Abstract
Cells utilize precise mechanisms to access genomic DNA with spatiotemporal accuracy. ATP-Dependent Chromatin Remodeling Enzymes (also known simply as “remodelers”) comprise a specialized class of enzymes that is intimately involved in genomic organization and accessibility. Remodelers selectively position nucleosomes to either alleviate chromatin compaction or achieve genomic condensation locally, based on a multitude of cellular signals. By dictating nucleosome position, remodelers control local euchromatic and heterochromatic states. These activities govern the accessibility of regulatory regions like promoters and enhancers to transcription factors, RNA polymerases, and co-activators or -repressors. As studies unravel the complexities of epigenetic topography, evidence points to a chromatin-based interactome where regulators interact competitively, cooperatively, and/or co-dependently through physical and functional means. These types of interactions, or crosstalk, between remodelers raise important questions for tissue development. Here, we briefly review the evidence for remodeler interactions and argue for additional studies examining crosstalk.
Introduction
The hypothesis that the epigenome contains a readable language of DNA methylation, post-translational modifications to unstructured histone tails, and multi-dimensional chromatin boundary elements has dramatically improved the understanding of chromatin regulation by stimulating careful analysis of previously unappreciated epigenetic details [1]. Regulation of gene subsets by epigenetic features can dictate cellular transitions (i.e. proliferation, potency, differentiation) and drive de novo tissue development with distinct epigenomic signatures [2]. Within the context of the epigenome, remodelers have emerged as critical regulators of developmental programs [3].
There are four major subfamilies of remodelers (SWI/SNF, INO80, ISWI, and CHD). All are considered derivatives of the SNF2 ATPase family (Figure 1). Each remodeling complex contains a single catalytic ATPase that utilizes ATP hydrolysis to achieve the energy-intensive modulation of chromatin (Figure 1, parentheses). Notably, several remodelers utilize multiple ATPases via mutually exclusive relationships (Figure 1; SWI/SNF, NURD). Mutual exclusivity is not limited to the ATPase subunit, as each remodeling family contains several positions within the complex that can be filled by mutually exclusive proteins. Additionally, remodelers can utilize the same accessory subunits even across family and subfamily classifications (Figure 1). The fact that remodeler components form distinct biochemical complexes on the basis of ATPase inclusion and accessory subunit incorporation is one of the most intriguing topics in remodeler biology. The compositional similarities between remodelers may implicate the complexes in direct relationships via cooperative or competitive means. Likewise, it is probable that variation in composition between remodelers is indicative of functional heterogeneity across remodeler subfamilies. However, limited knowledge on the biological functionality, locus-specific assembly, and regulatory crosstalk of remodelers exists to date. These topics are of central importance to the biomedical community given the broad implications for understanding gene regulation through epigenetic mechanisms.
Figure 1. SNF2 ATPase Chromatin Remodeling Enzymes.
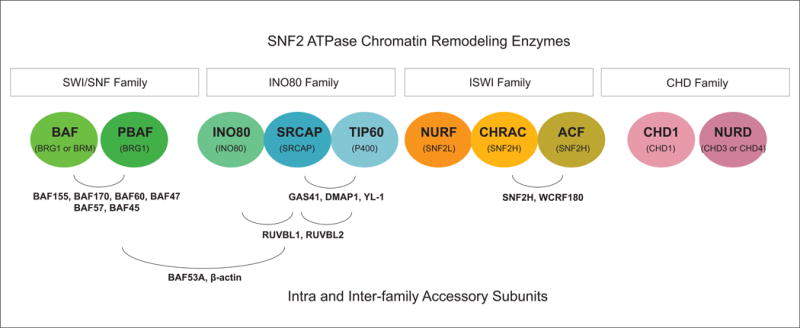
Representative diagram of ATP-Dependent Chromatin Remodeling Families. Families are designated by white boxes. Specific remodelers are represented as circles. Catalytic ATPases for each remodeling complex are indicated in parentheses. Subunits that incorporate into multiple complexes are identified by curved lines. Notably, the ISWI family includes additional complexes not depicted here (NoRC, RSF, WICH).
Remodeler ATPases are engines to the nuclear machines. They are archetypal subunits around which each remodeling complex is assembled, and are evolutionarily conserved across eukaryotes indicating their essential contribution to life. They hydrolyze hundreds of ATP molecules per minute to facilitate nucleosome sliding, histone variant exchange, and promoter clearance on a genome-wide scale [4]. They also contain additional functional domains that direct their function. For example, the predominant ATPase-containing subunit of SWI/SNF, BRG1 (encoded by the SMARCA4 gene in humans), contains a carboxy-terminal bromodomain that targets acetylated lysines on exposed histone tails, thereby promoting activation of transcription [5]. As each remodeler ATPase contains an essential SNF2 ATPase domain as well as conserved domains that promote specific activities of the remodelers, they are the subject of intense investigation.
In addition to the ATPase, each complex also contains multiple protein subunits that facilitate the assembly and action of each remodeling machine. The BPTF subunit of the NURF complex in the ISWI subfamily, for example, deposits trimethylation on lysine 4 residues of histone 3 (H3K4me3), a hallmark of chromatin priming that establishes local euchromatin and recruits transcriptional activators [6]. Deposition of H3K4me3 allows the NURF complex to dramatically alter the chromatin state of specific targets through both chromatin remodeling and post-translational modification of histones [7]. This bridging of chromatin remodeling and chromatin modifying activities is common in chromatin remodeling complexes. In comparison to the H3K4me3 deposition by the NURF complex, the NURD Complex in the CHD subfamily contains multiple histone deacetylases (HDAC1, HDAC2, HDAC3, HDAC8) that enable the complex to deacetylate histone tails and compact nucleosome arrays [8]. The counteracting activities of the NURF and NURD Complexes are representative of differing functions between remodelers as well as the importance of non-ATPase subunits in directing remodeler function. The canonical compositions and activities of each remodeler subfamily have been reviewed extensively and biochemically-derived maps of complex-members have been defined [9], [10], [11], [12], [13]. Yet we still know surprisingly little about the biological function of many proteins that co-purify with the complexes. Because each remodeler contains numerous accessory subunits that have undefined biological roles and it is likely that each confers a functional advantage to the complex(es) they associate with, we project that current knowledge on remodeler compositions and interactions underestimates the diversity of activities within remodeler-class epigenetic regulation in vivo.
New evidence indicates remodelers co-localize to a large proportion of the mammalian genome and directly compete or cooperate [14]. Studies on remodeler co-dependencies indicate remodelers may antagonize one another for promoter occupancy and locus control in yeast and mammals [15], similar to the counteracting remodeling exhibited by the NURF and NURD complexes. Additionally, extensive studies have revealed unexpected stochasticity in non-canonical assemblies of remodeler components in developmental and pathological contexts [16], [17], [18], [19], [20]. To gain a comprehensive understanding of remodeler-class regulatory activities, it is necessary to examine current data that implicate remodelers in an interdependent and dynamic network genome-wide. As we learn more about the abundance and significance of interactions between remodelers, the consequences for gene regulation, cell differentiation, and tissue development are immense. Such functional crosstalk between remodelers is the focus of this essay.
Remodeler Localization Genome-wide
Despite longstanding evidence from genetic experiments that remodelers perform specific functions throughout the genome [21], [22] there are new indications that remodelers engage in dynamic functional interactions. This unexpected result has shaped recent outlooks on chromatin biology in development by challenging biochemically-derived molecular structures and predictions on remodeler functionality. These insights reveal genomic crosstalk occurs on an unprecedented scale.
Extensive work has identified that the SWI/SNF complex localizes to active genomic elements [23], [24]. In one particular study, multiple subunits of the SWI/SNF complex were mapped genome-wide [24]. In addition, all interactions associated with the SWI/SNF complex were analyzed by mass spectrometry. Together, these data showed the full genome-wide and proteome-wide interactions carried out by the canonical SWI/SNF complex. While this work provides a valuable baseline for the genomic occupancy and the interacting partners of the SWI/SNF complex, variable subunits within the complex have not been mapped. Future work analyzing the various biochemically distinct forms of SWI/SNF and other chromatin remodeling complexes is needed to understand the functional and phenotypic diversity of these remodelers.
The localization of SWI/SNF components to regulatory positions throughout the genome aligned with previous findings that targeted mutations in remodelers cause dramatic phenotypes. Murine embryos lacking the Brg1 subunit of SWI/SNF fail to complete the perimplantation stage of development, for example [22]. However, investigations of remodelers from different families have confounded the assumption that mutagenic phenotypes arise in part by unique genomic localization. ChIP paired with high-throughouput sequencing (ChIP-seq) for the catalytic subunits of SWI/SNF, ISWI, and CHD subfamilies revealed that these distantly related remodelers co-localize across the genome with a high frequency [14]. Intriguingly, disruption of the complexes led to unique chromatin organization phenotypes according to nucleosome mapping techniques. Therefore despite co-localization, remodelers perform non-redundant roles in regulating genomic packaging. The regulation of this activity does not appear to be at the level of genomic occupancy, leaving a major unanswered question of how remodelers that bind similar regions and presumably modulate overlapping genes sets exhibit different phenotypes. Resolution of DNA binding by remodelers may obscure interpretations given the inability to accurately distinguish subtle variations using current technologies. One exciting alternative is that remodelers engage with local epigenetic cues after binding chromatin, resulting in dynamic regulation of remodelers despite similar targeting preferences.
One interesting scenario to evaluate crosstalk between remodelers would be to test the regulatory activities of closely-related complexes. Examining the genomic localization preferences and gene expression control by complexes that are biochemically-similar would clarify the biological functionality of biochemically-discrete remodelers. For example, the ARID1A and ARID2 subunits of SWI/SNF do not simultaneously appear in biochemical extractions of the SWI/SNF complex in vitro [25]. Each ARID (AT-rich interacting domain) associates with discrete configurations of SWI/SNF, known as BAF (ARID1A) and PBAF (ARID2), and is required for promoter targeting by the intact complex [26]. It is likely that ARID1A and ARID2 target discrete loci to facilitate unique cellular programs for the two SWI/SNF configurations. However, given the co-occupancy of genomic sites by remodelers from different families it is possible that ARID1A and ARID2 containing complexes also co-occupy binding sites. ARID1A and ARID2 may target the same loci at different times, utilizing SWI/SNF subunits to target similar features but unique ARID subunits to achieve different regulatory results. We hypothesize that given their overlapping, yet distinct subunit compositions, BAF and PBAF exhibit both competition and cooperation with one another in their function as regulators of gene expression (Figure 2).
Figure 2. SWI/SNF interactions genome-wide.
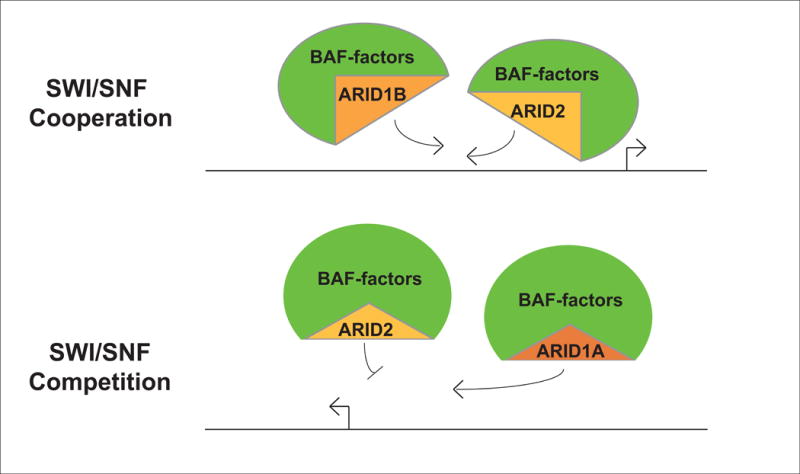
Putative functional interactions between SWI/SNF subcomplexes. Illustrations demonstrate competitive or cooperative genomic binding and gene regulation by SWI/SNF complexes that share regulatory components.
Currently, genomic co-localization by remodelers is best attributed to transient interactions at sites of nucleosome-depleted chromatin [24], [14], [27]. We propose that co-localization may serve a more specific purpose. Subunit-sharing between independent remodeling complexes could contribute to genomic targeting preferences by remodelers, where chromatin targeting by remodelers is dependent on direct cooperativity between counteracting or co-acting remodelers. Moreover, we predict that observed co-localization reflects that remodelers conduct separate regulatory functions throughout the genome and therefore allows cells to personalize their expression profiles based on local stimuli and epigenetic cues.
Accessory Subunits in Remodeler Activities
Given the mutual exclusivity of some remodeler factors and the similar localization patterns of evolutionarily divergent remodelers, the most elusive feature of remodelers to-date pertains to their functional composition in vivo. From glycerol gradient purifications and mass-spectrometry, each complex has been stringently purified to identify stable associates. SWI/SNF subfamily members are comprised of 8–15 subunits, INO80 subfamily members have up to 18 subunits, while ISWI and CHD have fewer ranging between 3–4 and 1–11 subunits, respectively [28]. Identifying if and when functional variations of these canonical complex compositions exist in vivo has been more challenging.
In addition to compositional heterogeneity, some remodelers like the INO80 and SWR complexes exhibit structural modularity [29]. Each complex exists in two states, one elongated form and a compressed form. Presumably, these different forms represent captured states of the complexes mid-remodeling, in accordance with the “hinge” hypothesis where the remodeler stretches to target the nucleosome then bends to embrace and mobilize it [30], [29]. The extent of structural modularity within remodeler families is not known, despite several crystal structures that indicate nucleosome targeting is a predominant feature of remodeling complexes [31], [32], [33], [34]. Likewise, there is currently limited knowledge on the functionality of biochemically-defined complexes. High-resolution genomic mapping of SWR and INO80 complexes in yeast identified nucleosomal targeting preferences of 20 subunits from the two complexes in total [35]. But each complex has been typically studied from the perspective of only one or two subunits in functional settings, leaving many of the accessory subunits with limited characterization. The gap in our understanding of the functional assemblage of complexes, particularly in higher eukaryotes or single cells, has left the roles of the accessory subunits largely undefined.
Several studies have recently provided intriguing hypotheses on the utility of lesser-known subunits. One well-supported hypothesis is that unique combinations of SWI/SNF subunits create distinct complexes, each with a different functional role. Namely, the degree of heterogeneity within SWI/SNF suggests accessory subunits perform essential and diverse functions. For example, SWI/SNF factor BAF53A (encoded by ACTL6A in humans) is replaced by BAF53B (encoded by ACTL6B) upon differentiation of neuronal progenitors to neurons in mammals [16]. The subunit “switch” is essential for cells to complete cell lineage specification. Genetic mutations in BAF53B lead mice to acquire defects in synaptic plasticity and long-term memory suggesting BAF53B is systematically essential for the activities of mature neurons [36]. Similar phenotypes have been observed in hematopoietic and embryonic tissue types where constitutive BAF53A expression is required for repopulating cells but insufficient for differentiation [37], [38].
Identifying the activities of accessory subunits could be key to unlocking remodeler specification in vivo. In several developmental systems, evidence suggests specific SWI/SNF assemblies drive cell lineage choices in development [23], [39], [18], [20], [17] or cell lineage abandonment in malignancy [40]. This evidence implicates accessory subunits as central players in remodeling activities. In development, subunit-exchanges by discrete remodelers are dictated by expression. The mechanism is likely more complex in cancer where remodeler components are ubiquitously expressed. Mutations in a single complex member likely allow chromatin remodeling activity by other forms of the complex in ways the are important for tumor formation and cancer proliferation. To understand physiology and pathophysiology, the roles of subunits that exist in multiple remodeling complexes and families should be clarified. We advocate for common subunits to be extensively characterized in order to clarify their role in remodeler-class crosstalk.
Intriguingly, BAF53A stably associates with many SNF2 remodelers. SWI/SNF, INO80, TIP60, and SRCAP Complexes all contain BAF53A as a core subunit [9]. Therefore assigning developmental phenotypes to any single BAF53A-containing remodeler is problematic because genetic manipulation of BAF53A would affect numerous activities of the other remodelers. Notably, BAF53A has been extensively studied in the context of the INO80 Complex. In vitro, BAF53A has been shown to be required for efficient DNA-dependent ATPase activity, histone targeting, and nucleosome mobility of the INO80 Complex [41], [42]. In addition, BAF53A forms a distinct structural module within the INO80 Complex composed of β-actin and the actin-related protein ACTR8. This module has a high affinity for the helicase-SANT-associated (HSA) domain within the INO80 ATPase indicative of a stable and evolutionarily conserved interaction [43]. SWI/SNF ATPase BRG1 also contains an HSA domain with BAF53A preference. Loss of either BAF53A alone or the BAF53A-β-actin-ACTR8 module is not required for the remodeling activities of the INO80 Complex, but greatly enhances them [41], [43]. Therefore it is likely that BAF53A acts in concert with the additional remodeler subunits to maximize the biological efficiency for nucleosome eviction and placement, perhaps by increasing the targeting efficiency of the complex to specific epigenetic features where remodeling is necessary. In such a scenario, the subunit “switch” to BAF53B may facilitate a different targeting preference for SWI/SNF thereby enabling expression of differentiation-specific genes (Figure 3). Little is known regarding the protein domains within BAF53A but clarification of its ability to be modified structurally or functionally may provide insight into its incorporation into specific remodeling complexes and its preference for histone epitopes. While the exact functions for BAF53A in the other SNF2 remodelers have not been clarified, it is probable that BAF53A serves a unifying feature among BAF53A-containing SNF2 remodelers that makes them functionally cooperative and competitive.
Figure 3. Role of Accessory Subunits in Remodeler Localization.
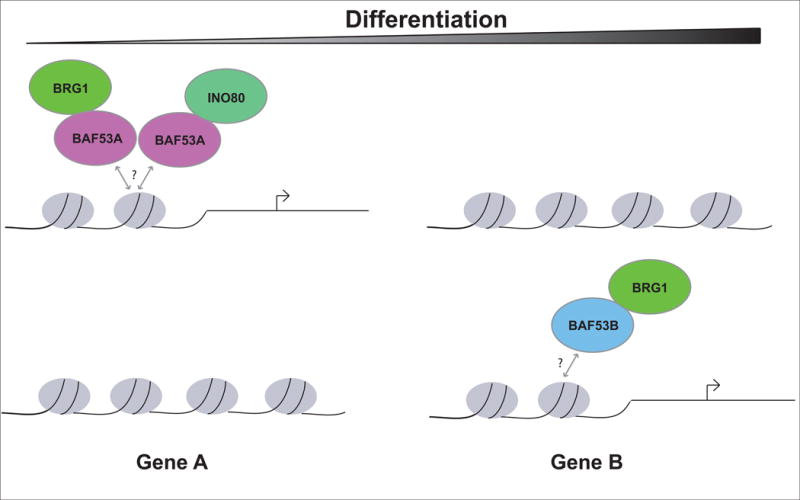
Predicted remodeler localization preferences based on subunit composition. We hypothesize that BAF53A-containing complexes utilize the histone-binding subunit to target similar epigenetic features. Similarly, we hypothesize that subunit switching from BAF53A to BAF53B in SWI/SNF in cell lineage specification serves to re-localize SWI/SNF to sites that require expression at specific cell-lineage timepoints.
Other subunits are shared between remodelers. The helicases RUVBL1 and RUVBL2 (RUVBLs) exist in INO80, TIP60, and SRCAP yet their remodeling-related functions are largely unknown, due in part to the fact that the RUVBLs promiscuously participate in non-remodeling activities through the Fanconi Anemia complex [44] and in snoRNP assembly [45]. Assembly of INO80 and TIP60 has been shown to be dependent on the RUVBLs [46], [47]. In addition, RUVBLs enhance the DNA binding ability of YY1, a transcription factor that canonically associates with INO80 [48]. This evidence suggests that the RUVBL proteins are required members of remodeler assemblies that exist to enhance binding efficiency and specification at epigenetic docking sites, similar to BAF53A. As with BAF53A, understanding the recruitment and action of accessory subunits like the RUVBLs within remodeling assemblies will be essential to clarifying their precise function in the context of chromatin remodeling and gene regulation.
Recent data on remodeler interactions, cooperation, and functional heterogeneity of remodeling complexes suggest there is likely a diversity of remodeler compositions in vivo. Emerging genome-wide studies of remodeler assemblies will improve our understanding of the range of functional compositions that remodelers form. We anticipate that intra-family and inter-complex variation exists to provide cells maximal plasticity to regulate gene expression rapidly and effectively. Biochemical purifications of remodelers inform our understanding of remodeler compositions, and are informative at the cell population level. However, at an individual locus and a single point of time, the combinatorial assembly of proteins from a diverse set of subunits may yield unique function on a locus-specific scale. Highlighting the different functions of unique compositions of chromatin remodeling complexes will be key to understanding the many roles of these complexes in gene regulation.
Conclusion
Faithful regulation of chromatin allows cells to maintain control over nucleosome position and gene expression. Because remodelers intimately interact with the genome to participate in chromatin regulation, their study is essential to understanding the dynamic epigenetic landscape as it pertains to genomic organization, integrity, and gene expression. As we currently understand only a fraction of the complexity that exists to modulate the epigenomic landscape in vivo, the study of remodelers, their interactions, and their components will provide important steps to clarify emergent questions of genomic regulation. The critical role of these complexes in development and disease highlight the necessity of understanding crosstalk within, and between, chromatin remodeling families.
References
- 1.Strahl B, Allis C. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller M, Amin V, Whitaker J, Schultz M, Ward L, Sarkar A, Quon G, Sandstrom R, Eaton M, Wu Y, Pfenning A, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris R, Shoresh N, Epstein C, Gjoneska E, Leung D, Xie W, Hawkins R, Lister R, Hong C, Gascard P, Mungall A, Moore R, Chuah E, Tam A, Canfield T, Hansen R, Kaul R, Sabo P, Bansal M, Carles A, Dixon J, Farh K, Feizi S, Karlic R, Kim A, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer T, Neph S, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari R, Siebenthall K, Sinnott-Armstrong N, Stevens M, Thurman R, Wu J, Zhang B, Zhou X, Beaudet A, Boyer L, De JP, Farnham P, Fisher S, Haussler D, Jones S, Li W, Marra M, McManus M, Sunyaev S, Thomson J, Tlsty T, Tsai L, Wang W, Waterland R, Zhang M, Chadwick L, Bernstein B, Costello J, Ecker J, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos J, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapier C, Cairns B. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 4.Holstege F, Jennings E, Wyrick J, Lee T, Hengartner C, Green M, Golub T, Lander E, Young R. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–28. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 5.Dutta A, Gogol M, Kim J, Smolle M, Venkatesh S, Gilmore J, Florens L, Washburn M, Workman J. Swi/Snf dynamics on stress-responsive genes is governed by competitive bromodomain interactions. Genes Dev. 2014;28:2314–30. doi: 10.1101/gad.243584.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenther M, Levine S, Boyer L, Jaenisch R, Young R. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wysocka J, Swigut T, Xiao H, Milne T, Kwon S, Landry J, Kauer M, Tackett A, Chait B, Badenhorst P, Wu C, Allis C. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 8.Denslow S, Wade P. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 9.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Science Advances. 2015;1(5):e1500447–e1500447. doi: 10.1126/sciadv.1500447. http://dx.doi.org/10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conaway R, Conaway J. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–7. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Gerhold C, Hauer M, Gasser S. INO80-C and SWR-C: guardians of the genome. J Mol Biol. 2015;427:637–51. doi: 10.1016/j.jmb.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Bartholomew B. ISWI chromatin remodeling: one primary actor or a coordinated effort? Curr Opin Struct Biol. 2014;24:150–5. doi: 10.1016/j.sbi.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koster M, Snel B, Timmers H. Genesis of chromatin and transcription dynamics in the origin of species. Cell. 2015;161:724–36. doi: 10.1016/j.cell.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 14.Morris S, Baek S, Sung M, John S, Wiench M, Johnson T, Schiltz R, Hager G. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parnell T, Schlichter A, Wilson B, Cairns B. The chromatin remodelers RSC and ISW1 display functional and chromatin-based promoter antagonism. Elife. 2015;4:e06073. doi: 10.7554/eLife.06073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lessard J, Wu J, Ranish J, Wan M, Winslow M, Staahl B, Wu H, Aebersold R, Graef I, Crabtree G. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–15. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staahl B, Tang J, Wu W, Sun A, Gitler A, Yoo A, Crabtree G. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J Neurosci. 2013;33:10348–61. doi: 10.1523/JNEUROSCI.1258-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lickert H, Takeuchi J, Von BI, Walls J, McAuliffe F, Adamson S, Henkelman R, Wrana J, Rossant J, Bruneau B. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 19.de I IS, Carlson K, Imbalzano A. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–90. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 20.Sawa H, Kouike H, Okano H. Components of the SWI/SNF complex are required for asymmetric cell division in C. elegans. Mol Cell. 2000;6:617–24. doi: 10.1016/s1097-2765(00)00060-5. [DOI] [PubMed] [Google Scholar]
- 21.Hargreaves D, Crabtree G. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bultman S, Gebuhr T, Yee D, La MC, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 23.Ho L, Jothi R, Ronan J, Cui K, Zhao K, Crabtree G. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–91. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Euskirchen G, Auerbach R, Davidov E, Gianoulis T, Zhong G, Rozowsky J, Bhardwaj N, Gerstein M, Snyder M. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011;7:e1002008. doi: 10.1371/journal.pgen.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nie Z, Xue Y, Yang D, Zhou S, Deroo B, Archer T, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–88. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandler R, Brennan J, Schisler J, Serber D, Patterson C, Magnuson T. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol. 2013;33:265–80. doi: 10.1128/MCB.01008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagaich A, Walker D, Wolford R, Hager G. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–74. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- 28.Clapier CR, Cairns BR. The Biology of Chromatin Remodeling Complexes. Annu Rev Biochem. 2009;78(1):273–304. doi: 10.1146/annurev.biochem.77.062706.153223. http://dx.doi.org/10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe S, Tan D, Lakshminarasimhan M, Washburn M, Hong E, Walz T, Peterson C. Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat Commun. 2015;6:7108. doi: 10.1038/ncomms8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold C, Lakomek K, Aebersold R, Beckmann R, Hopfner K. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–19. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Grüne T, Brzeski J, Eberharter A, Clapier C, Corona D, Becker P, Müller C. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–60. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 32.Kasten M, Clapier C, Cairns B. SnapShot: Chromatin remodeling: SWI/SNF. Cell. 2011;144:310.e1. doi: 10.1016/j.cell.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Dürr H, Flaus A, Owen-Hughes T, Hopfner K. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–7. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hauk G, McKnight J, Nodelman I, Bowman G. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–23. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yen K, Vinayachandran V, Pugh B. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013;154:1246–56. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel-Ciernia A, Matheos D, Barrett R, Kramár E, Azzawi S, Chen Y, Magnan C, Zeller M, Sylvain A, Haettig J, Jia Y, Tran A, Dang R, Post R, Chabrier M, Babayan A, Wu J, Crabtree G, Baldi P, Baram T, Lynch G, Wood M. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci. 2013;16:552–61. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krasteva V, Buscarlet M, Diaz-Tellez A, Bernard M, Crabtree G, Lessard J. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood. 2012;120:4720–32. doi: 10.1182/blood-2012-04-427047. [DOI] [PubMed] [Google Scholar]
- 38.Lu W, Fang L, Ouyang B, Zhang X, Zhan S, Feng X, Bai Y, Han X, Kim H, He Q, Wan M, Shi F, Feng X, Liu D, Huang J, Songyang Z. Actl6a protects embryonic stem cells from differentiating into primitive endoderm. Stem Cells. 2015;33:1782–93. doi: 10.1002/stem.2000. [DOI] [PubMed] [Google Scholar]
- 39.Ho L, Ronan J, Wu J, Staahl B, Chen L, Kuo A, Lessard J, Nesvizhskii A, Ranish J, Crabtree G. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–6. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadoch C, Crabtree G. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Cai Y, Jin J, Florens L, Swanson S, Washburn M, Conaway J, Conaway R. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J Biol Chem. 2011;286:11283–9. doi: 10.1074/jbc.M111.222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L, Conaway R, Conaway J. Multiple modes of regulation of the human Ino80 SNF2 ATPase by subunits of the INO80 chromatin-remodeling complex. Proc Natl Acad Sci U S A. 2013;110:20497–502. doi: 10.1073/pnas.1317092110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns B. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15:469–76. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajendra E, Garaycoechea J, Patel K, Passmore L. Abundance of the Fanconi anaemia core complex is regulated by the RuvBL1 and RuvBL2 AAA+ ATPases. Nucleic Acids Res. 2014;42:13736–48. doi: 10.1093/nar/gku1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bizarro J, Charron C, Boulon S, Westman B, Pradet-Balade B, Vandermoere F, Chagot M, Hallais M, Ahmad Y, Leonhardt H, Lamond A, Manival X, Branlant C, Charpentier B, Verheggen C, Bertrand E. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J Cell Biol. 2014;207:463–80. doi: 10.1083/jcb.201404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jónsson Z, Jha S, Wohlschlegel J, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16:465–77. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 47.Jha S, Gupta A, Dar A, Dutta A. RVBs are required for assembling a functional TIP60 complex. Mol Cell Biol. 2013;33:1164–74. doi: 10.1128/MCB.01567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.López-Perrote A, Alatwi H, Torreira E, Ismail A, Ayora S, Downs J, Llorca O. Structure of Yin Yang 1 oligomers that cooperate with RuvBL1-RuvBL2 ATPases. J Biol Chem. 2014;289:22614–29. doi: 10.1074/jbc.M114.567040. [DOI] [PMC free article] [PubMed] [Google Scholar]


