Abstract
We present a new method, Fine-Mapping of Adaptive Variation (FineMAV), which combines population differentiation, derived allele frequency, and molecular functionality to prioritize positively selected candidate variants for functional follow-up. We calibrate and test FineMAV using eight experimentally validated “gold standard” positively selected variants and simulations. FineMAV has good sensitivity and a low false discovery rate. Applying FineMAV to the 1000 Genomes Project Phase 3 SNP dataset, we report many novel selected variants, including ones in TGM3 and PRSS53 associated with hair phenotypes that we validate using available independent data. FineMAV is widely applicable to sequence data from both human and other species.
Electronic supplementary material
The online version of this article (10.1186/s13059-017-1380-2) contains supplementary material, which is available to authorized users.
Keywords: Human evolution, Positive selection, Selective sweep, Local adaptation, FineMAV
Background
The out-of-Africa expansion ~ 60,000 years ago exposed humans to a diverse range of new environments and selective pressures including new pathogens, climatic conditions, and diets [1–3]. Genetic drift and local adaptations in spatially distant populations consequently led to geographically structured phenotypic diversification, illustrated by the inter-population variation observed for numerous morphological and physiological traits, such as skin pigmentation [2–4]. Not only are the genetic variants underlying differences between populations crucial for understanding recent human evolution and present-day human diversity, but they may also be clinically relevant, as the prevalence and susceptibilities of some common diseases vary across regions (e.g. hypertension or type 2 diabetes) [4–6]. Medical implications of adaptive variation arise because natural selection can only act in a direct way on functionally important variants driving phenotypic variation [7, 8]; selected alleles usually confer protective effects, like pathogen resistance associated with CASP12 [9], CCR5 [10], and FUT2 [11] deficiency alleles, but paradoxically, may turn harmful in non-traditional environments or a homozygous state [5, 6, 12–14], e.g. sickle cell alleles [15], CPT1A [16, 17], and APOL1 [18, 19].
Selective episodes leave signatures in the human genome and thus can be recognized from the pattern of nucleotide polymorphisms in a population sample [2, 20, 21]. Most methods that have been developed to detect signals of recent and ongoing positive selection are based on the classical hard sweep model [2, 22]. This model assumes that a new advantageous mutation rapidly spreads to fixation or high frequency, purging nearby linked variation due to genetic hitchhiking [2, 20, 23]. Its genetic characteristics include high-frequency derived long-range haplotypes with a concomitant reduced level of genetic variation, large derived allele frequency differences between populations, and changes to the allele frequency spectrum (e.g. increased fraction of derived common and rare alleles, depletion of intermediate-frequency variation), although these features can also arise by genetic drift or purifying selection and are confounded by population demography [2–4, 7, 20, 22]. However, it has been argued that hard sweeps were rather rare in recent human evolution [2, 22] and that selection may more often operate on pre-existing variation that has evolved neutrally in the population until it becomes advantageous under certain conditions (“selection on standing variation”) [2, 4, 22]. Selection from standing variation is difficult to detect using most standard approaches, because the selected variant often exists on multiple haplotype backgrounds (a so-called “soft sweep”) and has weaker effects on closely linked sites, so does not produce the classical selective sweep signatures of extended linkage disequilibrium (LD) and site frequency spectrum (SFS) changes [2, 4, 21, 22, 24, 25].
Previous surveys have reported vast lists of putatively selected genomic segments, genes, and variants, which contrast sharply with the handful of functionally validated examples of genetic adaptations with both a strong population selection signal and a compelling explanation for the reasons for selection linked to a relevant phenotype in humans [2, 5, 22, 26]. This is because population-genetic-based methods are often imprecise, implicating large genomic regions harboring many genes and a myriad of single nucleotide polymorphisms (SNPs) that could potentially drive the selection signal, but which are mostly neutral [27]. Even if a selection statistic operates at the individual variant level, such as population-differentiation-based statistics (e.g. FST; difference in derived allele frequency [ΔDAF]) [28] or some composite likelihood approaches (e.g. composite of multiple signals [CMS]) [29], the highest scoring variant is not necessarily causal. High LD around the selected SNP often results in a stretch of highly differentiated variants with the same allele frequencies, further complicating the identification of the most likely causal variant. Similarly, for each potentially causal variant identified by CMS, there are on average 20 neutral proxies, all indistinguishable from the functional mutation [29]. As a result, the false discovery rate (FDR) of genome-wide selection scans is potentially high, which is reflected by the low concordance between such studies [2, 6, 7, 22, 26, 30–32]. The focus of this field now needs to move from candidate locus discovery to fine mapping of the signals of selection and biological understanding of their adaptive significance. However, population genetics alone is usually not sufficient to narrow down the signal of selection to a single causative SNP and the only way to distinguish true positives from artifacts or neutral passenger variation has been functional validation [2, 33]. Yet very few variants have been validated in this way, as current technology does not allow high-throughput functional validation, e.g. using genome editing in model systems [33]. Therefore, a useful step would be to subject candidate variants to rigorous evaluation and narrow down these extensive lists to a manageable subset of the strongest candidates for functional studies.
Despite these reservations, there are a few well-supported cases of local genetic adaptation that conform to the classical sweep model [22]. One example is the A allele at rs1426654 (within SLC24A5), which is nearly fixed in European populations, causing an amino acid (Thr to Ala) change and contributing to lighter skin pigmentation [34]. Such examples are not restricted to amino acid changes and have also been reported for cis-regulatory variants, such as the A allele at the rs4988235, an intronic regulatory variant in MCM6 which has been shown to increase the expression of the downstream lactase (LCT) gene in vitro enabling digestion of the milk sugar, lactose, as an adult in West Asian and European populations that traditionally practice pastoralism [35, 36].
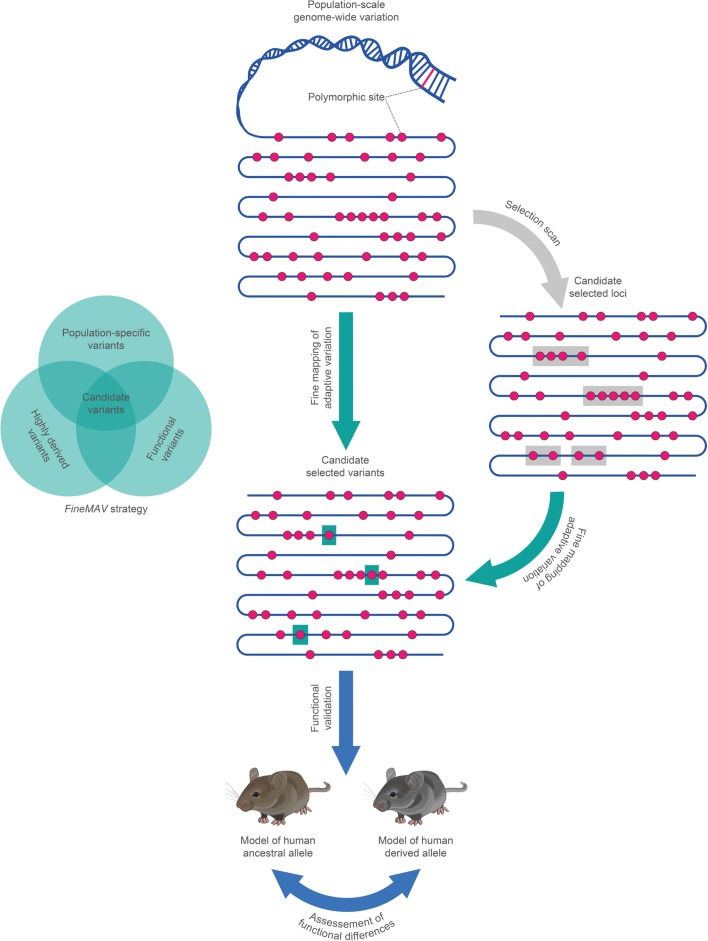
Here, we develop a new in silico framework to shortlist candidate positively selected variants for further functional follow-up (Fig. 1). In order to prioritize candidate variants, we need a starting list of variants, a protocol for prioritization, and a way of assessing whether or not the prioritization is effective. We use an integrative method that overlays population signatures of selection with functional annotation to produce a refined list of candidate variants in the 1000 Genomes Project Phase 3 SNP dataset [37]. We assessed the results using both eight “gold standard” examples where the evidence for positive selection acting on a particular variant is convincing; and also simulations, to explore the likely false positive and negative rates and further discuss some of the novel variants in our lists.
Fig. 1.
Workflow for prioritization of positively selected candidate variants for functional studies. The DNA molecule is represented as a blue line, with variants being red dots. Identification of the candidate positively selected variants from the genome-wide variation data, or the refinement of the known signal of selection to a functional SNP, is achieved by overlapping the statistical support from genetic analyses with functional annotation (implemented in FineMAV). A detailed follow-up functional study can then be performed (in vitro or in vivo experiments using model systems) to validate the implicated variant, quantify its phenotypic consequences, and clarify its relationship with reproductive fitness, e.g. by assessment of phenotypic differences between mouse models carrying the human-selected and non-selected alleles
Results
We performed a new analysis of 1000 Genomes Project Phase 3 whole-genome sequence data [37] focusing on identifying individual putatively selected SNPs. Our analysis overlays multiple lines of evidence for causality to prioritize the vast numbers of potential candidates in order to identify a small number for experimental follow up. FineMAV combines a new measure of population differentiation (derived allele purity [DAP]; see “Methods” Eq. 2), a measure of allele prevalence (DAF), and a measure of functionality (the Combined Annotation-Dependent Depletion [CADD] PHRED-scaled C-score [38]). We simply scaled and combined them to obtain a single measure giving high values to derived alleles that are common, population-specific, and functional (see “Methods” Eq. 1). FineMAV is designed to refine the location of a positive selection signal to a single variant and can be applied to a region of prior interest or to the whole genome for de novo discovery of selected variants, focusing on recent local adaptations, that arose after the out-of-Africa population expansion.
FineMAV power analyses using simulations
FineMAV’s power to detect selected variants depends on the strength of the selection coefficient. In simulations, it was unable to distinguish weak selection (s = 0.001) from neutrality since population differentiation under the scenarios tested was low (Additional file 1: Figure S1). In contrast, medium and strong selection coefficients (s = 0.007 and s = 0.01) produced FineMAV distributions that were different from the neutral variation (Additional file 1: Figure S1) and, assuming that CADD annotation is characterized by a low FDR, it was rare to find neutral variants in the extreme upper tail of the FineMAV distribution: FDR ~ 4%. The power to detect the selected variants that fall outside of the neutral FineMAV distribution was 46% and 77% for s = 0.007 and s = 0.01, respectively, although the real power, which depends on the accuracy of the functional annotation, might be lower (since functional annotation might be incomplete), these simulations demonstrated that FineMAV fits our aims, as we do not attempt to pick up all positive selection in the genome (accepting a high false negative rate), but rather try to minimize the FDR, which was < 5%.
FineMAV evaluation using 1000 Genomes Project data
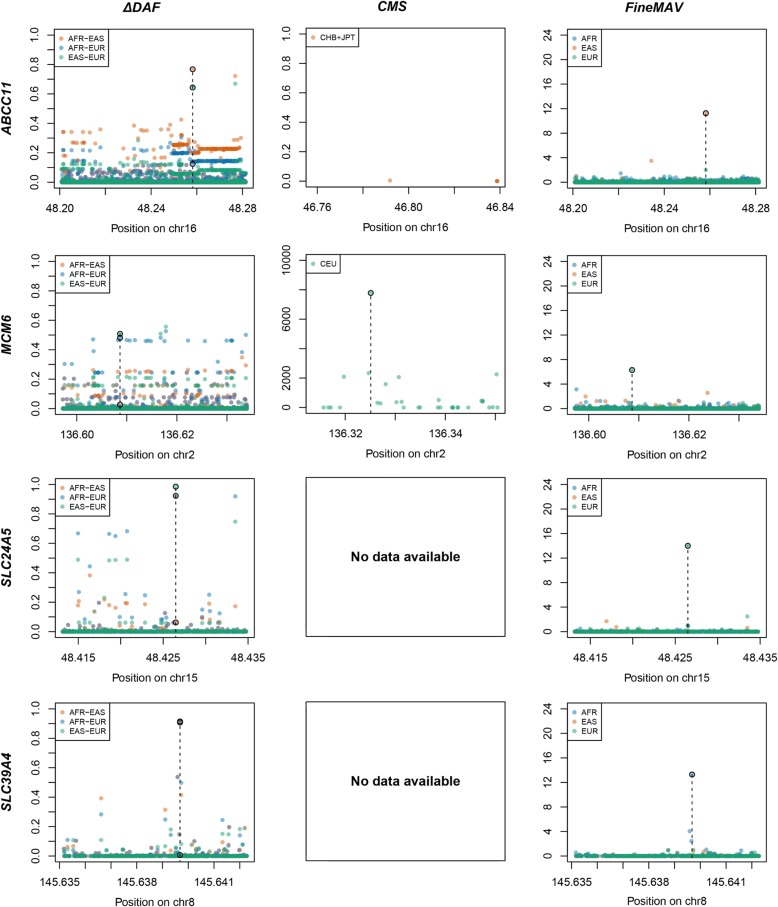
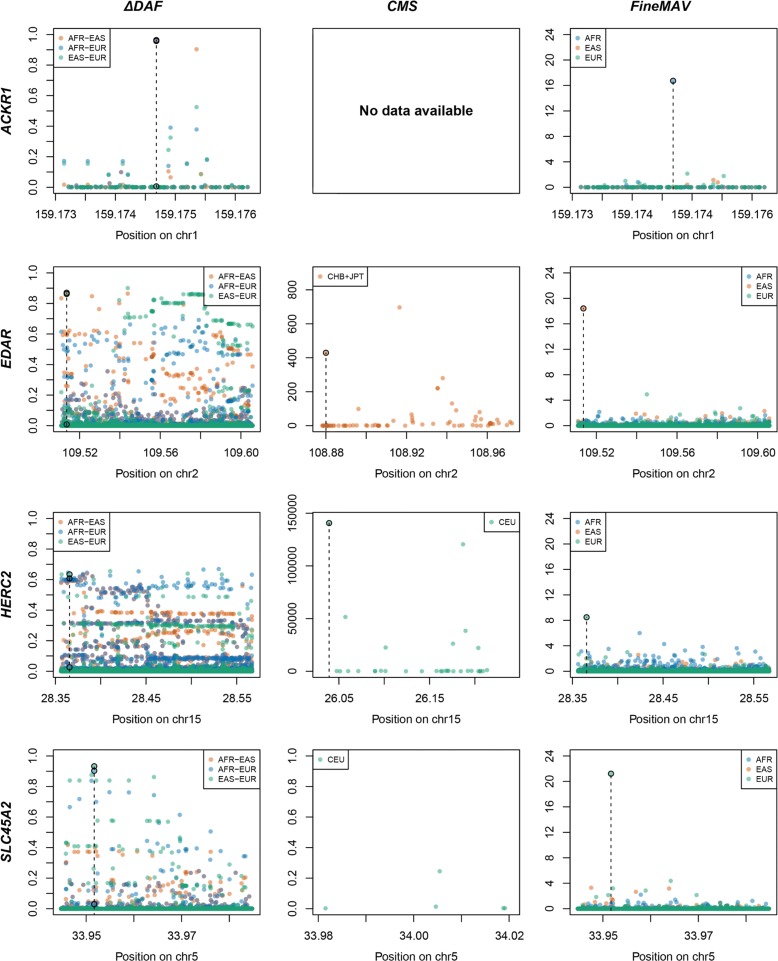
To calibrate FineMAV and evaluate its performance, we compiled a gold standard panel of the eight best examples of experimentally validated, positively selected variants underlying signals of positive selection that are linked to specific phenotypic consequences in the three well-characterized continental populations (Table 1). A key element was the value of the penalty for allele sharing between populations (parameter x). We first learned x from empirical data (subsets of the gold standards) and then tested it using simulations (100 simulated positive controls) to see if further increment of x increased FineMAV’s robustness in larger datasets (see “Methods”). The simulations showed that x deduced from empirical data was sufficient to pick up simulated selected variants and that its further increase did not affect FineMAV’s power. Calibration results were consistent across different combinations of gold standards used in the analysis (see “Methods”). We then applied FineMAV to genome-wide data from the 1000 Genomes Project (Phase 3) [37] to discover positive selection signals in Africa, East Asia, and Europe, and tested the results by examining whether or not: (1) our method was able to separate the other gold standard variants from the surrounding linked SNPs; (2) the gold standards as a group were found among the extreme outliers of the genome-wide distribution; and (3) FineMAV also enriched for genes identified in previous genome-wide selection scans with high Selection Support Index (SSI) values (Additional file 2).
Table 1.
List of “gold standard” selected variants used for FineMAV calibration and validation
| Gene | SNP | Population | Function |
|---|---|---|---|
| ACKR1 a | rs2814778 | AFR | Malaria resistance [115–118] |
| SLC39A4 | rs1871534 | AFR | Zinc level [119] |
| ABCC11 | rs17822931 | EAS | Earwax and sweat type [120, 121] |
| EDAR | rs3827760 | EAS | Hair shape and thickness [33, 122] |
| HERC2 | rs12913832 | EUR | Eye pigmentation [123–125] |
| MCM6 | rs4988235 | EUR | Lactose tolerance [35, 36] |
| SLC24A5 | rs1426654 | EUR | Skin pigmentation [34, 126] |
| SLC45A2 | rs16891982 | EUR | Skin pigmentation [126–128] |
aNote that ACKR1 is also known as DARC and the derived allele at rs2814778 is the Duffy O allele
AFR Africans, EAS East Asians, EUR Europeans
Results of the refinement of the signal of selection for the gold standard panel calibration and replication sets are shown in Figs. 2 and 3, respectively, together with the performance of methods relying on population-genetic data alone (ΔDAF – a standard measure of population differentiation [28] and CMS – a composite method [29, 39]). Our integrative approach successfully distinguished the positively selected variants from neutral background variation in all cases, whereas the standard methods were often unable to differentiate between the functional variant and its neutral proxies. Values of individual FineMAV components for each genomic window are shown in Additional file 1: Figure S2 and S3. Furthermore, we assessed how often the positively selected variant was the highest scoring one in a genomic window of 1000 SNPs in both the simulated and empirical data (1000 Genomes Project sequence data spanning the gold standard panel) according to three different tests (Table 2). In this comparison, we used two statistics relying on population genetic data alone (ΔDAF and DAPxDAF – population genetic component of FineMAV) and compared with our statistic FineMAV incorporating the measure of functionality. Inclusion of functionality improved the fine mapping of truly selected variants remarkably (Table 2). It is also worth noting that DAPxDAF is more sensitive to the signature of local adaptation than ΔDAF in the simulated data, especially for lower selection coefficients (Table 2 and Additional file 1: Figure S4 and S5).
Fig. 2.
Comparison of FineMAV with existing approaches for pinpointing positively selected variants in the calibration set. ΔDAF, CMS, and FineMAV scores are shown for the genomic windows spanning genes from the gold standard calibration panel. ΔDAF and FineMAV were calculated from the 1000 Genomes Project Phase3 dataset [37] for Africans (AFR, blue), East Asians (EAS, orange), and Europeans (EUR, green). CMS scores for localized regions were downloaded from an online repository [39] and included: region8new and region152new calculated using the pilot phase of 1000 Genomes Project [129]. Variants with CMS value set to “nan” were not plotted; thus, some CMS plots are missing. Genomic positions are given in Mb according to GRCh37 for ΔDAF and FineMAV, and build NCBI36 for CMS. The selected variant is marked with a dashed line. FineMAV notably reduced the noise of neutral background variation, so that the selected variant is always the highest scoring one in any given gene. Note that the y-axis scale in the CMS plots is not standardized
Fig. 3.
Comparison of FineMAV with existing approaches for pinpointing selected variants in the replication set. ΔDAF, CMS, and FineMAV scores are shown for the genomic windows spanning genes from the gold standard replication panel. ΔDAF and FineMAV were calculated from the 1000 Genomes Project Phase3 dataset [37] for Africans (AFR, blue), East Asians (EAS, orange), and Europeans (EUR, green). CMS scores for localized regions were downloaded from an online repository [39] and included: region34new, region104new, and SLC45A2old, all calculated using the pilot phase of 1000 Genomes Project [129]. Variants with CMS value set to “nan” were not plotted; thus, some CMS plots are missing. Genomic positions are given in Mb according to GRCh37 for ΔDAF and FineMAV, and build NCBI36 for CMS. The selected variant is marked with a dashed line. FineMAV notably reduced the noise of neutral background variation, so that the selected variant is always the highest scoring one in the given gene. Note that the y-axis scale in the CMS plots is not standardized
Table 2.
Different tests’ power to identify the selection driving SNP as the top scoring one in the genomic window of 1000 SNPs
| Scenario | ΔDAF | DAPxDAF | FineMAV |
|---|---|---|---|
| Empirical data | 0.75 | 0.75 | 1 |
| Simulation s = 0.001 | 0 | 0 | 0.01 |
| Simulation s = 0.007 | 0.23 | 0.44 | 0.75 |
| Simulation s = 0.01 | 0.72 | 0.84 | 0.92 |
“Empirical data” means 1000 Genomes Project sequence data of the gold standard panel. “Simulation” is given for three different selection coefficients (s). "DAPxDAF" specifies FineMAV without functional prediction
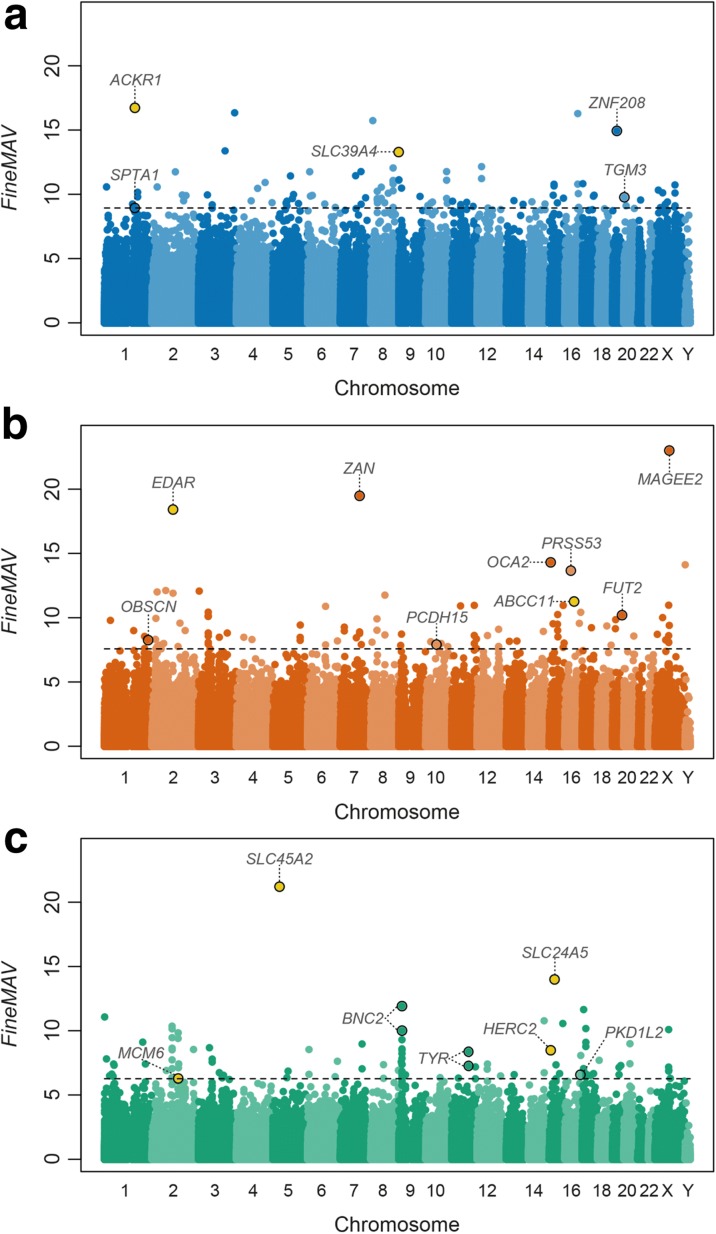
We then ranked all variants in the 1000 Genomes dataset according to their FineMAV value to identify extreme outliers in the upper tail of the empirical genome-wide distribution for each continent and examined whether or not the gold standard variants fell in the extreme tail. We indeed found all the gold standards to be high scoring (Fig. 4) (among the top 0.0004% of the whole-genome distribution [Additional file 1: Figure S6 and Additional file 3]) and set a conservative threshold to include the top 100 candidates per population (incorporating all gold standards and a total of 300 variants, out of more than 78 million derived alleles [Additional file 1: Figure S6 and Additional file 3]) for downstream analysis. Among those 300 FineMAV top-hits, we observed varying levels of allele frequency (DAF range of ~ 0.25–1) and allele sharing between populations (DAP range of ~ 0.38–1), all characterized by a functional CADD score prediction (in the range of ~ 11–47 with a mean of ~ 19). It is worth noting that although FineMAV prioritizes population-specific alleles, it also allows some degree of allele sharing between populations. The distribution of continental DAF, DAP, and CADD in the top FineMAV outliers in each population are shown in Additional file 1: Figure S7, S8, and S9, respectively.
Fig. 4.
Manhattan plot of genome-wide FineMAV scores. FineMAV scores were calculated for genome-wide SNPs from 1000 Genomes Project Phase 3 [37] in three continental populations: (a) Africans (AFR, blue); (b) East Asians (EAS, orange); (c) Europeans (EUR, green). Each dot in the Manhattan plots represents a single SNP plotted according to coordinates in GRCh37. The threshold (dashed lines) was set to include the top 100 variants (top ~ 0.0004% of the whole-genome distribution). All gold-standard SNPs (yellow dots found among the top outliers) and other interesting candidate variants are labeled with the name of the gene they fall into
Functional validation in silico
To further evaluate our top FineMAV hits, we performed an in silico validation by searching the available literature for relevant functional information. FineMAV’s performance is supported by several lines of evidence. The first verification comes from the “gold standard” replication set (the best examples of validated causal adaptive variants). Not only did FineMAV replicate the signals in these well-known cases of strong election, but it also narrowed it down to the known single functional SNP, even in high LD regions. Positive controls extend to other variants that were not included in the “gold standard” panel, but whose prior evidence of causality is also strong, potentially providing additional support for our method. FineMAV rediscovered additional known SNPs implicated in eye, hair, and skin pigmentation in non-Africans, such as rs1800414 in OCA2 (skin lightening in East Asians) [40–42], rs1042602 and rs1126809 in TYR (pigmentation and freckling in Europeans) [43–45], rs12350739 in BNC2 (freckling and color saturation of human skin pigmentation in Europeans) [46], and also rs1047781 in FUT2 (an enzyme-inactivating mutation conferring advantage in avoiding certain viral infections in East Asians) [11, 47], rs3211938 in CD36 selected in Yoruba (protection against malaria and/or the metabolic syndrome) [48–50], and rs1229984 in ADH1B (protection against alcohol dependence in East Asians) [51–54].
Finally, FineMAV also identified a variant with no prior implication of functionality that was experimentally validated while our study was in progress, thus providing additional evidence of its performance. We picked up the missense SNP rs11150606 as the sixth top-scoring FineMAV variant in East Asians and noticed that it fell in PRSS53, whose function was then largely unknown. PRSS53 encodes a polyserine protease called polyserase-3 (POL3S), which hydrolyzes peptide bonds. Subsequently, Adhikari et al. showed that PRSS53 is highly expressed in the hair follicle and rs11150606 was associated with hair shape in East Asians [55]. The authors confirmed the functionality of rs11150606 by in vitro assays, showing that it affects processing and secretion of the gene product, with the derived allele contributing to the straight hair phenotype (similarly to the well-established gold standard EDAR variant) [55]. It can thus be considered another gold standard example demonstrating the validity of our method in picking up true functional selected variants.
Furthermore, we looked at the enrichment of genome-wide association study (GWAS) hits and expression quantitative trait loci (eQTLs) more generally among our top outliers (Additional file 2). GWAS enrichment was carried out in an LD framework (Additional file 2) instead of a simple overlap between FineMAV outliers and GWAS hits, because many GWAS studies are based on array-genotyping, rather than whole-genome sequencing data and are confounded by LD between functional and linked variants. We saw enrichment in GWAS hits among FineMAV outliers in Europeans and Eurasians (especially in the high LD category r2 ≥ 0.9) (Additional file 2). As the majority of GWAS studies were conducted in populations with European ancestry, the lack of enrichment in Africans and East Asians is not surprising. The directionality of the association was not available for all variants, but most of the annotated selected alleles (or their tagging SNPs) conferred protective effect, except a few derived alleles associated with increased risk of schizophrenia, type 2 diabetes, increased adiposity, lupus erythematosus, and degeneration of lumbar disc (Additional file 4). We also saw selection on alleles associated with increased height and lower age of puberty in Europeans [56], delayed eruption of permanent teeth in Eurasians [57], facial morphology in Africans, and chin dimples in Europeans and Asians [58] (Additional file 4).
Similarly, we saw an enrichment in eQTLs among FineMAV top outliers in Europeans and Eurasians as compared to a random expectation (p values of 0.04 and 0.03, respectively), although with lower significance than for GWAS signals. In a similar fashion, non-European ancestries are under-represented in eQTL databases. Over half of the top 100 FineMAV outliers in Europeans and Eurasians were annotated as significant eQTLs (Additional file 3).
Novel candidate variants in Africa, East Asia, and Europe
Although we have thus far highlighted known variants replicated in our analysis, which serve as positive controls for evaluating our method’s performance, the vast majority of outliers discovered are novel and fall in non-coding regions (Additional file 1: Figure S10 and Additional file 2). We also identified variants on the X and Y chromosomes which have been under-represented in previous genomic scans [28, 29, 31, 39, 59–80], but further functional testing is needed to explore these findings. It is worth noting that the paucity of FineMAV hits on the Y chromosome (only one in the top 300) shows its strong dependence on the CADD score prediction.
We observed some high-scoring nonsense variants among our top candidates, suggesting pseudogenization of PKD1L2 (an endogenous fatty acid synthase in skeletal muscle) [81] in Europeans, ZNF208 (zinc finger and SRY-interacting protein) [82] in Africans, as well as ZAN, OBSCN (sacromeric signaling protein involved in myofibrillogenesis) [83] and MAGEE2 (melanoma-associated antigen expressed in the brain) [84] in East Asians. ZAN is particularly interesting as it encodes a zonadhesin protein located in the acrosome that mediates the species specificity of sperm binding to the extracellular coat of the egg (zona pellucida) [85]. We find a signal of selection at a nonsense mutation (rs2293766) present at 51% frequency in East Asians, but virtually absent elsewhere.
FineMAV also highlighted rs6048066, a missense variant in TGM3 in Africans. The TGM3 gene product’s deficiency in humans has been linked to Uncombable Hair Syndrome, characterized by dry, frizzy, and wiry hair [86], while the Tgm3 knockout mice exhibit rough-looking, curly, or brittle hair [87–89]. The missense variant we report here falls in the catalytic core of the protein, as does the mouse non-synonymous weBkr allele causing a wavy coat and curly whisker phenotype [89]. SNPs in TGM3 have been weakly associated with hair diameter in humans [90] and proteomic profiling of human hair shafts identified TGase 3 as a major component of the hair fiber and revealed considerable variation among samples of different ethnic origins, with the lowest levels in African Americans and Kenyans [91]. We propose that this missense variant (rs6048066) might cause enzyme deficiency and contribute to African hair texture, hypothesized to have experienced strong positive selection in equatorial climates due to body-temperature regulation [92, 93].
Finally, regulatory variants are particularly interesting as they form the most abundant functional category among FineMAV outliers (Additional file 1: Figure S10 and Additional file 2) and are responsible for the bulk of human phenotypic variation [21, 35, 94]. However, the functional effect of regulatory variants remains difficult to predict and interpret. We find a signal of selection on rs12881545—an intronic regulatory variant falling in a promoter-flanking region and transcription factor binding site that scores as the top sixth variant selected in Europeans. The region surrounding rs12881545, although non-coding, is characterized by high conservation across taxa and the presence of DNaseI hypersensitivity. Our GWAS analyses revealed that rs12881545 is tagged by rs7141210 (r2 = 0.96) associated with lower age at menarche [56]. rs12881545 is also a direct eQTL and the selected allele increases DLK1 expression (p value = 0.000015) [84]. DLK1 is an epidermal growth factor involved in differentiation of many tissues with strong links to adiposity and body growth. Furthermore, in accordance with the GWAS association [56, 95], aberrations in DLK1 has been linked to central precocious puberty (a condition where puberty starts too soon in children) [96]. Although potentially pleiotropic, it could be that this regulatory variant modulates the expression level of DLK1 and timing of menarche.
In-depth discussion of further novel alleles and speculation on the plausible selection pressures acting on them can be found in Additional file 2. The functional significance of these novel candidate variants presented here needs to be experimentally validated, but narrowing their signal of selection to a single most likely selected candidate SNP is a good starting point for such efforts.
FineMAV analysis in Admixed Americans and South Asians
After the calibration of our method and an assessment of its performance in African, East Asian, and European populations, we applied it to the remaining 1000 Genomes Project populations: Admixed Americans (AMR) and South Asians (SAS). FineMAV revealed less population-specific selection in these populations (Additional file 1: Figure S11) due to population admixture (AMR) or shared ancestry (SAS). Nevertheless, a single strong outlier was observed in the SAS, found at 0.54 frequency there but virtually absent elsewhere: the missense SNP rs201075024 in PRSS53 (Additional file 1: Figure S11.A). This is different from the non-synonymous variant in PRSS53 in East Asians (previous section), but lies in close proximity, only 10 bp away (Additional file 1: Figure S12), which might indicate a similar functional consequence and convergent evolution of a hair-related phenotype, especially as both mutations are non-synonymous and have a similar molecular nature. Besides PRSS53, we see several potential signals of convergent or parallel evolution [1], i.e. selection on the same gene in geographically distant populations but on a different SNP (Additional file 3); however, only in the case of PRSS53 do the similarities in locations and consequences of the SNPs provide a strong priori likelihood of convergent evolution.
In the AMR, even though admixture decreases the FineMAV signal, the gene flow into the Americas affects the frequency of derived Native American alleles, but not their purity (as private American alleles would still be found exclusively in Americas giving high DAP values). In the case of common derived alleles selected to high frequencies before the admixture event, a FineMAV signal should still be detectable (assuming their high functional prediction) in the extreme tail of the whole-genome distribution; the top three scores were missense variants: rs148608573 in MAP7D1, rs142326775 in ZNF438, and rs34890031 in LRGUK (where the mouse homologue is essential for multiple aspects of sperm assembly and function) [97] (Additional file 1: Figure S11.B).
Discussion
The aim of this study was not to perform another selection scan and it should not be interpreted in that way. Instead, it aims to refine a proportion of local adaptations to a single variant and prioritize positively selected candidates for further functional validation, as existing methods often do not pinpoint the selected SNPs. Furthermore, this paper does not focus on experimental follow-up of novel selection signals, but rather provides a decision-making algorithm for identifying high-priority causal variants for subsequent experimental work. To achieve these aims, we introduced the FineMAV statistic which combines measures of population differentiation, derived allele frequency, and molecular functionality. Incorporation of diverse functional annotations (such as predictors of deleteriousness) should improve the pinpointing of likely selected variants and lower FDR, as it has in the detection of disease-causing variants [98]. It is worth noting that variants classified as damaging alter the level or biochemical function of a gene product, but do not necessarily decrease the reproductive fitness of carriers [38, 99]. The functional consequence of the “damaging” change for a person depends on many factors and can be either negative or positive (as, for example, deficiency alleles might be either beneficial or detrimental) depending on the environmental context. For instance, variants disadvantageous in one environment can be favored under different conditions, e.g. CPT1A [16, 17].
FineMAV was calibrated and tested using a gold standard panel of the eight best examples of experimentally validated functional variants underlying signals of positive selection in humans and was able to identify the known functional candidate in all instances (Figs. 2 and 3). Using the 1000 Genomes Project Phase 3 dataset [37], we then ranked all genome-wide SNPs based on their FineMAV value and identified extreme outliers in the upper tail of the empirical genome-wide distribution in Africa, Europe, and East Asia (Additional file 3). FineMAV rediscovered many known variants with prior evidence for being causal of positive selection signals, which were not part of the calibration set, providing additional support for our method. We also identified potential functional variants in other genes reported to be under strong positive selection in the literature (with strong SSI scores; Additional file 5) where the specific positively selected variant had not been confirmed, including LPP, PCDH15, and PRSS53. The selection signal in PCDH15 and PRSS53 was attributed to a single missense variant per population (rs4935502, rs11150606, and rs201075024, respectively), replicating and extending the results obtained by CMS [39, 55].
The signal in BNC2 was particularly strong in Europeans, as reflected by a cluster of 12 SNPs found among the top 100 hits in the FineMAV distribution (Fig. 4c). The hypothesized functional SNP (the intergenic rs12350739) was the second highest-scoring BNC2 variant in our analysis and has been reported to be a functional eQTL as it falls in a highly conserved melanocyte-specific enhancer and regulates BNC2 transcription [46]. The highest-scoring BCN2 variant (rs10962600) might also contribute to the differential expression of BNC2 isoforms as several regions inside and outside of the BNC2 gene contain enhancer features [46]. Interestingly, BNC2 has been highlighted as present in a region of the human genome that shows Neanderthal ancestry (Additional file 1: Figure S13), suggesting that Neanderthal introgression might have provided modern humans with adaptive variation for skin phenotypes involving BNC2 [46, 100–102]. Furthermore, a cluster of high-scoring SNPs in FineMAV analysis might more generally be indicative of introgression as a source of adaptive variation, as opposed to advantageous de novo mutations that usually arise individually. Although we cannot exclude the possibility of more than one causal SNP in regions introgressed from archaic hominins (especially those falling in regulatory elements), it seems that FineMAV may have low resolution in cases of adaptive introgression. We also found other FineMAV outliers in regions proposed to be adaptively introgressed from an archaic source (27 SNPs in total) in GNAI2, GPATCH1, IRF6, POU2F3, RASSF1, SEMA3F, and SLC38A3 (Additional file 1: Figure S13) [100–103], suggesting that some of the candidates might be of archaic rather than de novo origin. However, the origin of the adaptive mutations is not the focus of this study and has been considered elsewhere [100–103]. Apart from BNC2, several other introgressed SNPs also showed GWAS associations, including IRF6 (cleft lip), GPATCH1 (bone density), and, most interestingly, a high-LD eQTL region on chromosome 3 spanning GNAI2, HYAL1, HYAL2, RASSF1, SEMA3F, and SLC38A3 in East Asians associated with keloid scar formation resulting from dysfunction of the wound-healing processes [104]. It has been shown that keloid susceptibility varies across ethnicities with higher incidence in Africans and East Asians, and darker-skinned populations in general [105, 106].
Finally, FineMAV picked up variants with modest to high derived allele frequency in the range of ~ 0.25–1 within continental populations (Additional file 1: Figure S7). Most classical methods detect only extreme allele frequency differences between populations, which are less likely to arise by chance [22]. On the other hand, highly functional alleles are less likely to be subjected to random changes in their frequency; thus, it seems that filtering out neutral variation by applying functional information might allow more examples of weaker sweeps (potentially including selection on standing variation) to be discovered, which are characterized by more modest allele frequency shifts [4, 22], although our method has no power to detect low selection coefficients that do not produce population differentiation patterns.
Functional validation of candidate signals of selection is a current roadblock in the field, limiting both our understanding of the modes and importance of positive selection and the independent evaluation of methods to detect it. Modeling of non-pathological human genetic variation in cell or animal systems, however, has received only limited attention to date [107]. Our study misses some genuine selected variants, but our prioritization aims to enrich for true positives, which is what matters for studies that may spend years examining individual candidates in cellular or animal models. For example, the reason for selection of the TRPV6 haplotype containing three derived non-synonymous substitutions observed in non-African populations remains enigmatic despite detailed functional characterization of selected and non-selected forms at the cellular level [108]. Although it remains possible that the ancestral and derived forms differ in aspects that were not tested or can only be observed at the whole-organism level [108], none of the three candidate sites was supported by our FineMAV analysis (in both selection scenarios n = 3 [AFR, EAS, EUR] and n = 2 [AFR, EAS + EUR]; see “Methods”) since their score was low (FineMAV ~ 1 for each variant in the EAS + EUR scenario). Therefore, we see them as weak candidates for causality and would not suggest a high priority for modeling them.
Conclusions
Modeling human selection in cell or animal systems is challenging since relevant phenotypic consequences (often very subtle) might be overlooked. Some phenotypes may be manifest only in certain conditions, such as the presence of specific pathogens or environmental stresses, and might be missed even by association studies in humans [7, 33]. The inability to directly demonstrate phenotypic consequences in a limited set-up, therefore, does not entirely rule out the possibility that a variant has been selected [21]. Nonetheless, regardless of challenges like these, cell and animal models often provide the best way to test hypotheses regarding recent human evolution [33]. FineMAV now offers an improved way to identify specific variants for these tests and paves the way for systematic identification of selected alleles driving phenotypic differences among human populations, future functional studies of individual loci, and more general understanding of the circumstances in which local adaptations occur.
Methods
Fine-mapping of adaptive variation
Fine-Mapping of Adaptive Variation (FineMAV) is designed to refine a signal of selection to a single most likely selected variant and thus to differentiate it from the passenger variants for functional follow-up studies. FineMAV is most relevant for targets of recent or ongoing local positive selection underlying local adaptations in humans following the out-of-Africa migration (within the last ~ 60,000 years) and can be applied to a region of prior interest, or to the whole genome, for discovering novel positively selected variants. It could also potentially address old selection in the human lineage (preceding the out-of-Africa expansion; see “FineMAV calibration” below), but this is not the main focus of this study.
A FineMAV score was calculated for the derived allele of each SNP by combining its DAP, DAF, and functional prediction score (the CADD PHRED-scaled C-score) [38] (Eq. 1). The rationale behind doing so is that variants predicted to be non-functional are (within the limitations of the prediction) likely to be neutral, since natural selection can only act directly on variants that confer a phenotypic effect. If an allele is predicted to be highly functional and rare, it will often be deleterious; but it cannot be harmful if it is both functional and common, and may potentially be adaptive. Importantly, all three metrics are allele-specific (rather than site- or gene-specific) and consequently allow direct evaluation of individual alleles. We simply scaled and combined the metrics to obtain a single measure giving high values to derived alleles that are common, population-specific, and functional (Eq. 1). Individual components are introduced in the following sections. Although FineMAV can be also applied to ancestral alleles by calculating their allele frequency and purity, detection of selection on segregating ancestral alleles would be limited by extensive sharing of ancestral alleles worldwide (across different populations) and their low purity scores. Therefore, it is unlikely to detect selection on segregating ancestral alleles that do not produce a high population differentiation signature.
Equation 1. Fine-Mapping of Adaptive Variation. To compute FineMAV per derived allele across n populations, suppose i ∈ {1, 2, …, n} and let DAFi be derived allele frequency in population i.
Measure of population differentiation
We used an allele frequency differentiation method as a signature of local selection in FineMAV. We chose a measure of population structure differing somewhat from existing methods, as it: (1) operates at the variant level; (2) does not rely on the hard sweep assumptions of strong LD and SFS signatures (which can be erased by recombination); (3) is sensitive to many types of selection including classic sweeps and selection from standing variation; and (4) detects recent human adaptations [4, 21, 22, 24, 25]. Alternative methods based on extended LD or distortion of SFS are characterized by low genomic resolution (summary statistics are calculated for large genomic windows) [109] and do not allow single candidate variants to be pinpointed, which is the key aim of this manuscript. Furthermore, such methods lead to signals mainly in cases of strong hard sweeps (known to be rare in human evolution). Therefore, such tests were not incorporated into the FineMAV score.
Any selection event, regardless of its mode, will eventually produce an excess of allele frequency differentiation between populations as long as: (1) it has taken place in one population but not in another, and the allele was at low frequency when first favored; (2) there is variation in selection coefficient over space; (3) migration and gene flow between the populations have been restricted; and (4) there has been enough time for selection to act [4, 22]. Even if an allele is equally advantageous in all environments, but its selection happened in a regionally restricted manner, the selected variant will be concentrated around its geographic origin due to limited dispersal [4, 7].
We proposed and applied a new measure of population differentiation called DAP. DAP is related to ΔDAF [28] and other pairwise comparison-based methods, but is able to summarize population differentiation (spatial pattern of the derived allele) across many populations in a single measure for each variant. DAP is a measure of derived allele entropy based on Gini impurity [110] and describes how unequally the derived allele is distributed among diverse populations. DAP operates on derived allele counts in a population sample when distinct groups are equally represented and is calculated according to Eq. 2. When population groups are not equally represented, derived allele count can be estimated from derived allele frequency. DAP counts derived allele occurrences across populations and describes their spatial distribution, reaching its maximum of 1 when all cases (derived alleles) fall into a single population category and penalizes allele sharing between different populations. The magnitude of the penalty can be controlled by the x parameter (“penalty parameter”) depending on the user’s purposes and the number of populations being compared (n) (see “FineMAV calibration”). x is an inherent parameter of the population differentiation test, as without it DAP would not measure entropy and would remain constant (equal to 1) for all alleles. We calibrated x using a subset of our gold standards (see “FineMAV calibration”). It is worth noting that DAP is a measure of derived allele purity (or “privateness”) and scores highly for both rare and common alleles found exclusively in a single population (characterized by high population differentiation) and therefore needs to be combined with a measure of allele abundance (DAF) in order to detect local adaptation.
Equation 2. Derived allele purity. To compute derived allele purity per site (DAP) across n equally represented populations, suppose i ∈ {1, 2, …, n} and let di be derived allele count in population i.
Measure of allele prevalence
We estimated allele abundance using two alternative approaches: (1) global derived allele frequency; and (2) continental derived allele frequency. In both cases, DAF is in the range of 0–1. We obtained the continental DAF by averaging DAF across all populations within each continent and calculated global DAF for each variant by averaging continental DAFs. Both approaches yielded similar results (almost identical lists of top 100 extreme outliers). The main difference between these two measures of allele prevalence is that incorporation of global DAF results in a single FineMAV score for each derived allele (which is then assigned to a single population based on the difference in derived allele frequency between examined populations), while application of continental DAF leads to calculation of FineMAV scores for each population separately. Global DAF is n-dependent, while continental DAF remains constant regardless of n, thereby making FineMAV values comparable across different values of n. Here, we report results incorporating continental DAF. The combined measure of DAPxDAF is the population genetic component of the FineMAV test that detects the signature of local adaptation.
Measure of functionality
It is crucial that variant-level functional inferences are based on whole-genome measures to ensure that all potentially selected variants are treated equally. We needed a measure of functionality to be allele-specific and applicable to all variation, both coding and non-coding, since many signals of selection localize in regulatory elements or intergenic regions [21, 29]. As proteins are usually involved in multiple processes through complicated interaction pathways with other proteins, amino acid change in one protein may affect diverse traits, i.e. pleiotropic phenotypes [33]. In general, pleiotropic changes are thought to be disadvantageous [94], so it is believed that a great deal of human phenotypic variation is based in regulatory variation [21, 35, 94]. Thus, different sets of annotations for coding and non-coding variation would make it challenging to compare these distinct variant categories and consensus methods combining multiple annotations, each with its own strengths and weaknesses, are especially needed here for unbiased prioritization of variants [38]. In our analysis, we used the CADD (v1.2 PHRED-scaled C-score), which integrates 63 diverse genome annotations into a single measure for each variant and in theory can take a value in the range of 0–99 [38].
FineMAV calibration
We compiled a gold standard panel of the eight best examples of experimentally validated functional variants underlying signals of positive selection which are linked to specific phenotypic consequences (Table 1) and calibrated FineMAV using population-scale sequence data (1000 Genomes Project Phase 3) of genomic windows spanning half of the gold standards (randomly chosen from each population). We then examined if the calibration results are sensitive to different combinations of gold standards in a quantitative and reproducible manner.
Increment of the penalty for allele sharing (x) increases the difference between the FineMAV score of the differentiated selected SNPs and less differentiated nearby neutral variants. The magnitude of this difference increases with increasing x, reaches a plateau, and then decreases for larger values of x. For each gold standard gene (empirical data), we calculated the fold change between the FineMAV scores of the selected variant and the highest scoring neutral background SNP using different values of x (1 ≤ x ≤ 4). We then selected the optimal values of x for each gold standard gene that maximizes the difference between the selected and neutral variants (Additional file 1: Figure S14). Based on our calibration set, we decided to set the penalty parameter x to a consensus value of 3.5.
If our calibration analysis relied on a single gold standard gene, the optimal x would fall in the rage of 2.5 ≤ x ≤ 4 (Additional file 1: Figure S14). The same holds true for different combinations of gold standards. Although some gold standards reached a plateau earlier than others, and some of them did not reach a plateau in the examined interval at all, it seems that the minimal value of 2.5 is large enough to differentiate between selected and neutral SNPs in all cases (Additional file 1: Figure S14). Although further increase of x (>4) improves the fold change between some of the gold standards and neutral variants, we also noticed a decrease in the fold change in the examined range of x and decided not to extend this range in our calibration analyses.
Furthermore, we examined the overall rank of gold standards in the empirical whole-genome distributions of FineMAV and DAPxDAF. The average rank improves dramatically with increasing x until 2.5, and then plateaus (with further decrease above 4 in case of DAPxDAF distribution) (Additional file 1: Figure S15). The optimal value of x for highest ranks is seen at x = 3.5 and 4 (all gold standards among top 100 genome-wide outliers). Similarly, FineMAV’s and DAPxDAF’s power to detect the selection-driving SNP as the highest scoring in a genomic window of 1000 nearby SNPs does not increase substantially with x > 2 (using both simulated and empirical data; Tables 3 and 4). Finally, we performed overlap analyses of the top 100 FineMAV outliers from the empirical whole-genome distribution across different values of x (from 1 to 4) (Additional file 1: Figure S16). We conclude that a value of x from ~ 3 to 4 is optimal and further increment does not improve FineMAV analyses usefully (very similar set of top 100 outliers) (Additional file 1: Figure S16). We recommend x = 3.5 (in three populations comparison; n = 3) as an optimal penalty in FineMAV analyses, although higher penalties would yield very similar results. Note that this calibration was carried out using gold standards across three continental populations (n = 3) and x is sensitive to n. To see that: for maximally differentiated derived alleles (observed in one population only) DAP is constant (DAPmax = 1) and insensitive to n, while at the other extreme, minimally differentiated derived alleles (with the same frequency in all populations), DAP depends on n and DAPi > DAPi+1 (1 < i ≤ n). To adjust for this and keep the FineMAV value insensitive to n, the x parameter for lower n values needs to be higher. Additional file 1: Figure S17 specifies the values of x for different values of n that make FineMAV values comparable across different values of n.
Table 3.
FineMAV’s power to identify the selection-driving SNP as the top scoring one in a genomic window of 1000 SNPs across different values of x parameter
| Scenario | x = 1 | x = 1.5 | x = 2 | x = 2.5 | x = 3 | x = 3.5 | x = 4 |
|---|---|---|---|---|---|---|---|
| Empirical data | 0.5 | 0.88 | 1 | 1 | 1 | 1 | 1 |
| Simulation s = 0.007 | 0.6 | 0.7 | 0.71 | 0.72 | 0.74 | 0.75 | 0.76 |
| Simulation s = 0.01 | 0.82 | 0.88 | 0.9 | 0.92 | 0.92 | 0.92 | 0.92 |
“Empirical data” means 1000 Genomes Project sequence data of the gold standard panel. “Simulation” is given for two different selection coefficients (s)
Table 4.
DAPxDAF’s power to identify the selection-driving SNP as the top scoring one in a genomic window of 1000 SNPs across different values of x parameter
| Scenario | x = 1 | x = 1.5 | x = 2 | x = 2.5 | x = 3 | x = 3.5 | x = 4 |
|---|---|---|---|---|---|---|---|
| Empirical data | 0 | 0.75 | 1 | 1 | 1 | 1 | 1 |
| Simulation s = 0.007 | 0.08 | 0.39 | 0.44 | 0.43 | 0.44 | 0.44 | 0.45 |
| Simulation s = 0.01 | 0.41 | 0.84 | 0.84 | 0.84 | 0.84 | 0.84 | 0.85 |
“Empirical data” means 1000 Genomes Project sequence data of the gold standard panel. “Simulation” is given for two different selection coefficients (s)
This calibration is robust to different combinations and number of gold standards used in the analysis and is supported by both empirical and simulated data. We encourage modified values of x if users wish to apply FineMAV to different species characterized by different levels of population differentiation or to different modes of selection, following the calibration framework presented here.
Balance between differentiation and functionality
The penalty parameter x also tunes DAP in relation to DAF and CADD and controls the balance between population differentiation and the prediction of functionality. The magnitude of the penalty for allele sharing in our population differentiation test (DAPxDAF) needs to fit the purpose of detecting selected alleles. We aim to pick up highly functional variants among the most differentiated variants (not the other way around) with the minimal cut-off for functionality at ~ 10 (variants with CADD scores below this threshold are considered non-functional).
When the x parameter is set to 1, population differentiation is not taken into account (DAP is constant and equals 1 for all variants) and FineMAV (or rather DAFxCADD) picks up derived alleles of high frequencies (often nearly fixed in the human lineage) with a strong prediction of functionality that are not differentiated between populations (e.g. the stop mutation in CASP12 - rs497116) [9] (green tail in Additional file 1: Figure S18, S19 and S20; x = 1). We provide a list of such variants in Additional file 6. 77 of the top 100 such outliers were shared between Africans, East Asians, and Europeans. They could represent old selection events in the human lineage (presumably preceding the out-of-Africa expansion) and are potentially interesting, although beyond the scope of this study which focuses on recent selection and population diversification.
In the calibration stage, we needed to find the value of the x penalty parameter that assigns low scores to the background variation and highly functional derived alleles nearly fixed on the human lineage in the window around the selected variant. Imagine two scenarios. In scenario 1: a maximally differentiated derived allele that is exclusively fixed in population i but absent elsewhere (DAPmax = 1), which implies a maximal frequency (DAFi = 1), and is predicted to be functional (CADD = 20). In this scenario, FineMAV = 20 and would be constant regardless of n (number of populations used in the analysis). Alternatively, in scenario 2, for a derived mutation that is fixed in all populations (DAFi = 1) and is highly functional (CADD = 45) we need to penalize for allele sharing between populations to keep DAP (and consequently the FineMAV value) at a low level relative to scenario 1. x set to ~ 3 (and above) fits all above criteria (Additional file 1: Figures S18–S20). Increment of x removes the tail of nearly fixed derived alleles of high CADD prediction which disappears around x = 3 and 3.5 and leaves the most differentiated variants that are predicted to be functional (with CADD scores > 10) (Additional file 1: Figures S18–S20). Further increase of x (within the interval examined) has little effect on the results (see “FineMAV calibration” above and Additional file 1: Figures S18–S20). The penalty parameter x set according to Additional file 1: Figure S17 (3.5 and > 3 population comparison) is sufficient to give low scores to highly functional nearly fixed alleles (scenario 2: DAP ~ 0.064 and FineMAV ~ 2.88, which is at least seven times lower than the gold standard calibration set).
FineMAV calculation in 1000 Genomes Project samples
DAF and DAP values were calculated from the 1000 Genomes Project Phase 3 data release [37] using a custom script; CADD PHRED-scaled C-scores v1.2 were obtained from an online repository [38]. We ran our analysis for both autosomes and sex chromosomes, focusing initially on three continental populations: Africans (AFR), East Asians (EAS), and Europeans (EUR). We ran it in two contexts: (1) to re-discover continent-specific positive selection signals in Africa, East Asia, and Europe (n = 3; x = 3.5); and (2) to analyze selection that happened outside of Africa by pooling East Asians and Europeans together (n = 2; x = 4.96). Although our study focuses on local adaptation driving population differentiation at the continental scale, FineMAV can be also applied to study signals of selection within continents. It is also possible to investigate signals of selection shared between populations by relevant population grouping depending on the user’s purposes, e.g. selection outside Africa by pooling East Asians and Europeans together (Additional file 3).
FineMAV was calculated for derived alleles (annotated according to Ensembl) [111, 112] using a custom script (SNPs only). We applied a conservative FineMAV cut-off to include only the top 100 candidate variants in each continental population (which incorporated all gold standards and gave a total of 300 variants corresponding to the top ~ 0.0004% of the whole-genome distribution) for our downstream enrichment analysis (Additional file 2).
Subsequently, we also ran FineMAV in AMR and SAS from the 1000 Genomes Project Phase 3 data release [37], together with the three main continental populations, as follows: AFR, AMR, EAS, EUR; n = 4; x = 2.98 and AFR, EAS, EUR, SAS; n = 4; x = 2.98, to investigate population-specific local adaptation in those populations.
Simulation analyses
Simulations assessing FineMAV’s performance were limited by the unknown relationship between the prediction of functionality (CADD score) and the selection coefficient. Although the functional range of CADD scores has been estimated, its FDR and sensitivity are poorly understood, while FineMAV’s performance is closely tied to the accuracy of the functional annotation. Nevertheless, we performed simulation analyses using individual-based forward-time simulations implemented in simuPOP v1.1.7 [113] to assess the power (true positive rate) and FDR of the FineMAV algorithm. We simulated three populations with a set of demographic parameters (starting effective population size, migration rate, and time of divergence) similar to estimates in European, African, and East Asian populations accordingly to published values [114]. We simulated a genomic window of 1000 SNPs with one SNP per window under selection in one population. The probability of recombination between two SNPs was set to increase with increasing physical distance between sites. The starting derived allele frequency for the selected marker was set to 0.01, while the allele frequencies of the remaining neutral SNPs were drawn from a beta distribution. Each SNP was assigned a CADD score value as follows:
Neutral SNPs were randomly assigned a CADD score value drawn from the genome-wide CADD distribution of derived alleles seen at ≥ 2% frequency in the 1000 Genomes Project Phase 3. Our simulation does not include purifying selection against pathogenic variants with high CADD values, so the derived allele frequency cutoff was set to 2% (approximately the minimal frequency at which a neutral derived allele should be seen at least once in a homozygous state in a population of the Phase 3 size) to remove rare deleterious variants from the CADD distribution.
We assumed that the CADD score distribution of selected variants is high and corresponds to known functional variants (which is supported by the CADD predictions of the gold standard panel). Based on this assumption, the CADD score for the selected SNP was drawn from the outlier distribution in the range of 10.78–47 (see below and “Results” section).
We then simulated four scenarios under the additive selection model with different selection coefficients: s = 0.001, s = 0.007, s = 0.01, and s = 0 (no selection); and a sample size of 500 individuals in each population. The populations were sampled after 1000 generations of selection and drift. Each scenario was replicated 100 times. The FineMAV algorithm was subsequently applied to each output dataset. We then checked how often the selected variants fall outside of the neutral FineMAV distribution. To determine the upper end of the neutral distribution we bootstrapped 1000 FineMAV values from the simulated neutral variation 100 times and took the maximum sampled value as our cut-off (set to FineMAV of 10.7).
Additional files
Description of meta-analysis, enrichment analyses, and novel candidates found in this study. (DOCX 182 kb)
Top 100 FineMAV SNPs in each continental population (x = 3.5). (XLSX 92 kb)
GWAS hits among the top 100 FineMAV SNPs within each continental population. GWAS_SNP column specifies GWAS hits linked to given FineMAV SNPs. r2 column specifies the level of LD between the FineMAV SNP and the closest GWAS SNP. Genomic positions and the direction of genome-wide association are given for FineMAV SNPs. DER, derived allele of FineMAV SNP; ANC, ancestral allele of FineMAV SNP. Grayed variants might underline the same selection event. (XLSX 59 kb)
Top SSI protein-coding genes. (XLSX 60 kb)
Top 100 FineMAV SNPs in each continental population (x = 1). (XLSX 77 kb)
Acknowledgements
The authors would like to thank Dr Graham Ritchie for his help with setting up the CADD score mining script.
Funding
Our work was funded by The Wellcome Trust (098051).
Availability of data and materials
The 1000 Genomes data are available from http://www.internationalgenome.org/home [37]. Datasets generated and analyzed during the current study, as well as custom scripts are available at https://figshare.com/projects/FineMAV/26845 under an open source license (CC BY 4.0 and MIT) [132–136].
List of relevant urls
1000 Genomes Project, http://www.internationalgenome.org/, CADD, http://cadd.gs.washington.edu/, CMS, https://www.broadinstitute.org/cms/results, ENSEMBL, http://www.ensembl.org/, fitCons, http://compgen.cshl.edu/fitCons/, GEUVADIS, http://www.geuvadis.org/, GTEx, http://www.gtexportal.org/, GWAVA, http://www.sanger.ac.uk/sanger/StatGen_Gwava/, IMPC, http://mousephenotype.org/, MGI, http://informatics.jax.org/, OMIM, http://www.omim.org/, PUBMED, http://www.ncbi.nlm.nih.gov/pubmed/.
Abbreviations
- AFR
Africans
- AMR
Admixed Americans
- CADD
Combined annotation-dependent depletion
- CMS
Composite of multiple signals
- DAF
Derived allele frequency
- DAP
Derived allele purity
- EAS
East Asians
- eQTL
Expression quantitative trait loci
- EUR
Europeans
- FDR
False discovery rate
- FineMAV
Fine-mapping of adaptive variation
- FST
Wright’s fixation index
- GTEx
Genotype-Tissue Expression project
- LD
Linkage disequilibrium
- s
Selection coefficient
- SAS
South Asians
- SFS
Site frequency spectrum
- SNP
Single nucleotide polymorphism
- SSI
Selection support index
- ΔDAF
Difference in derived allele frequency
Authors’ contributions
MS participated in the project design and coordination, performed analyses and drafted the manuscript. MM and YC performed analyses. QA, YX, and CTS participated in project design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13059-017-1380-2) contains supplementary material, which is available to authorized users.
Contributor Information
Michał Szpak, Email: ms30@sanger.ac.uk.
Massimo Mezzavilla, Email: mm25@sanger.ac.uk.
Qasim Ayub, Email: qasim.ayub@monash.edu.
Yuan Chen, Email: yuan@sanger.ac.uk.
Yali Xue, Email: ylx@sanger.ac.uk.
Chris Tyler-Smith, Email: cts@sanger.ac.uk.
References
- 1.Jobling MA, Hollox E, Hurles M, Kivisild T, Tyler-Smith C. Human evolutionary genetics. 2. New York: Garland Science; 2013. [Google Scholar]
- 2.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong C, Di Rienzo A. Adaptations to local environments in modern human populations. Curr Opin Genet Dev. 2014;29:1–8. doi: 10.1016/j.gde.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat Rev Genet. 2009;10:745–55. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez JA, Marigorta UM, Navarro A. Integrating genomics into evolutionary medicine. Curr Opin Genet Dev. 2014;29:97–102. doi: 10.1016/j.gde.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–46. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8:857–68. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura M. The neutral theory of molecular evolution. New York, Cambridge: Cambridge University Press; 1983. [Google Scholar]
- 9.Xue Y, Daly A, Yngvadottir B, Liu M, Coop G, Kim Y, et al. Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am J Hum Genet. 2006;78:659–70. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novembre J, Galvani AP, Slatkin M. The geographic spread of the CCR5 Delta32 HIV-resistance allele. PLoS Biol. 2005;3:e339. doi: 10.1371/journal.pbio.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–53. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 12.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353–62. [PMC free article] [PubMed] [Google Scholar]
- 13.Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21:596–601. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Raj T, Kuchroo M, Replogle JM, Raychaudhuri S, Stranger BE, De Jager PL. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am J Hum Genet. 2013;92:517–29. doi: 10.1016/j.ajhg.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allison AC. Protection afforded by sickle-cell trait against subtertian malareal infection. Br Med J. 1954;1:290–4. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardona A, Pagani L, Antao T, Lawson DJ, Eichstaedt CA, Yngvadottir B, et al. Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS One. 2014;9:e98076. doi: 10.1371/journal.pone.0098076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemente FJ, Cardona A, Inchley CE, Peter BM, Jacobs G, Pagani L, et al. A selective sweep on a deleterious mutation in CPT1A in Arctic populations. Am J Hum Genet. 2014;95:584–9. doi: 10.1016/j.ajhg.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko WY, Rajan P, Gomez F, Scheinfeldt L, An P, Winkler CA, et al. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am J Hum Genet. 2013;93:54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–5. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–20. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 21.Scheinfeldt LB, Tishkoff SA. Recent human adaptation: genomic approaches, interpretation and insights. Nat Rev Genet. 2013;14:692–702. doi: 10.1038/nrg3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez RD, Kelley JL, Elyashiv E, Melton SC, Auton A, McVean G, et al. Classic selective sweeps were rare in recent human evolution. Science. 2011;331:920–4. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 24.Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–23. [PubMed] [Google Scholar]
- 25.Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–52. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlidis P, Jensen JD, Stephan W, Stamatakis A. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol Biol Evol. 2012;29:3237–48. doi: 10.1093/molbev/mss136. [DOI] [PubMed] [Google Scholar]
- 27.Vitti JJ, Grossman SR, Sabeti PC. Detecting natural selection in genomic data. Annu Rev Genet. 2013;47:97–120. doi: 10.1146/annurev-genet-111212-133526. [DOI] [PubMed] [Google Scholar]
- 28.Colonna V, Ayub Q, Chen Y, Pagani L, Luisi P, Pybus M, et al. Human genomic regions with exceptionally high levels of population differentiation identified from 911 whole-genome sequences. Genome Biol. 2014;15:R88. doi: 10.1186/gb-2014-15-6-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman SR, Shlyakhter I, Karlsson EK, Byrne EH, Morales S, Frieden G, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–6. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 30.Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 2009;19:711–22. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–9. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oleksyk TK, Smith MW, O’Brien SJ. Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci. 2010;365:185–205. doi: 10.1098/rstb.2009.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamberov YG, Wang S, Tan J, Gerbault P, Wark A, Tan L, et al. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell. 2013;152:691–702. doi: 10.1016/j.cell.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–6. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 35.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–7. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 36.Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet. 2003;12:2333–40. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 37.Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–13. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards M, Bigham A, Tan J, Li S, Gozdzik A, Ross K, et al. Association of the OCA2 polymorphism His615Arg with melanin content in east Asian populations: further evidence of convergent evolution of skin pigmentation. PLoS Genet. 2010;6:e1000867. doi: 10.1371/journal.pgen.1000867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eaton K, Edwards M, Krithika S, Cook G, Norton H, Parra EJ. Association study confirms the role of two OCA2 polymorphisms in normal skin pigmentation variation in East Asian populations. Am J Hum Biol. 2015;27:520–5. doi: 10.1002/ajhb.22678. [DOI] [PubMed] [Google Scholar]
- 42.Donnelly MP, Paschou P, Grigorenko E, Gurwitz D, Barta C, Lu RB, et al. A global view of the OCA2-HERC2 region and pigmentation. Hum Genet. 2012;131:683–96. doi: 10.1007/s00439-011-1110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, Jarman C, et al. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 2007;81:1119–32. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutton SM, Spritz RA. A comprehensive genetic study of autosomal recessive ocular albinism in Caucasian patients. Invest Ophthalmol Vis Sci. 2008;49:868–72. doi: 10.1167/iovs.07-0791. [DOI] [PubMed] [Google Scholar]
- 45.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 46.Visser M, Palstra RJ, Kayser M. Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum Mol Genet. 2014;23:5750–62. doi: 10.1093/hmg/ddu289. [DOI] [PubMed] [Google Scholar]
- 47.Kudo T, Iwasaki H, Nishihara S, Shinya N, Ando T, Narimatsu I, et al. Molecular genetic analysis of the human Lewis histo-blood group system. II. Secretor gene inactivation by a novel single missense mutation A385T in Japanese nonsecretor individuals. J Biol Chem. 1996;271:9830–7. doi: 10.1074/jbc.271.16.9830. [DOI] [PubMed] [Google Scholar]
- 48.Fry AE, Ghansa A, Small KS, Palma A, Auburn S, Diakite M, et al. Positive selection of a CD36 nonsense variant in sub-Saharan Africa, but no association with severe malaria phenotypes. Hum Mol Genet. 2009;18:2683–92. doi: 10.1093/hmg/ddp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayodo G, Price AL, Keinan A, Ajwang A, Otieno MF, Orago AS, et al. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am J Hum Genet. 2007;81:234–42. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love-Gregory L, Sherva R, Sun L, Wasson J, Schappe T, Doria A, et al. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum Mol Genet. 2008;17:1695–704. doi: 10.1093/hmg/ddn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuo Y, Yokoyama R, Yokoyama S. The genes for human alcohol dehydrogenases beta 1 and beta 2 differ by only one nucleotide. Eur J Biochem. 1989;183:317–20. doi: 10.1111/j.1432-1033.1989.tb14931.x. [DOI] [PubMed] [Google Scholar]
- 52.Jornvall H, Hempel J, Vallee BL, Bosron WF, Li TK. Human liver alcohol dehydrogenase: amino acid substitution in the beta 2 beta 2 Oriental isozyme explains functional properties, establishes an active site structure, and parallels mutational exchanges in the yeast enzyme. Proc Natl Acad Sci U S A. 1984;81:3024–8. doi: 10.1073/pnas.81.10.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hurley TD, Edenberg HJ, Bosron WF. Expression and kinetic characterization of variants of human beta 1 beta 1 alcohol dehydrogenase containing substitutions at amino acid 47. J Biol Chem. 1990;265:16366–72. [PubMed] [Google Scholar]
- 54.Han Y, Gu S, Oota H, Osier MV, Pakstis AJ, Speed WC, et al. Evidence of positive selection on a class I ADH locus. Am J Hum Genet. 2007;80:441–56. doi: 10.1086/512485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adhikari K, Fontanil T, Cal S, Mendoza-Revilla J, Fuentes-Guajardo M, Chacon-Duque JC, et al. A genome-wide association scan in admixed Latin Americans identifies loci influencing facial and scalp hair features. Nat Commun. 2016;7:10815. doi: 10.1038/ncomms10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–7. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geller F, Feenstra B, Zhang H, Shaffer JR, Hansen T, Esserlind AL, et al. Genome-wide association study identifies four loci associated with eruption of permanent teeth. PLoS Genet. 2011;7:e1002275. doi: 10.1371/journal.pgen.1002275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48:709–17. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–14. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X, Ong RT, Pillai EN, Elzein AM, Small KS, Clark TG, et al. Detecting and characterizing genomic signatures of positive selection in global populations. Am J Hum Genet. 2013;92:866–81. doi: 10.1016/j.ajhg.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–5. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 62.Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, Rieder MJ, et al. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–65. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fagny M, Patin E, Enard D, Barreiro LB, Quintana-Murci L, Laval G. Exploring the occurrence of classic selective sweeps in humans using whole-genome sequencing data sets. Mol Biol Evol. 2014;31:1850–68. doi: 10.1093/molbev/msu118. [DOI] [PubMed] [Google Scholar]
- 66.Hofer T, Foll M, Excoffier L. Evolutionary forces shaping genomic islands of population differentiation in humans. BMC Genomics. 2012;13:107. doi: 10.1186/1471-2164-13-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tennessen JA, Akey JM. Parallel adaptive divergence among geographically diverse human populations. PLoS Genet. 2011;7:e1002127. doi: 10.1371/journal.pgen.1002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johansson A, Gyllensten U. Identification of local selective sweeps in human populations since the exodus from Africa. Hereditas. 2008;145:126–37. doi: 10.1111/j.0018-0661.2008.02054.x. [DOI] [PubMed] [Google Scholar]
- 69.Kimura R, Fujimoto A, Tokunaga K, Ohashi J. A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS One. 2007;2:e286. doi: 10.1371/journal.pone.0000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez Herraez D, Bauchet M, Tang K, Theunert C, Pugach I, Li J, et al. Genetic variation and recent positive selection in worldwide human populations: evidence from nearly 1 million SNPs. PLoS One. 2009;4:e7888. doi: 10.1371/journal.pone.0007888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–37. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rafajlovic M, Klassmann A, Eriksson A, Wiehe T, Mehlig B. Demography-adjusted tests of neutrality based on genome-wide SNP data. Theor Popul Biol. 2014;95:1–12. doi: 10.1016/j.tpb.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 73.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–8. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai Z, Camp NJ, Cannon-Albright L, Thomas A. Identification of regions of positive selection using Shared Genomic Segment analysis. Eur J Hum Genet. 2011;19:667–71. doi: 10.1038/ejhg.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong M, Lange K, Papp JC, Fan R. A powerful score test to detect positive selection in genome-wide scans. Eur J Hum Genet. 2010;18:1148–59. doi: 10.1038/ejhg.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang K, Thornton KR, Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci U S A. 2006;103:135–40. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C, Bailey DK, Awad T, Liu G, Xing G, Cao M, et al. A whole genome long-range haplotype (WGLRH) test for detecting imprints of positive selection in human populations. Bioinformatics. 2006;22:2122–8. doi: 10.1093/bioinformatics/btl365. [DOI] [PubMed] [Google Scholar]
- 81.Mackenzie FE, Romero R, Williams D, Gillingwater T, Hilton H, Dick J, et al. Upregulation of PKD1L2 provokes a complex neuromuscular disease in the mouse. Hum Mol Genet. 2009;18:3553–66. doi: 10.1093/hmg/ddp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oh HJ, Li Y, Lau YF. Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol Reprod. 2005;72:407–15. doi: 10.1095/biolreprod.104.034447. [DOI] [PubMed] [Google Scholar]
- 83.Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J Cell Biol. 2001;154:123–36. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wassarman PM, Jovine L, Litscher ES. A profile of fertilization in mammals. Nat Cell Biol. 2001;3:E59–64. doi: 10.1038/35055178. [DOI] [PubMed] [Google Scholar]
- 86.U Basmanav FB, Cau L, Tafazzoli A, Mechin MC, Wolf S, Romano MT, et al. Mutations in three genes encoding proteins involved in hair shaft formation cause uncombable hair syndrome. Am J Hum Genet. 2016;99:1292–304. doi: 10.1016/j.ajhg.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.John S, Thiebach L, Frie C, Mokkapati S, Bechtel M, Nischt R, et al. Epidermal transglutaminase (TGase 3) is required for proper hair development, but not the formation of the epidermal barrier. PLoS One. 2012;7:e34252. doi: 10.1371/journal.pone.0034252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bognar P, Nemeth I, Mayer B, Haluszka D, Wikonkal N, Ostorhazi E, et al. Reduced inflammatory threshold indicates skin barrier defect in transglutaminase 3 knockout mice. J Invest Dermatol. 2014;134:105–11. doi: 10.1038/jid.2013.307. [DOI] [PubMed] [Google Scholar]
- 89.Brennan BM, Huynh MT, Rabah MA, Shaw HE, Bisaillon JJ, Radden LA, 2nd, et al. The mouse wellhaarig (we) mutations result from defects in epidermal-type transglutaminase 3 (Tgm3) Mol Genet Metab. 2015;116:187–91. doi: 10.1016/j.ymgme.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fujimoto A, Nishida N, Kimura R, Miyagawa T, Yuliwulandari R, Batubara L, et al. FGFR2 is associated with hair thickness in Asian populations. J Hum Genet. 2009;54:461–5. doi: 10.1038/jhg.2009.61. [DOI] [PubMed] [Google Scholar]
- 91.Laatsch CN, Durbin-Johnson BP, Rocke DM, Mukwana S, Newland AB, Flagler MJ, et al. Human hair shaft proteomic profiling: individual differences, site specificity and cuticle analysis. PeerJ. 2014;2:e506. doi: 10.7717/peerj.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jablonski NG, Chaplin G. The evolution of skin pigmentation and hair texture in people of African ancestry. Dermatol Clin. 2014;32:113–21. doi: 10.1016/j.det.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 93.Robbins CR. Chemical and physical behavior of human hair. 4. New York: Springer; 2002. [Google Scholar]
- 94.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 95.Day FR, Thompson DJ, Helgason H, Chasman DI, Finucane H, Sulem P, et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet. 2017;49:834–41. doi: 10.1038/ng.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dauber A, Cunha-Silva M, Macedo DB, Brito VN, Abreu AP, Roberts SA, et al. Paternally inherited DLK1 deletion associated with familial central precocious puberty. J Clin Endocrinol Metab. 2017;102:1557–67. doi: 10.1210/jc.2016-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Y, DeBoer K, de Kretser DM, O’Donnell L, O’Connor AE, Merriner DJ, et al. LRGUK-1 is required for basal body and manchette function during spermatogenesis and male fertility. PLoS Genet. 2015;11:e1005090. doi: 10.1371/journal.pgen.1005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ionita-Laza I, Capanu M, De Rubeis S, McCallum K, Buxbaum JD. Identification of rare causal variants in sequence-based studies: methods and applications to VPS13B, a gene involved in Cohen syndrome and autism. PLoS Genet. 2014;10:e1004729. doi: 10.1371/journal.pgen.1004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–76. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vernot B, Akey JM. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–21. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 101.Sankararaman S, Mallick S, Dannemann M, Prufer K, Kelso J, Paabo S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–7. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Racimo F, Sankararaman S, Nielsen R, Huerta-Sanchez E. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 2015;16:359–71. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova JL, et al. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet. 2016;98:5–21. doi: 10.1016/j.ajhg.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakashima M, Chung S, Takahashi A, Kamatani N, Kawaguchi T, Tsunoda T, et al. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42:768–71. doi: 10.1038/ng.645. [DOI] [PubMed] [Google Scholar]
- 105.Alhady SM, Sivanantharajah K. Keloids in various races. A review of 175 cases. Plast Reconstr Surg. 1969;44:564–6. doi: 10.1097/00006534-196912000-00006. [DOI] [PubMed] [Google Scholar]
- 106.Marneros AG, Norris JE, Olsen BR, Reichenberger E. Clinical genetics of familial keloids. Arch Dermatol. 2001;137:1429–34. doi: 10.1001/archderm.137.11.1429. [DOI] [PubMed] [Google Scholar]
- 107.Enard W. Mouse models of human evolution. Curr Opin Genet Dev. 2014;29:75–80. doi: 10.1016/j.gde.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 108.Hughes DA, Tang K, Strotmann R, Schoneberg T, Prenen J, Nilius B, et al. Parallel selection on TRPV6 in human populations. PLoS One. 2008;3:e1686. doi: 10.1371/journal.pone.0001686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu M, Ayub Q, Guerra-Assuncao JA, Long Q, Ning Z, Huang N, et al. Exploration of signals of positive selection derived from genotype-based human genome scans using re-sequencing data. Hum Genet. 2012;131:665–74. doi: 10.1007/s00439-011-1111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Breiman L. Classification and regression trees. Belmont, CA: Wadsworth International Group; 1984. [Google Scholar]
- 111.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Paten B, Herrero J, Fitzgerald S, Beal K, Flicek P, Holmes I, et al. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008;18:1829–43. doi: 10.1101/gr.076521.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peng B, Kimmel M. simuPOP: a forward-time population genetics simulation environment. Bioinformatics. 2005;21:3686–7. doi: 10.1093/bioinformatics/bti584. [DOI] [PubMed] [Google Scholar]
- 114.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–8. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 116.Iwamoto S, Li J, Omi T, Ikemoto S, Kajii E. Identification of a novel exon and spliced form of Duffy mRNA that is the predominant transcript in both erythroid and postcapillary venule endothelium. Blood. 1996;87:378–85. [PubMed] [Google Scholar]
- 117.Iwamoto S, Li J, Sugimoto N, Okuda H, Kajii E. Characterization of the Duffy gene promoter: evidence for tissue-specific abolishment of expression in Fy(a-b-) of black individuals. Biochem Biophys Res Commun. 1996;222:852–9. doi: 10.1006/bbrc.1996.0833. [DOI] [PubMed] [Google Scholar]
- 118.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 119.Engelken J, Carnero-Montoro E, Pybus M, Andrews GK, Lalueza-Fox C, Comas D, et al. Extreme population differences in the human zinc transporter ZIP4 (SLC39A4) are explained by positive selection in Sub-Saharan Africa. PLoS Genet. 2014;10:e1004128. doi: 10.1371/journal.pgen.1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yoshiura K, Kinoshita A, Ishida T, Ninokata A, Ishikawa T, Kaname T, et al. A SNP in the ABCC11 gene is the determinant of human earwax type. Nat Genet. 2006;38:324–30. doi: 10.1038/ng1733. [DOI] [PubMed] [Google Scholar]
- 121.Martin A, Saathoff M, Kuhn F, Max H, Terstegen L, Natsch A. A functional ABCC11 allele is essential in the biochemical formation of human axillary odor. J Invest Dermatol. 2010;130:529–40. doi: 10.1038/jid.2009.254. [DOI] [PubMed] [Google Scholar]
- 122.Mou C, Thomason HA, Willan PM, Clowes C, Harris WE, Drew CF, et al. Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum Mutat. 2008;29:1405–11. doi: 10.1002/humu.20795. [DOI] [PubMed] [Google Scholar]
- 123.Eiberg H, Troelsen J, Nielsen M, Mikkelsen A, Mengel-From J, Kjaer KW, et al. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet. 2008;123:177–87. doi: 10.1007/s00439-007-0460-x. [DOI] [PubMed] [Google Scholar]
- 124.Sturm RA, Duffy DL, Zhao ZZ, Leite FP, Stark MS, Hayward NK, et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet. 2008;82:424–31. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–55. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tsetskhladze ZR, Canfield VA, Ang KC, Wentzel SM, Reid KP, Berg AS, et al. Functional assessment of human coding mutations affecting skin pigmentation using zebrafish. PLoS One. 2012;7:e47398. doi: 10.1371/journal.pone.0047398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 2005;25:278–84. doi: 10.1002/humu.20143. [DOI] [PubMed] [Google Scholar]
- 128.Cook AL, Chen W, Thurber AE, Smit DJ, Smith AG, Bladen TG, et al. Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J Invest Dermatol. 2009;129:392–405. doi: 10.1038/jid.2008.211. [DOI] [PubMed] [Google Scholar]
- 129.Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–9. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–6. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Szpak M, Mezzavilla M, Ayub Q, Chen Y, Xue Y, Tyler-Smith C. FineMAV: custom script code. figshare. 2017. http://doi.org/10.6084/m9.figshare.5632021.
- 133.Szpak M, Mezzavilla M, Ayub Q, Chen Y, Xue Y, Tyler-Smith C. FineMAV: Simulations data set. figshare. 2017. http://doi.org/10.6084/m9.figshare.5631859.
- 134.Szpak M, Mezzavilla M, Ayub Q, Chen Y, Xue Y, Tyler-Smith C. FineMAV: AFR + EUREAS data set. figshare. 2017. http://doi.org/10.6084/m9.figshare.5630215.
- 135.Szpak M, Mezzavilla M, Ayub Q, Chen Y, Xue Y, Tyler-Smith C. FineMAV: AFR + EAS + EUR + SAS data set. figshare 2017. http://doi.org/10.6084/m9.figshare.5630200.
- 136.Szpak M, Mezzavilla M, Ayub Q, Chen Y, Xue Y, Tyler-Smith C. FineMAV: AFR + EAS + EUR data set. figshare 2017. http://doi.org/10.6084/m9.figshare.5625088.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of meta-analysis, enrichment analyses, and novel candidates found in this study. (DOCX 182 kb)
Top 100 FineMAV SNPs in each continental population (x = 3.5). (XLSX 92 kb)
GWAS hits among the top 100 FineMAV SNPs within each continental population. GWAS_SNP column specifies GWAS hits linked to given FineMAV SNPs. r2 column specifies the level of LD between the FineMAV SNP and the closest GWAS SNP. Genomic positions and the direction of genome-wide association are given for FineMAV SNPs. DER, derived allele of FineMAV SNP; ANC, ancestral allele of FineMAV SNP. Grayed variants might underline the same selection event. (XLSX 59 kb)
Top SSI protein-coding genes. (XLSX 60 kb)
Top 100 FineMAV SNPs in each continental population (x = 1). (XLSX 77 kb)
Data Availability Statement
The 1000 Genomes data are available from http://www.internationalgenome.org/home [37]. Datasets generated and analyzed during the current study, as well as custom scripts are available at https://figshare.com/projects/FineMAV/26845 under an open source license (CC BY 4.0 and MIT) [132–136].