Abstract
Cryptosporidium ubiquitum is an emerging zoonotic pathogen in humans. Recently, a subtyping tool targeting the 60-kDa glycoprotein (gp60) gene was developed for C. ubiquitum, and identified six subtype families (XIIa–XIIf). In this study, we selected five genetic loci known to be polymorphic in C. hominis and C. parvum for the development of a multilocus subtyping tool for C. ubiquitum, including CP47 (cgd6_1590), MSC6-5 (cgd6_4290), cgd6_60, cgd2_3690, and cgd4_370. PCR primers for these targets were designed based on whole genome sequence data from C. ubiquitum. DNA sequence analyses of 24 C. ubiquitum specimens showed the presence of 18, 1, 5, 4, and 5 subtypes at the CP47, MSC6-5, cgd6_60, cgd2_3690, and cgd4_370 loci, respectively. Altogether, 18 multilocus sequence typing (MLST) subtypes were detected among the 19 specimens successfully sequenced at all polymorphic loci. Phylogenetic analyses of the MLST data indicated that the rodent subtype families of XIIe and XIIf were highly divergent from others, and the ruminant XIIa subtype family formed a monophyletic group genetically distant from other rodent subtype families XIIb, XIIc, and XIId. The latter showed no consistent grouping of specimens and formed one large cluster in phylogenetic analysis of concatenated multilocus sequences. This was supported by results of STRUCTURE and FST analyses, which further suggested that XIIa originated from one common ancestor whereas XIIb, XIIc, and XIId contained mixed ancestral types, reflecting a close relatedness of the three subtype families and the likely occurrence of genetic recombination among them. Thus, an MLST tool was developed for high-resolution subtyping of C. ubiquitum and results of preliminary characterizations of specimens from humans and animals supported the conclusion on the existence of ruminant and rodent-adapted C. ubiquitum groups.
Keywords: Cryptosporidium ubiquitum, Multilocus sequence typing, Zoonotic transmission, Host adaptation
1. Introduction
Cryptosporidium ubiquitum, previously known as the Cryptosporidium cervine genotype, infects a broad range of host species (Fayer et al., 2010). It has been found in human cases worldwide, especially in industrialized countries (Blackburn et al., 2006; Chalmers et al., 2009; Cieloszyk et al., 2012; Davies et al., 2009; Elwin et al., 2012; Feltus et al., 2006; Molloy et al., 2010; Ong et al., 2002; Trotz-Williams et al., 2006). In addition to causing human disease, C. ubiquitum is detected in a variety of animals, especially ruminants and rodents (Diaz et al., 2015; Fayer et al., 2010; Feng et al., 2007; Li et al., 2016; Mirhashemi et al., 2016). Thus, C. ubiquitum has emerged as an important zoonotic species in humans. Nevertheless, little is known on the transmission route of C. ubiquitum in humans and animals and the significance of zoonotic infection in its epidemiology.
To characterize the transmission of human-pathogenic Cryptosporidium spp., various molecular diagnostic tools have been developed. For C. ubiquitum, a subtyping tool targeting the 60-kDa glycoprotein (gp60) gene has identified six subtype families (XIIa-XIIf) (Li et al., 2014). These subtype families differ in host range and geographic distribution. However, this observation is yet to be supported by multilocus sequence typing (MLST). Thus far, MLST tools are available for five Cryptosporidium species, including C. hominis, C. parvum, C. meleagridis, C. muris, and C. andersoni, and have been shown to be useful in genetic characterizations of these species (Cama et al., 2006; Feng et al., 2011; Gatei et al., 2007; Gatei et al., 2006; Wang et al., 2014).
In this study, we selected five genetic loci known to be polymorphic in C. hominis and C. parvum, developed an MLST tool for high-resolution subtyping of C. ubiquitum from humans and animals.
2. Materials and methods
2.1. Specimens
DNA extractions from 24 C. ubiquitum specimens were used in this study. The specimens were from humans, one Verreaux’s sifaka (Propithecus verreauxi coquereli), and various species of ruminants and rodents in the United States, Spain, the Slovak Republic, South Africa, China, and Nepal (Table 1). The specimens were assigned to XIIa–XIIf subtype families (Table 1) by gp60 sequence analysis in a previous study (Li et al., 2014).
Table 1.
Specimens of Cryptosporidium ubiquitum used in the study and their subtype designations at the five selected loci.
| Specimen | Host | Source location | gp60 subtype familya | MLST subtype | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| CP47 | MSC6-5 | cgd6_60 | cgd2_3690 | cgd4_370 | ||||
| 38664 | Sheep | China | XIIa | 4 | 1 | 1 | 1 | 3 |
| 38665 | Sheep | China | XIIa | 4 | + | 1 | 1 | 3 |
| 32563 | Impala | South Africa | XIIa | 4 | 1 | 1 | 1 | 4 |
| 32560 | Buffalo | South Africa | XIIa | 5 | 1 | 1 | 1 | 4 |
| 30238 | Swamp deer | Nepal | XIIa | 2 | − | 1 | 1 | 4 |
| 30241 | Swamp deer | Nepal | XIIa | 3 | − | 1 | 1 | 4 |
| 31570 | Sheep | Spain | XIIa | 1 | 1 | 1 | 1 | 4 |
| 34141 | Human | USA | XIIb | 7 | 1 | 2 | 2 | 1 |
| 33456 | Verreaux’s sifaka | USA | XIIb | 6 | 1 | 2 | 2 | 1 |
| 18868 | Chipmunk | USA | XIIb | 18 | − | − | − | − |
| 37192 | Human | USA | XIIb | 6 | 1 | 2 | 3 | 5 |
| 37202 | Human | USA | XIIb | 8 | − | 2 | 3 | 5 |
| 17574 | Human | USA | XIIc | 9 | 1 | 2 | 2 | 1 |
| 36638 | Porcupine | USA | XIIc | 10 | 1 | 2 | 2 | 1 |
| 37211 | Human | USA | XIIc | 9 | + | 2 | 3 | 5 |
| 17577 | Human | USA | XIIc | 11 | 1 | 2 | 2 | 1 |
| 29361 | Human | USA | XIId | 13 | 1 | 3 | 2 | 1 |
| 37204 | Human | USA | XIId | 14 | 1 | 2 | 2 | 1 |
| 14223 | Beaver | USA | XIId | 15 | − | − | 2 | 1 |
| 18370 | Eastern gray squirrel | USA | XIId | − | − | − | 2 | 2 |
| 14886 | Eastern chipmunk | USA | XIId | 12 | 1 | 2 | 2 | 1 |
| 37193 | Human | USA | XIId | 13 | 1 | 2 | 3 | 5 |
| 37646 | Field mouse | Slovak Republic | XIIe | 16 | + | 4 | 4 | + |
| 37644 | Field mouse | Slovak Republic | XIIf | 17 | − | 5 | + | + |
PCR positive but produced noisy signals at sequencing; −, PCR negative.
The gp60 subtype family data of these specimens are obtained from a previous report (Li et al., 2014).
2.2. MLST markers
For multilocus subtyping of C. ubiquitum, we selected five genetic loci based on their polymorphic nature in C. parvum and C. hominis. Three polymorphic markers of cgd6_60, CP47 (cgd6_1590) and MSC6-5 (cdg6_4290) were previously used in several MLST and population genetic analyses of C. hominis and C. parvum (Feng et al., 2014; Feng et al., 2013; Li et al., 2013), and two other loci of cgd2_3690 and cgd4_370 were found to be polymorphic in a recent comparison of C. parvum and C. hominis genomes (Guo et al., 2015). Their orthologs in C. ubiquitum were identified by alignment of whole genome sequence of C. ubiquitum obtained in a previous study (Li et al., 2014) with the reference sequence from the C. parvum IOWA genome (Abrahamsen et al., 2004). Primers for nested PCR were designed based on semi-conserved sequence of each locus with expected PCR products ranging from 619 bp to 1055 bp (Table 2).
Table 2.
Primer sequences of five multilocus sequence typing loci used in the study.
| Locus | Nature of polymorphism | Primer | Sequence (5′ to 3′) | Annealing temp (°C) | Expected PCR product (bp) |
|---|---|---|---|---|---|
| CP47 (cgd6_1590) | Microsatellite-TAC/TAA | F1 | TACCGAGTGGGTGGTACATATTATA | 58 | ~937 |
| R1 | AGCTAACAGGCCCTGTATCAG | ||||
| F2 | GCTCAAGAATTTATTCCCAGAGG | 55 | ~752 | ||
| R2 | TCATCTAATACTTCATTTTTGGTATC | ||||
| MSC6-5 (cgd6_4290) | Microsatellite-TCT/TCC | F1 | CTGATAGCATCCCTTCCTGG | 55 | ~1118 |
| R1 | GAATTTACACTGAAGTTCC | ||||
| F2 | GGATTTGGACTTACTCCATTTCT | 55 | ~970 | ||
| R2 | CCATCGTTATTACCTTCAGATG | ||||
| cgd6_60 | SNP | F1 | GTTACTCTGTTTGGCCAACTAGG | 55 | ~1217 |
| R1 | CTTTAATAGTACCAGTACAAGGAGTT | ||||
| F2 | ATTTCTCTTCAGATGTGGGAA | 52 | ~1055 | ||
| R2 | AAGCAACTTCATCTCCAGC | ||||
| cgd2_3690 | SNP | F1 | AGCTCTGGCAGTATACCTCA | 55 | ~870 |
| R1 | ATTAGTCCAAGTAAGCTCATGGT | ||||
| F2 | ATGGTGAGTATTCATCCCTTACA | 52 | ~780 | ||
| R2 | AGCATCCTTCAATACAAAGTAG | ||||
| cgd4_370 | SNP | F1 | TGCATCTAATATACCACTATCATC | 52 | ~1009 |
| R1 | CCAGAGTATTCTGAAGGATATA | ||||
| F2 | TACTAAGGTAACATTGGCGCCAT | 52 | ~619 | ||
| R2 | TTGCCCATAATGACTTGCATTTC |
SNP, single nucleotide polymorphisms.
2.3. MLST PCR
Nested PCR was used for the amplification of MLST markers. For each nested PCR, the total volume of PCR was 50 μl, which consisted of 1 μl of DNA (primary PCR) or 2 μl of the primary PCR product (secondary PCR), primers at a concentration of 0.25 μM (primary PCR using F1 and R1) or 0.5 μM (secondary PCR using F2 and R2), 200 mM deoxyribonuleotide triphosphate mix (Promega, Madison, WI), 3 mM MgCl2 (Promega), 1 × GeneAmp PCR buffer (Applied Biosystems, Foster City, CA), and 1.25 U of Taq DNA polymerase (Promega). The primary PCR reactions also contained 400 ng/μl of non-acetylated bovine serum albumin (Sigma, Louis, MO). PCR amplification consisted of an initial denaturation at 94 °C for 5 min; 35 cycles of 94 °C for 45 s, the specified annealing temperature for each primer set (Table 2) for 45 s, and 72 °C for 1 min; and a final 7-min extension at 72 °C. Two negative controls (reagent-grade water) for primary PCR and secondary PCR were used in each PCR run. The secondary PCR products were detected by agarose gel electrophoresis and ethidium bromide staining. Two PCR replicates were used to analyze each DNA extraction.
2.4. DNA sequencing
The secondary PCR products of the expected size were sequenced in both directions using an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster city, CA). A second PCR product was sequenced if the initial sequence was unreadable. The sequences obtained were assembled using ChromasPro version 1.5 (http://www.technelysium.com.au/ChromasPro.html), edited using BioEdit version 7.1 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and aligned using ClustalX version 1.81 (http://www.clustal.org/).
2.5. Data analysis
To assess the phylogenetic relationships among various gp60 subtype families of C. ubiquitum, neighbor-joining trees of nucleotide sequences from polymorphic loci were constructed using genetic distances from the Kimura-2 parameter model and the software Mega version 6.06 (http://www.megasoftware.net/). Thereafter, we used the Bayesian analysis tool STRUCTURE version 2.3.4 (http://pritch.bsd.uchicago.edu/structure.html) by K-means clustering and the admixture model to identify subpopulations of C. ubiquitum. To assess the robustness of the substructuring, population differentiation (FST) between gp60 subtype families was also calculated using Arlequin version 3.5 (http://cmpg.unibe.ch/software/arlequin35).
2.6. Nucleotide sequence accession numbers
Representative nucleotide sequences generated in this study were deposited in GenBank under accession numbers KX286354 to KX286386.
3. Results
3.1. Sequence polymorphisms at MLST loci
PCR analyses of 24 C. ubiquitum specimens produced the expected products for 23, 17, 21, 23, and 23 specimens at the CP47, MSC6-5, cgd6_60, cgd2_3690, and cgd4_370 loci, respectively (Table 1). Sequences were obtained for the majority of PCR amplicons with the exception of seven specimens. One specimen (37644) at cgd2_3690, two specimens (37644 and 37646) at cgd4_370, and three specimens (38665, 37211, and 37646) at MSC6-5, produced noisy signals. Sequence polymorphism was observed at CP47, cgd6_60, cgd2_3690, and cgd4_370, including variations in copy numbers of simple tandem repeats, single nucleotide substitutions, and insertion and deletion in the non-repeat regions. In contrast, all specimens at the MSC6-5 locus produced the same nucleotide sequence. Thus, further MLST data analyses include only the four polymorphic loci. Altogether, there were 18, 5, 4, and 5 subtypes at the CP47, cgd6_60, cgd2_3690, and cgd4_370 loci, respectively (Table 1). A total of 19 specimens were subtyped successfully at all four polymorphic loci, forming 18 MLST subtypes. Most of the MLST subtypes had only one specimen, with the exception of one MLST subtype, which had two specimens (38664 and 38665).
3.2. Phylogenetic relationships among gp60 subtype families
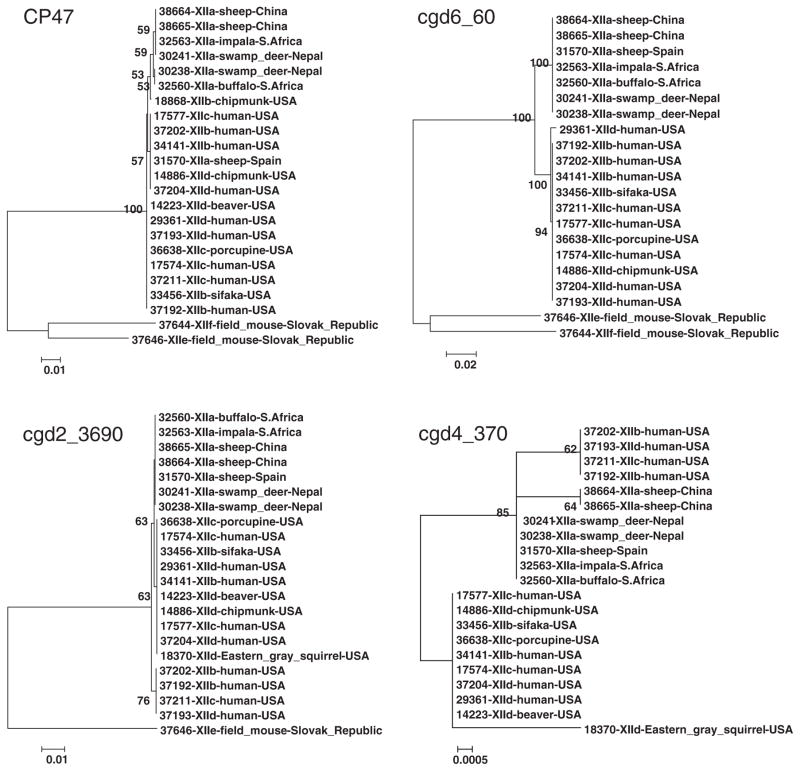
To establish phylogenetic relationships among gp60 subtype families (XIIa–XIIf) of C. ubiquitum, a neighbor-joining analysis was carried out for each of the four polymorphic loci (Fig. 1). At the CP47 locus, two subtype families of XIIe and XIIf formed a cluster highly divergent from the major cluster of the four subtype families of XIIa, XIIb, XIIc, and XIId. Within the latter, most XIIa specimens (6/7) formed one sub-cluster and were relatively distant from sub-clusters by the remaining specimens. These were no consistent separation of XIIb, XIIc, and XIId specimens in the phylogenetic tree. At the cgd6_60 locus, XIIe and XIIf also formed a cluster highly distinct from the major cluster. Within the latter, XIIa formed a monophyletic group with 100% bootstrap support and was distant from the group formed by XIIb, XIIc, and XIId. At the cgd2_3690 locus, XIIe was distant from the major cluster formed by other subtype families, in which XIIa formed a mono-phyletic group. At the cgd4_370 locus, none of XIIe and XIIf specimens were sequenced successfully, sequences from XIIa formed two clusters, and there was no consistent grouping among XIIb, XIIc, and XIId specimens.
Fig. 1.
Phylogenetic relationships among gp60 subtype families of Cryptosporidium ubiquitum. Four polymorphic loci of CP47, cgd6_60, cgd2_3690, and cgd4_370 were selected in constructing trees by a neighbor-joining analysis of nucleotide sequences using distance calculated by the Kimura 2-parameter model. Bootstrap values greater than 50% from 1000 replicates are shown.
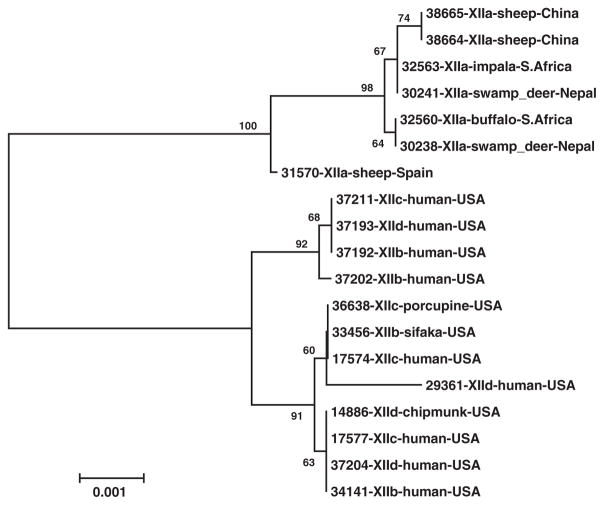
The genetic relationship among gp60 subtype families was further assessed by a neighbor-joining tree constructed using concatenated multilocus sequences from the four MLST loci (Fig. 2). Two genetically related major groups were seen. One group consisted of all XIIa specimens, and the other group consisted of XIIb, XIIc, and XIId specimens with no consistent clustering of members of the three gp60 subtype families.
Fig. 2.
Phylogenetic relationships among gp60 subtype families of Cryptosporidium ubiquitum. A neighbor-joining tree was constructed by analysis of concatenated nucleotide sequences of four polymorphic loci (CP47, cgd6_60, cgd2_3690, and cgd4_370) using distance calculated by the Kimura 2-parameter model.
3.3. Substructure in C. ubiquitum
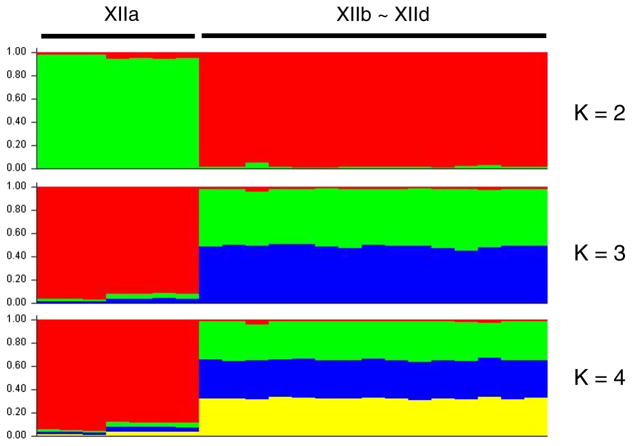
The evolutionary relationship and existence of substructuring among gp60 subtype families of C. ubiquitum were assessed by using STUCTURE analysis (Fig. 3). When K = 2 was used in the analysis, two subpopulations were generated. All seven XIIa specimens formed a sub-population, and 15 specimens of XIIb, XIIc, and XIId formed the other subpopulation. When K > 2, all XIIa specimens still formed one subpopulation and XIIb, XIIc, and XIId formed the other, with admixed colors in the latter. Thus, the result of STUCTURE analysis was in agreement with the grouping in the neighbor-joining analysis of concatenated multilocus sequences, and supported the conclusion on the genetic homogeneity of the XIIa subtype family. Further pairwise FST analysis also revealed highly significant population differentiation between XIIa and the other three subtype families (FST = 0.489 to 0.564; P < 0.00001), and did not detect significant differentiation (FST = −0.020 to 0.046; P = 0.247 to 0.619) among the XIIb, XIIc, and XIId subtype families (Table 3).
Fig. 3.
Relationships among various gp60 subtype families of Cryptosporidium ubiquitum by substructuring analysis. MLST data including the gp60 locus from 22 specimens of the XIIa, XIIb, XIIc and XIId subtype families were used by K-means clustering and the admixture model to identify subpopulations. The expected populations K = 2, 3, and 4 were used in the STRUCTURE analysis.
Table 3.
Population differentiation among gp60 subtype families of Cryptosporidium ubiquitum in FST analysis of sequences at five genetic loci.
| Parameter and subtype | XIIa | XIIb | XIIc |
|---|---|---|---|
| Population pairwise FST | |||
| XIIb | 0.48930 | ||
| XIIc | 0.56385 | 0.04278 | |
| XIId | 0.49324 | 0.04622 | −0.01996 |
| FST P values | |||
| XIIb | 0.00000 ± 0.0000* | ||
| XIIc | 0.00000 ± 0.0000* | 0.25000 ± 0.0134 | |
| XIId | 0.00000 ± 0.0000* | 0.24707 ± 0.0153 | 0.61914 ± 0.0177 |
P < 0.00001, highly significant population differentiation.
4. Discussion
In this study, we took advantage of the recent whole genome sequence data of C. ubiquitum (Li et al., 2014), and identified four genetic loci that showed high sequence polymorphism among the C. ubiquitum specimens examined. With the inclusion of the gp60 locus previously characterized (Li et al., 2014), an MLST tool targeting the five polymorphic loci (gp60, CP47, cgd6_60, cgd2_3690, and cgd4_370) was developed for high-resolution subtyping of C. ubiquitum in humans and animals. This tool allowed the identification of 18 MLST subtypes in the 19 C. ubiquitum specimens with complete data for all polymorphic loci. Compared with gp60 subtyping alone, this MLST tool revealed a greater genetic diversity in C. ubiquitum.
Phylogenetic analyses of the MLST data generated from this study showed that two gp60 subtype families of XIIe and XIIf formed a cluster highly divergent from the dominant cluster of XIIa, XIIb, XIIc, and XIId. This is in agreement with the previous observation at the gp60 locus (Li et al., 2014), indicating the presence of a unique C. ubiquitum group in rodents in the Slovak Republic. Within the major cluster, XIIa formed a monophyletic group distinct from other subtype families, and genetic homogeneity was also observed within XIIa in STRUCTURE analysis, providing strong support for XIIa being a separate group with a unique ancestry. In contrast, within the group formed by XIIb, XIIc, and XIId, the pattern of combinations in STRUCTURE suggested a mixture of ancestral types, reflecting a close relatedness of the three subtype families and the likely occurrence of genetic recombination among them. Thus, gp60 subtype family XIIa apparently represents a ruminant-adapted C. ubiquitum group and XIIb, XIIc and XIId subtype families represent a rodent-adapted group. These two groups of C. ubiquitum likely have different population genetic structures.
The identification of broad host-adapted groups improves our understanding of the transmission of C. ubiquitum. Previously, it was shown by sequence analysis of the gp60 locus that all human C. ubiquitum infections in the United States were caused by subtype families XIIb, XIIc and XIId, whereas those in the United Kingdom were mostly caused by XIIa (Li et al., 2014). The new data on genetic differences between ruminant-adapted (XIIa) and rodent-adapted (XIIb/XIIc/XIId) C. ubiquitum groups re-affirm the likely roles of rodents in the transmission of human infections in the United States and sheep in the United Kingdom. The broad rodent host range of XIIb, XIIc and XIId subtype families seemingly allows the occurrence of a sylvatic transmission cycle of C. ubiquitum, with occasional spillover of infections to humans. As C. ubiquitum XIIb, XIIc and XIId subtypes are among the most frequently detected Cryptosporidium species in protected drinking source watershed in the United States (Jiang et al., 2005; Li et al., 2014), further studies using the newly established MLST tool are needed to examine the role of drinking water in the transmission of C. ubiquitum infection in humans.
In conclusion, four polymorphic genetic markers were identified in this study for genetic characterizations of C. ubiquitum, and an MLST tool was developed for high-resolution subtyping of C. ubiquitum. With validation studies of additional specimens from diverse animals and the inclusion of the gp60 locus, the MLST tool should be useful in studies of C. ubiquitum transmission in different areas and population genetic characterizations of isolates from different host sources, especially the potential role of host species or geography in genetic structuring of parasite populations.
Acknowledgments
In memory of Ying Tang, who conducted bulk of the bench work in this study and passed away in 2015. We thank the authors of the previous study on C. ubiquitum gp60 subtyping for providing DNA used in this study.
This work was supported by the National Natural Science Foundation of China (grant numbers 31425025, 31229005, 3110103901, and 31302078); Open Funding Project of the State Key Laboratory of Veterinary Etiological Biology, Lanzhou, China (grant number SKLVEB2014KFKT008); Fundamental Research Funds for the Central Universities, China; and the U.S. Centers for Disease Control and Prevention.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Blackburn BG, Mazurek JM, Hlavsa M, Park J, Tillapaw M, Parrish M, Salehi E, Franks W, Koch E, Smith F, Xiao L, Arrowood M, Hill V, da Silva A, Johnston S, Jones JL. Cryptosporidiosis associated with ozonated apple cider. Emerg Infect Dis. 2006;12:684–686. doi: 10.3201/eid1204.050796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cama VA, Arrowood MJ, Ortega YR, Xiao L. Molecular characterization of the Cryptosporidium parvum IOWA isolate kept in different laboratories. J Eukaryot Microbiol. 2006;53(Suppl 1):S40–S42. doi: 10.1111/j.1550-7408.2006.00168.x. [DOI] [PubMed] [Google Scholar]
- Chalmers RM, Elwin K, Thomas AL, Guy EC, Mason B. Long-term Cryptosporidium typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Euro Surveill. 2009;14:19086. doi: 10.2807/ese.14.02.19086-en. [DOI] [PubMed] [Google Scholar]
- Cieloszyk J, Goni P, Garcia A, Remacha MA, Sanchez E, Clavel A. Two cases of zoonotic cryptosporidiosis in Spain by the unusual species Cryptosporidium ubiquitum and Cryptosporidium felis. Enferm Infecc Microbiol Clin. 2012;30:549–551. doi: 10.1016/j.eimc.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Davies AP, Campbell B, Evans MR, Bone A, Roche A, Chalmers RM. Asymptomatic carriage of protozoan parasites in children in day care centers in the United Kingdom. Pediatr Infect Dis J. 2009;28:838–840. doi: 10.1097/INF.0b013e31819d646d. [DOI] [PubMed] [Google Scholar]
- Diaz P, Quilez J, Prieto A, Navarro E, Perez-Creo A, Fernandez G, Panadero R, Lopez C, Diez-Banos P, Morrondo P. Cryptosporidium species and subtype analysis in diarrhoeic pre-weaned lambs and goat kids from north-western Spain. Parasitol Res. 2015;114:4099–4105. doi: 10.1007/s00436-015-4639-0. [DOI] [PubMed] [Google Scholar]
- Elwin K, Hadfield SJ, Robinson G, Chalmers RM. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000–2008. Epidemiol Infect. 2012;140:673–683. doi: 10.1017/S0950268811000860. [DOI] [PubMed] [Google Scholar]
- Fayer R, Santin M, Macarisin D. Cryptosporidium ubiquitum n. sp in animals and humans. Vet Parasitol. 2010;172:23–32. doi: 10.1016/j.vetpar.2010.04.028. [DOI] [PubMed] [Google Scholar]
- Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, McEvoy JM. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbiol. 2006;44:4303–4308. doi: 10.1128/JCM.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, Reid M, Xiao L. Cryptosporidium genotypes in wildlife from a New York watershed. Appl Environ Microbiol. 2007;73:6475–6483. doi: 10.1128/AEM.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Tiao N, Li N, Hlavsa M, Xiao L. Multilocus sequence typing of an emerging Cryptosporidium hominis subtype in the United States. J Clin Microbiol. 2014;52:524–530. doi: 10.1128/JCM.02973-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Torres E, Li N, Wang L, Bowman D, Xiao L. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int J Parasitol. 2013;43:1141–1147. doi: 10.1016/j.ijpara.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Feng Y, Yang W, Ryan U, Zhang L, Kvac M, Koudela B, Modry D, Li N, Fayer R, Xiao L. Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni. J Clin Microbiol. 2011;49:34–41. doi: 10.1128/JCM.01329-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatei W, Das P, Dutta P, Sen A, Cama V, Lal AA, Xiao L. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect Genet Evol. 2007;7:197–205. doi: 10.1016/j.meegid.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Gatei W, Hart CA, Gilman RH, Das P, Cama V, Xiao L. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J Eukaryot Microbiol. 2006;53(Suppl 1):S43–S48. doi: 10.1111/j.1550-7408.2006.00169.x. [DOI] [PubMed] [Google Scholar]
- Guo Y, Tang K, Rowe LA, Li N, Roellig DM, Knipe K, Frace M, Yang C, Feng Y, Xiao L. Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. BMC Genomics. 2015;16:320. doi: 10.1186/s12864-015-1517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Alderisio KA, Xiao L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol. 2005;71:4446–4454. doi: 10.1128/AEM.71.8.4446-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Cai J, Cai M, Wu W, Li C, Lei M, Xu H, Feng L, Ma J, Feng Y, Xiao L. Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet Parasitol. 2016;215:58–62. doi: 10.1016/j.vetpar.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, Santin M, Fayer R, Kvac M, Ryan U, Sak B, Stanko M, Guo Y, Wang L, Zhang L, Cai J, Roellig D, Feng Y. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis. 2014;20:217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, Feng Y. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg Infect Dis. 2013;19:1573–1582. doi: 10.3201/eid1910.121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirhashemi ME, Zintl A, Grant T, Lucy F, Mulcahy G, Waal TD. Molecular epidemiology of Cryptosporidium species in livestock in Ireland. Vet Parasitol. 2016;216:18–22. doi: 10.1016/j.vetpar.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy SF, Smith HV, Kirwan P, Nichols RA, Asaolu SO, Connelly L, Holland CV. Identification of a high diversity of Cryptosporidium species genotypes and subtypes in a pediatric population in Nigeria. Am J Trop Med Hyg. 2010;82:608–613. doi: 10.4269/ajtmh.2010.09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CS, Eisler DL, Alikhani A, Fung VW, Tomblin J, Bowie WR, Isaac-Renton JL. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg Infect Dis. 2002;8:263–268. doi: 10.3201/eid0803.010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotz-Williams LA, Martin DS, Gatei W, Cama V, Peregrine AS, Martin SW, Nydam DV, Jamieson F, Xiao L. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol Res. 2006;99:346–352. doi: 10.1007/s00436-006-0157-4. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang W, Cama V, Wang L, Cabrera L, Ortega Y, Bern C, Feng Y, Gilman R, Xiao L. Population genetics of Cryptosporidium meleagridis in humans and birds: evidence for cross-species transmission. Int J Parasitol. 2014;44:515–521. doi: 10.1016/j.ijpara.2014.03.003. [DOI] [PubMed] [Google Scholar]