Abstract
Atomic force microscopy (AFM)-based single-molecule force spectroscopy (SMFS) is a powerful yet accessible means to characterize the unfolding/refolding dynamics of individual molecules and resolve closely spaced, transiently occupied folding intermediates. On a modern commercial AFM, these applications and others are now limited by the mechanical properties of the cantilever. Specifically, AFM-based SMFS data quality is degraded by a commercial cantilever’s limited combination of temporal resolution, force precision, and force stability. Recently, we modified commercial cantilevers with a focused ion beam to optimize their properties for SMFS. Here, we extend this capability by modifying a 40 × 18 μm2 cantilever into one terminated with a gold-coated, 4 × 4 μm2 reflective region connected to an uncoated 2-μm-wide central shaft. This “Warhammer” geometry achieved 8.5-μs resolution coupled with improved force precision and sub-pN stability over 100 s when measured on a commercial AFM. We highlighted this cantilever’s biological utility by first resolving a calmodulin unfolding intermediate previously undetected by AFM and then measuring the stabilization of calmodulin by myosin light chain kinase at dramatically higher unfolding velocities than in previous AFM studies. More generally, enhancing data quality via an improved combination of time resolution, force precision, and force stability will broadly benefit biological applications of AFM.
Main Text
Single-molecule force spectroscopy (SMFS) provides valuable insights into diverse biophysical systems (1). One particularly exciting application is studying the unfolding and refolding of nucleic acid structures (2) and proteins (3, 4). Detecting closely spaced and/or transiently occupied intermediate states yields insights into a molecule’s folding pathway (5, 6, 7, 8, 9). Such studies require a technically challenging triumvirate of experimental capabilities: temporal resolution, force precision, and force stability. Temporal resolution and force precision are needed to distinguish closely spaced and briefly occupied states. Force stability enables equilibrium assays, where individual molecules repeatedly unfold and refold (2) and thereby also enables reconstruction of a one-dimensional free-energy landscape along the stretching axis (10). Dual-beam optical traps have emerged as the SMFS modality of choice for such studies (6, 7, 8, 10) due to their combination of force stability and precision. For instance, studies of calmodulin with an optical trap resolved additional folding intermediates (7) that were previously undetected by highly stable, custom atomic force microscopy (AFM) (11).
Historically, AFMs have had poor force precision and stability in comparison to custom-built optical traps (1). However, commercial AFMs are much more user accessible. SMFS on a modern AFM is now limited by the mechanical properties of commercial cantilevers rather than the rigidity of the AFM frame (12, 13, 14). For example, we achieved sub-pN stability over 100 s by removing the gold coating from long, soft cantilevers (L = 100 μm; k ≈ 7 pN/nm) (12). Yet these cantilevers still suffered from relatively poor force precision and time resolution (450 ms) in comparison to the best optical trapping results (15).
A high-speed AFM using ultrashort cantilevers (L = 9 μm) (16), on the other hand, has exceeded the time resolution of advanced optical traps (15) (0.7 versus 6–10 μs, respectively). However, this benefit comes at the expense of force precision due to underdamped motion (quality factor (Q) > 0.5) and stability due to low-frequency (low-ƒ) noise (14). Recently, we optimized ultrashort cantilevers for SMFS by modifying them with a focused ion beam (FIB), achieving 1-μs resolution and improved force precision (14). Yet these cantilevers had only moderate stability, achieving sub-pN performance over ∼1–3 s. Further, detecting such modified, ultrashort cantilevers required retrofitting our commercial AFM with a home-built detection system (14). Thus, there is an exciting opportunity in AFM-based SMFS to combine excellent time resolution and extended force stability with the ease of use provided by an unmodified commercial AFM.
Here, we extend our earlier efforts in modifying cantilevers with an FIB (13, 14) to achieve 8.5-μs resolution coupled with sub-pN stability over 100 s on an unmodified commercial AFM. We demonstrated the utility of this, to our knowledge, new cantilever geometry by unfolding a single protein domain embedded in a polyprotein (Fig. 1 A), and thereby resolved a calmodulin unfolding intermediate previously undetected by AFM (11). We also measured calmodulin stabilization by myosin light chain kinase (MLCK) at much higher unfolding velocities than in earlier AFM studies (11).
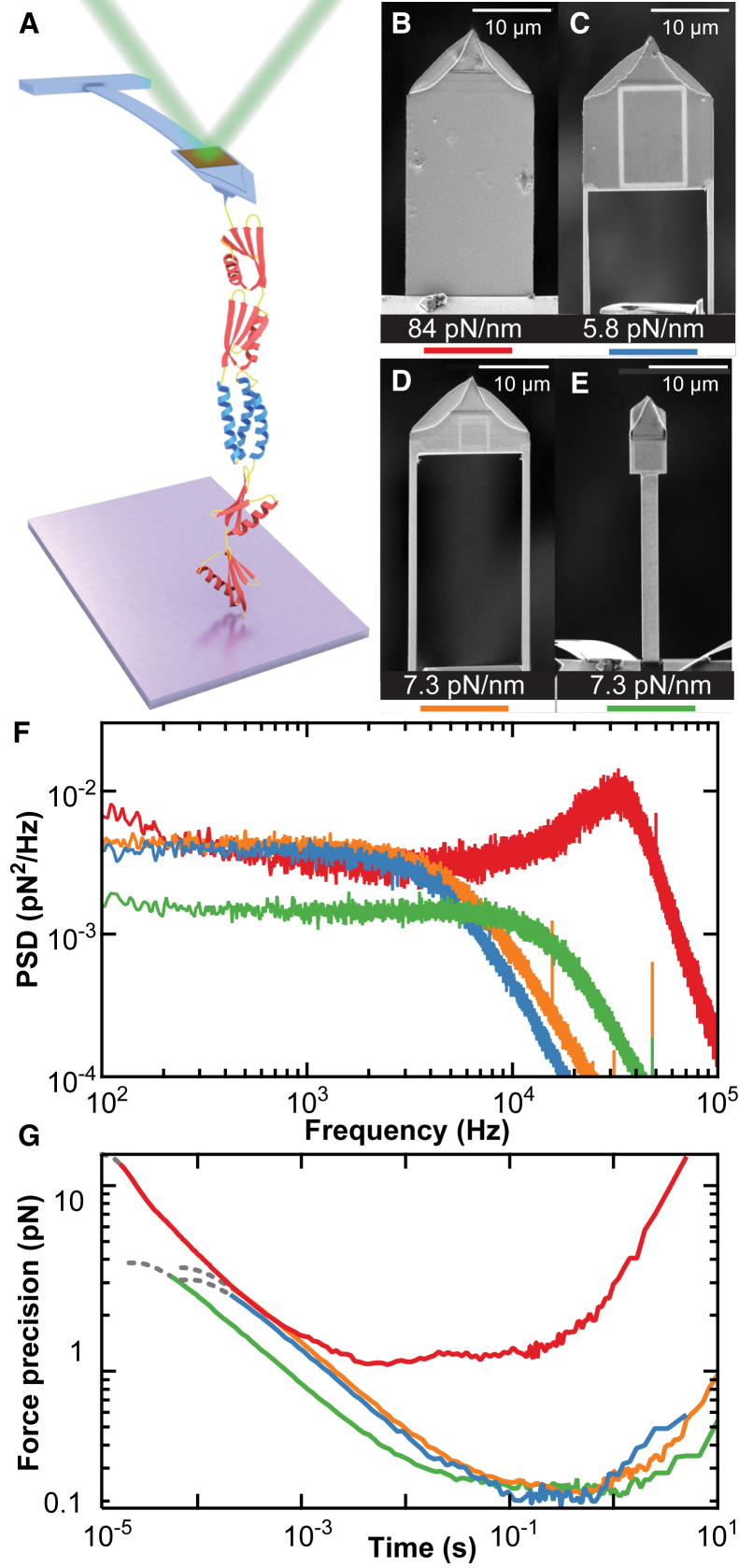
Figure 1.
Improved performance of modified AFM cantilevers. (A) Schematic of the assay showing a polyprotein consisting of four domains of NuG2 (red) and one domain of α3D (blue) being unfolded with a Warhammer cantilever. (B–E) Images of cantilevers prior to gold removal: an unmodified BioLever Mini (B), a standard Mod Mini (C), a Long-cut Mod Mini (D), and a Warhammer (E). The cantilever’s spring constant is noted below each image. (F) Comparison of the force PSD for each cantilever using the color code denoted in (B)–(E). (G) Force precision over a given averaging time, technically the Allan deviation (17). At the very shortest times, the motion of the cantilever becomes correlated, distorting the force precision calculation. This region of the curve is de-emphasized using a dashed line.
Our new “Warhammer” cantilever is best understood if we briefly review the process and benefits of FIB-modifying cantilevers. To achieve detection with our commercial AFM (Cypher ES; Asylum Research, Goleta, CA), we started with a cantilever of intermediate length, a BioLever Mini (L = 40 μm; k ≈ 100 pN/nm; Olympus) (Fig. 1 B). Relative to the oft-used BioLever Long (L = 100 μm; k ≈ 7 pN/nm; Olympus), this shorter and stiffer cantilever offered improved short-term force precision due to its lower hydrodynamic drag (β) (17) and improved time resolution (τ ≈ k/β in the overdamped limit [Q < 0.5]). However, these benefits come at the expense of force stability due to low-ƒ noise that increases with k. Our original FIB-modifying process yielded a soft yet short cantilever (L = 40 μm; k ≈ 7 pN/nm) (Fig. 1 C) (13). In that work, we simultaneously reduced k and β by removing a rectangular region at the base of the cantilever and thinning the remaining supporting beams. We then removed the majority of the cantilever’s gold coating to improve force stability, and retained high reflectivity by preserving a small gold patch at the end of the cantilever. For brevity, we refer to these cantilevers as a “Mod Mini.” In our original work (13), they exhibited a good response time (76 μs) by SMFS standards coupled with sub-pN force stability over five decades of time (0.001–100 s).
Using this basic FIB-modification process, we hypothesized that reducing β via reduced surface area at the end of the cantilever would further improve performance. Such reduction could be accommodated while still efficiently detecting the resulting cantilever by using the small spot-size (9 × 3 μm2) detection module available for our commercial AFM. In particular, we tested two new cantilever geometries, referred to as “Long-cut Mini” and Warhammer (Fig. 1, D and E). Although the Long-cut Mini was an extension of our original Mod Mini (Fig. 1 C) (13), the Warhammer used a small (4 × 4 μm2) yet highly reflective region supported by a central uncoated shaft. We modified all cantilevers to have a ∼10-fold reduction in k. We did not investigate very soft cantilevers (k < 4 pN/nm), which tend to irreversibly fold when immersed in liquid.
To compare our set of four cantilever geometries, we measured their thermal motion in liquid positioned 50 nm over the surface and thereby deduced their force power spectral density (PSD) (Fig. 1 F) and force precision (Fig. 1 G). Specifically, we computed the mean force precision over a given averaging time, technically the Allan deviation (18). Importantly, these metrics reflect performance in typical SMFS assays because they account for the increased β when a cantilever is positioned near a surface.
Analysis of the PSD reveals several benefits arising from FIB modification. First, a standard BioLever Mini remained resonant even near the surface (Q = 1.9), as illustrated by the peak in its PSD at ∼32 kHz (Fig. 1 F, red). Yet, standard SMFS theory assumes the force probe is overdamped (Q < 0.5) (19). All three modified cantilevers exhibited PSDs with no peak. Unexpectedly, the Long-cut Mini exhibited a PSD essentially identical to a standard Mod Mini (Fig. 1 F, orange versus blue) despite removing an extra 30% of the cantilever’s planar surface area. In contrast, the Warhammer geometry had a ∼3-fold higher characteristic frequency (ƒc) and better force precision in the thermally limited regime (flat portion of the PSD). Hence, both time resolution and force precision improved despite constant k. Our results, therefore, suggest that a single supporting shaft has substantially reduced β relative to two widely spaced supports. Finite-element modeling may provide for further enhancements.
Computing force precision as a function of averaging time highlights that averaging Brownian motion over short timescales improved data quality (Fig. 1 G). Over longer timescales, force precision was degraded due to low-ƒ noise. For these four cantilevers, the Warhammer had the best stability, with similar performance by the other two modified cantilevers. The Warhammer also exhibited the best short-term force precision due to its lower β. More quantitatively, the Warhammer had ∼40% less force noise than a Mod Mini in the thermally limited regime (0.2–10 ms).
We next compared the performance of the three modified cantilevers (Fig. 2 A) when applied to the unfolding of a polyprotein, a widely used assay (Fig. 1 A). To do so, we used a polyprotein containing a single copy of α3D centered within four repeats of NuG2 (20). NuG2 is a fast-folding variant of GB1 (21) that has been well studied by AFM (22), and acts as an internal standard to assure individual polyproteins were stretched. α3D is a computationally designed, three-helix bundle (23), and is the most mechanically labile protein probed to date by AFM-based SMFS (20). For improved data quality, we site-specifically anchored one end to a polyethylene glycol-coated cover slip via a copper-free click chemistry, and the other end to a polyethylene glycol-coated AFM tip via a streptavidin-biotin linkage. This scheme enabled a strong but reversible coupling to the AFM tip (20).
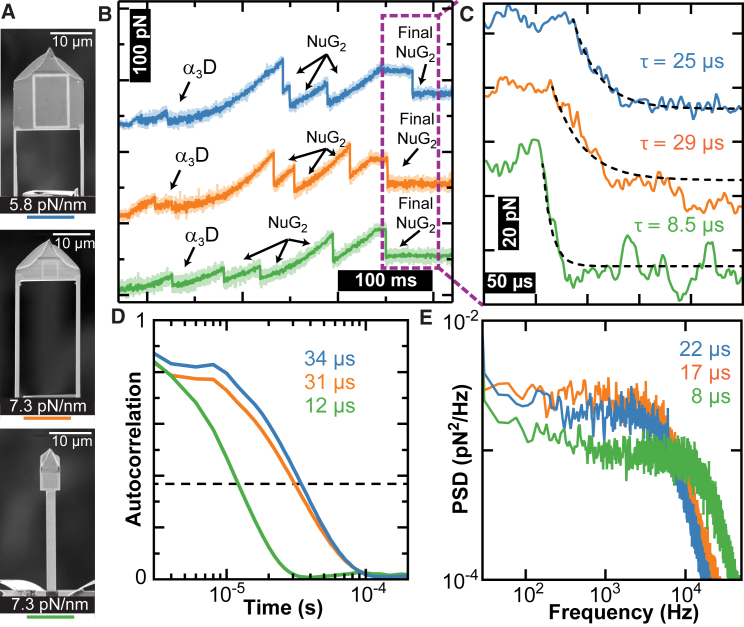
Figure 2.
Temporal resolution of different cantilever geometries. (A) Scanning electron microscopy images of the modified cantilevers. (B) Force-versus-time traces show unfolding of the (NuG2)2-α3D-(NuG2)2 construct. In this assay, the construct was stretched until the α3D and three NuG2 domains unfolded. The stage was further retracted until the polyprotein was held at ∼80 pN. The stage retraction was then stopped and the last folded NuG2 domain unfolded. Data smoothed to 2 kHz. (C) High-bandwidth force-versus-time traces from (B) showing the unfolding of the fourth NuG2 domain at v = 0 nm/s. Time constants determined from exponential fits. Data acquired at 500 kHz. (D) Autocorrelation of the cantilever motion after the final NuG2 domain unfolded but was still attached to the polyprotein. Time constants shown were determined from the 1/e point of the autocorrelation (dashed line). (E) Force PSDs of the 500-kHz data after unfolding of the final NuG2 domain. Time constants estimated from τ ≈ Q/(πƒc) based on the characteristic frequency, ƒc.
To directly measure the cantilever response time during a SMFS assay, we used a stretching protocol in which we initially unfolded α3D and the first three NuG2 domains at v = 400 nm/s, and then stopped retraction at ∼80 pN (Fig. 2 B) to measure the resulting force decay after the rupture of the final NuG2 domain (Fig. 2 C). This mechanical response to a step change in force was well described by a single exponential. Comparison among the modified cantilevers showed that the Warhammer had a decay time of 8.5 μs, a 3-fold improvement over the Long-cut Mini and the Mod Mini (29 and 25 μs, respectively). Additionally, all three cantilevers clearly resolved the low-force unfolding of α3D.
Although this force decay is our preferred metric, the cantilever response time is also encoded in the Brownian motion of the cantilever. In particular, we computed two alternative metrics by analyzing the cantilever’s thermal motion when pulling on the fully unfolded polyprotein, because the taut polyprotein contributes added stiffness to the full system during a SMFS assay. In the first alternative, we estimated the cantilever response time from the 1/e point in the cantilever’s autocorrelation curve (Fig. 2 D), yielding a characteristic time similar to our preferred metric. The second metric was based on analysis of force PSDs (Fig. 2 E), similar to Fig. 1 D, but when pulling on the polyprotein. Based on a traditional AFM analysis (16), the response time was estimated using τ ≈ Q/(πfc). Although this estimate is accurate in the underdamped limit (Q ≫ 1), the resulting response times were nevertheless similar to the other metrics. Finally, we note that careful inspection of Fig. 2 C leads to an apparent anomaly: the Warhammer has larger force fluctuations and hence larger force noise than the other two modified cantilevers. In actuality, the Warhammer has better force precision (Fig. 1, G and F); this discrepancy arises from the temporal filtering by the slower responding cantilevers given the depicted 500-kHz data (Fig. 2 C). When the data from all three cantilevers were filtered to 5 kHz (Fig. S1), the Warhammer exhibited better force precision (1.7 pN) than either the Mod Mini (2.4 pN) or the Long-cut Mini (2.6 pN).
To demonstrate the Warhammer’s improved performance in AFM-based SMFS, we revisited a pioneering AFM study that resolved the unfolding and refolding of calmodulin (11). To review, this prior work used a custom AFM to pull at very low velocities (v = 1 nm/s) and thereby revealed two unfolding steps: the N-terminal and C-terminal domains, each of which is composed of two EF-hand motifs. However, a subsequent study using a dual-beam optical trap revealed an additional intermediate in which one of the EF-hand motifs in the C-terminal domain unfolds (7). With the Warhammer, we now observed this additional unfolding intermediate, and did so at a comparatively high stretching velocity (100 nm/s) relative to the original AFM work (1 nm/s) (Fig. 3, A and B). For clarity, we color coded the three unfolding states, fully folded (blue), partial unfolding of the C-terminal EF hand (green), full unfolding of the C-terminal domain (orange), and the fully unfolded calmodulin (purple). As expected, the total change in contour length (53.0 nm) agreed with the previously measured value (52.2 nm) (7).
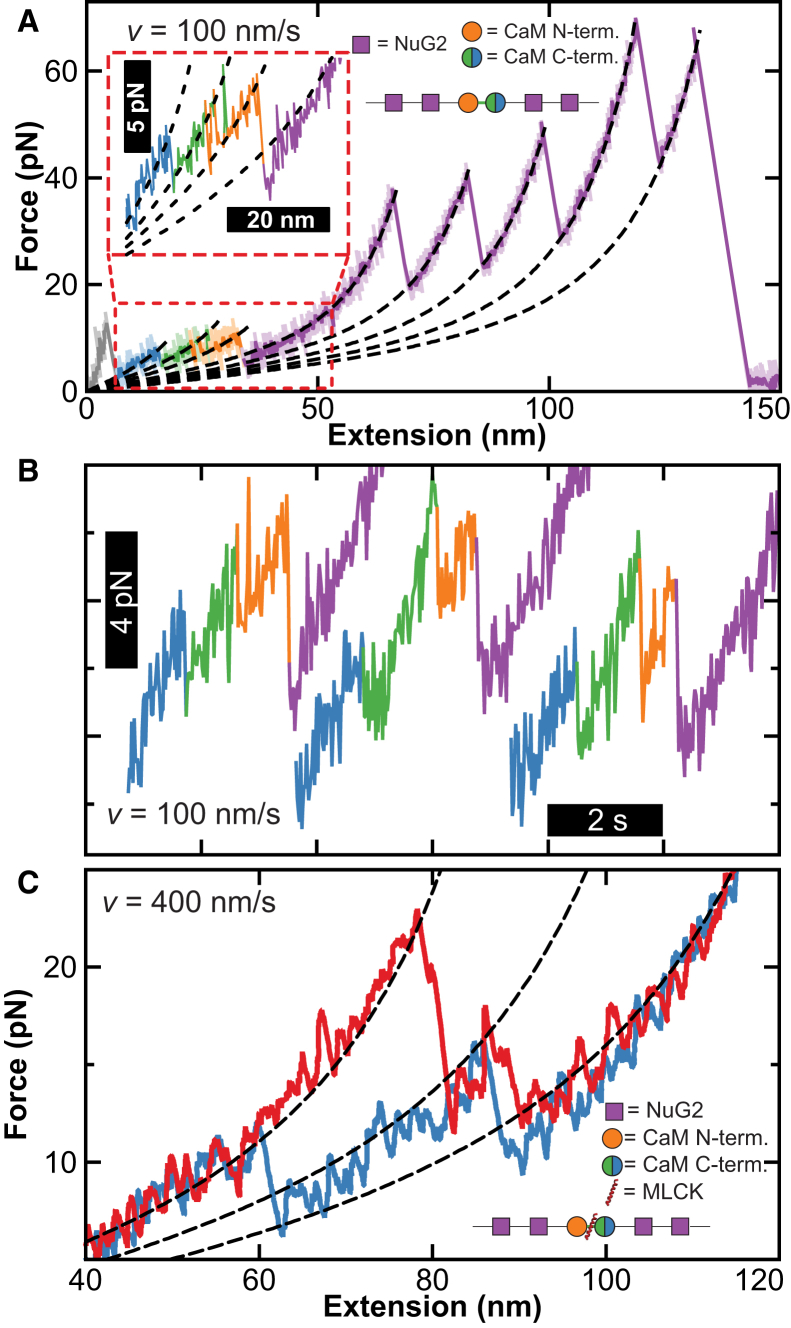
Figure 3.
Improved AFM-based SMFS studies of calmodulin unfolding when using a Warhammer cantilever. (A) Force-extension curves show the unfolding of a polyprotein containing calmodulin and four repeats of NuG2 at v = 100 nm/s. Trace color coding indicates fully folded calmodulin (blue), partial unfolding of the N-terminal domain (green), followed by the full unfolding of the C- and N-terminal domains (orange and purple, respectively). The unfolding of the NuG2 domains at higher force is also color coded purple. Dashed lines represent worm-like chain fits. (Inset) Calmodulin unfolding at low force. (B) Three force-versus-time traces at v = 100 nm/s highlight three-step unfolding of calmodulin. (C) Force-extension curves comparing the unfolding of calmodulin bound and unbound to MLCK at v = 400 nm/s (red and blue, respectively). Dark traces filtered to 250 Hz.
We next recapitulated the mechanical stabilization of calmodulin when bound to one of its target ligands, MLCK (11). In that earlier study, MLCK stabilized the N-terminal domain. When we added MLCK to the buffer, we also clearly observed MLCK-induced stabilization (Fig. 3 C, red versus blue) despite pulling at much higher velocity (400 versus 1 nm/s). Thus, the Warhammer provides for significantly enhanced signal/noise ratio and rapid characterization of low-force unfolding events by AFM standards (1, 11).
From a practical point of view, we emphasize that FIB-modified cantilevers were straightforward to fabricate and were reusable. Until recently (13), our laboratory had no prior expertise with an FIB. After initial training in FIB operation, fabrication of the Warhammer geometry was not technically challenging, but rather a modification of a previously published, step-by-step protocol (24). The change in geometry, on the other hand, was the key to improved performance. Fabrication remained efficient; we produced 2–3 cantilevers/h. Unlike FIB-modified, ultrashort cantilevers (14), we detected the Warhammer when using the standard small spot-size module of our commercial AFM with no loss in precision over all measured frequencies (Fig. S2 A). That said, we preferred to use a home-built module that featured a 3-μm diameter circular spot (14) because it reduced an optical interference artifact (Fig. S2 B). Finally, Warhammer cantilevers were robust and reusable. After functionalization (20), a Warhammer could be reused over multiple days and refunctionalized after plasma cleaning. Handling or bending of the cantilever during plasma cleaning was typically the limiting factor.
In summary, Warhammer cantilevers offer an excellent combination of 8.5-μs resolution coupled with sub-pN force stability over 100 s. We expect this combination to enable equilibrium folding studies of proteins and nucleic acid structures over long periods on a commercial AFM. The advances in data quality demonstrated here for SMFS are immediately applicable to a wide range of biological AFM applications, including rapid nanomechanical mapping of live cells (25).
Author Contributions
T.T.P. designed the research. M.-A.L. purified labeled polyprotein constructs. D.T.E. and J.K.F. fabricated the cantilevers. D.T.E. acquired the data, and D.T.E. and T.T.P. analyzed the data and wrote the manuscript.
Acknowledgments
We thank M. Sousa for scientific discussions.
This work was supported by a National Institutes of Health Molecular Biophysics Training grant awarded to M.-A.L. (T32 GM-065103), by the National Science Foundation (grants DBI-1353987, MCB-1716033, and Phy-1734006), and by the National Institute of Standards and Technology (NIST). Mention of commercial products is for information only; it does not imply NIST’s recommendation or endorsement. T.T.P. is a staff member of NIST’s Quantum Physics Division.
Editor: Keir Neuman.
Footnotes
Supporting Materials and Methods and seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31141-4.
Supporting Citations
References (26, 27, 28, 29) appear in the Supporting Material.
Supporting Material
References
- 1.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liphardt J., Onoa B., Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 3.Rief M., Gautel M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 4.Cecconi C., Shank E.A., Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 5.Onoa B., Dumont S., Bustamante C. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenleaf W.J., Frieda K.L., Block S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stigler J., Ziegler F., Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 8.Yu H., Liu X., Woodside M.T. Direct observation of multiple misfolding pathways in a single prion protein molecule. Proc. Natl. Acad. Sci. USA. 2012;109:5283–5288. doi: 10.1073/pnas.1107736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H., Siewny M.G., Perkins T.T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science. 2017;355:945–950. doi: 10.1126/science.aah7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodside M.T., Anthony P.C., Block S.M. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junker J.P., Ziegler F., Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 12.Churnside A.B., Sullan R.M., Perkins T.T. Routine and timely sub-picoNewton force stability and precision for biological applications of atomic force microscopy. Nano Lett. 2012;12:3557–3561. doi: 10.1021/nl301166w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bull M.S., Sullan R.M., Perkins T.T. Improved single molecule force spectroscopy using micromachined cantilevers. ACS Nano. 2014;8:4984–4995. doi: 10.1021/nn5010588. [DOI] [PubMed] [Google Scholar]
- 14.Edwards D.T., Faulk J.K., Perkins T.T. Optimizing 1-μs-resolution single-molecule force spectroscopy on a commercial atomic force microscope. Nano Lett. 2015;15:7091–7098. doi: 10.1021/acs.nanolett.5b03166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neupane K., Foster D.A., Woodside M.T. Direct observation of transition paths during the folding of proteins and nucleic acids. Science. 2016;352:239–242. doi: 10.1126/science.aad0637. [DOI] [PubMed] [Google Scholar]
- 16.Rico F., Gonzalez L., Scheuring S. High-speed force spectroscopy unfolds titin at the velocity of molecular dynamics simulations. Science. 2013;342:741–743. doi: 10.1126/science.1239764. [DOI] [PubMed] [Google Scholar]
- 17.Viani M.B., Schaffer T.E., Hansma P.K. Small cantilevers for force spectroscopy of single molecules. J. Appl. Phys. 1999;86:2258–2262. [Google Scholar]
- 18.Sullivan D.B., Allan D.W., Walls E.L. U.S. Government Printing Office; Washington: 1990. Characterization of Clocks and Oscillators. [Google Scholar]
- 19.Evans E., Ritchie K. Strength of a weak bond connecting flexible polymer chains. Biophys. J. 1999;76:2439–2447. doi: 10.1016/S0006-3495(99)77399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walder R., LeBlanc M.-A., Perkins T.T. Rapid characterization of a mechanically labile α-helical protein enabled by efficient site-specific bioconjugation. J. Am. Chem. Soc. 2017;139:9867–9875. doi: 10.1021/jacs.7b02958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nauli S., Kuhlman B., Baker D. Computer-based redesign of a protein folding pathway. Nat. Struct. Biol. 2001;8:602–605. doi: 10.1038/89638. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y., Kuske R., Li H. Direct observation of markovian behavior of the mechanical unfolding of individual proteins. Biophys. J. 2008;95:782–788. doi: 10.1529/biophysj.107.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y., Alonso D.O., Gai F. Ultrafast folding of alpha3D: a de novo designed three-helix bundle protein. Proc. Natl. Acad. Sci. USA. 2003;100:15486–15491. doi: 10.1073/pnas.2136623100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faulk J.K., Edwards D.T., Perkins T.T. Improved force spectroscopy using focused-ion-beam-modified cantilevers. Methods Enzymol. 2017;582:321–351. doi: 10.1016/bs.mie.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Alsteens D., Dupres V., Dufrêne Y.F. High-resolution imaging of chemical and biological sites on living cells using peak force tapping atomic force microscopy. Langmuir. 2012;28:16738–16744. doi: 10.1021/la303891j. [DOI] [PubMed] [Google Scholar]
- 26.Proksch R., Schaffer T.E., Viani M.B. Finite optical spot size and position corrections in thermal spring constant calibration. Nanotechnology. 2004;15:1344–1350. [Google Scholar]
- 27.Cao Y., Balamurali M.M., Li H. A functional single-molecule binding assay via force spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:15677–15681. doi: 10.1073/pnas.0705367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He C., Hu C., Li H. Direct observation of the reversible two-state unfolding and refolding of an alpha/beta protein by single-molecule atomic force microscopy. Angew. Chem. Int. Ed. Engl. 2015;54:9921–9925. doi: 10.1002/anie.201502938. [DOI] [PubMed] [Google Scholar]
- 29.Bouchiat C., Wang M.D., Croquette V. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.