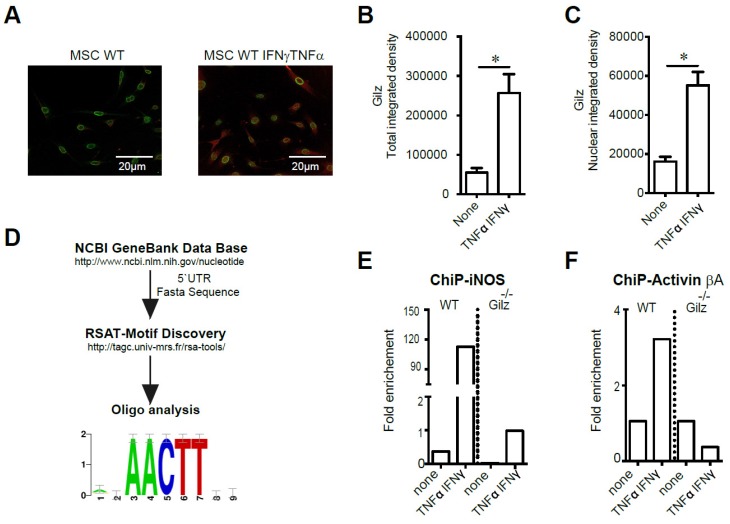
Figure 4.
Study of Gilz DNA binding motifs in primed MSC. (A-C) Immunodetection of Gilz localization in MSC by confocal microscopy. The cells were visualized and photographed using a confocal microscope (Leica TCS SP5; Leica, Heidelberg, Germany). All the analyses were performed using the ImageJ program. For quantification, the fluorescence intensity reflecting Gilz expression profile (either total or nuclear) was quantified in different fields of each image for each condition. (D) The fasta sequences of the promoters of iNOS and Activin were used to look for overrepresented motif enrichment through the Motif Discovery oligo-analysis from Regulatory Sequence Analysis Tools (RSAT) that could be a possible target for Gilz binding, selecting oligomer lengths up to 5 bases and count on single strand parameters. (E-F) WT and Gilz-/- MSC were treated at 80% confluence either with 20 ng/mL IFN-γ and 10 ng/mL TNF-α. Then, the cells were harvested to perform chromatin immunoprecipitation (ChIP) experiments and the binding of Gilz on inos or Activin βa promoter regions was analyzed. Mean values of n≥3 independent experiments. * P < 0.05. All error bars indicate SEM.