Abstract
In cryogenic correlated light and electron microscopy (cryo-CLEM), frozen targets of interest are identified and located on EM grids by fluorescence microscopy and then imaged at higher resolution by cryo-EM. Whilst working with these methods, we discovered that a variety of mammalian cells exhibit strong punctate autofluorescence when imaged under cryogenic conditions (80 K). Autofluorescence originated from multilamellar bodies (MLBs) and secretory granules. Here we describe a method to distinguish fluorescent protein tags from these autofluorescent sources based on the narrower emission spectrum of the former. The method is first tested on mitochondria and then applied to examine the ultrastructural variability of secretory granules within insulin-secreting pancreatic beta-cell-derived INS-1E cells.
Keywords: Cryo-CLEM, Autofluorescence, Mammalian cells, Electron cryo-tomography, Fluorescent proteins
1. Introduction
Electron microscopy (EM) is an essential tool in the study of cell ultrastructure, with resolving power several orders of magnitude greater than that of light microscopy (LM). Frequently, however, it is difficult to identify objects of interest in EM images when their ultrastructure is unknown. In conventional “thin-section” transmission electron microscopy (TEM), this challenge was originally addressed by the development of immuno-gold labeling (Faulk and Taylor, 1971). This method allows direct labeling of specific targets, therefore permitting their visualization within the cell. However, the fixation, dehydration and/or resin embedding steps can result in poor cell and antigen preservation, leading to an accompanying loss of information (Pilhofer et al., 2010).
Alternatively, in correlated light and electron microscopy (CLEM), each target can be specifically labelled with a fluorophore; typically, genetically-encoded fluorescent protein tags or dyes have been used for labelling (Briegel et al., 2010). The target is located first by fluorescence light microscopy, and then imaged at higher magnification by electron microscopy. CLEM can be conducted either at room- or cryogenic-temperatures (“cryo-CLEM”). Like immuno-EM, room-temperature CLEM also requires chemically fixed and dehydrated cells, which can distort or obscure important structural features (Lucic et al., 2013, Afzelius and Maunsbach, 2004). Nevertheless, room-temperature CLEM has been instrumental in the visualization of numerous bacterial and mammalian cellular events that would otherwise have been challenging or impossible to capture prior to the advent of this method (Grabenbauer et al., 2005, Muller-Reichert et al., 2007, Bertipaglia et al., 2016, Darcy et al., 2006, Schorb et al., 2016, Avinoam et al., 2015, Kukulski et al., 2011, Kukulski et al., 2012, Redemann and Muller-Reichert, 2013, Kapoor et al., 2006).
In cryo-CLEM, samples are preserved in a near-native, “frozen-hydrated” state. To visualize fluorescence inside frozen-hydrated cells, cryogenic LM (cryo-LM) stages are used (Schlimpert et al., 2012, Bertipaglia et al., 2016, Schorb and Briggs, 2014, Briegel et al., 2010, Schellenberger et al., 2014, Schorb et al., 2016). Unfortunately, because the sample has to be kept frozen, long-working-distance air-objective lenses with low numerical apertures are used instead of oil-immersion lenses. Therefore, to increase the resolution of light microscopy under cryogenic conditions, several “super-resolution” cryo-CLEM studies have now been performed (Chang et al., 2014, Liu et al., 2015, Kaufmann et al., 2014).
Cryo-CLEM experiments on mammalian cells have been previously performed using organic fluorescent dyes or proteins tagged with fluorophores (Kaufmann et al., 2014, Liu et al., 2015, Schorb and Briggs, 2014, Schorb et al., 2016, Bykov et al., 2016, Schellenberger et al., 2014, Jun et al., 2011). Though organic dyes are very bright, it can be challenging to use these compounds to efficiently label specific proteins inside cells. Membrane permeabilization is necessary for most organic dyes to gain access to their relevant intracellular targets. However, permeabilization creates structural alterations which may introduce potential artifacts (Lucic et al., 2013, Pilhofer et al., 2010). To address this issue, genetically-encoded fluorescent proteins can be used for intracellular labelling. Unlike dyes, genetically-encoded fluorophores are not limited by permeabilization issues. Genetically-encoded fluorescent proteins can also be easily fused with most proteins and offer a rich repertoire of colors (Shaner et al., 2005). One disadvantage is, however, fluorescent proteins’ limited brightness (Tinnefeld et al., 2015).
During the course of our work, we began imaging an established insulin-secreting pancreatic beta cell line, INS-1E (Merglen et al., 2004). Insulin-secreting pancreatic beta cells are a well-studied model for regulated secretion. Besides insulin, these cells release other diffusible molecules including peptide hormones and neurotransmitters critical in signaling (Merglen et al., 2004). INS-1E cells are characterized by their ability to rapidly secrete large amounts of protein through exocytic fusion of secretory granules. The advent of genetically-encoded fluorophores and the ability to selectively express them in specific compartments enabled the observation of intracellular traffic through the secretory pathway with great temporal and spatial specificity. Direct visualization of secretory granules by EM has complemented these studies by providing higher resolution detail (Burgess and Kelly, 1987, Kelly, 1985). EM revealed the electron dense core of mature secretory granules, termed dense core secretory granule (DCSGs). These cores are known to contain matrix proteins such as chromogranins including chromogranin A (CgA), which aggregate in the vesicle lumen during secretory granule maturation (Gerdes et al., 1989, Taupenot et al., 2005, Gorr et al., 1989, Chanat and Huttner, 1991, Yoo and Albanesi, 1991, Videen et al., 1992, Lemaire et al., 2009).
In an attempt to further characterize secretory granules in a native state, we tagged CgA with GFP (CgA-GFP) and performed cryo-CLEM. Unfortunately, in the course of these studies, we discovered strong autofluorescence in frozen mammalian cells. To overcome this problem, we developed a method to distinguish fluorescent protein tags from autofluorescent sources.
2. Results
2.1. INS-1E cells exhibit bright, punctate autofluorescence under cryogenic conditions
Initially, we set out to image the secretory pathway of the pancreatic beta cell-derived INS-1E cell line, which has long been used as a model system for regulated cell secretion (Merglen et al., 2004, Farino et al., 2016). Though the secretory machinery of these cells has been extensively studied by both LM and conventional EM (Rubi et al., 2005, Giordano et al., 2008), we sought to advance those earlier efforts by imaging these cells in a near-native, frozen-hydrated state in 3-D via electron cryo-tomography (ECT) (Oikonomou and Jensen, 2016). We were particularly interested in dense core secretory granules (DCSGs), intracellular compartments which are central to the efficient secretion of hormones including insulin (Kim et al., 2001). To identify DCSGs in cryo-tomograms, we tagged chromogranin A (CgA), a granin protein widely used as a marker for DCSGs, given its almost exclusive localization to this intracellular compartment (Huh et al., 2005).
We transfected INS-1E cells with CgA that was C-terminally tagged with GFP (CgA-GFP), and grew the cells to ~30–40% confluency on EM finder grids. To facilitate image correlation, we added 500 nm blue fluorospheres to the samples before plunge-freezing [as in (Bykov et al., 2016, Schorb and Briggs, 2014, Chang et al., 2014, Liu et al., 2015, Schellenberger et al., 2014)]. Significantly, these fluorospheres were visible in both cryo-LM and cryo-EM modalities. Moreover, owing to the broad excitation/emission spectrum of the 500 nm blue fluorophores, we used their signals to simultaneously align all fluorescence (DAPI, FITC and YFP) channels.
We first performed cryo-LM to identify targets of interest in thin ice at the periphery of cells using CgA-GFP. We observed a punctate cytosolic distribution (Fig. 1A, Supplementary Fig. 1A) consistent with CgA’s expected intracellular localization to secretory granules (Huh et al., 2005). However, the CgA-GFP signal was not bright either due to the low temperature or the low numerical aperture (NA) of our long-working-distance air-objective (NA 0.7), or both. We therefore used exposure times of up to 2s and applied 2D real-time deblur deconvolution (Nikon).
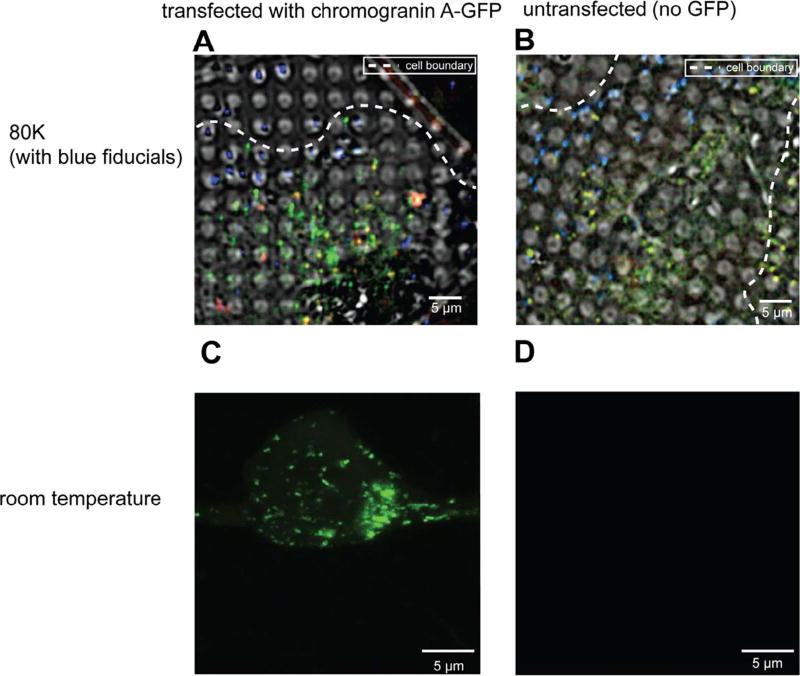
Fig. 1.
INS-1E cells exhibit strong autofluorescence at 80 K. (A, B) Deconvolved cryo-LM images (composite of bright field and epifluorescence in FITC, mCherry and DAPI channels) of INS-1E cells transfected with CgA-GFP (A) or untransfected (B). (C, D) Room-temperature light microscopy images of epifluorescence in FITC channel of INS-1E cells transfected with CgA-GFP (C) or untransfected (D).
As a negative control, we also imaged untransfected INS-1E cells by cryo-LM (Fig. 1B, Supplementary Fig. 1B) using the same methods. Surprisingly, unlike images of untransfected cells at room-temperature, which display little autofluorescence (Fig. 1C–D), images of untransfected cells at ~80 K (cooled by liquid nitrogen) exhibited clear puncta. To determine whether the autofluorescence observed at 80 K was broad-spectrum, as is typical at room-temperature (Billinton and Knight, 2001), we also imaged cells using an mCherry filter. In transfected cells, there were two populations of puncta: one emitting both green and red fluorescence, and another emitting primarily green fluorescence (Fig. 1A, Supplementary Fig. 1A). In untransfected cells, all the puncta emitted both green and red fluorescence (Fig. 1B, Supplementary Fig. 1B), indicating that the autofluorescence in the sample was broad spectrum.
2.2. Other mammalian cells also exhibit bright autofluorescence at ~80 K
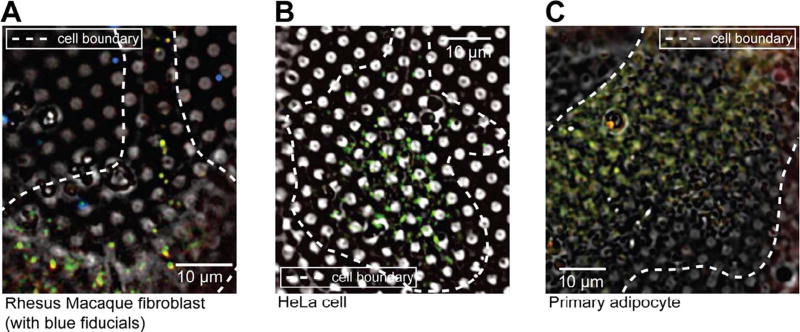
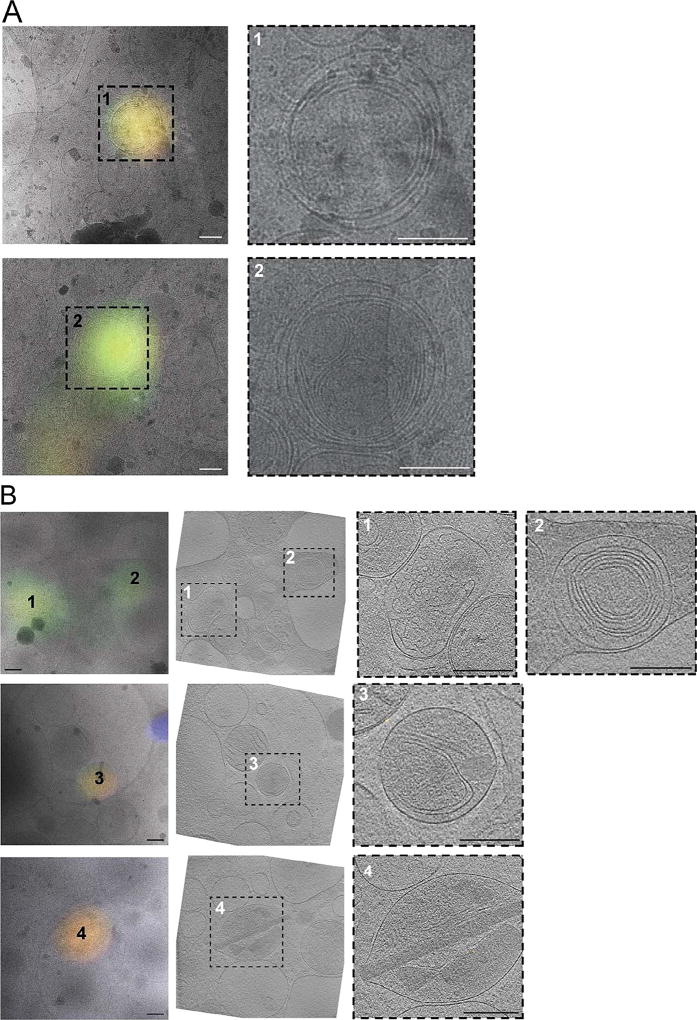
We next tested whether bright autofluorescence at 80 K was unique to INS-1E cells. We applied the same cryogenic imaging techniques, including deconvolution to three other untransfected mammalian cell lines: rhesus macaque fibroblasts, HeLa cells and human primary adipocytes. All three cell lines exhibited numerous puncta distributed throughout the cell volume with broad fluorescence spectra ranging from green (FITC) to red (mCherry) (Fig. 2, Supplementary Fig. 1C–D). To identify the source of the observed autofluorescence, we imaged several puncta at high-resolution by electron cryo-tomography (ECT). All of the autofluorescent puncta in rhesus macaque fibroblasts (Fig. 3A) and HeLa cells (data not shown) correlated to multi-membranous structures, some resembling multilamellar bodies (MLBs) (Hariri et al., 2000, Lajoie et al., 2005). Similarly, many autofluorescent puncta in untransfected INS-1E cells correlated to multi-membranous structures in cryo-tomograms (Fig. 3B, panel 1–3). Other autofluorescent puncta in INS-1E cells correlated to membrane-enclosed crystalline structures (Fig. 3B, panel 4).
Fig. 2.
Autofluorescence at 80 K is a general feature of mammalian cells. Deconvolved cryo-LM images (bright field and epifluorescence in FITC, mCherry and DAPI channels) of (A) rhesus macaque fibroblasts, (B) HeLa cells and (C) primary adipocytes.
Fig. 3.
Cryo-CLEM reveals sources of autofluorescence in untransfected rhesus macaque fibroblasts (A) and INS-1E cells (B). Left panels show deconvolved epifluorescence images overlaid on high-magnification cryo-EM projection images. Middle panel in (B) shows the corresponding tomographic slice. Right panels show zoomed-in views of the boxed areas in corresponding panels at left. Numbers indicate corresponding locations. Scale bars = 200 nm.
2.3. Distinguishing fluorescent protein tags from autofluorescence
Because at room-temperature background autofluorescence sources tend to bleach faster than fluorescent proteins (Billinton and Knight, 2001), one approach that has proven useful to reduce autofluorescence is photobleaching. We therefore compared the rates of bleaching of autofluorescence sources and our fluorescent protein tags. It was previously shown that fluorescent protein tags are remarkably resistant to photobleaching at 80 K (Schwartz et al., 2007). Unfortunately, the autofluorescence did not bleach away quickly either (data not shown), so we did not pursue this strategy further.
Instead, we tried to identify autofluorescence by its broader emission spectrum, a strategy which has also been used previously at room-temperature (Szollosi et al., 1995, Mansfield et al., 2005).
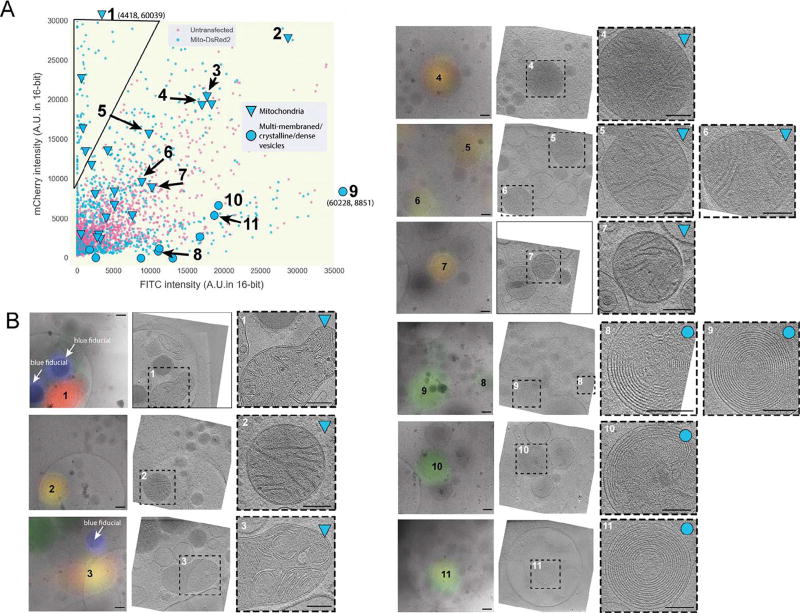
To test the method, we chose a “positive control” that could be unambiguously identified in cryo-tomograms with or without a fluorescent tag. Mitochondria proved to be a good choice due to their well-defined ultrastructure including easily-recognizable cristae (Davies et al., 2014, Zick et al., 2009, Rabl et al., 2009). We therefore transfected INS-1E cells with the mitochondrial marker Mito-DsRed2, which is targeted to the intermembrane space between the inner and outer mitochondrial membranes by the signal sequence of cytochrome c oxidase (Park et al., 2013). We again imaged both untransfected and transfected cells by cryo-CLEM, measured the fluorescence of a large number of deconvolved puncta in both the green (FITC) and red (mCherry) channels, and plotted their intensities. While both transfected and untransfected cells exhibited puncta with both green and red signals, there was a clear population of puncta in the transfected cells with considerably purer red emission (top left corner of Fig. 4A).
Fig. 4.
Relative fluorescence intensity can distinguish target fluorescent signal from autofluorescence at 80 K. (A) Scatter plot of mCherry and FITC channel deconvolved intensity values of fluorescent puncta in INS-1E cells untransfected (magenta) or transfected with Mito-DsRed2 (blue) [n > 875 for transfected, 2500 untransfected]. The black line indicates the area of the scatter plot with no puncta in untransfected INS-1E cells and is described in the text. Larger symbols denote structures observed by cryo-CLEM of selected puncta in transfected cells, corresponding to mitochondria (triangles) or multi-membraned structures, dense vesicles, crystalline structures or structures with a combination of these features (circles). (B) Examples of structures correlated to fluorescent puncta in INS-1E cells transfected with Mito-DsRed2 (32 spots were correlated in total). Left panels show deconvolved epi-fluorescence images overlaid on high-magnification cryo-EM projection images. Middle panels show tomographic slices of the same areas. Right panels show zoomed-in views of the boxed areas in the middle panels. Numbers indicate corresponding locations. Scale bars = 200 nm.
Next, we used ECT to image 32 randomly chosen deconvolved mCherry puncta in the transfected cells. These puncta were located along the thin edges of the cell. Many of the puncta correlated to mitochondria, and the centroid of fluorescence fell within the boundaries of the organelle, validating the accuracy of our correlation procedure (Fig. 4B, targets 1–7). Other deconvolved puncta correlated to multi-membranous structures (Fig. 4B, targets 8–11). As expected, puncta correlating to mitochondria exhibited a greater ratio of red fluorescence to green fluorescence, and were therefore positioned near the upper left corner of the fluorescence plot (Fig. 4A). The multi-membranous structures corresponded to puncta near the opposite (lower right) corner. To distinguish fluorescent protein tags from cellular autofluorescence, one can therefore record fluorescence images of both tagged and untagged frozen cells and plot the fluorescence in two channels. Puncta in the overlap region near the middle of the plot may be from either fluorescent protein tags or autofluorescence, and cannot be distinguished with great confidence. Puncta in the extreme corner towards the pure color of the fluorescent protein tag where autofluorescent puncta are not seen (triangle in the upper left corner of Fig. 4A), however, are very likely to contain the tag.
2.4. Variations on the method
Given the heterogeneous nature of vitreous ice in which the sample is embedded, it is likely the point spread function (PSF) of different puncta is non-uniform (Hell et al., 1993). The fluorescence from identical fluorophores at different depths within the ice is attenuated and spread more or less. We therefore tested whether using raw images (before deconvolution) would give better results. Unfortunately, the distribution of fluorescence in unprocessed images shifted towards the lower portion of the scatter plot, making the signals from fluorescent protein tags more difficult to distinguish from autofluorescence (Supplementary Fig. 2).
In all the work described to this point, the intensity of the brightest pixel within each punctum was used in the scatter plots. As another variation, we tested whether using integrated values over a 3 × 3 pixel square area around the peak gave better results. For this procedure, the centroid pixel and its neighboring pixels were first fitted using a 2DGaussian curve. No significant differences were seen (Supplementary Fig. 3A), so there appears to be no reason for a more sophisticated calculation.
Finally, we tested whether exciting with different lasers and/or measuring entire spectra (instead of just two colors with a dichroic filter cube) would be more effective, hoping that it could perhaps even allow different sources of autofluorescence to be distinguished. To test this, we transfected INS-1E cells with an EYFP-tagged ER marker, calreticulin (calreticulin-EYFP) and imaged transfected and untransfected cells with a confocal microscope (see Methods for details). Autofluorescent puncta in untransfected cells exhibited broad emission spectra without distinctive peaks (14 in total; Supplementary Fig. 4C–D). In contrast, many puncta in transfected cells exhibited narrow emission when excited by 488 nm light and almost no emission when excited by 560 nm light (Supplementary Fig. 4A–B, puncta #3 and #4), consistent with fluorescence from the fluorescent protein tag. Unfortunately distinct classes of autofluorescent puncta in untransfected cells were not seen, so the simpler, faster, and cheaper two-color method is more practical.
2.5. Cryo-CLEM of INS-1E cells transfected with chromogranin A-GFP
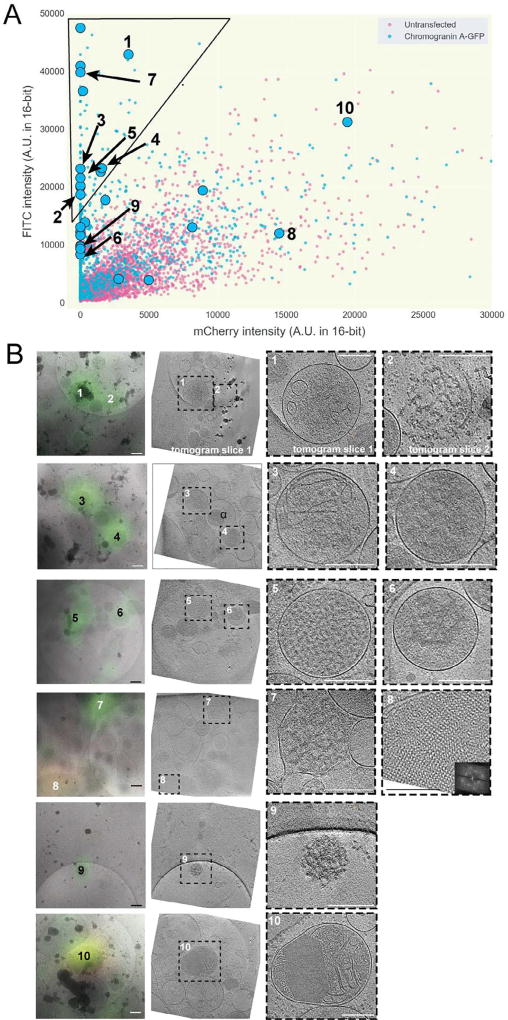
Having established a technique to distinguish desired fluorescent signals from autofluorescence, we returned to INS-1E cells expressing CgA-GFP. Once again, we imaged both untransfected and transfected cells by cryo-LM, recorded their deconvolved intensity values in the green (FITC) and red (mCherry) channels, and plotted these values in two dimensions (Fig. 5A). As expected, we observed a broad overlap region in the center of the plot with puncta from both the transfected (tagged) and untransfected (untagged) cells. We also observed a region in the upper left corner of the plot devoid of puncta from untransfected cells which very likely contained CgA-GFP-specific signal.
Fig. 5.
Application of method enables cryo-CLEM of CgA-GFP in INS-1E cells. (A) Scatter plot of FITC and mCherry channel deconvolved intensity values of fluorescent puncta in INS-1E cells untransfected (magenta) or transfected with CgA-GFP (blue) [n > 1025 for transfected, 4600 untransfected]. The black line indicates the area of the scatter plot with no puncta in untransfected INS-1E cells and is described in the text. Larger symbols denote structures observed by cryo-CLEM of selected puncta in transfected cells (circles). (B) Examples of structures correlated to fluorescent puncta (27 spots were correlated in total). Left panels show deconvolved epi-fluorescence images overlaid on high-magnification cryo-EM projection images. Middle panels show tomographic slices of the same areas. Right panels show zoomed-in views of the boxed areas in the middle panels. Numbers indicate corresponding locations. Scale bars = 200 nm.
Twenty-seven deconvolved puncta were then imaged by ECT, including 15 in the upper left corner of the plot and 12 in the ambiguous overlap region. Cryo-tomographic slices through ten examples are shown in Fig. 5B (see also Supplementary Movie 1). Among the deconvolved puncta from the upper left corner of the plot (very likely to contain CgA-GFP), we observed vesicles with a dense aggregate core (#’s 4 and 6, for example), vesicles with a dense granular core (#5 for example), vesicles with a dense aggregate or granular core and internal smaller vesicles (#’s 1 and 3, for example), and clusters of dense aggregated material partially surrounded by membrane fragments (#’s 2 and 7). Among the puncta in the ambiguous overlap region, we saw two vesicles with crystalline cores (#’s 8 and 10) and a cluster of dense aggregated material (# 9).
3. Discussion
3.1. Approach
Here we showed that many mammalian cell lines exhibit strong punctate autofluorescence at ~80 K. To distinguish fluorescent protein tags from this background, we recommend the following approach: (1) Record cryo-LM images of both non-transfected (unlabeled) and transfected (labelled) cells in two fluorescent channels, one channel corresponding to the color of the fluorescent protein tag and the other from an unrelated channel (e.g. red and green). (2) Deconvolve the images (Agard, 1984). (3) Plot the peak pixel intensities of the puncta from the two fluorescent channels on a two-dimensional plot. (4) Note which regions of the plot contain puncta from unlabeled cells, and which contain puncta from only the labelled cells. Puncta in the latter region will very likely correspond to objects containing the labelled protein, puncta in the former may or may not.
One advantage of this method is that it is simple, fast, and can be done with any standard fluorescence microscope equipped with a cryostage and long-working-distance air-objective lens. We found that deconvolving the fluorescence images was very helpful, but measuring emission in more than two channels was unnecessary. We also found that integrating intensities over areas larger than the single peak pixel was unnecessary.
3.2. Potential sources of autofluorescence in mammalian cells under cryogenic conditions
Previous work has identified the following sources of cellular autofluorescence at room-temperature: (1) biomolecules such as amino acids containing aromatic rings, (2) the three-ring system of flavins (producing green spectra) (Jackson et al., 2004, Benson et al., 1979), (3) the reduced form of pyridine nucleotides (NAD(P)H, producing blue/green spectra) (Galeotti et al., 1970, Chance and Thorell, 1959), (4) lipid pigments (orange/yellow spectra) (Dayan and Wolman, 1993, Billinton and Knight, 2001), (5) porphyrins (red spectra), and (6) chlorophyll (Sheen et al., 1995). In our work on frozen insulin-secreting INS-1E cells, here we found that a variety of multi-membranous structures fluoresced. These structures exhibited up to 14 tightly nested concentric spherical vesicles and resembled MLBs (Lajoie et al., 2005, Hariri et al., 2000). Other autofluorescent structures were bounded by a single outer membrane and contained partially degraded membranes, consistent with earlier descriptions of autolysosomes (Klionsky et al., 2014, Klumperman and Raposo, 2014). Additionally, we observed autofluorescent signals originating from crystalline structures enclosed within the lumen of secretory vesicles. Previous studies have already reported fluorescence from protein crystals, particularly under cryogenic temperatures, which enhanced their intrinsic fluorescence intensity up to ten-fold (Lukk et al., 2016). More generally, work by König et al. also provided evidence that intracellular autofluorescence was more pronounced at 80 K than at room-temperature. They attributed this effect to the increase in quantum yield of the fluorophores due to the reduction in thermal relaxation processes at lower temperatures (Konig et al., 2014).
3.3. Application of the method to characterize secretory vesicles
Pancreatic INS-1E cells have long been used to study insulin release through secretory granules. To date, the majority of light microscopy studies characterizing these secretory granules have been constrained by limited resolution in vivo and the conventional electron microscopy studies have been limited by potential artifacts brought on by sample preparation. Here we applied cryo-CLEM to image secretory granules in situ under near-native conditions.
We used a GFP tag on a standard secretory granule marker, chromogranin A, to identify relevant structures of interest. Using our method to distinguish GFP signal from background autofluorescence, we found that CgA-GFP was present in a diverse set of objects. Some were vesicles possessing dense aggregate or granular cores consistent with previous descriptions of secretory vesicles at different stages of maturation (Novikoff et al., 1977). We also observed CgA-GFP-positive signal within clusters of dense aggregated material partially surrounded by membrane fragments, and a cluster of very dense aggregated material in the cytoplasm with no membrane fragments. These were likely lysed vesicles. We also found CgA-GFP in vesicles incorporating smaller vesicles within their lumen. Previous work describing such structures has identified them as autolysosomes, which are autophagosomes after fusion with lysosomes (Klionsky et al., 2014). In the context of INS-1E cells, these structures are likely involved in degrading vesicle content including insulin and may therefore act as a regulatory control for insulin secretion, though they were not clearly surrounded by two membranes as expected for autophagosomes (Goginashvili et al., 2015, Marsh et al., 2007, Liu et al., 2015). The lysed vesicles and probable autolysosomes may have formed here simply because of the unnaturally high levels of CgA expression following transfection.
We also observed vesicles by ECT with dense aggregated cores that were not fluorescent (example denoted by α in Fig. 5B; see also Supplementary Movie 1). One possible explanation for this is that the variable pH within DCSGs modifies the fluorescence of GFP. Indeed, in pancreatic beta cells, granule acidification is a critical step for proper maturation of pro-insulin to the mature form, ultimately leading to crystallization and exocytosis (Orci et al., 1986, Paroutis et al., 2004). Consequently, a more acidic pH in mature vesicles may quench GFP fluorescence.
This could lead to more difficultly in identifying puncta within these acidic structures, and ultimately bias our approach towards less mature granules. Additionally, such a bias highlights a caveat to interpreting fluorescence images: some cellular objects of a given type may not fluoresce or even incorporate tagged protein, even if it is expressed at levels above those present endogenously. Thus, the previously unrecognized diversity of structures that did contain CgA-GFP and the observation of dense core secretory granules that did not fluoresce both illustrate the power of cryo-CLEM to more completely characterize cellular pathways and objects.
4. Online methods
4.1. Cell growth and transfection
Rat insulinoma INS-1E cells (gift of P. Maechler, Université de Genève) were maintained in a humidified 37 °C incubator with 5% CO2. INS-1E cells were cultured in RPMI 1640 media with L-glutamine (Life Technologies, Grand Island, NY), supplemented with 5% fetal bovine serum (FBS) (heat inactivated), 10 mM HEPES, 100 units/mL penicillin, 100 µg/mL streptomycin, 1 mM sodium pyruvate, and 50 µM 2-Mercaptoethanol. HeLa cells and rhesus macaque fibroblasts where cultured in DMEM media with no phenol red (Gibco), containing 10% FBS, 100 units/mL penicillin, 100 µg/mL streptomycin. Primary adipocyte cells were cultured in human preadipocyte growth medium (Sigma-Aldrich). For cryo-FM and cryo-ET, cells were plated onto fibronectin-coated 200 mesh gold R2/2 London finder Quantifoil grids (Quantifoil Micro Tools GmbH, Jena, Germany) at a density of 2 × 105 cells/mL. We did not observe significant autofluorescence from the support film at 80 K. After 48 h incubation, cultures were plunge frozen in liquid ethane/propane mixture using a Vitrobot Mark IV (FEI, Hillsboro, OR) (Iancu et al., 2006). For cell transfections, INS-1E cells were plated onto fibronectin-coated 200 mesh gold R2/2 Quantifoil grids at a 2 × 105 cells/mL density and cultured for 24–48 h (37 °C, 5% CO2). The cells were then transfected with 2 µg DNA constructs in serum-free RPMI media (5 h, 37 °C, 5% CO2) using Lipofectamine 2000 (Life Technologies, Carlsbad, CA). Transfected constructs included CgA-GFP (Taupenot et al., 2002), calreticulin-EYFP (pEYFP-ER plasmid; Clontech Laboratories, Inc., Mountain View, CA) and Mito-dsRed2 (pDsRed2-Mito plasmid; Clontech Laboratories, Inc.). Following 16 h incubation in serum-containing RPMI media, cells were briefly rinsed in PBS and plunge-frozen for subsequent imaging. Immediately prior to plunge-freezing, 3 µl of a suspension of beads was applied to grids. The bead suspension was made by diluting 500 nm blue (345/435 nm) polystyrene fluorospheres (Phosphorex) with a colloidal solution of 20 nm gold fiducials (Sigma Aldrich) pretreated with bovine serum albumin. The gold served as fiducial markers for tomogram reconstruction while the blue fluorospheres served as landmarks for registering fluorescence light microscopy (FLM) images from different channels as well as EM images. In addition, the blue fluorospheres helped locate target areas in phase contrast light microscopy and low-magnification EM images containing thin ice suitable for high-resolution ECT. Plunge-frozen grids were subsequently loaded into Polara EM cartridges (FEI). EM cartridges containing frozen grids were stored in liquid nitrogen and maintained at ≤−150 °C throughout the experiment including cryo-FLM imaging, cryo-EM imaging, storage and transfer.
4.2. Fluorescence imaging and image processing
The EM cartridges were transferred into a cryo-FLM stage (FEI Cryostage) modified to hold Polara EM cartridges (Nickell et al., 2006, Briegel et al., 2010) and mounted on a Nikon Ti inverted microscope. The grids were imaged using a 60X extra-long-working-distance air-objective (Nikon CFI S Plan Fluor ELWD 60X NA 0.7 WD 2.62–1.8 mm). Images were recorded using a Neo 5.5 sCMOS camera (Andor Technology, South Windsor, CT) using a 2D real-time deblur deconvolution module in the NIS Elements software from AutoQuant (Nikon Instruments Inc., Melville, NY). The 2D real-time deconvolution algorithm estimates a PSF using several factors such as sample thickness, noise levels in the image, background subtraction and contrast enhancement. The pixel size corresponding to the objective lens was ~108 nm (at the sample level). All fluorescence images (individual channels) were saved in 16-bit grayscale format. CgA-GFP was visualized with a FITC filter. Mito-dsRed2 was visualized with an mCherry filter. Blue fluorospheres were visualized with a DAPI filter.
Following cryo-FLM imaging, images from different channels were aligned using a custom written Python alignment script. The 500 nm blue fluorospheres have a broad excitation/emission spectrum that encompassed the DAPI, FITC and YFP channels (sometimes faintly in the mCherry channel) and were therefore used to align these channels. When the blue beads were not visible in the mCherry channel, other broad-spectrum sources of fluorescence (autofluorescence from cells or ice particles) were used to align the mCherry channel with the rest, in particular, the FITC and YFP channels. By using these various broad-spectrum fluorescence signals we were able to align all fluorescence channels. By choosing fairly small point sources of fluorescence, we were even able to align the channels to sub-pixel accuracy by fitting a 2D-Gaussian for the point sources in each of the channels.
Before the cryo-FLM images were further analyzed, background fluorescence in each channel was subtracted from the respective images. This background fluorescence was uniform throughout each image (even outside cellular areas) and likely originates from the grid/ice. Fluorescent puncta were identified in the channel of interest using a Python script and their peak fluorescence intensities measured. In addition, intensities of the same pixels in other channels were also recorded. For example, in the case of CgA-GFP dataset, peak intensities of puncta in the FITC channel and their corresponding pixel intensities in the mCherry channel were recorded. Peak intensity values in both channels of both untransfected and transfected cells were plotted on scatter plots.
4.3. Cumulative intensity calculations
To calculate cumulative intensities for each of the fluorescence puncta, we first identified the centroid pixel using a 2D-Gaussian estimation. To this end, we first identified fluorescence puncta and their peak intensity pixel in the primary fluorescence channel of interest (e.g. RFP channel in the case of Mito-DsRed dataset). Subsequently, we used 9 pixels in-and-around each of the peak intensity pixels to accurately estimate a centroid for individual puncta. When these sub-pixel shifts along both x and y directions were applied to the original image using spline interpolation, we were able to reconstruct images for each of the fluorescence puncta perfectly centered around a theoretically estimated peak. Using these reconstructed images for each of the fluorescence puncta, we calculated the cumulative value for 3×3 pixels in-and-around the center pixel. We applied the same shifts calculated for one channel to the other fluorescence channels as well.
4.4. Confocal microscopy and emission spectra analysis
Lambda stacks were collected using a 60X objective lens on a Zeiss LSM 710 microscope equipped with a 32-channel spectral detector with sequential 561 nm and 488 nm laser excitations.
Conditions were imaged under the same laser power and gain settings. Spectral curves were then extracted from the lambda stacks using ImageJ for comparisons. All the imaging parameters within each channel were standardized.
4.5. Cryo-CLEM and ECT
Grids previously imaged by cryo-FLM were subsequently imaged by ECT using an FEI G2 Polara 300 kV FEG TEM equipped with an energy filter (slit width 20 eV for higher magnifications; Gatan, Inc.). Images were recorded using a 4 k × 4 k K2 Summit direct detector (Gatan, Inc.) operating in the electron counting mode. First, areas containing the fluorescent puncta of interest were located in the TEM. Tilt series were then recorded of these areas using UCSF Tomography (Zheng et al., 2007) or SerialEM (Mastronarde, 2005) software at a magnification of 18,000×. This corresponds to a pixel size of 6 Å (at the specimen level) and was found to be sufficient for this study. Each tilt series was collected from −60° to+60° with an increment of 1° in an automated fashion at 8–10 µm underfocus. The cumulative dose of one tilt-series was between 80 and 200 e−/Å2.
Areas of interest were located by TEM in a stepwise manner. First, the grid square/cell of interest on the finder grid was located by using large location markers or other features visible at a low magnification (100×). Second, a smaller area containing the fluorescent punctum of interest was located by mapping cryo-FLM images to intermediate-magnification EM images (typically 3000× or 1200×) by eye using various local features within the identified grid square. These features included (1) clusters of 500 nm microspheres that were arranged in a uniquely identifiable pattern, (2) cracks and regularly spaced 2 μm holes in the carbon film and (3) ice contamination. This was done with either UCSF Tomography (Zheng et al., 2007) or SerialEM (Mastronarde, 2005). With UCSF Tomography, the area of interest first had to be mentally mapped on the low/intermediate magnification EM image using the local features described above before being identified again at 18,000× magnification by the same features (if available within the field of view). Correlation with SerialEM was more streamlined. cryo-FLM images were registered with EM images of the grid square of interest using local features (described above) as control points. These EM images could be either single projection images at a low enough magnification (360× or 1200×) to contain the area of interest and enough control points to enable tilt-series collection or a montage assembled from higher-magnification (3000×) EM images. Once the cryo-FLM images were registered, areas of interest were marked using the “anchor maps” feature. Using this feature, marked areas could be revisited and tilt-series collected in an automated fashion. Once acquired, tilt-series were aligned and binned fourfold into 1 k × 1 k arrays before reconstruction into 3D tomograms with the IMOD software package (Kremer et al., 1996). In addition to the tilt-series, projection images of the location at various magnifications (360×, 1200×, 3000×, 9300× and 18,000×) were saved and used for high-precision post-data collection correlation.
4.6. High-precision post-data collection correlation of FLM images and tomographic slices
RGM fluorescence images were correlated to EM projection images using an in-house image registration script written in Python. This entailed stepwise registration of images recorded at various magnifications, starting with the cryo-FLM image, recorded at the lowest magnification (60X) (2056 × 2054 pixels), all the way up to the 18,000× EM image. First, the cryo-FLM image (60×) was registered with a low/intermediate-magnification EM image (1200×). The centroid positions of 500 nm blue microspheres were estimated to sub-pixel accuracy and used as control points for this registration. The magnification of the EM image used for this registration was chosen such that there were at least 4 control points available in the field of view. The microspheres were clearly visible at magnifications above 1200× and less reliably visible at magnifications as low as 360×. Therefore the registration process was more accurate when using >1200× EM images. The registration parameters (affine transformation) were saved. Successive steps involved similar calculation of affine transformation parameters between EM projection images of various magnifications up to 18,000×. Gold fiducials, surface ice contaminations, and cellular features visible at these magnifications were used as control points for these registration steps. In general, we found that high defocus values at lower magnifications (~50 µm at 3000× and ~1 mm at 1200×) enhanced the visibility of control points and thus resulted in better registration. Also, having more control points (at least 5) resulted in better registration. Precise registration at lower magnifications is particularly important because of the relatively larger pixel sizes involved. To produce the final registration of the cryo-FLM images with the 18,000× projection EM images, the transformations calculated in each of the previous steps were successively applied to the cryo-FLM images. The resulting FLM images were overlaid on the 18,000× EM projection images using Adobe Photoshop CC (San Jose, CA), setting the visibility of the upper cryo-FLM image layer to linear dodge.
Supplementary Material
Acknowledgments
This work was supported by the NIH (grant GM082545 to G.J.J., grant GM082545-6935 to T.J.H., grant K08 DA031241 to Z.F.), the Department of Defense (grant PR141292 to Z.F.), and the John F. and Nancy A. Emmerling Fund of The Pittsburgh Foundation (to Z.F.). We thank Dr. Joachim Frank, Dr. Maïté Courel, Dr. Hans Breunig, Dr. Karsten König, Dr. Estela Area-Gomez, Robert Grassucci and Stephanie Siegmund for guidance, suggestions and reagents. We thank Dr. Pierre Maechler for generously providing INS-1E cells for our studies. We would like to thank Steven Wilbert for technical assistance. Confocal imaging was performed in the Biological Imaging Facility, with the support of the Caltech Beckman Institute and the Arnold and Mabel Beckman Foundation.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2017.10.009.
References
- Afzelius BA, Maunsbach AB. Biological ultrastructure research; the first 50 years. Tissue Cell. 2004;36:83–94. doi: 10.1016/j.tice.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Agard DA. Optical sectioning microscopy: cellular architecture in three dimensions. Annu. Rev. Biophys. Bioeng. 1984;13:191–219. doi: 10.1146/annurev.bb.13.060184.001203. [DOI] [PubMed] [Google Scholar]
- Avinoam O, Schorb M, Beese CJ, Briggs JA, Kaksonen M. ENDOCYTOSIS. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science. 2015;348:1369–1372. doi: 10.1126/science.aaa9555. [DOI] [PubMed] [Google Scholar]
- Benson RC, Meyer RA, Zaruba ME, McKhann GM. Cellular autofluorescence-is it due to flavins? J. Histochem. Cytochem. 1979;27:44–48. doi: 10.1177/27.1.438504. [DOI] [PubMed] [Google Scholar]
- Bertipaglia C, Schneider S, Jakobi AJ, Tarafder AK, Bykov YS, Picco A, Kukulski W, Kosinski J, Hagen WJ, Ravichandran AC, Wilmanns M, Kaksonen M, Briggs JA, Sachse C. Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle. EMBO Rep. 2016;17:1044–1060. doi: 10.15252/embr.201541960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billinton N, Knight AW. Seeing the wood through the trees: a review of techniques for distinguishing green fluorescent protein from endogenous autofluorescence. Anal. Biochem. 2001;291:175–197. doi: 10.1006/abio.2000.5006. [DOI] [PubMed] [Google Scholar]
- Briegel A, Chen S, Koster AJ, Plitzko JM, Schwartz CL, Jensen GJ. Correlated light and electron cryo-microscopy. Methods Enzymol. 2010;481:317–341. doi: 10.1016/S0076-6879(10)81013-4. [DOI] [PubMed] [Google Scholar]
- Burgess TL, Kelly RB. Constitutive and regulated secretion of proteins. Annu. Rev. Cell Biol. 1987;3:243–293. doi: 10.1146/annurev.cb.03.110187.001331. [DOI] [PubMed] [Google Scholar]
- Bykov YS, Cortese M, Briggs JA, Bartenschlager R. Correlative light and electron microscopy methods for the study of virus-cell interactions. FEBS Lett. 2016;590:1877–1895. doi: 10.1002/1873-3468.12153. [DOI] [PubMed] [Google Scholar]
- Chanat E, Huttner WB. Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J. Cell Biol. 1991;115:1505–1519. doi: 10.1083/jcb.115.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B, Thorell B. Localization and kinetics of reduced pyridine nucleotide in living cells by microfluorometry. J. Biol. Chem. 1959;234:3044–3050. [PubMed] [Google Scholar]
- Chang YW, Chen S, Tocheva EI, Treuner-Lange A, Lobach S, Sogaard-Andersen L, Jensen GJ. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat. Methods. 2014;11:737–739. doi: 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcy KJ, Staras K, Collinson LM, Goda Y. An ultrastructural readout of fluorescence recovery after photobleaching using correlative light and electron microscopy. Nat. Protoc. 2006;1:988–994. doi: 10.1038/nprot.2006.146. [DOI] [PubMed] [Google Scholar]
- Davies KM, Daum B, Gold VA, Muhleip AW, Brandt T, Blum TB, Mills DJ, Kuhlbrandt W. Visualization of ATP synthase dimers in mitochondria by electron cryo-tomography. J. Vis Exp. 2014:51228. doi: 10.3791/51228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan D, Wolman M. Lipid pigments. Prog. Histochem. Cytochem. 1993;25:1–74. doi: 10.1016/s0079-6336(11)80029-x. [DOI] [PubMed] [Google Scholar]
- Farino ZJ, Morgenstern TJ, Vallaghe J, Gregor N, Donthamsetti P, Harris PE, Pierre N, Freyberg R, Charrier-Savournin F, Javitch JA, Freyberg Z. Development of a rapid insulin assay by homogenous time-resolved fluorescence. PLoS One. 2016;11:e0148684. doi: 10.1371/journal.pone.0148684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk WP, Taylor GM. An immunocolloid method for the electron microscope. Immunochemistry. 1971;8:1081–1083. doi: 10.1016/0019-2791(71)90496-4. [DOI] [PubMed] [Google Scholar]
- Galeotti T, van Rossum GD, Mayer D, Chance B. Fluorescence studies of NAD (P)H binding in intact cells. Hoppe Seylers Z. Physiol. Chem. 1970;351:274–275. [PubMed] [Google Scholar]
- Gerdes HH, Rosa P, Phillips E, Baeuerle PA, Frank R, Argos P, Huttner WB. The primary structure of human secretogranin II, a widespread tyrosine-sulfated secretory granule protein that exhibits low pH- and calcium-induced aggregation. J. Biol. Chem. 1989;264:12009–12015. [PubMed] [Google Scholar]
- Giordano T, Brigatti C, Podini P, Bonifacio E, Meldolesi J, Malosio ML. Beta cell chromogranin B is partially segregated in distinct granules and can be released separately from insulin in response to stimulation. Diabetologia. 2008;51:997–1007. doi: 10.1007/s00125-008-0980-5. [DOI] [PubMed] [Google Scholar]
- Goginashvili A, Zhang Z, Erbs E, Spiegelhalter C, Kessler P, Mihlan M, Pasquier A, Krupina K, Schieber N, Cinque L, Morvan J, Sumara I, Schwab Y, Settembre C, Ricci R. Insulin granules. Insulin secretory granules control autophagy in pancreatic beta cells. Science. 2015;347:878–882. doi: 10.1126/science.aaa2628. [DOI] [PubMed] [Google Scholar]
- Gorr SU, Shioi J, Cohn DV. Interaction of calcium with porcine adrenal chromogranin A (secretory protein-I) and chromogranin B (secretogranin I) Am. J. Physiol. 1989;257:E247–E254. doi: 10.1152/ajpendo.1989.257.2.E247. [DOI] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat. Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- Hariri M, Millane G, Guimond MP, Guay G, Dennis JW, Nabi IR. Biogenesis of multilamellar bodies via autophagy. Mol. Biol. Cell. 2000;11:255–268. doi: 10.1091/mbc.11.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell S, Reiner G, Cremer C, Stelzer EHK. Aberrations in confocal fluorescence microscopy induced by mismatches in refractive index. J. Microsc. 1993;169:391–405. [Google Scholar]
- Huh YH, Bahk SJ, Ghee JY, Yoo SH. Subcellular distribution of chromogranins A and B in bovine adrenal chromaffin cells. FEBS Lett. 2005;579:5145–5151. doi: 10.1016/j.febslet.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A, Gan L, He Y, Jensen GJ. Electron cryotomography sample preparation using the Vitrobot. Nat. Protoc. 2006;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- Jackson KA, Snyder DS, Goodell MA. Skeletal muscle fiber-specific green autofluorescence: potential for stem cell engraftment artifacts. Stem Cells. 2004;22:180–187. doi: 10.1634/stemcells.22-2-180. [DOI] [PubMed] [Google Scholar]
- Jun S, Ke D, Debiec K, Zhao G, Meng X, Ambrose Z, Gibson GA, Watkins SC, Zhang P. Direct visualization of HIV-1 with correlative live-cell microscopy and cryo-electron tomography. Structure. 2011;19:1573–1581. doi: 10.1016/j.str.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann R, Schellenberger P, Seiradake E, Dobbie IM, Jones EY, Davis I, Hagen C, Grunewald K. Super-resolution microscopy using standard fluorescent proteins in intact cells under cryo-conditions. Nano Lett. 2014;14:4171–4175. doi: 10.1021/nl501870p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RB. Pathways of protein secretion in eukaryotes. Science. 1985;230:25–32. doi: 10.1126/science.2994224. [DOI] [PubMed] [Google Scholar]
- Kim T, Tao-Cheng JH, Eiden LE, Loh YP. Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell. 2001;106:499–509. doi: 10.1016/s0092-8674(01)00459-7. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Eskelinen EL, Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I'm confused. Autophagy. 2014;10:549–551. doi: 10.4161/auto.28448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Raposo G. The complex ultrastructure of the endolysosomal system. Cold Spring Harb. Perspect. Biol. 2014;6:a016857. doi: 10.1101/cshperspect.a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig K, Uchugonova A, Breunig HG. High-resolution multiphoton cryomicroscopy. Methods. 2014;66:230–236. doi: 10.1016/j.ymeth.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Kaksonen M, Briggs JA. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie P, Guay G, Dennis JW, Nabi IR. The lipid composition of autophagic vacuoles regulates expression of multilamellar bodies. J. Cell Sci. 2005;118:1991–2003. doi: 10.1242/jcs.02324. [DOI] [PubMed] [Google Scholar]
- Lemaire K, Ravier MA, Schraenen A, Creemers JW, van de Plas R, Granvik M, van Lommel L, Waelkens E, Chimienti F, Rutter GA, Gilon P, IN'T VELD PA, SCHUIT FC. Insulin crystallization depends on zinc transporter ZnT8 expression, but is not required for normal glucose homeostasis in mice. Proc. Natl. Acad. Sci. USA. 2009;106:14872–14877. doi: 10.1073/pnas.0906587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Xue Y, Zhao W, Chen Y, Fan C, Gu L, Zhang Y, Zhang X, Sun L, Huang X, Ding W, Sun F, Ji W, Xu T. Three-dimensional super-resolution protein localization correlated with vitrified cellular context. Sci. Rep. 2015;5:13017. doi: 10.1038/srep13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Rigort A, Baumeister W. Cryo-electron tomography: the challenge of doing structural biology in situ. J. Cell Biol. 2013;202:407–419. doi: 10.1083/jcb.201304193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk T, Gillilan RE, Szebenyi DM, Zipfel WR. A visible-light-excited fluorescence method for imaging protein crystals without added dyes. J. Appl. Crystallogr. 2016;49:234–240. doi: 10.1107/S160057671502419X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JR, Gossage KW, Hoyt CC, Levenson RM. Autofluorescence removal, multiplexing, and automated analysis methods for in-vivo fluorescence imaging. J. Biomed. Opt. 2005;10:41207. doi: 10.1117/1.2032458. [DOI] [PubMed] [Google Scholar]
- Marsh BJ, Soden C, Alarcon C, Wicksteed BL, Yaekura K, Costin AJ, Morgan GP, Rhodes CJ. Regulated autophagy controls hormone content in secretory-deficient pancreatic endocrine beta-cells. Mol. Endocrinol. 2007;21:2255–2269. doi: 10.1210/me.2007-0077. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Merglen A, Theander S, Rubi B, Chaffard G, Wollheim CB, Maechler P. Glucose sensitivity and metabolism-secretion coupling studied during two-year continuous culture in INS-1E insulinoma cells. Endocrinology. 2004;145:667–678. doi: 10.1210/en.2003-1099. [DOI] [PubMed] [Google Scholar]
- Muller-Reichert T, Srayko M, Hyman A, O'Toole ET, McDonald K. Correlative light and electron microscopy of early Caenorhabditis elegans embryos in mitosis. Methods Cell Biol. 2007;79:101–119. doi: 10.1016/S0091-679X(06)79004-5. [DOI] [PubMed] [Google Scholar]
- Nickell S, Kofler C, Leis AP, Baumeister W. A visual approach to proteomics. Nat. Rev. Mol. Cell Biol. 2006;7:225–230. doi: 10.1038/nrm1861. [DOI] [PubMed] [Google Scholar]
- Novikoff AB, Mori M, Quintana N, Yam A. Studies of the secretory process in the mammalian exocrine pancreas. I. The condensing vacuoles. J. Cell. Biol. 1977;75:148–165. doi: 10.1083/jcb.75.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou CM, Jensen GJ. A new view into prokaryotic cell biology from electron cryotomography. Nat. Rev. Microbiol. 2016;14:205–220. doi: 10.1038/nrmicro.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Amherdt M, Madsen O, Perrelet A, Vassalli JD, Anderson RG. Conversion of proinsulin to insulin occurs coordinately with acidification of maturing secretory vesicles. J. Cell Biol. 1986;103:2273–2281. doi: 10.1083/jcb.103.6.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Choi H, Min JS, Park SJ, Kim JH, Park HJ, Kim B, Chae JI, Yim M, Lee DS. Mitochondrial dynamics modulate the expression of pro-inflammatory mediators in microglial cells. J. Neurochem. 2013;127:221–232. doi: 10.1111/jnc.12361. [DOI] [PubMed] [Google Scholar]
- Paroutis P, Touret N, Grinstein S. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology (Bethesda) 2004;19:207–215. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ. Bacterial TEM: new insights from cryo-microscopy. Methods Cell Biol. 2010;96:21–45. doi: 10.1016/S0091-679X(10)96002-0. [DOI] [PubMed] [Google Scholar]
- Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Korner C, Jagasia R, Keil T, Baumeister W, Cyrklaff M, Neupert W, Reichert AS. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell. Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S, Muller-Reichert T. Correlative light and electron microscopy for the analysis of cell division. J. Microsc. 2013;251:109–112. doi: 10.1111/jmi.12056. [DOI] [PubMed] [Google Scholar]
- Rubi B, Ljubicic S, Pournourmohammadi S, Carobbio S, Armanet M, Bartley C, Maechler P. Dopamine D2-like receptors are expressed in pancreatic beta cells and mediate inhibition of insulin secretion. J. Biol. Chem. 2005;280:36824–36832. doi: 10.1074/jbc.M505560200. [DOI] [PubMed] [Google Scholar]
- Schellenberger P, Kaufmann R, Siebert CA, Hagen C, Wodrich H, Grunewald K. High-precision correlative fluorescence and electron cryo microscopy using two independent alignment markers. Ultramicroscopy. 2014;143:41–51. doi: 10.1016/j.ultramic.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlimpert S, Klein EA, Briegel A, Hughes V, Kahnt J, Bolte K, Maier UG, Brun YV, Jensen GJ, Gitai Z, Thanbichler M. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151:1270–1282. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorb M, Briggs JA. Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy. 2014;143:24–32. doi: 10.1016/j.ultramic.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorb M, Gaechter L, Avinoam O, Sieckmann F, Clarke M, Bebeacua C, Bykov YS, Sonnen AF, Lihl R, Briggs JA. New hardware and workflows for semi-automated correlative cryo-fluorescence and cryo-electron microscopy/tomography. J. Struct Biol. 2016 doi: 10.1016/j.jsb.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J. Microsc. 2007;227:98–109. doi: 10.1111/j.1365-2818.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW. Green-fluorescent protein as a new vital marker in plant cells. Plant J. 1995;8:777–784. doi: 10.1046/j.1365-313x.1995.08050777.x. [DOI] [PubMed] [Google Scholar]
- Szollosi J, Lockett SJ, Balazs M, Waldman FM. Autofluorescence correction for fluorescence in situ hybridization. Cytometry. 1995;20:356–361. doi: 10.1002/cyto.990200412. [DOI] [PubMed] [Google Scholar]
- Taupenot L, Harper KL, Mahapatra NR, Parmer RJ, Mahata SK, O'Connor DT. Identification of a novel sorting determinant for the regulated pathway in the secretory protein chromogranin A. J. Cell Sci. 2002;115:4827–4841. doi: 10.1242/jcs.00140. [DOI] [PubMed] [Google Scholar]
- Taupenot L, Harper KL, O'Connor DT. Role of H+-ATPase-mediated acidification in sorting and release of the regulated secretory protein chromogranin A: evidence for a vesiculogenic function. J. Biol. Chem. 2005;280:3885–3897. doi: 10.1074/jbc.M408197200. [DOI] [PubMed] [Google Scholar]
- Tinnefeld P, Eggeling C, Hell S. Far-field optical nanoscopy. Springer; Berlin Heidelberg, New York, NY: 2015. [Google Scholar]
- Videen JS, Mezger MS, Chang YM, O'Connor DT. Calcium and catecholamine interactions with adrenal chromogranins. Comparison of driving forces in binding and aggregation. J. Biol. Chem. 1992;267:3066–3073. [PubMed] [Google Scholar]
- Yoo SH, Albanesi JP. High capacity, low affinity Ca2+ binding of chromogranin A. Relationship between the pH-induced conformational change and Ca2+ binding property. J. Biol. Chem. 1991;266:7740–7745. [PubMed] [Google Scholar]
- Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J. Struct. Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793:5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.