Abstract
Metabolite profiling of novel psychoactive substances (NPS) is critical for documenting drug consumption. N-(1-amino-3,3-dimethyl-1-oxobutan-2-yl)-1-(4-fluorobenzyl)-1H-indazole-3-carboxamide (ADB-FUBINACA) is an emerging synthetic cannabinoid whose toxicological and metabolic data are currently unavailable. We aimed to determine optimal markers for identifying ADB-FUBINACA intake. Metabolic stability was evaluated with human liver microsome incubations. Metabolites were identified after 1 and 3 h incubation with pooled human hepatocytes, liquid chromatography- high resolution mass spectrometry in positive-ion mode (5600+ TripleTOF®, Sciex) and several data mining approaches (MetabolitePilot™, Sciex). Metabolite separation was achieved on an Ultra Biphenyl column (Restek®); full-scan TOF-MS and information-dependent acquisition MS/MS data were acquired. ADB-FUBINACA microsomal half-life was 39.7 min, with a predicted hepatic clearance of 9.0 mL/min/kg and a 0.5 extraction ratio (intermediate-clearance drug). Twenty-three metabolites were identified. Major metabolic pathways were alkyl and indazole hydroxylation, terminal amide hydrolysis, subsequent glucuronide conjugations, and dehydrogenation. We recommend ADB-FUBINACA hydroxyalkyl, hydroxydehydroalkyl and hydroxylindazole metabolites as ADB-FUBINACA intake markers. N-dealkylated metabolites are not specific ADB-FUBINACA metabolites and should not be used as definitive markers of consumption. This is the first ADB-FUBINACA in vitro metabolism study; in vivo experiments enabling pharmacokinetic and pharmacodynamics studies or urine from authentic clinical/forensic cases are needed to confirm our results.
Keywords: ADB-FUBINACA, synthetic cannabinoid, novel psychoactive substances, metabolism, hepatocytes, LC-HRMS
1. INTRODUCTION
Synthetic cannabinoids (SCs) are novel psychoactive substances (NPS) eliciting cannabimimetic psychoactive effects [1]. SCs were originally produced for biomedical research but now are abused, leading many countries to schedule these drugs as controlled substances [2-5]. Clandestine laboratories continue producing new compounds that mimic effects of scheduled NPS to circumvent legislation. More than 130 SCs were identified to date [6]. Little pharmacological or toxicological data are available when substances first appear. Analytical methods require constant updating, and pharmacodynamic and pharmacokinetic studies must be conducted for the continual emergence of NPS. With its patent filed only in 2009, ADB-FUBINACA, N-(1-amino-3,3-dimethyl-1-oxobutan-2-yl)-1-(4-fluorobenzyl)-1H-indazole-3-carboxamide (Fig. 1) is one of the newest SC [7]. In 2013, the drug was identified for the first time in illegal herbal blends in Japan, in association with two other SCs ADBICA and XLR-11 [8]. The drug was reported for the first time in Europe in the same year in Hungary [6] as Facebook logo shaped tablets, and in biological samples from patients who swallowed or crushed and insufflated the product [9, 10].
Fig. (1).
Structure of ADB-FUBINACA and Schedule I controlled analogs.
There are currently no in vivo human or animal ADB-FUBINACA pharmacodynamic data. In vitro experiments demonstrated potent CB1 cannabinoid receptor binding (Ki = 0.36 nM, EC50 = 0.98 – 1.2 nM) [7, 11] and CB2 (EC50 = 3.5 nM) [11]. CB1 binding affinity that correlates with cannabinoid psychoactive effects [12] is 140 times stronger than for THC, comparable to other indole and indazole SCs with an aminomethylbutanamide or aminodimethylbutanamide structure [11]. Affinity for CB1 receptors is slightly lower for the desmethyl and indole analogs, AB-FUBINACA (scheduled) [13] and ADB-FUBICA, and higher with the N-pentyl and N-fluoropentyl analogs, ADB-PINACA (scheduled) [13] and 5F-ADB-PINACA [11]. ADB-FUBINACA reportedly produces drowsiness, respiratory deprivation, CO2 retention, hypotonia, hypothermia, vomiting, sinus tachycardia, mydriasis, extrapyramidal movement disorder, hyperkinesis, acoustic and visual hallucinations, agitation and mixed symptoms [2, 10]. Structural analogs were involved in several intoxication and fatalities [14-19], with the first ADB-FUBINACA death reported in 2016 [20].
ADB-FUBINACA pharmacodynamic and pharmaco-kinetic studies are still lacking. Many SCs are active at low doses and extensively metabolized with little to no parent compound found in urine, making urinary metabolite detection critical for documenting consumption in forensic and clinical cases [21-25]. One ADB-FUBINACA human liver microsome (HLM) in vitro metabolism study identified a single hydroxyalkyl metabolite [26]. Identifying the SC responsible for resultant toxicities also is important for educating the public on the drug’s dangers.
Our aim was to determine ADB-FUBINACA human metabolism to improve analytical identification of this new SC in biological samples. In vitro HLM [27-29] and human hepatocytes incubations [30-33] with high resolution mass spectrometry (HRMS) analysis proved useful for predicting human SC metabolism. Human hepatocytes contain all required hepatic metabolic enzymes and endogenous cofactors and provide the natural orientation of membrane enzymes, better predicting metabolite production in in vivo conditions than enzymes alone or HLM [34].
2. EXPERIMENTAL
2.1. Chemicals and Reagents
ADB-FUBINACA and diclofenac standards were purchased from Cayman Chemical (Ann Arbor, MI, USA) and Toronto Research Chemicals (Toronto, Canada) respectively. LC-MS grade water, methanol, and formic acid (Optima™ LC/MS) were acquired from Fisher Scientific (Fair Lawn, NJ, USA), and trypan blue and LC-MS grade acetonitrile from Sigma-Aldrich® (St. Louis, MT, USA). Distilled water was produced by an ELGA PURELAB® Ultra Analytic purifier (Siemens Water Technologies, Lowell, MA, USA). Fifty-donor pooled HLM, ten-donor-pooled cryopreserved human hepatocytes, InVitroGRO™ hepatocyte thawing (HT) medium, and InVitroGRO™ Krebs-Henseleit buffer (KHB) were obtained from BioreclamationIVT (Westbury, NY, USA). Solutions A (NADPH regeneration system) and B (glucose-6-phosphate dehydrogenase) were purchased from BD Biosciences (San Jose, CA, USA).
2.2. Metabolic Stability Assessment with Human Liver Microsomes (HLM)
2.2.1. Incubation
The reaction mixture included 780 µL distilled water, 100 µL 0.5 M potassium phosphate buffer pH 7.4, 10 µL solution B, and 10 µL 100 µmol/L ADB-FUBINACA in methanol. After vortexing, HLM suspensions (20 mg/mL) were thawed at 37ºC, 50 µL added to the reaction mixture and gently mixed. The suspension was pre-incubated at 37ºC for 3 min, and the reaction initiated by adding 50 µL solution A. Samples (100 µL) were collected after 0, 3, 8, 13, 20, 45 and 60 min incubation and the reaction quenched with an equal volume of ice-cold acetonitrile. Samples were stored at – 80ºC until analysis.
HLM samples were thawed and diluted 100-fold with mobile phase A before injection onto the LC-HRMS. Liquid chromatography (LC)-HRMS parameters for HLM incubations are described below.
2.2.2. LC-MS Parameters
HLM LC-MS analysis was performed on a 3200 QTRAP® mass spectrometer (triple quadrupole-linear ion trap) equipped with an TurboV ion source operated in positive electrospray mode from Sciex (Foster City, CA, USA) and coupled with an LC-20ADXR HPLC system from Shimadzu (Columbia, MD, USA). Data were acquired with analyst TF software version 1.5 (Sciex).
Separation was performed using a Kinetex® C18 column (100 x 2.1 mm, 2.6 µm) combined with a 50 x 2.1 mm guard column of identical mobile phase from Phenomenex (Torrance, CA, USA). Elution was achieved within 15 min with a gradient mobile phase composed of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) at a flow rate of 0.3 mL/min. Injection volume was 5 µL. Autosampler and column oven temperatures were 4 and 40ºC, respectively. Gradient conditions started at 5% B for 0.5 min, increased to 95% B in 9.5 min, were held for 2.5 min, returned to initial conditions in 0.1 min and re-equilibrated for 2.4 min.
The mass spectrometer was operated in multiple reaction monitoring (MRM) mode. Three transitions were monitored between 6.5 and 8.5 min (accumulation time, 0.5 s): transition m/z 383.2 > 253.3 with a 33 V collision energy (CE) and a 4 V collision cell exit potential (CXP), transition m/z 383.2 > 109.0 with a 57 V CE and a 4 V CXP, and transition m/z 383.2 > 366.3 with a 17 V CE and a 6 V CXP. The first transition was for quantification and the last two for identification confirmation. Source parameters were: gas 1, 45 psi; gas 2, 70 psi; curtain gas, 30 psi; temperature, 500ºC; ion spray voltage, 4 kV; declustering potential, 31 V.
2.2.3. Calculations
In vitro microsomal half-life (T1/2) and clearance (CLint, micr), predicted intrinsic clearance (CLint), predicted human hepatic clearance (CLhep), and extraction ratio (ER) were calculated based on Baranczewski [35] and McNaney’s [36] models, without consideration of plasma protein binding. T1/2 was calculated based on the percentage of ADB-FUBINACA MS signal remaining at each time point.
2.3. Metabolite Profiling with Human Hepatocytes
2.3.1. Incubation
Hepatocytes were thawed at 37ºC, washed twice with InVitroGRO™ HT medium and KHB, and centrifuged at 100 g for 5 min. Supernatant was removed and the pellet re-suspended in 2 mL KHB, yielding a 2 x 106 cells/min concentration. Cell viability, assessed with trypan blue exclusion dye (0.4%, v/v), was ≥80%. ADB-FUBINACA was dissolved in methanol and diluted in KHB to a final 20 µmol/L concentration. The reaction mixture was prepared with 250 µL hepatocyte suspension and 250 µL drug solution and incubated in a Forma™ Steri-Cycle™ CO2 incubator at 37ºC (Thermo Scientific). Samples (500 µL) were incubated for 0, 1 and 3 h and the reaction quenched with an equal volume of ice-cold acetonitrile. Diclofenac was incubated in a separate mixture under the same conditions and 4’-hydroxydiclofenac and acyl-β-D-glucuronide diclofenac monitored to ensure hepatocyte metabolic activity. In addition, a negative control with ADB-FUBINACA in buffer without hepatocytes was incubated to rule out non-enzymatic reactions. Samples were stored at – 80ºC until analysis.
Hepatocyte samples were thawed and centrifuged at 4ºC, 15,000 g for 10 min, to remove cell debris. Supernatants were diluted 5-fold with 0.1% formic acid in water (mobile phase A) before injection.
2.3.2. LC-HRMS Parameters
LC-HRMS analysis of hepatocyte incubations was performed on a 5600+ TripleTOF® quadrupole-time of flight mass spectrometer equipped with a DuoSpray source (Sciex) operated in positive electrospray mode, coupled with an LC-20ADXR HPLC system from Shimadzu. Data were acquired with Analyst TF software version 1.6 (Sciex).
Separation was performed on an Ultra Biphenyl column (100 x 2.1 mm, 3 µm) combined with a 10 x 2.1 mm guard column of identical phase from Restek® (Bellefonte, PA, USA). Elution was achieved within 15 min with a gradient mobile phase composed of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) at a flow rate of 0.5 mL/min. Injection volume was 15 µL. Autosampler and column oven temperatures were set at 4 and 30ºC respectively. Gradient conditions started with 20% B for 0.5 min, increased to 95% B in 10.5 min and then held for 2 min, returned to initial conditions in 0.1 min and re-equilibrated for 1.9 min. Efflux was diverted to waste before 2 and after 13 min.
The mass spectrometer operated in full-scan TOF-MS and information-dependent acquisition (IDA) MS/MS modes. One MS cycle was composed of one full-scan TOF-MS survey scan followed by up to eight IDA MS/MS scans (accumulation time, 0.5 s). The eight ions detected with the most intense signal in the survey scan (intensity threshold, 100 cps), triggered an MS/MS event with fragmentation (isolation window, m/z 1.5). IDA mode included a list of expected metabolites (Supplementary material, Table S1 (5.3MB, pdf) ) that were fragmented in priority, regardless of their intensity (mass tolerance, 50 mDa). Full MS scans were acquired from m/z 100 – 1000 with high resolution and a 0.1 s accumulation time. IDA MS/MS scans were acquired from m/z 30 – 1000 with high resolution; collision energy was 35 ± 15 V and accumulation time 0.05 s. Source parameters were: gas 1, 60 psi; gas 2, 75 psi; curtain gas, 45 psi; temperature, 650ºC; ion spray voltage, 4 kV; declustering potential, 80 V.
2.3.3. Metabolite Identification
Data from hepatocyte incubations were analyzed with MetabolitePilot™ software (v. 1.6; Sciex) for ADB-FUBINACA metabolites identification. Processing settings are described in Table 1.
Table 1.
MetabolitePilot™ processing settings. MS(/MS), (tandem) mass spectrometry.
| Parent Compound | ADB-FUBINACA |
|---|---|
| Formula | C21H23N4O2F |
| Theoretical m/z (positive mode) | 383.1878 |
| Biotransformations | |
| C7H5F loss, C6H11NO loss, C6H12N2O loss, NH loss, amide hydrolysis, carboxylation, defluorination, defluorobenzylation, desaturation, glucuronidation, glutathione conjugation, hydrogenation, amide hydrolysis to carboxylic acid, internal hydrolysis, ketone formation, hydroxylation, sulfation, and combinations | |
| Chromatographic parameters | |
| Min peak width | 2.5 s |
| Min chromatographic intensity | 500 cps |
| Retention time window | 1 to 13 min |
| MS parameters | |
| m/z tolerance | 50 ppm |
| Min MS peak intensity | 100 cps |
| Mass range window | 150 to 800 m/z |
| Isotopic pattern intensity tolerance | 20% |
| Isotopic pattern m/z tolerance | 25 ppm |
| MS/MS parameters | |
| m/z tolerance | 50 ppm |
| Min MS/MS peak intensity | 50 cps |
| Characteristic product ions | Find at least 1 |
| Neutral loss | Find at least 1 |
3. RESULTS
3.1. Metabolic Stability Assessment
ADB-FUBINACA had a 39.7 ± 0.1 min half-life (T1/2) and an in vitro microsomal intrinsic clearance (CLint micr) of 17.5 ± 0.1 µL/min/mg in HLM incubations. Intrinsic (CLint) and hepatic (CLhep) clearances were estimated at 16.5 and 9.0 mL/min/kg, respectively, with a 0.5 extraction ratio (ER).
3.2. Metabolite Profiling
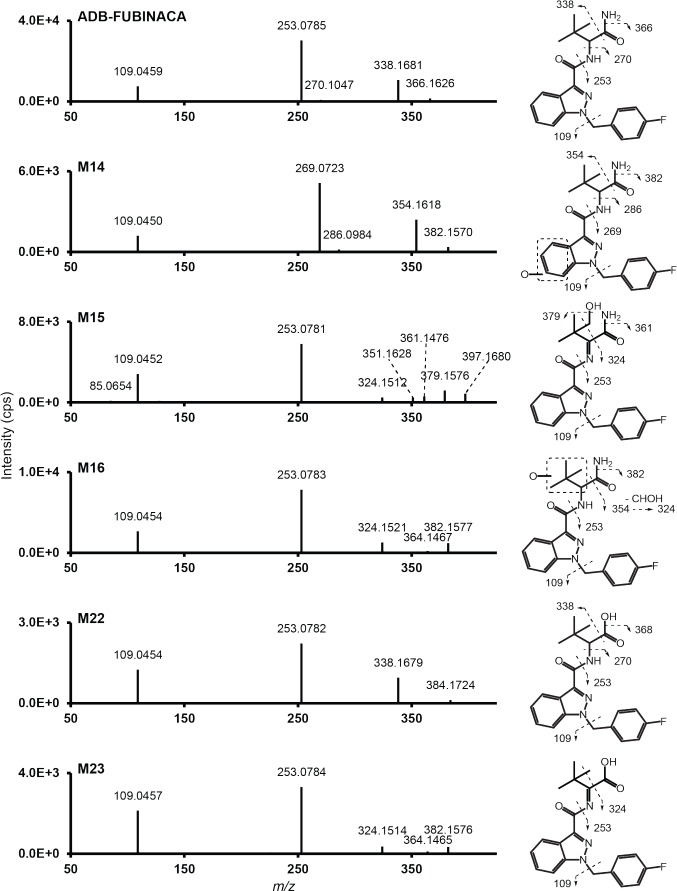
ADB-FUBINACA peak area was 7.8 x 106, 4.8 x 106, and 2.3 x 106 respectively after 0, 1, and 3 h incubation with human hepatocytes. Parent MS/MS spectrum (m/z 383.1878, retention time: 6.58 min) presented major characteristic fragments produced by aminodimethylbutanamide loss (m/z 253.0772), carboxamide loss (m/z 338.1663) or fluorobenzylium ion formation (m/z 109.0448) (Fig. 2). Two minor fragments also were formed by loss of dimethylbutanamide (m/z 270.1037) and amine (m/z 366.1612). ADB-FUBINACA fragments m/z 109, 253, and 338 were previously observed by Uchiyama et al. and Takayama et al. by LC-ESI-MS/MS (QTOF and triple quadrupole, respectively) [8, 26].
Fig. (2).
ADB-FUBINACA and major metabolites’ MS/MS spectrum and assigned fragmentation patterns. Metabolites are presented in ascending retention time order.
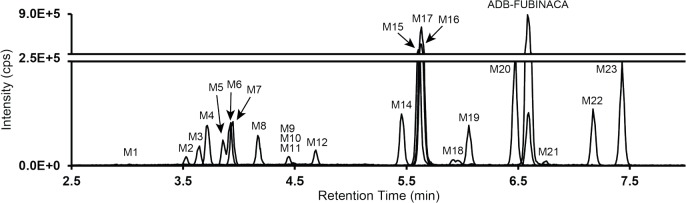
Twenty-one and twenty-two ADB-FUBINACA metabolites were identified in hepatocyte samples after 1 and 3 h incubation, respectively, totaling twenty-three different metabolites (labeled M1 to M23 in ascending retention time (RT) order)
Accurate mass for diagnostic product ions are given in Fig. (2) and S1 (5.3MB, pdf) (Supplementary material). Peak area for ADB-FUBINACA at 0 h was 7.8 x 106. Metabolites are listed by ascending RT and ranked depending on signal intensity after 1 and 3 h incubation. MS, mass spectrometry; ND, not detected; RT: retention time; *, no MS/MS.
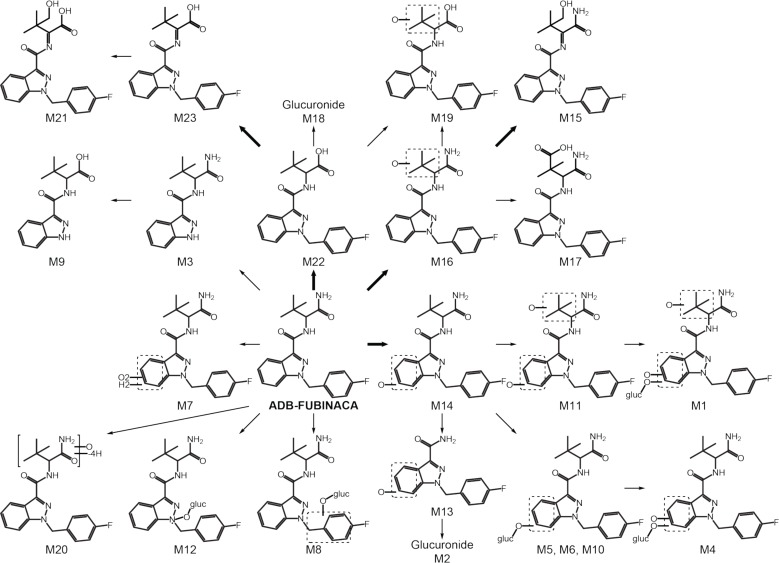
(Fig. 3). Biotransformations, retention times, accurate mass molecular ion, nominal mass product ions, and MS peak areas for parent and metabolites are listed in Table 2. MS/MS spectra and fragmentation patterns are reported in Fig. (2) (major metabolites) and S1 (5.3MB, pdf) (Supplementary material). Accurate mass errors were within 3.6 ppm. The main biotransformations included hydroxylation at the dimethylpropane chain (M1, M11, M15 to M17, M19, and M21) or the indazole ring (M1, M4, M11, M13, and M14), amide hydrolysis (M9, M18, M19, and M21 to M23), subsequent corresponding glucuronide conjugations (M1, M2, M4 to M6, M10, M12, and M18), and dehydrogenation (M15, M21, and M23). Other reactions included epoxidation followed by epoxide hydrolysis (M7), methylene-fluorophenyl hydroxylation then glucuronidation (M8), and N-dealkylations (M2, M3, M9, and M13). All metabolites were absent from the control 0 h incubation. No metabolite was the result of a non-enzymatic transformation as none was detected after 3 h incubation without hepatocytes. Structure elucidation of the top ten metabolites is described below.
Fig. (3).
Combined extracted ion chromatogram of ADB-FUBINACA and metabolites obtained from hepatocyte incubation after 3 h. ADB-FUBINACA metabolites are numbered M1 to M23 in ascending order of retention time.
Table 2.
Elemental composition, accurate mass molecular ion, nominal mass for diagnostic product ions, MS peak areas, and RT in hepatocyte incubations.
| ID | Biotransformation | [M+H]+ | RT (min) | Elemental Composition | Mass Error (ppm) |
Diagnostic
Product Ions (m/z) |
Incubation (1 h) | Incubation (3 h) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Peak Area | Rank | Peak Area | Rank | |||||||
| ADB-FUBINACA | 383.1890 | 6.58 | C21H23N4O2F | 3.3 | 109, 253, 270, 338, 366 | 4.8 x 106 | 2.3 x 106 | |||
| M1 | Aliphatic hydroxylation + indazole dihydroxylidation + glucuronidation | 607.2061 | 2.91 | C27H31N4O11F | 2.5 | 109, 285, 461, 590 | N.D. | 7.9 x 103 | 21 | |
| M2 | N-Dealkylation + indazole hydroxylidation + glucuronidation | 462.1314 | 3.53 | C21H20N3O8F | 1.5 | 109, 269, 286, 445 | 2.3 x 104 | 12 | 4.9 x 104 | 16 |
| M3 | Methylenefluorophenyl loss | 275.1509 | 3.65 | C14H18N4O2 | 2.5 | 145, 162, 230, 258 | 8.4 x 104 | 8 | 1.3 x 105 | 13 |
| M4 | Indazole dihydroxylation + glucuronidation | 591.2107 | 3.72 | C27H31N4O10F | 1.8 | 109, 267, 285, 370, 461, 546, 574 |
5.7 x 104 | 10 | 2.3 x 105 | 8 |
| M5 | Indazole hydroxylation + glucuronidation | 575.2160 | 3.86 | C27H31N4O9F | 2.1 | 109, 269, 354, 445, 530, 558 |
1.6 x 104 | 15 | 1.3 x 105 | 12 |
| M6 | Indazole hydroxylation + glucuronidation | 575.2159 | 3.93 | C27H31N4O9F | 2.0 | 109, 269, 354, 445, 530, 558 |
1.2 x 105 | 6 | 2.3 x 105 | 9 |
| M7 | Dihydrodiol formation | 417.1946 | 3.95 | C21H25N4O4F | 3.2 | 109, 241, 287, 372, 400 | 9.3 x 104 | 7 | 2.6 x 105 | 6 |
| M8 | Methylbenzene hydroxylation + glucuronidation | 575.2160 | 4.17 | C27H31N4O9F | 2.1 | 125, 145, 269, 354, 530, 558 |
8.4 x 104 | 9 | 1.7 x 105 | 11 |
| M9 | Methylenefluorophenyl loss + amide hydrolysis | 276.1350 | 4.42 | C14H17N3O3 | 2.5 | 145, 162, 230 | N.D. | 7.9 x 103 | 22 | |
| M10 | Indazole hydroxylation + glucuronidation | 575.2156 | 4.44 | C27H31N4O9F | 1.4 | 109, 269, 354, 445, 530, 558 |
2.0 x 104 | 14 | 5.2 x 104 | 15 |
| M11 | Aliphatic hydroxylation + indazole hydroxylation | 415.1791 | 4.48 | C21H23N4O4F | 3.6 | 109, 269, 340, 398 | 7.4 x 103 | 19 | 2.0 x 104 | 20 |
| M12 | N-hydroxylation + glucuronidation | 575.2154 | 4.69 | C27H31N4O9F | 1.0 | 109, 253, 324, 338, 382, 558 |
2.1 x 104 | 13 | 8.4 x 104 | 14 |
| M13 | N-Dealkylation + indazole hydroxylation | 286.0986 | 4.76 | C15H12N3O2F | 0.0 | 109, 269 | 6.1 x 103 | 20 | N.D. | |
| M14 | Indazole hydroxylation | 399.1836 | 5.45 | C21H23N4O3F | 2.2 | 109, 269, 286, 354 | 1.7 x 105 | 5 | 3.4 x 105 | 5 |
| M15 | Aliphatic hydroxylation + dehydrogenation | 397.1678 | 5.60 | C21H21N4O3F | 1.9 | 109, 253, 270, 324, 361 | 4.5 x 105 | 2 | 8.5 x 105 | 2 |
| M16 | Aliphatic hydroxylation | 399.1838 | 5.63 | C21H23N4O3F | 2.9 | 109, 253, 324, 382 | 1.6 x 106 | 1 | 1.7 x 106 | 1 |
| M17 | Aliphatic carboxylation | 413.1635 | 5.65 | C21H21N4O4F | 3.7 | 109, 253, 324, 368, 396 | 1.1 x 104 | 18 | 4.2 x 104 | 17 |
| M18 | Amide hydrolysis + glucuronidation | 560.2041 | 5.92 | C27H30N3O9F | 0.3 | 109, 253, 270, 338, 366, 384 |
1.5 x 104 | 16 | 3.2 x 104 | 19 |
| M19 | Amide hydrolysis + aliphatic hydroxylation | 400.1677 | 6.06 | C21H22N3O4F | 2.5 | 109, 253, 324 | 1.5 x 104 | 17 | 2.6 x 105 | 7 |
| M20 | Not identified (+O, -4H) | 395.1525 | 6.48 | C21H19N4O3F | 2.9 | 109, 253 | 5.4 x 104 | 11 | 2.0 x 105 | 10 |
| M21 | Amide hydrolysis + aliphatic hydroxylation + dehydrogenation | 398.1513 | 6.75 | C21H20N3O4F | 0.7 | 109, 253, 324, 352 | 3.1 x 103* | 21 | 3.3 x 104 | 18 |
| M22 | Amide hydrolysis | 384.1726 | 7.17 | C21H22N3O3F | 2.1 | 109, 253, 338 | 1.8 x 105 | 4 | 3.7 x 105 | 4 |
| M23 | Amide hydrolysis + dehydrogenation | 382.1572 | 7.43 | C21H20N3O3F | 2.7 | 109, 253, 324 | 1.8 x 105 | 3 | 6.6 x 105 | 3 |
3.2.1. Hydroxylated Metabolites
M14 and M16 (m/z 399.1827, 5.45 and 5.63 min respectively) were hydroxyl metabolites of ADB-FUBINACA as suggested by the 15.9949 Da increase from parent mass (+O).
M14 MS/MS spectrum included the fragments m/z 269.0721, 286.0986, 354.1612, and 382.1561 that were also 15.9949 Da larger and also included the fluorobenzylium ion (m/z 109.0448), indicating that the hydroxylation occurred on the benzene part of the indazole ring. It is noteworthy that no water loss was detected during M14 fragmentation as the delocalized electrons of the benzene ring strengthened the hydroxyl bond. M6 (m/z 575.2148, 3.93 min) likely is the corresponding glucuronide of M14. M4 (m/z 591.2097, 3.72 min) was formed by further hydroxylation of ADB-FUBINA hydroxyindazole-glucuronide on the benzene ring.
M16 MS/MS spectrum included fragments m/z 354.1612 and 382.1561 that were 15.9949 Da larger than those of the parent. Fragments m/z 109.0448 and 253.0772, also present in the parent MS/MS spectrum, further suggested that M16 hydroxylation occurred on the dimethylpropane chain. Fragment m/z 324.1507 was produced by a carboxamide and CHOH loss, perhaps the result of a hydroxylation on a methyl group of the dimethylpropane chain. Further hydroxylation of M16 generated M17 (m/z 413.1619, 5.65 min).
3.2.2. Metabolites Generated by Amide Hydrolysis
Metabolites generated by amide hydrolysis eluted later than non-hydrolyzed metabolites, as observed with other SCs with the same terminal carboxamide group [37].
M22 (m/z 384.1718, 7.17 min) was the result of amide hydrolysis as suggested by a 0.9840 Da increase (-NH2, +OH). The MS/MS spectrum showed fragments m/z 109.0448, 253.0772, 338.1663, and 366.1612, also present in parent spectrum, demonstrating that ADB-FUBINACA was not transformed except amide hydrolysis on the terminal amine function. Similarly, M19 (m/z 400.1667, 6.06 min) presented a 0.9840 Da increase from M16 mass (ADB-FUBINACA hydroxydimethylpropyl) and the same fragmentation pattern, indicating that both amide hydrolysis and dimethylpropane hydroxylation occurred.
3.2.3. Dehydrogenated Metabolites
M23 (m/z 382.1561, 5.92 min) was formed by amide hydrolysis and dehydrogenation as suggested by a 1.0317 Da decrease (-NH2, +OH, -H2). Given the late RT, M23 could not possibly be an artifact derived from a water loss of a hydroxylated metabolite. MS/MS spectrum presented fragments m/z 109.0448 and 253.0772, indicating that the dehydrogenation occurred on the aminodimethylbutanamide portion of ABD-FUBINACA, leading to an imine formation. Fragment m/z 324.1507 was produced by the loss of the carboxylic acid and one methyl group (– 58.0054 Da).
M15 (m/z 397.1670, 5.60 min) was formed by ADB-FUBINACA hydroxylation and dehydrogenation as suggested by the 13.9792 Da decrease (+O, -H2). Fragments m/z 109.0448, 253.0772, 270.1037, and 324.1507 indicate imine formation and methyl hydroxylation. Fragment m/z 379.1565 confirmed hydroxylation (water loss).
M20 (m/z 395.1514, 6.48 min) was generated by hydroxylation and a di-dehydrogenation, as suggested by the 11.9636 Da increase (+O, -2H2). Fragments m/z 109.0448 and 253.0772, also detected in ADB-FUBINACA’s mass spectrum, indicated that reactions occurred on the amino-dimethylbutanamide moiety. Fragment m/z 324.1507 indicated dehydrogenation to the imine function.
3.2.4. Dealkylated Metabolites
M13 (m/z 286.0986, 4.76 min) was formed by dimethylbutanamide cleavage and a hydroxylation, as suggested by the 97.0892 Da decrease (+O, -C6H11NO). Few fragments were produced as M13 signal intensity was low in the Full-MS scan after 1 h incubation and not detectable after 3 h incubation. Fragment m/z 109.0448 corresponds to unchanged methylenefluorophenyl and indicated that the reaction occurred on the indazole ring benzene group. Fragment m/z 269.0720 was formed by amine loss.
3.2.5. Epoxidized Metabolite
M7 (m/z 417.1933, 3.95 min) was a dihydrodiol, as suggested by a 34.0055 Da increase (+2O, +2H). Fragments m/z 287.0826, 304.1092, 372.1718, and 400.1667, were 34.0055 Da larger than the corresponding ADB-FUBINACA fragments while fragment m/z 109.0448 was still present, indicating that the reactions did only affect the indazole ring. We hypothesized that M7 was formed by epoxidation of the benzene moiety of ADB-FUBINACA’s indazole ring, followed by hydrolysis of the newly formed epoxide. This metabolic reaction was previously observed in several SCs containing indole or indazole ring metabolic pathways [33, 37].
4. DISCUSSION
4.1. Metabolic Stability Assessment
According to McNaney’s [36] and Lavé’s [38] classifications based on intrinsic clearance and extraction ratio estimations, respectively, ADB-FUBINCA is considered an intermediate-clearance drug (15<CLint<40 and 0.3<ER<0.7). As a result, ADB-FUBINCA metabolites are expected to be found in urine several days after intake. First-pass hepatic elimination is expected to be significant, and hepatic clearance sensitive to changes in plasma protein binding, drug metabolism, and hepatic blood flow [39]. However, the extent of plasma protein binding can play an important role in a drug’s phar-macokinetics. If ADB-FUBINACA was a highly protein-bound drug, this would lower the hepatic clearance and extend the detection window.
4.2. Metabolic Profiling
4.2.1. Method
HRMS is a powerful tool in metabolite identification studies as it theoretically allows capturing every compound in a single injection, and facilitates molecular formula determination of metabolites and fragments. Nitrogen atoms of the indazole core and the two amide functions are easily charged in acidic conditions, making positive-ion mode ionization suitable for ADB-FUBINACA metabolites’ detection. A 50 ppm MS mass tolerance was chosen during preliminary automated metabolite identification as it is broad enough that no possible metabolite with a high mass error could be missed, but narrow enough to predict elemental composition. MS/MS IDA mode scan speed allowed monitoring eight compounds at each MS cycle with a ramped collision energy fragmentation to maximize the production of different fragments and facilitate identification.
Samples were prepared by simple dilution as opposed to extraction before injection to increase detection of potential metabolites. However, matrix suppression could impede detection of metabolites with low signal intensity. Similarly, relative metabolite rank based on MS peak intensity could be confounded by matrix effect and ionization efficiency.
4.2.2. Metabolite Identification
The suggested ADB-FUBINACA metabolic pathway is described in Fig. (4). M16 (ADB-FUBINACA hydroxy-alkyl), M15 (ADB-FUBINACA hydroxydehydroalkyl) and M14 (ADB-FUBINACA hydroxylindazole) are the three metabolites recommended as ADB-FUBINACA intake markers. Several glucuronidated metabolites (M1, M2, M4 to M6, M8, M10, M12, and M18) suggest that hydrolysis of biological samples (e.g. urine, blood) prior to extraction could increase non-glucuronidated metabolites’ concentrations and facilitate their detection.
Fig. (4).
ADB-FUBINACA suggested metabolic pathway. M20 accurate mass and fragmentation pattern suggest the loss of four hydrogen atoms and the addition of an oxygen on ADB-FUBINACA dimethylbutanamide moiety but its structure was not fully elucidated. Bold arrow, major pathway; gluc, glucuronide.
Major metabolic pathways of AB-FUBINACA, ADB-FUBINACA’s analog (Fig. 1), in hepatocyte incubations included amide hydrolysis and alkyl and indazole hydroxylations [37]. Alkyl dehydrogenations, dihydrodiol formations, and glucuronidations also were observed to a lesser extent. ADB-FUBINACA metabolism in the present experiments was consistent with these results. ADB-FUBINACA N-dealkylated metabolites formed by dimethylbutanamide cleavage (M2 and M13) also were detected in AB-FUBINACA metabolism [37]. The same two metabolites could theoretically be formed after MDMB-FUBINACA metabolism as it presents the same indazole- methylenefluorophenyl structure. Similarly, metabolites formed by N-dealkylation (M3 and M9) could theoretically be formed after MAB-CHMINACA, ADB-CHMINACA, ADB-PINACA and 5-F-ADB-PINACA metabolism, as they present the same indazole-dimethylbutanamide structure. Metabolites generated by amide hydrolysis, M21, M22 and M23, could theoretically be formed after MDMB-FUBINACA O-demethylation. However, MDMB-FUBINACA, MAB-CHMINACA, ADB-CHMINACA, ADB-PINACA and 5-F-ADB-PINACA metabolism has yet to be elucidated. Given the constant emergence of new substances and depending on the legislation, being able to precisely distinguish one synthetic cannabinoid from another is critical. As M2, M3, M9, M13, M21, M22 and M23 may not be specific for ADB-FUBINACA intake, they are not recommended as ADB-FUBINACA consumption markers. Noteworthy, AB-FUBINACA carboxylic acid, the corresponding M22 ADB-FUBINACA metabolite, is specific for AB-FUBINACA metabolism, as there are no other compounds known that can produce it [37].
This in vitro hepatocyte marker profile should be confirmed with human urine samples after ADB-FUBINACA intake, as post-liver metabolism processes also can occur (e.g. re-absorption, extrahepatic metabolism, enterohepatic circulation). Unfortunately, authentic urine or blood specimens following ADB-FUBINACA consumption were not available despite our efforts to obtain them. It is important to verify in vitro hepatocyte metabolism results with human urine samples, although we have observed good correlation with authentic urine specimens and hepatocyte incubations for 10 µM FDU-PB-22, FUB-PB-22 [40], AB-PINACA [32] and AB-FUBINACA [37]. Castaneto et al. observed good agreement between metabolites identified during AB-FUBINACA hepatocyte incubations and authentic urine specimens following AB-FUBINACA intake, including the most intense signals for AB-FUBINACA carboxylic acid, AB-FUBINACA carboxylic acid-glucuronide, and AB-FUBINACA carboxylic acid-hydroxyalkyl metabolites [37].
CONCLUSION
The first ADB-FUBINACA metabolic profile is presented. ADB-FUBINACA pharmacokinetics were determined in human liver microsomes: ADB-FUBINACA was predicted as an intermediate-clearance compound with a 37.9 min microsomal half-life. ADB-FUBINACA metabolism was studied following human hepatocyte incubations: 23 meta-bolites were detected; M16 (ADB-FUBINACA hydroxyalkyl), M15 (ADB-FUBINACA hydroxydehydroalkyl) and M14 (ADB-FUBINACA hydroxylindazole) were detected with the most intense MS signals and are suggested as ADB-FUBINACA markers. Urine hydrolysis should be performed to improve detection capability.
Experiments were conducted to improve ADB-FUBINACA metabolite identification in human matrices for forensic or clinical cases. Identification of major metabolite markers is critical to guide synthesis efforts of manufacturers to provide suitable analytical standards and for further pharmacodynamic and pharmacokinetic studies. In the past, HLM and hepatocyte incubation approaches proved useful to predict human metabolism [27-33]. However, in vivo data are necessary to confirm results.
ACKNOWLEDGEMENTS
The authors would like to thank Tim Moeller of Bioreclamation IVT for his assistance with the incubations. This research was supported by the Intramural Research Program of the National Institute on Drug Abuse, National Institutes of Health.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
CONFLICT OF INTEREST
The authors confirm that they do not have any conflicts of interest with this article contents.
REFERENCES
- 1.Gurney S.M., Scott K.S., Kacinko S.L., Presley B.C., Logan B.K. Pharmacology, toxicology, and adverse effects of synthetic cannabinoid drugs. Forensic Sci. Rev. 2014;26(1):53–78. [PMID: 26226970]. [PubMed] [Google Scholar]
- 2.U.S. Department of Justice, Drug Enforcement Administration, Office of Diversion Control http://www.deadiversion.usdoj.gov. (Accessed February 1, 2016)
- 3.Government of Canada. Justice Laws Website. http://www.laws-lois.justice.gc.ca. (Accessed February 1, 2016)
- 4.European Monitoring Centre for Drugs and Drug Addiction http://www.ednd.emcdda.europa.eu. (Accessed February 1, 2016)
- 5.Australian Government, Department of Health, Therapeutic Goods Administration http://www.tga.gov.au. (Accessed February 1, 2016)
- 6.European Monitoring Centre for Drugs and Drug Addiction. European Drug Report – Trends and Developments. Lisbon: Portugal: 2015. [Google Scholar]
- 7.Buchler I.P., Hayes M.J., Hegde S.G., Hockerman S.L., Jones D.E., Kortum S.W., Rico J.G., Tenbrink R.E., Wu K.K. Indazole derivatives. U.S. Patent CA 2,714,573 A1, 2009 September 3. . 2009 [Google Scholar]
- 8.Uchiyama N., Matsuda S., Kawamura M., Kikura-Hanajiri R., Goda Y. Two new-type cannabimimetic quinolinyl carboxylates, QUPIC and QUCHIC, two new cannabimimetic carboxamide derivatives, ADB-FUBINACA and ADBICA, and five synthetic cannabinoids detected with a thiophene derivative α-PVT and an opioid receptor agonist AH-7921 identified in illegal products. Forensic Toxicol. 2013;31(2):223–240. [http://dx.doi.org/10. 1007/s11419-013-0182-9]. [Google Scholar]
- 9. http://www2.aekwien.at/news_pdf/8781_1.pdf. (Accessed February 1, 2016)
- 10. Scottish Families Affected by Alcohol & Drugs http://www.sfad.org.uk/media-centre/news/130/15-non-fatal-intoxications-associated-with-tablets-containing-ADB-FUBINACA-in-Hungary. (Accessed February 1, 2016)
- 11.Banister S.D., Moir M., Stuart J., Kevin R.C., Wood K.E., Longworth M., Wilkinson S.M., Beinat C., Buchanan A.S., Glass M., Connor M., McGregor I.S., Kassiou M. Pharmacology of Indole and Indazole Synthetic Cannabinoid Designer Drugs AB-FUBINACA, ADB-FUBINACA, AB-PINACA, ADB-PINACA, 5F-AB-PINACA, 5F-ADB-PINACA, ADBICA, and 5F-ADBICA. ACS Chem. Neurosci. 2015;6(9):1546–1559. doi: 10.1021/acschemneuro.5b00112. [http://dx.doi.org/10. 1021/acschemneuro.5b00112]. [PMID: 26134475]. [DOI] [PubMed] [Google Scholar]
- 12.Huestis M.A., Gorelick D.A., Heishman S.J., Preston K.L., Nelson R.A., Moolchan E.T., Frank R.A. Blockade of effects of smoked marijuana by the CB1-selective cannabinoid receptor antagonist SR141716. Arch. Gen. Psychiatry. 2001;58(4):322–328. doi: 10.1001/archpsyc.58.4.322. [http://dx.doi.org/10.1001/archpsyc.58.4.322]. [PMID: 11296091]. [DOI] [PubMed] [Google Scholar]
- 13.Schedules of controlled substances: temporary placement of four synthetic cannabinoids into Schedule I. Final order. Fed. Regist. 2014;79(27):7577–7582. [PMID: 24605391]. [PubMed] [Google Scholar]
- 14.Notes from the field: severe illness associated with reported use of synthetic marijuana - Colorado, August-September 2013. MMWR Morb. Mortal. Wkly. Rep. 2013;62(49):1016–1017. [PMID: 24336136]. [PMC free article] [PubMed] [Google Scholar]
- 15.European Monitoring Centre for Drugs and Drug Addiction. EMCDDA-Europol 2013 Annual Report on the implementation of Council Decision 2005/387/JHA. 2013 [Google Scholar]
- 16. Federal Drug Control Service of Russia http://fskn.gov.ru/ includes/periodics/speeches_fskn/2014/1006/124332682/detail.shtml. (Accessed February 1, 2016).
- 17.Monte A.A., Bronstein A.C., Cao D.J., Heard K.J., Hoppe J.A., Hoyte C.O., Iwanicki J.L., Lavonas E.J. An outbreak of exposure to a novel synthetic cannabinoid. N. Engl. J. Med. 2014;370(4):389–390. doi: 10.1056/NEJMc1313655. [http://dx.doi.org/10.1056/NEJMc1313655]. [PMID: 24450915]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa K., Wurita A., Minakata K., Gonmori K., Nozawa H., Yamagishi I., Watanabe K., Suzuki O. Postmortem distribution of AB-CHMINACA, 5-fluoro-AMB, and diphenidine in body fluids and solid tissues in a fatal poisoning case: usefulness of adipose tissue for detection of the drugs in unchanged forms. Forensic Toxicol. 2015;33(1):45–53. [http://dx.doi.org/10.1007/ s11419-014-0245-6]. [Google Scholar]
- 19.Schwartz M.D., Trecki J., Edison L.A., Steck A.R., Arnold J.K., Gerona R.R. A common source outbreak of severe delirium associated with exposure to the novel synthetic cannabinoid ADB-PINACA. J. Emerg. Med. 2015;48(5):573–580. doi: 10.1016/j.jemermed.2014.12.038. [http://dx.doi.org/ 10.1016/j.jemermed.2014.12.038]. [PMID: 25726258]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanks K.G., Clark W., Behonick G. Death associated with the use of the synthetic cannabinoid ADB-FUBINACA. J. Anal. Toxicol. 2016;40(3):236–239. doi: 10.1093/jat/bkv142. [http://dx.doi.org/10.1093/jat/ bkv142]. [PMID: 26755539]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adamowicz P., Zuba D., Sekuła K. Analysis of UR-144 and its pyrolysis product in blood and their metabolites in urine. Forensic Sci. Int. 2013;233(1-3):320–327. doi: 10.1016/j.forsciint.2013.10.005. [http://dx.doi.org/10.1016/j. forsciint.2013.10.005]. [PMID: 24314536]. [DOI] [PubMed] [Google Scholar]
- 22.Jang M., Yang W., Choi H., Chang H., Lee S., Kim E., Chung H. Monitoring of urinary metabolites of JWH-018 and JWH-073 in legal cases. Forensic Sci. Int. 2013;231(1-3):13–19. doi: 10.1016/j.forsciint.2013.03.053. [http://dx.doi. org/10.1016/j.forsciint.2013.03.053]. [PMID: 23890611]. [DOI] [PubMed] [Google Scholar]
- 23.Jang M., Yang W., Shin I., Choi H., Chang H., Kim E., Kim E. Determination of AM-2201 metabolites in urine and comparison with JWH-018 abuse. Int. J. Legal Med. 2014;128(2):285–294. doi: 10.1007/s00414-013-0884-x. [http://dx.doi.org/10.1007/s00414-013-0884-x]. [PMID: 23884698]. [DOI] [PubMed] [Google Scholar]
- 24.Bertol E., Vaiano F., Di Milia M.G., Mari F. In vivo detection of the new psychoactive substance AM-694 and its metabolites. Forensic Sci. Int. 2015;256:21–27. doi: 10.1016/j.forsciint.2015.07.018. [http://dx.doi.org/10.1016/ j.forsciint.2015.07.018]. [PMID: 26295909]. [DOI] [PubMed] [Google Scholar]
- 25.Jang M., Kim I.S., Park Y.N., Kim J., Han I., Baeck S., Yang W., Yoo H.H. Determination of urinary metabolites of XLR-11 by liquid chromatography-quadrupole time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2015;408:503–516. doi: 10.1007/s00216-015-9116-1. [PMID: 26514671]. [DOI] [PubMed] [Google Scholar]
- 26.Takayama T., Suzuki M., Todoroki K., Inoue K., Min J.Z., Kikura-Hanajiri R., Goda Y., Toyooka T. UPLC/ESI-MS/MS-based determination of metabolism of several new illicit drugs, ADB-FUBINACA, AB-FUBINACA, AB-PINACA, QUPIC, 5F-QUPIC and α-PVT, by human liver microsome. Biomed. Chromatogr. 2014;28(6):831–838. doi: 10.1002/bmc.3155. [http://dx.doi.org/10.1002/ bmc.3155]. [PMID: 24861751]. [DOI] [PubMed] [Google Scholar]
- 27.Sobolevsky T., Prasolov I., Rodchenkov G. Detection of urinary metabolites of AM-2201 and UR-144, two novel synthetic cannabinoids. Drug Test. Anal. 2012;4(10):745–753. doi: 10.1002/dta.1418. [http://dx. doi.org/10.1002/dta.1418]. [PMID: 23042760]. [DOI] [PubMed] [Google Scholar]
- 28.De Brabanter N., Esposito S., Tudela E., Lootens L., Meuleman P., Leroux-Roels G., Deventer K., Van Eenoo P. In vivo and in vitro metabolism of the synthetic cannabinoid JWH-200. Rapid Commun. Mass Spectrom. 2013;27(18):2115–2126. doi: 10.1002/rcm.6673. [http://dx.doi. org/10.1002/rcm.6673]. [PMID: 23943333]. [DOI] [PubMed] [Google Scholar]
- 29.Sobolevsky T., Prasolov I., Rodchenkov G. Study on the phase I metabolism of novel synthetic cannabinoids, APICA and its fluorinated analogue. Drug Test. Anal. 2015;7(2):131–142. doi: 10.1002/dta.1756. [http://dx.doi.org/10.1002/dta.1756]. [PMID: 25428705]. [DOI] [PubMed] [Google Scholar]
- 30.Gandhi A.S., Zhu M., Pang S., Wohlfarth A., Scheidweiler K.B., Huestis M.A. Metabolite profiling of RCS-4, a novel synthetic cannabinoid designer drug, using human hepatocyte metabolism and TOF-MS. Bioanalysis. 2014;6(11):1471–1485. doi: 10.4155/bio.14.13. [http://dx.doi.org/10.4155/bio.14.13]. [PMID: 25046048]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wohlfarth A., Gandhi A.S., Pang S., Zhu M., Scheidweiler K.B., Huestis M.A. Metabolism of synthetic cannabinoids PB-22 and its 5-fluoro analog, 5F-PB-22, by human hepatocyte incubation and high-resolution mass spectrometry. Anal. Bioanal. Chem. 2014;406(6):1763–1780. doi: 10.1007/s00216-014-7668-0. [http://dx.doi.org/10.1007/s00216-014-7668-0]. [PMID: 24518903]. [DOI] [PubMed] [Google Scholar]
- 32.Wohlfarth A., Castaneto M.S., Zhu M., Pang S., Scheidweiler K.B., Kronstrand R., Huestis M.A. Pentylindole/pentylindazole synthetic cannabinoids and their 5-fluoro analogs produce different primary metabolites: Metabolite profiling for AB-PINACA and 5F-AB-PINACA. AAPS J. 2015;17(3):660–677. doi: 10.1208/s12248-015-9721-0. [http://dx.doi. org/10.1208/s12248-015-9721-0]. [PMID: 25721194]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diao X., Wohlfarth A., Pang S., Scheidweiler K.B., Huestis M.A. High-resolution mass spectrometry for characterizing the metabolism of synthetic cannabinoid THJ-018 and Its 5-Fluoro analog THJ-2201 after incubation in human hepatocytes. Clin. Chem. 2015;62:157–169. doi: 10.1373/clinchem.2015.243535. [PMID: 26430074]. [DOI] [PubMed] [Google Scholar]
- 34.Li A.P. Human hepatocytes: isolation, cryopreservation and applications in drug development. Chem. Biol. Interact. 2007;168(1):16–29. doi: 10.1016/j.cbi.2007.01.001. [http://dx.doi.org/10.1016/j.cbi.2007.01.001]. [PMID: 17270162]. [DOI] [PubMed] [Google Scholar]
- 35.Baranczewski P., Stańczak A., Sundberg K., Svensson R., Wallin A., Jansson J., Garberg P., Postlind H. Introduction to in vitro estimation of metabolic stability and drug interactions of new chemical entities in drug discovery and development. Pharmacol. Rep. 2006;58(4):453–472. [PMID: 16963792]. [PubMed] [Google Scholar]
- 36.McNaney C.A., Drexler D.M., Hnatyshyn S.Y., Zvyaga T.A., Knipe J.O., Belcastro J.V., Sanders M. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay Drug Dev. Technol. 2008;6(1):121–129. doi: 10.1089/adt.2007.103. [http://dx.doi.org/10.1089/adt.2007.103]. [PMID: 18336089]. [DOI] [PubMed] [Google Scholar]
- 37.Castaneto M., Wohlfarth A., Pang S., Zhu M., Scheidweiler K., Kronstrand R., Huestis M. Identification of AB-FUBINACA metabolites in human hepatocytes and urine using high-resolution mass spectrometry. Forensic Toxicol. 2015;39(9):707–713. [Google Scholar]
- 38.Lavé T., Dupin S., Schmitt C., Valles B., Ubeaud G., Chou R.C., Jaeck D., Coassolo P. The use of human hepatocytes to select compounds based on their expected hepatic extraction ratios in humans. Pharm. Res. 1997;14(2):152–155. doi: 10.1023/a:1012036324237. [http://dx.doi.org/ 10.1023/A:1012036324237]. [PMID: 9090701]. [DOI] [PubMed] [Google Scholar]
- 39.Atkinson A.J., Jr, Kushner W. Clinical pharmacokinetics. Annu. Rev. Pharmacol. Toxicol. 1979;19:105–127. doi: 10.1146/annurev.pa.19.040179.000541. [http://dx.doi.org/10. 1146/annurev.pa.19.040179.000541]. [PMID: 378099]. [DOI] [PubMed] [Google Scholar]
- 40.Diao X., Scheidweiler K.B., Wohlfarth A., Pang S., Kronstrand R., Huestis M.A. In vitro and in vivo human metabolism of synthetic cannabinoids FDU-PB-22 and FUB-PB-22. AAPS J. 2016;18(2):455–464. doi: 10.1208/s12248-016-9867-4. [http://dx.doi.org/10.1208/s12248-016-9867-4]. [PMID: 26810398]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.