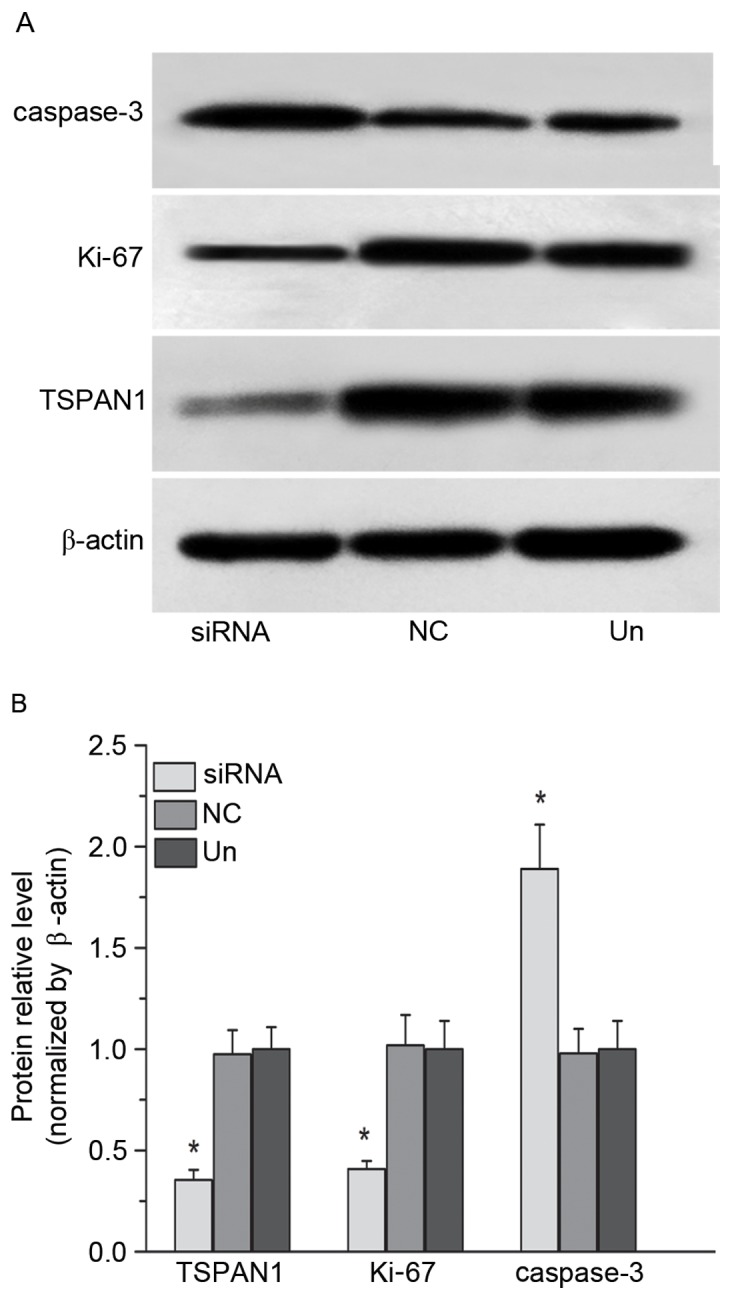
Expression levels of TSPAN1, Ki-67 and caspase-3 protein in Eca-109 cells were measured by (A) western blot analysis and (B) densitometric analysis. Compared with the negative control group, the TSPAN1 protein level was significantly decreased in the TSPAN1 siRNA group (0.35±0.05 vs. 0.97±0.12), the Ki-67 protein level was highly reduced in the TSPAN1 siRNA group (0.41±0.04 vs. 1.02±0.15) and the caspase-3 protein level was increased in the TSPAN1 siRNA group (1.89±0.22 vs. 0.98±0.12). All data are expressed as a percentage of the negative control group volume density. *P<0.05. TSPAN1, tetraspanin 1; siRNA, small interfering RNA; NC, negative control; Un, untreated.
TSPAN1 silencing inhibits the expression of Ki-67 and promotes the expression of caspase-3
The optical density value for the bands for each of the three groups was compared with β-actin. The expression of the cell proliferation-associated protein Ki-67 in the TSPAN1 siRNA group was significantly decreased (up to 61.0%) compared with the negative control group (0.41±0.04 vs. 1.02±0.15; P<0.05; Fig. 4). The expression of the apoptosis-associated protein caspase-3 was significantly increased (up to 91.0%) in the TSPAN1 siRNA group compared with the negative control group (1.89±0.22 vs. 0.98±0.12; P<0.05; Fig. 4). These results demonstrated that silencing the TSPAN1 gene could inhibit the expression of Ki-67 and promote the expression of caspase-3.
Knockdown of the TSPAN1 gene inhibits cell proliferation
The effect of silencing of TSPAN1 on proliferation of Eca-109 cells was examined. The absorbance values of Eca-109 cells at 48 and 72 h after transfection with TSPAN1 siRNA were significantly decreased compared with the negative control group (1.15±0.10 vs. 1.54±0.09 at 48 h; 1.65±0.13 vs. 2.43±0.11 at 72 h; P<0.05; Fig. 5).
Figure 5.
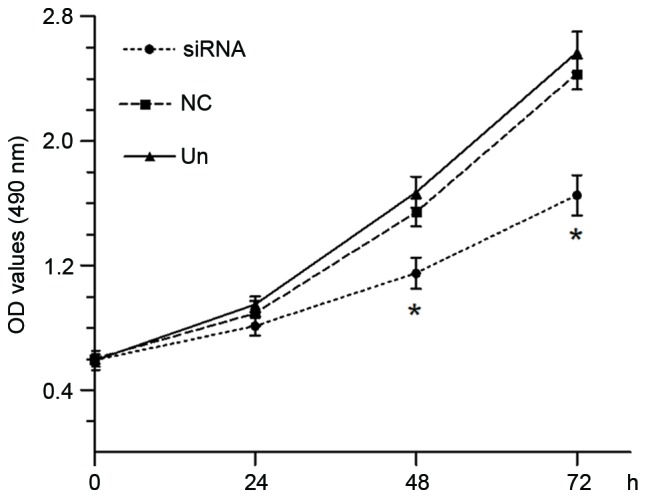
The proliferation of Eca-109 cells was assayed using MTT at 0, 24, 48 and 72 h post-transfection. Growth curve showed that the OD values (490 nm) obviously decreased in TSPAN1 siRNA group at 48 (1.15±0.10 vs. 1.54±0.09) and 72 h (1.65±0.13 vs. 2.43±0.11). *P<0.05 compared with the negative control group at the same time point. TSPAN1, tetraspanin 1; siRNA, small interfering RNA; OD, optical density; NC, negative control; Un, untreated.
TSPAN1 gene silencing induces cell apoptosis
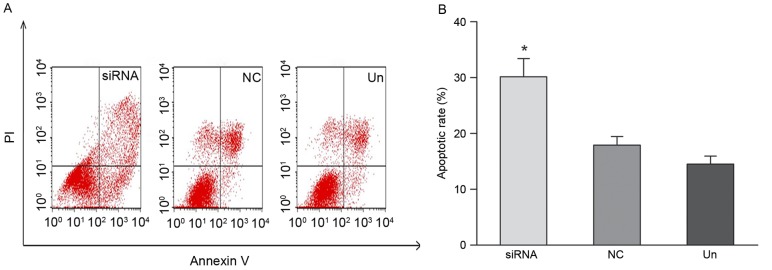
Apoptosis induced by silencing the TSPAN1 gene was evaluated using the Annexin V-FITC/PI staining assay and flow cytometry analysis. As demonstrated in Fig. 6, apoptotic cells were significantly increased by TSPAN1 siRNA treatment as compared with the negative control group (30.16±3.24 vs. 17.92±1.52%; P<0.05).
Figure 6.
Eca-109 cell apoptosis was detected using Annexin V-fluorescein isothiocyanate/PI staining assay by flow cytometry. (A) Cells in the lower left quadrant represent survivals; lower right quadrant represent early apoptosis; the upper right quadrant represent post-apoptotic and the upper left quadrant represent necrosis. (B) Data are expressed as the early apoptosis rate percentage and the result demonstrated that compared with the negative control group, the early apoptosis significantly increased in the tetraspanin 1 siRNA group (30.16±3.24 vs. 17.92±1.52%; *P<0.05). PI, propidium iodide; siRNA, small interfering RNA; NC, negative control; Un, untreated.
Discussion
As a new member of the tetraspanin group, TSPAN1 is a recently discovered tumor-associated gene (7,9–13). Previous studies have demonstrated that TSPAN1 is associated with the occurrence and development of numerous cancers, including hepatocellular carcinoma (7), gastric carcinoma (9), colorectal carcinoma (10), lung cancer (11), skin cancer (12) and pancreatic cancer (13). RNA interference (RNAi) technology is an effective approach to silence tumor gene expression for studies of gene function, and has the potential for gene therapy (14–17). Synthetic siRNA can trigger RNAi responses in mammalian cells and induce inhibition of gene expression (14). The specific siRNA used in the current study has been used in a previous study and confirmed to be effective (15). He et al (16) reported that silencing TSPAN1 using siRNA in liver carcinoma MHCC97H cells suppressed cell proliferation and induced apoptosis. Chen et al (17) reported that silencing TSPAN1 depressed the proliferation and growth of skin carcinoma A431 cells. These previous studies are in agreement with the present study, in which TSPAN1 siRNA was designed to investigate the effects of TSPAN1 in the proliferation and apoptosis of EC cells, and to disclose the role of TSPAN1 in EC.
Esophageal carcinogenesis is a complex process, which involves a variety of molecular events associated with fundamental alterations in cell physiological processes (18). Elucidation of the underlying molecular mechanisms of esophageal carcinogenesis is required to develop novel treatments. In the present study, 60 EC and matched adjacent normal tissue cases were initially studied to identify the association between TSPAN1 and clinicopathological parameters of patients with EC. In adjacent normal tissues, TSPAN1 was weakly stained. In EC tissues, TSPAN1 expressed strongly and was particularly enhanced in metastatic emboli in blood vessels, filtrating depth muscle and in poorly differentiated cells. The positive ratio of TSPAN1 expression was significantly increased in EC compared with adjacent normal tissue. The results supported the hypothesis that TSPAN1 over expression may be an important molecular event in the progression of EC. The present study demonstrated that lower differentiation EC tissue exhibited higher expression of TSPAN1, which indicated that TSPAN1 may be a marker for cell differentiation phenotype. In addition, TSPAN1 over expression was associated with histological differentiation, depth of tumor infiltration and lymph node metastasis, which indicated that the accumulation of TSPAN1 in cancer cells is associated with the malignant progression of EC. These results are similar to the studies by Zhang et al (12) and Wollscheid et al (19). TSPAN1 has been demonstrated to accumulate at the cell surface in EC, and may transduce signals of cell proliferation, synergistically promoting EC development (3). Evaluate cancer cell proliferation and detecting the expression level of Ki-67 in tumors demonstrated that TSPAN1 may function with Ki-67 to increase tumor cell proliferation. Similarly, several studies have reported that the development of cancers is associated with the activity and expression of TSPAN1 in a variety of cancers, including hepatocellular carcinoma, skin squamous carcinoma and osteoclastogenesis (4,7,17).
With the development of RNAi technology, a siRNA technique was used to down regulate TSPAN1 in the Eca-109 cell line. Compared with the negative group and the untreated group, RT-qPCR and western blot analysis results revealed that the mRNA expression level of the TSPAN1 decreased (up to 67.7%) and the protein expression level of the TSPAN1 (up to 62.0%) decreased within 48 h post-transfection of TSPAN1 siRNA in Eca-109 cells. MTT assay revealed that cell proliferation was inhibited (up to 39% at 48 h; 78% at 72 h) and flow cytometry showed that apoptosis was induced (30.16±3.24 vs. 17.92±1.52%). The present study demonstrated that TSPAN1 siRNA effectively knocked down the expression of TSPAN1 in Eca-109 cells and reduced the cell growth and proliferation. By contrast, to elucidate the molecular function of TSPAN1 on promoting proliferation and inhibiting apoptosis, the expression of the key regulators Ki-67 and caspase-3 was examined. To the best of our knowledge, this is the first study to demonstrate that the expression level of Ki-67 is significantly decreased (up to 61.0%) and caspase-3 is significantly increased (up to 91.0%) in Eca-109 cells transfected with TSPAN1 siRNA. Ki-67 was known to be over expressed in EC and in the early events of esophageal carcinogenesis, and is also a useful biomarker for early detection (20). By contrast, caspase-3 is a member of the caspase family of enzymes, which are the major inducers of apoptosis (21). This apoptotic protein is identified as one of the mechanisms by which cancer cells accelerate apoptosis, and is activated in the apoptotic cell by extrinsic (death ligand) and intrinsic (mitochondrial) pathways (22). The present study revealed that silencing the TSPAN1 gene by siRNA resulted in an increase in caspase-3 expression, consequently promoting the apoptosis of EC.
In summary, the results of the present study demonstrated that TSPAN1 is highly expressed in EC cases, which may be regarded as one of the independent favorable indicators for the initiation and development of EC. The results in Eca-109 cells demonstrated that the main function of the TSPAN1 gene may be to promote cell proliferation by increasing levels of Ki-67, and to decrease apoptosis by down regulating caspase-3 expression. Therefore, TSPAN1 maybe a candidate gene for diagnosis and a target gene for therapy of EC.
Acknowledgements
The present study was supported by the foundation of the production-study-research prospective joint research programs of Jiangsu Province, China (grant no. BY 2013042-06), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, and from the Science Foundation of Nantong City, Jiangsu, China (grant no. BK2014001).
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen SB, Weng HR, Wang G, Yang JS, Yang WP, Liu DT, Chen YP, Zhang H. Surgical treatment for early esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. 2013;14:3825–3830. doi: 10.7314/APJCP.2013.14.6.3825. [DOI] [PubMed] [Google Scholar]
- 3.Serru V, Dessen P, Boucheix C, Rubinstein E. Sequence and expression of seven new tetraspans. Biochim Biophys Acta. 2000;1478:159–163. doi: 10.1016/S0167-4838(00)00022-4. [DOI] [PubMed] [Google Scholar]
- 4.Iwai K, Ishii M, Ohshima S, Miyatake K, Saeki Y. Expression and function of transmembrane-4 superfamily (tetraspanin) proteins in osteoclasts: Reciprocal roles of Tspan-5 and NET-6 during osteoclastogenesis. Allergol Int. 2007;56:457–463. doi: 10.2332/allergolint.O-07-488. [DOI] [PubMed] [Google Scholar]
- 5.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: Molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 6.Claas C, Wahl J, Orlicky DJ, Karaduman H, Schnölzer M, Kempf T, Zöller M. The tetraspanin D6.1A and its molecular partners on rat carcinoma cells. Biochem J. 2005;389:99–110. doi: 10.1042/BJ20041287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Yuan D, Wang GL, Wang Y, Wu YY, Zhu J. Clinicopathological significance of expression of Tspan-1, Jab1 and p27 in human hepatocellular carcinoma. J Korean Med Sci. 2010;25:1438–1442. doi: 10.3346/jkms.2010.25.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 9.Lu Z, Luo T, Nie M, Pang T, Zhang X, Shen X, Ma L, Bi J, Wei G, Fang G, Xue X. TSPAN1 functions as an oncogene in gastric cancer and is downregulated by miR-573. FEBS Lett. 2015;589:1988–1994. doi: 10.1016/j.febslet.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Fei B, Wang Q, Song M, Yin Y, Zhang B, Ni S, Guo W, Bian Z, Quan C, et al. MicroRNA-638 inhibits cell proliferation, invasion and regulates cell cycle by targeting tetraspanin 1 in human colorectal carcinoma. Oncotarget. 2014;5:12083–12096. doi: 10.18632/oncotarget.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Peng W, Lu Y, Chen J, Zhu YY, Xi T. MiR-200a enhances the migrations of A549 and SK-MES-1 cells by regulating the expression of TSPAN1. J Biosci. 2013;38:523–532. doi: 10.1007/s12038-013-9351-6. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Wang J, Chen L, Wang G, Qin J, Xu Y, Li X. Expression and function of NET-1 in human skin squamous cell carcinoma. Arch Dermatol Res. 2014;306:385–397. doi: 10.1007/s00403-013-1423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou FQ, Lei XF, Yao JL, Wang YJ, Zhang W. Tetraspanin 1 is involved in survival, proliferation and carcinogenesis of pancreatic cancer. Oncol Rep. 2015;34:3068–3076. doi: 10.3892/or.2015.4272. [DOI] [PubMed] [Google Scholar]
- 14.Li T, Zhu YY, Chen L, Sun Y, Yuan J, Graham M, French P. Size unbiased representative enzymatically generated RNAi (SURER) library and application for RNAi therapeutic screens. Nucleic Acid Ther. 2015;25:35–46. doi: 10.1089/nat.2014.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang GL, Chen L, Wei YZ, Zhou JM, Wu YY, Zhang YX, Qin J, Zhu YY. The effect of NET-1 on the proliferation, migration and endocytosis of the SMMC-7721 HCC cell line. Oncol Rep. 2012;27:1944–1952. doi: 10.3892/or.2012.1698. [DOI] [PubMed] [Google Scholar]
- 16.He S, Wei YZ, Wang GL, Xu YY, Zhou JM, Zhang YX, Chen L. Study of RNA interference targeting NET-1 combination with sorafenib for hepatocellular carcinoma therapy in vitro and in vivo. Gastroenterol Res Pract. 2013;2013:685150. doi: 10.1155/2013/685150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Zhu Y, Li H, Wang GL, Wu YY, Lu YX, Qin J, Tuo J, Wang JL, Zhu J. Knockdown of TSPAN1 by RNA silencing and antisense technique inhibits proliferation and infiltration of human skin squamous carcinoma cells. Tumori. 2010;96:289–295. doi: 10.1177/030089161009600217. [DOI] [PubMed] [Google Scholar]
- 18.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 19.Wollscheid V, Kühne-Heid R, Stein I, Jansen L, Köllner S, Schneider A, Dürst M. Identification of a new proliferation-associated protein NET-1/C4.8 characteristic for a subset of high-grading cervical intraepithelial neoplasia and cervical carcinoma. Int J Cancer. 2002;99:771–775. doi: 10.1002/ijc.10442. [DOI] [PubMed] [Google Scholar]
- 20.Jin Y, Zhang W, Liu B. Abnormal expression of p53, Ki67 and iNOS in human esophageal carcinoma in situ and pre-malignant lesions. Zhonghua Zhong Liu Za Zhi. 2001;23:129–131. (In Chinese) [PubMed] [Google Scholar]
- 21.Cohen GM. Caspases: The executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, Yang X, Yao S, LwinOo T, He H, Wang A, Li C, He L. Reactive oxygen species burst induced by aluminum stress triggers mitochondria-dependent programmed cell death in peanut root tip cells. Plant Physiol Biochem. 2014;82:76–84. doi: 10.1016/j.plaphy.2014.03.037. [DOI] [PubMed] [Google Scholar]