Abstract
Objective
To study the genetic cause of Mayer-Rokitansky-Kuster-Hauser (MRKH) syndrome. Although a few candidate genes and genomic domains for have been reported for MRKH, the genetic underpinnings remain largely unknown. Some of the top candidate genes are WNT4, HNF1B, and LHX1. The goals of this study were to: 1) determine the prevalence of WNT4, HNF1B, and LHX1 point mutations, as well as new copy number variants (CNVs) in people with MRKH; and 2) identify and characterize MRKH cohorts.
Design
Laboratory and community based study
Setting
Academic medical centers
Patients
147 MRKH probands and available family members
Interventions
DNA sequencing of WNT4, HNF1B, and LHX1 in 100 MRKH patients; chromosomal microarray analysis in 31 North American MRKH patients; and ascertainment and sample collection of 147 North American and Turkish MRKH probands and their families
Main Outcome Measure(s)
DNA sequence variants and CNVs; pedigree structural analysis
Results
We report finding CNVs in 6/31 (~19%) people with MRKH, but no point mutations or small indels in WNT4, HNF1B, or LHX1 in 100 MRKH patients. Our MRKH families included 43 quads, 26 trios, and 30 duos. Of our MRKH probands, 87/147 (59%) had MRKH type 1 and 60/147 (41%) had type 2 with additional anomalies.
Conclusions
Although the prevalence of WNT4, HNF1B, and LHX1 point mutations is low in people with MRKH, the prevalence of CNVs was about 19%. Further analysis of our large familial cohort of patients will facilitate gene discovery to better understand the complex etiology of MRKH.
Introduction
Mayer-Rokitanksy-Kuster-Hauser (MRKH) syndrome (MIM #27700), also known as Müllerian aplasia, consists of congenital absence of the uterus and vagina. MRKH, which is the name patients prefer, accounts for 10% of the cases of primary amenorrhea (1), and it affects ~1/5,000 women (2). This anomaly constitutes the most severe malformation of the female reproductive tract, and occurs in isolation in two-thirds of patients, often referred to as Type 1 (2). A subset of patients (Type 2) presents with associated structural abnormalities such as unilateral renal agenesis (30%), skeletal defects (10–15%), cardiac anomalies (2–3%), and deafness (2–3%). These patients have a normal 46,XX karyotype and typically exhibit normal ovarian function with normal development of breasts and external genitalia (2, 3).
The molecular pathways of Müllerian development have been well studied in animal models, and document the importance of WNT signaling, as mice with null or mutant alleles for Wnt9b, Wnt4, Wnt5a, and Wnt7a manifest varying degrees of Müllerian hypoplasia (4). In addition, other genes such as Pbx1, Pax2, Lhx1, and Emx2, have also been implicated in normal Müllerian development in the mouse (5, 6). However, the genetic underpinnings of human Müllerian developmental abnormalities are largely unknown. While many cases are sporadic, some familial cases have also been reported, suggesting a genetic role in the pathogenesis in some patients (7). Genetic transmission is difficult to ascertain since affected families are often small and affected individuals are unable to have children unless they undergo surrogacy or a uterine transplant (3, 8).
A number of chromosomal regions and candidate genes have been studied in humans with MRKH, and several have been observed by different investigators, but conclusive evidence for causation is lacking (3) except for causative mutations in WNT4 (9) and HNF1B (10). An LHX1 nonsense variant has been reported, but the segregation within families and in vitro confirmation have not been documented (11). However, studies in mice suggest that mutations in Lhx1 could lead to an MRKH-like phenotype (12–14). Chromosomal microarrays have suggested numerous copy number variants (CNVs) associated with MRKH, with 17q12 and 16p11 being two more commonly affected regions (3, 15). These CNVs contain multiple genes, and it is currently not clear if MRKH is a genomic disorder or if one or a few genes within these regions could be involved in its pathophysiology (3).
The genetic component of MRKH may be complex and its complete understanding is hindered by the lack of large collections of MRKH families. Herlin et al (16) reported one family with two MRKH probands and reviewed the literature, reporting 67 families with at least two MRKH patients or one MRKH and one with MRKH-associated anomalies. However, these families were ascertained from multiple publications in the literature, and there has not been any large characterization of unselected MRKH patients from a single research team. Importantly and in addition, the prevalence of gene mutations in WNT4, HNF1B, and LHX1 has not been substantiated in a relatively large sample of MRKH women. The purpose of the present study was to: 1) collect and obtain clinical information and blood samples from a large cohort of MRKH families containing at least one MRKH patient; 2) determine the prevalence of variants in two accepted MRKH genes—WNT4 and HNF1B, as well as the candidate gene—LHX1; and 3) to determine if CNVs are present in a subset of our North American MRKH patients, which are absent in their unaffected parents.
Methods
Cohort characterization
Patients and families were recruited to participate via ascertainment of MRKH probands through our Developmental Gene Discovery Project (DGDP). Many probands were collected by authors (LCL and OMA), but a substantial number were ascertained from the Beautiful You MRKH Foundation (author ACL). MRKH was defined as a female with normal breast development, Tanner 5 pubic hair, and an absent vagina and uterus based upon physical exam, supported by imaging (ultrasound and/or an MRI) and/or surgery (2, 3). All patients had a 46,XX karyotype except our one previously reported patient with a chromosomal translocation involving chromosomes 3 and 16 (17). MRKH associated anomalies were identified by reviewing medical records and obtaining family history. Every attempt was made to collect available family members. Peripheral blood was collected for creating lymphoblastoid cell lines and extracting DNA as described previously (18). Lymphoblastoid cells were created so that a long-term supply of DNA, RNA, and protein could be available for in vitro analyses on identified genetic variants for confirmation. This study was approved by the Institutional Review Board at Augusta University; and all participating patients and available family members signed a consent form.
Sanger DNA Sequencing for WNT4, LHX1, HNF1B
DNA was extracted from peripheral blood leukocytes as described previously (18, 19). Sanger sequencing was performed on a cohort of 100 MRKH patients (79 with type 1 and 21 with type 2) for the protein coding regions and splice junctions for 5 exons of WNT4 (NM_030761.4), 5 exons of LHX1 (NM_005568.3), and 9 exons of HNF1B (NM_000458.3). Each fragment was amplified by PCR for 30 cycles consisting of a 5 minute denaturation step at 95° C followed by 30 cycles of the following: one minute at 95° C, 30–60 seconds at 55° C, and 30–60 seconds at 72° C followed by a 7 minute 72° C extension step. Each fragment was resolved by agarose gel electrophoresis, and then an aliquot of the sample was subjected to dideoxy sequencing on an ABI 310 Automated DNA Sequencer as described previously (20–22). Each fragment was sequenced in the forward and reverse directions. If a variant was identified, two additional sequencing reactions were performed. The obtained sequence was blasted to the wild type sequence.
Chromosomal Microarrays
DNA from 31 unrelated North American MRKH probands (10 with type 1 and 21 with type 2) was subjected to chromosomal microarrays. Copy number variant analysis was performed at Harvard utilizing an Affymetrix Cytoscan HD array, which consisted of 750,000 SNP probes and 1.9 million copy number probes to detect CNVs. The lower limit of detection for CNVs was 50 kb. 100 ng of genomic DNA was labeled with Cytoscan reagent kit according to the manufacturer's instructions. The array data was analyzed with Chromosome Analysis Suite (ChAS) Software as described previously (22). Human genome hg19 assembly was used to map genomic coordinates.
Results
Cohort characterization
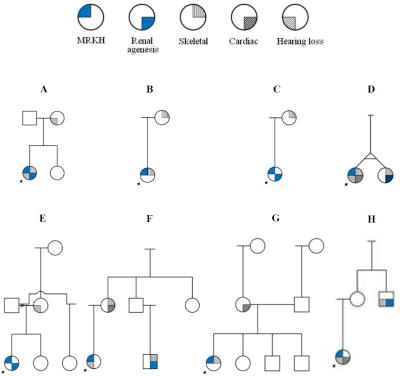
We acquired DNA (and lymphoblastoid cell lines on most) from a cohort of 147 MRKH female probands. These patients include 80 North American probands, 58 for which we have other family members (shown in Supplemental Figure 1a) and 22 singletons (not shown). We also have 67 Turkish probands, 41 with family members (shown in Supplemental Figure 1b) and 26, which are singletons. We collected 43 quads (proband, 1–2 parents, 1–2 siblings), 26 trios (proband, 1–2 parents, 1 sibling), and 30 duos (proband and 1 first degree relative). We have characterized phenotypes for the patients and family members in pedigrees, including Müllerian duct agenesis and associated anomalies (Figure 1; Table 1). Overall, 87 have type 1 MRKH, and 60 are type 2. Of our affected type 2 probands, anomalies include: renal (n=24), skeletal (n = 34), hearing impairment (n = 25), and cardiac (n = 13).
Figure 1.
(A–H) Eight MRKH pedigrees from North America with at least one affected family member with an MRKH-associated anomaly are shown. Arrows indicated MRKH probands.
Table 1.
The frequency that an associated anomaly was observed in our cohort of 147 MRKH patients.
| MRKH (n) | Renal | Skeletal | Cardiac | Hearing Deficits |
|---|---|---|---|---|
| 147 | 24 | 34 | 13 | 25 |
| (16%) | (23%) | (9%) | (17%) |
Examination of family members indicates that none of the probands (58 families from North America and 41 from Turkey) in this study has a family member who is also affected by MRKH. However, we observed that 8/58 (14%) of the families from North America (Figure 1) and 0/41 of the families from Turkey had a relative with an associated anomaly (Supplemental Figure 1).
Sanger DNA Sequencing for WNT4, LHX1, HNF1B and Chromosomal Microarrays
For the 100 patients with MRKH (79 type 1 and 21 type 2) in our cohort, we performed PCR on all protein coding exons and splice sites of WNT4, HNF1B, and LHX1. Agarose gel electrophoresis and Sanger sequencing failed to reveal evidence of deletions, small indels, or likely pathogenic variants in WNT4, HNF1B, or LHX1.
Chromosomal microarray studies performed on 31 North American patients revealed that 6 of 31 (~19%) had CNVs of potential clinical significance: 2/10 were found in people with MRKH type 1and 4/21 were found in individuals with MRKH type 2 (Table 2). Two MRKH type 1 patients had deletions involving 17q12—DGDP156 had a de novo CNV that was 1.9 Mb and DGDP287 had a 1.4 Mb deletion. The other four CNVs were found in individuals with MRKH type 2. Another MRKH patient (DGDP284) had a 16p11 deletion (746 kb) and a duplication (456kb) of 11p11. MRKH patient DGDP149 had three different CNVs (a deletion involving 2q11 and two duplications—one at 8p23 and another at 12p12-12p13), but only the 12p CNV was de novo (Table 2). Both parents of DGDP149 had CNVs involving 8p23 and 2q11. A 4 Mb deletion at 1q21-1q22 was identified in DGDP273; and a 2q13 deletion of 840 kb was found in patient DGDP155. We could only document that one 17q12 deletion (in DGDP156) and the 12p12-12p13 duplication (DGDP149) were de novo---parents were not available for the other four patients with CNVs. Both HNF1B and LHX1 reside within 17q12, while WNT4 is localized to chromosome 1p36.12. Except for the 16p11 and 17q12 deletions, all other CNVs were not found in DECIPHER or ClinVar. Only 14 MRKH patients had both DNA sequencing and chromosomal microarrays.
Table 2.
Copy number variants found in six MRKH patients. Parents were studied in two (DGDP156 and DGDP149). Del = deletion; Dup = duplication; kb = kilobases; Mb = megabases; NS = not studied. Human genome hg19 assembly was used for coordinates.
| Patient | Location | CNV | Size | Coordinates | Associated anomalies | De novo |
|---|---|---|---|---|---|---|
| DGDP155 | 2q13 | Del | 840kb | chr2:110,520-111,360 | Type 2: Renal, | NA |
| Cardiac, Skeletal | ||||||
| DGDP156 | 17q12 | Del | 1.9Mb | chr17:34,455,782-36,307,773 | Type 1 | De novo |
| Mother | Normal | Normal | ||||
| Father | Normal | Normal | ||||
| DGDP273 | 1q21-1q22 | Del | 4 Mb | chr1:144,823,069-148,832,359 | Type 2: Skeletal | NA |
| DGDP149 | 2q11 | Del | 1.5 Mb | chr2:96,712,139-98,249,638 | Type 2: Renal, | |
| 8p23 | Dup | 1.9 Mb | chr8:4,322,747-6,214,120 | hearing, skeletal | ||
| 12p12-12p13 | Dup | 1 Mb | chr12:13,938,281-14,954,514 | De novo | ||
| Mother | 2q11 | Del | 962kb | chr2:96,766,402-97,728,447 | Normal | |
| 8p23 | Del | 455 kb | chr8:3,926,021-4,381,311 | |||
| Father | 8p23 | Dup | 2 Mb | chr8:4,300,484-6,290,708 | Normal | |
| DGDP284 | 16p11 | Del | 746 kb | chr16:29,432,212-30,178,406 | Type 2: Hearing | NA |
| 11p11 | Dup | 456 kb | chr11:48,065,462-48,521,382 | |||
| DGDP287 | 17q12 | Del | 1.4Mb | chr17:34,815,077-36,249,799 | Type 1 | NA |
Discussion
The molecular basis of MRKH remains largely unknown. A number of prior studies, most of which had small sample sizes, failed to show mutations in reasonable candidate genes including WT1, CFTR, WNT7A, GALT, HOXA7, PBX1, HOXA13, PAX2, HOXA10, AMH, AMHR, RARG, RXRA, CTNNB1, LAMC1, DLGH1, and SHOX (reviewed in Layman (3)). The best supportive evidence for a causative gene is WNT4, which was the first genetic cause identified and corroborated by functional studies (9). Mouse studies also provide additional support for WNT signaling in normal Müllerian development (4). The four MRKH patients described have heterozygous WNT4 mutations that impair WNT4 function in vitro, but there was no evidence of genetic heritability, making inheritance ascertainment difficult (9, 23–25). Other affected family members have shown wild type WNT4 sequences in some of these families, which suggests de novo autosomal dominant inheritance with variable expressivity. Interestingly, MRKH patients with WNT4 mutations may also demonstrate hyperandrogenism (9), but hyperandrogenism is common in reproductive aged women (26), so some caution should be exercised. These investigators found that 4/37 (10.8%) of MRKH patients had WNT4 mutations (9, 23–25), so we expected to identify ~10 patients with WNT4 mutations. However, none of our 100 MRKH patients had WNT4 mutations detected by DNA sequencing. These findings indicate that perhaps the prevalence of WNT4 mutations is much less than previously reported in an unselected group of MRKH probands.
A second causative MRKH gene is HNF1B, mutations of which result in maturity onset diabetes of the young type 5 (MODY5) with nondiabetic renal disease (10). Interestingly, two affected females with heterozygous intragenic HNF1B deletions impairing function in vitro, also had Müllerian aplasia (10). Two studies examined the prevalence of HNF1B mutations in small numbers of patients. Bernardini et al (27) studied 20 MRKH patients and Ledig et al (28) studied 56 patients, and found no point mutations. Our DNA sequencing results also failed to show any HNF1B point mutations or small indels in 100 patients, again indicating low prevalence. Interestingly, HNF1B is located within the common 17q12 deletion CNV. To date these findings suggest that HNF1B mutations are causative, but since the 17q12 CNV has been reported more than 10–15 times, it suggests other genes within this region could play some role (3). Thus, a different gene within the interval or a contiguous gene deletion syndrome cannot be excluded at this time.
LHX1 is a promising candidate gene for MRKH for several reasons. First, LHX1 resides within the relatively common 17q12 CNV, which has been associated with MRKH in several studies (3, 15), including ours. Second, conditional deletion of Lhx1 in Wolffian-derived (12) or Mullerian-derived (14) tissues recapitulates an MRKH-like phenotype in female mice. One heterozygous human nonsense variant in LHX1 has been described, but no other family members were available and no in vitro analyses were performed (11). Two other studies found LHX1 missense variants, but no family studies or in vitro analyses were performed (28, 29). We found neither LHX1 point mutations nor small intragenic indels in 100 MRKH patients indicating that these types of mutations in this gene are uncommon in MRKH. This gene also resides within the 17q12 CNV deletion interval, but appears to be uncommonly involved in MRKH by itself. Therefore, there is no conclusive evidence that LHX1 variants cause MRKH. A variety of other gene variants in TBX6 (29, 30), WNT9B (31, 32), and RBM8A (30) have been described, but all reports suffer from a lack of in vitro confirmation and family studies, clouding their true role in MRKH causality at this time.
Our chromosomal microarray analysis revealed potentially pathogenic CNVs in 6 of 31 (~19%) patients, including two patients with 17q12 deletions, both of whom had MRKH type 1. The 17q12 region is one of the most commonly identified CNVs in MRKH, and it has been found in both type 1 and type 2 MRKH (3, 15). We also found one proband with a 16p11 deletion, which has also been reported by multiple different investigators (3, 15). All other CNVs were absent in DECIPHER and ClinVar. Of interest, our proband with the 16p11 deletion also had an 11p11 duplication. We identified a deletion in the 1q21.1-1q21.2 region that includes the region for TAR (thrombocytopenia absent radius) syndrome, marked by absent radii and early onset thrombocytopenia due to RBM8A gene mutations (30). Some patients with TAR syndrome have Mullerian aplasia, but our patient did not have the TAR phenotype. CNVs of the long arm of chromosome 2 are associated with developmental delay, intellectual disability, microcephaly, skeletal abnormalities, short stature, and cleft lip and palate (33). A patient with type 2 MRKH was found to have a microdeletion at 2q11.2, as we have confirmed here. Of interest, our patient with the 2q13 deletion presented with renal agenesis, cardiac, and skeletal defects. The 2q13 region includes PAX8, which has been shown to coordinate with PAX2 to regulate branching morphogenesis and nephron differentiation during kidney development, a process closely linked to Müllerian duct development (34).
We could only document two patients with de novo CNVs (we did not have DNA on parents of other probands with CNVs)—one with a 17q12 deletion and the other with a 12p12-12p13 duplication. The patient with the 1 Mb duplication at 12p had two other CNVs involving 2q11 and 8p23 that originated from the parents, suggesting they could be polymorphisms. However the sizes and breakpoints are different from the proband. Our MRKH proband had a 1.5Mb deletion at 2q11, which encompasses the 962 kb deletion identified in her mother. This same MRKH proband also had a 1.9 Mb duplication at 8p23, which is similar in size to the 2 Mb duplication found in her father. Intriguingly, the proband’s mother had an 8p23 deletion (rather than duplication) of 455 kb, which is partially encompassed in the deleted region of the affected daughter and husband (DGDP149 in Table 2). This dynamic change of CNV breakpoints on the same chromosomal regions in this family is worthy of future investigation. Perhaps these parental CNVs of 8p23 and 2q11 deletions in her mother and the 8p23 duplication in the father had an interchromosomal effect contributing to the generation of the 12p12-12p13 duplication in the MRKH proband. Although the CNVs we have identified are possibly pathogenic, they do not provide convincing evidence of causation.
The molecular basis of MRKH is difficult to characterize. The major barrier is the lack of a large familial cohort of MRKH families to systematically study both clinically and at the molecular level. Any identified variant should be subjected to family studies to determine if the variant segregates with the phenotype, which cannot be easily done without a large cohort. Then, in vitro analysis is required for confirmation. In the current study, we sought to gather a large sample of MRKH families, enlisting the assistance of the patient support foundation—Beautiful You MRKH (https://www.beautifulyoumrkh.org/). We collected a sample of 147 MRKH probands, including 99 with available family members (Supplemental Figure 1). From our large familial MRKH cohort, no proband had an additional affected relative with MRKH. Only 14% of North American and no Turkish probands had a relative with an MRKH-associated anomaly. This, along with unknown phenotypic effects in males, adds to the complexity of MRKH genetics, and it must be considered when studying families. Even in small families, segregation analyses can be performed, and we would not expect unaffected individuals to have the same variant, as exemplified for WNT4 (9) and HNF1B (10) mutations.
Herlin et al (16) described a familial case of MRKH and ascertained MRKH probands in the literature who had at least one more person affected with MRKH or MRKH associated anomalies. They found that 36 of the 67 families had two or more MRKH patients, while the remainder had at least one associated anomaly. Our findings are more unselected since we collect all MRKH patients and families we can recruit, which perhaps gives a more reasonable estimate of MRKH family structure, avoiding publication bias as in Herlin et al (16). The family structures of the reported multiplex MRKH families argue against autosomal recessive inheritance, although this seems more possible in our Turkish patients because of potential consanguinity. Instead de novo autosomal dominant, or polygenic/multifactorial modes of inheritance are more likely, which has been proposed by others (16). As recommended by Schaffer (35), true digenic/polygenic inheritance is extremely challenging to substantiate since human families, cellular studies, and mouse models are needed for corroboration.
Clearly, our understanding of the molecular basis of MRKH is in its infancy, unlike other reproductive disorders such as hypogonadotropic hypogonadism and gonadal failure for which the genetic basis is known for a significant proportion of affected individuals (36–38). Our cohort of MRKH probands and family members, as well as our collection and usage of lymphoblastoid cell lines, will allow gene segregation analysis to test for true de novo variants, determine effects upon RNA and protein, and examine for inheritance patterns. Because we have a large number of quads and trios, determination of inheritance of identified variants should be more easily accomplished. Future efforts will include whole exome sequencing and whole genome sequencing in our cohort to generate comprehensive data via high throughput next generation sequencing methods. In vitro analyses will be needed to complement the effect of any genetic variant, particularly for missense variants, which are commonly variants of undetermined significance unless they are studied in vitro (39). However, newer bioinformatic programs, such as pVAAST (pedigree variant annotation, analysis and search tool), are able to mine next generation DNA sequencing data from quads, trios, or any family structure. pVAAST incorporates linkage, association, and variant severity. This method has been shown to be superior to other methods in analyzing genome sequencing data for autosomal dominant, autosomal recessive, and particularly relevant to the MRKH phenotype, dominant inheritance resulting from de novo mutations (40). Digenic/polygenic disease is also amenable to pVAAST analysis (40).
In summary, the genetic basis for MRKH remains largely unknown, and nongenetic causes such as epigenetic changes and somatic anomalies could play a role in some patients. The prevalence of mutations in the two known genes WNT4 (n = 4) and HNF1B (n = 1 family) is exceedingly low. There is no solid evidence at this time for causation in other genes, although there are promising candidate genes, such as LHX1, TBX6, WNT9B, and RMB8A that require future study. The repetitive identification of CNVs in regions such as 17q12, 16p11, and 1q21 suggests their involvement, as well as a potential explanation for the etiology of multiple cases that would be missed by DNA sequencing of exons. However, further work is needed to prove causation. Large well-characterized MRKH families with lymphoblastoid cell lines, such as our large international cohort, along with significant public support from the MRKH community, will be essential to unravel its complex genetics.
Supplementary Material
Acknowledgments
L.C.L. was supported from NIH grant HD33004
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: The authors have nothing to disclose.
References
- 1.Reindollar RH, Byrd JR, McDonough PG. Delayed sexual development:study of 252 patients. Am J Obstet Gynecol. 1981;140:371–80. doi: 10.1016/0002-9378(81)90029-6. [DOI] [PubMed] [Google Scholar]
- 2.Oppelt PG, Lermann J, Strick R, Dittrich R, Strissel P, Rettig I, et al. Malformations in a cohort of 284 women with Mayer-Rokitansky-Kuster-Hauser syndrome (MRKH) Reprod Biol Endocrinol. 2012;10:57. doi: 10.1186/1477-7827-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Layman LC. The genetics of mullerian aplasia. Expert Rev Endocrinol Metab. 2014;9:411–9. doi: 10.1586/17446651.2014.914433. [DOI] [PubMed] [Google Scholar]
- 4.Mericskay M, Kitajewski J, Sassoon D. Wnt5a is required for proper epithelial-mesenchymal interactions in the uterus. Development. 2004;131:2061–72. doi: 10.1242/dev.01090. [DOI] [PubMed] [Google Scholar]
- 5.Masse J, Watrin T, Laurent A, Deschamps S, Guerrier D, Pellerin I. The developing female genital tract: from genetics to epigenetics. Int J Dev Biol. 2009;53:411–24. doi: 10.1387/ijdb.082680jm. [DOI] [PubMed] [Google Scholar]
- 6.Kurita T. Normal and abnormal epithelial differentiation in the female reproductive tract. Differentiation. 2011;82:117–26. doi: 10.1016/j.diff.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson JL. Genetics of female infertility due to anomalies of the ovary and mullerian ducts. Methods in molecular biology. 2014;1154:39–73. doi: 10.1007/978-1-4939-0659-8_3. [DOI] [PubMed] [Google Scholar]
- 8.Petrozza JC, Gray MR, Davis AJ, Reindollar RH. Congenital absence of the uterus and vagina is not commonly transmitted as a dominant genetic trait: outcomes of surrogate pregnancies. Fertility and sterility. 1997;67:387–9. doi: 10.1016/S0015-0282(97)81927-9. [DOI] [PubMed] [Google Scholar]
- 9.Biason-Lauber A, Konrad D, Navratil F, Schoenle EJ. A WNT4 mutation associated with Mullerian-duct regression and virilization in a 46,XX woman. The New England journal of medicine. 2004;351:792–8. doi: 10.1056/NEJMoa040533. [DOI] [PubMed] [Google Scholar]
- 10.Lindner TH, Njolstad PR, Horikawa Y, Bostad L, Bell GI, Sovik O. A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1beta. Hum Mol Genet. 1999;8:2001–8. doi: 10.1093/hmg/8.11.2001. [DOI] [PubMed] [Google Scholar]
- 11.Ledig S, Brucker S, Barresi G, Schomburg J, Rall K, Wieacker P. Frame shift mutation of LHX1 is associated with Mayer-Rokitansky-Kuster-Hauser (MRKH) syndrome. Hum Reprod. 2012;27:2872–5. doi: 10.1093/humrep/des206. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005;132:2809–23. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi A, Shawlot W, Kania A, Behringer RR. Requirement of Lim1 for female reproductive tract development. Development. 2004;131:539–49. doi: 10.1242/dev.00951. [DOI] [PubMed] [Google Scholar]
- 14.Huang CC, Orvis GD, Kwan KM, Behringer RR. Lhx1 is required in Mullerian duct epithelium for uterine development. Dev Biol. 2014;389:124–36. doi: 10.1016/j.ydbio.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nik-Zainal S, Strick R, Storer M, Huang N, Rad R, Willatt L, et al. High incidence of recurrent copy number variants in patients with isolated and syndromic Mullerian aplasia. J Med Genet. 2011;48:197–204. doi: 10.1136/jmg.2010.082412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herlin M, Hojland AT, Petersen MB. Familial occurrence of Mayer-Rokitansky-Kuster-Hauser syndrome: a case report and review of the literature. Am J Med Genet A. 2014;164A:2276–86. doi: 10.1002/ajmg.a.36652. [DOI] [PubMed] [Google Scholar]
- 17.Williams LS, Kim HG, Kalscheuer VM, Tuck JM, Chorich LP, Sullivan ME, et al. A balanced chromosomal translocation involving chromosomes 3 and 16 in a patient with Mayer-Rokitansky-Kuster-Hauser syndrome reveals new candidate genes at 3p22.3 and 16p13.3. Molecular cytogenetics. 2016;9:57. doi: 10.1186/s13039-016-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhagavath B, Podolsky RH, Ozata M, Bolu E, Bick DP, Kulharya A, et al. Clinical and molecular characterization of a large sample of patients with hypogonadotropic hypogonadism. Fertility and sterility. 2006;85:706–13. doi: 10.1016/j.fertnstert.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 19.Layman LC, Roach DJ, Plouffe L, Jr, McDonough PG, Wilson JT. The detection of Hind III restriction fragment length polymorphisms (RFLP's) using a DNA probe for the β-subunit of follicle-stimulating hormone (FSH) Fertility and sterility. 1990;53:261–5. doi: 10.1016/s0015-0282(16)53278-6. [DOI] [PubMed] [Google Scholar]
- 20.Layman LC, Cohen DP, Jin M, Xie J, Li Z, Reindollar RH, et al. Mutations in gonadotropin-releasing hormone receptor gene cause hypogonadotropic hypogonadism. Nat Genet. 1998;18:14–5. doi: 10.1038/ng0198-14. [DOI] [PubMed] [Google Scholar]
- 21.Layman LC, Lee EJ, Peak DB, Namnoum AB, Vu KV, van Lingen BL, et al. Delayed puberty and hypogonadism caused by mutations in the follicle-stimulating hormone beta-subunit gene. The New England journal of medicine. 1997;337:607–11. doi: 10.1056/NEJM199708283370905. [DOI] [PubMed] [Google Scholar]
- 22.Quaynor SD, Stradtman EW, Jr, Kim HG, Shen Y, Chorich LP, Schreihofer DA, et al. Delayed puberty and estrogen resistance in a woman with estrogen receptor alpha variant. The New England journal of medicine. 2013;369:164–71. doi: 10.1056/NEJMoa1303611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biason-Lauber A, De Filippo G, Konrad D, Scarano G, Nazzaro A, Schoenle EJ. WNT4 deficiency--a clinical phenotype distinct from the classic Mayer-Rokitansky-Kuster-Hauser syndrome: a case report. Hum Reprod. 2007;22:224–9. doi: 10.1093/humrep/del360. [DOI] [PubMed] [Google Scholar]
- 24.Philibert P, Biason-Lauber A, Gueorguieva I, Stuckens C, Pienkowski C, Lebon-Labich B, et al. Molecular analysis of WNT4 gene in four adolescent girls with mullerian duct abnormality and hyperandrogenism (atypical Mayer-Rokitansky-Kuster-Hauser syndrome) Fertility and sterility. 2011;95:2683–6. doi: 10.1016/j.fertnstert.2011.01.152. [DOI] [PubMed] [Google Scholar]
- 25.Philibert P, Biason-Lauber A, Rouzier R, Pienkowski C, Paris F, Konrad D, et al. Identification and functional analysis of a new WNT4 gene mutation among 28 adolescent girls with primary amenorrhea and mullerian duct abnormalities: a French collaborative study. The Journal of clinical endocrinology and metabolism. 2008;93:895–900. doi: 10.1210/jc.2007-2023. [DOI] [PubMed] [Google Scholar]
- 26.Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, et al. Positions statement: criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: an Androgen Excess Society guideline. The Journal of clinical endocrinology and metabolism. 2006;91:4237–45. doi: 10.1210/jc.2006-0178. [DOI] [PubMed] [Google Scholar]
- 27.Bernardini L, Gimelli S, Gervasini C, Carella M, Baban A, Frontino G, et al. Recurrent microdeletion at 17q12 as a cause of Mayer-Rokitansky-Kuster-Hauser (MRKH) syndrome: two case reports. Orphanet J Rare Dis. 2009;4:25. doi: 10.1186/1750-1172-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ledig S, Schippert C, Strick R, Beckmann MW, Oppelt PG, Wieacker P. Recurrent aberrations identified by array-CGH in patients with Mayer-Rokitansky-Kuster-Hauser syndrome. Fertility and sterility. 2011;95:1589–94. doi: 10.1016/j.fertnstert.2010.07.1062. [DOI] [PubMed] [Google Scholar]
- 29.Sandbacka M, Laivuori H, Freitas E, Halttunen M, Jokimaa V, Morin-Papunen L, et al. TBX6, LHX1 and copy number variations in the complex genetics of Mullerian aplasia. Orphanet J Rare Dis. 2013;8:125. doi: 10.1186/1750-1172-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tewes AC, Rall KK, Romer T, Hucke J, Kapczuk K, Brucker S, et al. Variations in RBM8A and TBX6 are associated with disorders of the mullerian ducts. Fertility and sterility. 2015;103:1313–8. doi: 10.1016/j.fertnstert.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Waschk DE, Tewes AC, Romer T, Hucke J, Kapczuk K, Schippert C, et al. Mutations in WNT9B are associated with Mayer-Rokitansky-Kuster-Hauser syndrome. Clinical genetics. 2016;89:590–6. doi: 10.1111/cge.12701. [DOI] [PubMed] [Google Scholar]
- 32.Ma W, Li Y, Wang M, Li H, Su T, Li Y, et al. Associations of Polymorphisms in WNT9B and PBX1 with Mayer-Rokitansky-Kuster-Hauser Syndrome in Chinese Han. PLoS One. 2015;10:e0130202. doi: 10.1371/journal.pone.0130202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley KN, Catalano LM, Bernat JA, Adams SD, Martin DM, Lalani SR, et al. Recurrent deletions and duplications of chromosome 2q11.2 and 2q13 are associated with variable outcomes. Am J Med Genet A. 2015;167A:2664–73. doi: 10.1002/ajmg.a.37269. [DOI] [PubMed] [Google Scholar]
- 34.Narlis M, Grote D, Gaitan Y, Boualia SK, Bouchard M. Pax2 and pax8 regulate branching morphogenesis and nephron differentiation in the developing kidney. J Am Soc Nephrol. 2007;18:1121–9. doi: 10.1681/ASN.2006070739. [DOI] [PubMed] [Google Scholar]
- 35.Schaffer AA. Digenic inheritance in medical genetics. J Med Genet. 2013;50:641–52. doi: 10.1136/jmedgenet-2013-101713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Layman LC. The genetic basis of female reproductive disorders: Etiology and clinical testing. Mol Cell Endocrinol. 2013;370:138–48. doi: 10.1016/j.mce.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Layman LC. Clinical genetic testing for kallmann syndrome. The Journal of clinical endocrinology and metabolism. 2013;98:1860–2. doi: 10.1210/jc.2013-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.AlAsiri S, Basit S, Wood-Trageser MA, Yatsenko SA, Jeffries EP, Surti U, et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. The Journal of clinical investigation. 2015;125:258–62. doi: 10.1172/JCI78473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu H, Roach JC, Coon H, Guthery SL, Voelkerding KV, Margraf RL, et al. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat Biotechnol. 2014;32:663–9. doi: 10.1038/nbt.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.